Abstract
Background
Understanding of iron oxide nanoparticles (IONP) interaction with the body milieu is crucial to guarantee their efficiency and biocompatibility in nanomedicine. Polymer coating to IONP, with polyethyleneglycol (PEG) and polyvinylpyrrolidone (PVP), is an accepted strategy to prevent toxicity and excessive protein binding.
Aim
The aim of this study was to investigate the feature of IONP adsorption of complement proteins, their activation and consequent inflammatory response as a strategy to further elucidate their biocompatibility.
Methods
Three types of IONP with different surface characteristics were used: bare (IONP-bare), coated with PVP (IONP-PVP) and PEG-coated (IONP-PEG). IONPs were incubated with human plasma and adsorbed proteins were identified. BALB/c mice were intravenously exposed to IONP to evaluate complement activation and proinflammatory response.
Results
Protein corona fingerprinting showed that PEG surface around IONP promoted a selective adsorption of complement recognition molecules which would be responsible for the complement system activation. Furthermore, IONP-PEG activated in vitro, the complement system and induced a substantial increment of C3a and C4a anaphylatoxins while IONP-bare and IONP-PVP did not. In vivo IONP-PEG induced an increment in complement activation markers (C5a and C5b-9), and proinflammatory cytokines (IL-1β, IL-6, TNF-α).
Conclusion
The engineering of nanoparticles must incorporate the association between complement proteins and nanomedicines, which will regulate the immunostimulatory effects through a selective adsorption of plasma proteins and will enable a safer application of IONP in human therapy.
Keywords: iron oxide nanoparticles, complement, inflammatory response, protein corona
Introduction
Iron oxide nanoparticles (IONP) have been subject of intensive research for many years due to their intrinsic properties for which they are used in nanomedicine as contrast agents,1 hyperthermia inductors,2 and drug-delivery carriers.3 However, despite the initial enthusiasm to popularize and spread their use in biomedical areas, promising candidates previously approved by the Food and Drug Administration have been withdrawn due to hypersensitivity and toxicity concerns.4,5 One of the major pitfalls of IONP-based nanomedicines is the lack of biocompatibility with blood components and the immune system. When IONP and other nanomedicines are injected into the bloodstream, proteins and other biomolecules are rapidly adsorbed on their surface creating a novel biological entity which will would be dependent of nanoparticle (NP) physicochemical properties.6,7 This newly formed complex, known as “protein corona” (PC), shapes the biological interaction between the IONP with other biomolecules, cells, and physical barriers.8
Moreover, the interaction of IONP with components of the immune system such as phagocytic cells, and the complement system are key modulators for their efficacy distribution and toxicity.9,10 The complement system is a group of ~30 proteins (distributed as soluble elements in plasma and as extracellular receptors in immune cells) that provides critical immunoprotective and immunoregulatory functions; it opsonizes and induces a series of inflammatory processes against pathogens and nanostructured materials, which are perceived as foreign agents.11 The complement system can be triggered through three different pathways: 1) the classic pathway, activated by the immune complexes (antigen–antibody) and by others molecules such as reactive protein C, 2) the lectin pathway, activated by the union of the mannan-binding lectin (MBL) or ficolins to the mannose-containing carbohydrates or N-acetylglucosamine, and 3) the alternative pathway that can be spontaneously initiated when the complement component C3 binds to a reactive surface.12 Although complement pathways depend on different molecules for their initiation, they converge to generate the same set of effector molecules. Anaphylatoxins (C3a, C4a, and C5a) are soluble byproducts of the complement activation that are potent inflammatory inductors. When these peptides are produced in an uncontrolled condition, they can induce a wide spectrum of side effects such as cardiac and respiratory complications.13 Some nanomedicines, such as Doxil,14 nanomaterials such as carbon nanotubes (CNT),15 and IONP16 have shown the potential to interact with the complement system resulting in a significant activation. Consequently, evaluation of the complement system has recently gained attention in nanomedicine development since its activation has been linked to numerous adverse effects in animal models and patients.12,13,17
In this study, we aimed to test whether the use of polymeric coatings on IONP is capable of adsorbing specific proteins to modulate the complement activation and if this interaction would be translated to an inflammatory response. Through characterization of the human PC, we demonstrate that polyethyleneglycol (PEG) coating on IONP promoted a selective adsorption of complement recognition molecules, which would be responsible for the complement system activation. These observations will help to gain insight on how nanomedicines interact with the proteins of the immune system and to develop potential strategic interventions to modulate (inhibiting or stimulating) complement activation in vivo to increase therapeutic applications of IONP.
Materials and methods
Fe3O4 NP and physicochemical characterization
Three Fe3O4 NP were used in this study; bare Fe3O4 NP (IONP-bare) were a kind gift from Dr Jaime Santoyo (Physics Department, Cinvestav-IPN), and were synthesized as previously described.18 Polyvinylpyrrolidone- (PVP) and PEG-coated IONP were purchased from Sigma Aldrich (St Louis, MO, USA). IONP were examined by transmission electron microscope (TEM), JEM2010 (JEOL Ltd.) for particle morphology and size distribution in TEM mode and elemental mapping by energy-dispersive X-ray spectroscopy. Hydrodynamic particle size distributions were measured by centrifugal liquid sedimentation in a DC24000 system (CPS Instruments Inc.) A certified polyvinyl chloride particle calibration standard provided by the instrument supplier, was used to calibrate all measurements. Zeta potential of IONP was analyzed by Laser Doppler microelectrophoresis using the Zetasizer Nano ZS90 size analyzer (Malvern Instruments Ltd.). The endotoxin content was assessed by the endpoint chromogenic Limulus Amebocyte Lysate. The three IONP were negative for endotoxin contamination (<0.1 EU/mL).
PC formation and protein identification
IONP were incubated with pooled human plasma (2.5 mg/mL) at 37°C for 30 minutes. This temperature was chosen to emulate the physiological conditions in the bloodstream and to functionally evaluate complement in vitro.19 Unbound proteins were removed after centrifugation at 22,000× g for 30 minutes followed by three washing steps with PBS-EDTA. The resulting IONP–protein complex is considered as the “hard corona,” which consists of those proteins adsorbed on the NP surface for enough time to influence the NPs interactions with living system.20 Proteins were desorbed from IONP by incubation with LDS Sample Buffer (Novex) at 70°C for 10 minutes. Then, the total recovered protein of each IONP was separated by one-dimensional SDS-PAGE 4%–12% Bis–Tris polyacrylamide gels.21,22 The lanes were cut into fractions23 (Figure S1) and prepared for further analysis by reduction, alkylation, and tryptic digestion (trypsin-LysC, 37°C, overnight).24 The peptide extracts were analyzed on a nanoliquid chromatography system (Acquity UPLC, Waters), coupled by an electrospray ionization interface to a linear ion trap mass analyzer (LTQ values, Thermo). The raw files were converted to mzML HUPO standard archives using the ProteoWizard converter.25 The protein search was performed with Comet26 against a Uniprot fasta database for Homo sapiens, employing a target-decoy strategy.27 The resulting peptide and protein hits were validated using PeptideProphet/ProteinProphet,28,29 and converted to the final result files. The complete bioinformatic workflow was implemented in taverna (https://taverna.incubator.apache.org/),30 running on massypup64.30 The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/)31 via the PRoteomics IDEntifications (PRIDE) partner repository (http://www.ebi.ac.uk/pride/archive/)32 with the dataset identifier PXD004441. The complete list of identified proteins is listed in Table S1.
PC biological analysis using ClueGO
ClueGO, a Cytoscape plug-in (version 2.2.5), was employed to analyze, classify, and visualize the network of biological process related to each PC.33 Data were analyzed by hypergeometric test and P-value correction with the Bonferroni step down. Global Homo sapiens was used as a background annotation database and Gene Ontology terms were visualized as nodes linked base on their kappa score level (0.5).
Complement activation in human plasma
Blood plasma samples were obtained from 30 healthy volunteer donors by venipuncture of whole blood. Samples were pooled, aliquoted, and stored at −80°C until use. IONP were dispersed at 1 mg/mL in plasma and incubated for 30 minutes at 37°C. After that, cold EDTA was added (at 10 mM final concentration) to stop all the complement activation pathways. Protein–IONP complexes were separated by centrifugation at 15,000× g for 30 minutes at 4°C. After that, C3a, C4a, and C5a concentrations were determined in plasma samples with the Cytometric Bead Array Human Anaphylatoxin Kit (Cat No 561418, Becton Dickinson), which measures both C3a, C4a, and C5a, and their desArg forms (C3adesArg, C4adesArg, and C5adesArg). Informed consent was obtained in writing from each participant prior to inclusion in the study, which was performed in accordance with the Declaration of Helsinki and according to institutionalized bioethics code. The procedure followed for extracting plasma proteins from human blood is classified as research with minimal risk according to the current Regulation of the General Health Law in the Field of Health Research, Art 17, and does not require approval from an institutional research ethics committee.
In vivo complement activation
Male BALB/c mice (7–8 weeks old) were randomly assigned into four experimentation groups (five mice per group): 1) control (sterile 0.9% NaCl solution); 2) exposed to IONP-bare; 3) exposed to IONP-PVP; and 4) exposed to IONP-PEG. Then, mice were anesthetized under 3% isoflurane/oxygen mixture. Mice were intravenously injected by the lateral tail vein at a dose of 5 mg NP/kg body weight. The selected dose of 5 mg/kg of body weight is in agreement with a high dose achieved in Phase II clinical studies of IONP used for MR angiography.34,35 A pilot study with Zymosan A, a known activator of the complement system, was performed to determine a time point where a significant complement activation occurs. Based on our results, the order and timeline of the hypothesized biological events (complement activation – increment of cytokines), and previous literature,17,36 we chose 90 minutes as exposure time in order to evidence an increment of both the complement markers and cytokines. After exposure, mice were deeply anesthetized and euthanized by terminal exsanguination (intracardiac puncture). Liver, spleen, kidney, heart, and brain were extensively washed in saline solution and collected for Fe biodistribution analysis. Plasma concentrations of sC5a and sC5b-9 were determined by ELISA with commercial kits (Mouse Complement Component C5a assay kit Cat; DY2150, R&D Systems Inc., and Mouse Terminal Complement Complex C5b-9 Kit Cat; CSB-E08710m Cusa-bio Biotech Co. Ltd., respectively). Levels of TFN-α, IL-1β, and IL-6 were measured with a multiplexed cytokine assay (Mouse Magnetic Panel catalog no. LMC0001M, Novex; Life Technologies), following the manufacturer’s instructions. Quantification of Fe in the tissues was performed by particle-induced X-ray emission (PIXE) following a methodology previously described.37 PIXE measurements were validated with two standards from the International Atomic Energy Agency (IAEA 153 and IAEA 155). All animal experimental procedures were approved by the Institutional Committee for the Care and Use of Laboratory Animals at CINVESTAV-IPN that follows the regulations established by the Mexican Official Norm for the Use and Welfare of Laboratory Animals (NOM-062-ZOO-1999), which is in accordance with the Guide for the Care and Use of Laboratory Animals, USA.
Statistical analysis
Data analysis was performed using GraphPad Prism 6 (GraphPad Software Inc.). Normal distribution of the data was tested with Shaphiro–Wilk test. Comparison between experimentation groups was analyzed by one-way ANOVA followed by Dunnett’s post hoc test. Differences with P<0.05 were considered statistically significant. Data are presented as the mean ± standard error of the mean (SEM).
Results
Physicochemical characterization of IONP
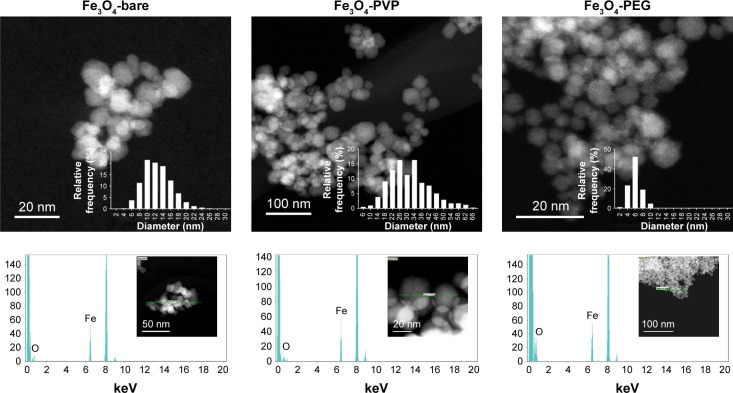
IONP were characterized as pristine material TEM (Figure 1). The three IONP nanoparticles showed a diameter range below the 100 nm and a semispherical shape. EDX analysis showed the absence of contamination with other elements rather than iron and oxygen. The IONP were characterized suspended in physiological sterile saline solution since this was the vehicle used for corona formation and complement activation assays. The hydrodynamic diameter measurements showed that three IONP tend to spread within two size populations, the smaller (few nanometers) corresponding to primary particles and a larger one corresponding to agglomerates of primary particles (Table 1). The percentages of primary particles that remain after IONP suspension were: 71.2% for IONP-bare, 83.3% for IONP-PVP, and 65.1% for IONP-PEG. Compared to IONP-bare and IONP-bare, the IONP-PVP particles suspended in saline media were less susceptible to form low-sized agglomerates from primary particles.
Figure 1.
Characterization of IONP by TEM.
Notes: Micrographs of three different IONP and their size distribution (upper panel). Elemental mapping by energy-dispersive X-ray spectroscopy (lower panel). Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone; TEM, transmission electron microscope.
Table 1.
Physicochemical characterization of IONP
| Particle | Primary size range (nm)a | Hydrodynamic size (nm)b (% of total population) | Zeta potential (mV)c | |
|---|---|---|---|---|
| Primary | Agglomerates | |||
| IONP-bare | 4.9–25.5 | 6.2±0.5 (71.21) | 75±14 (19.01) | −14.1±1.8 |
| IONP-PVP | 4.4–65.7 | 6.5±1.0 (83.36) | 75.6±10.8 (0.024) | −13.3±2.5 |
| IONP-PEG | 1.8–11.21 | 6.4±0.6 (65.1) | 31.0±2.80 (31.8) | −46.3±3.8 |
Notes:
Size distribution of pristine materials reported in Escamilla Rivera et al.69
Size distribution of IONP suspended in saline solution obtained by centrifugal liquid sedimentation (distribution based on particle number). Values are expressed as mean ± SD, and those in parenthesis represent the percentage from the total measured population.
Laser Doppler microelectrophoresis of IONP suspended in saline solution. Values are expressed as mean ± SD.
Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone.
Protein adsorption to IONP surface
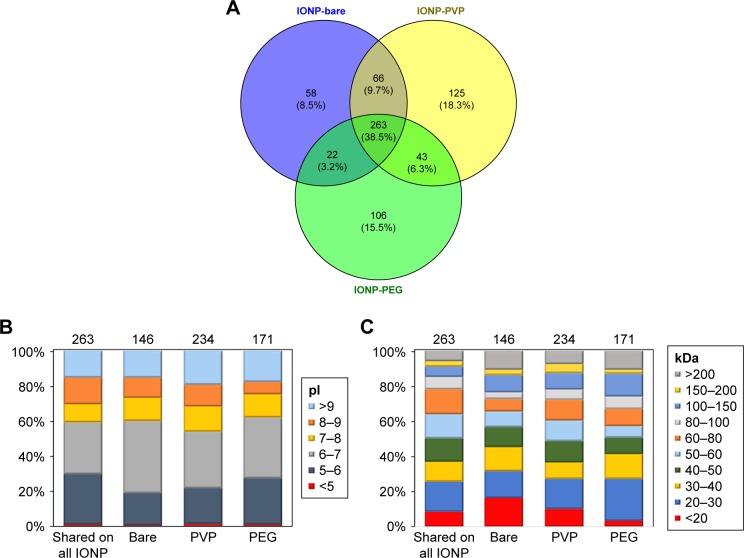
In order to identify the adsorbed proteins onto the three IONP and to determine how surface coatings influence the activation of the immune response, we obtained a pro-teomic profile of the PC that surrounds the three IONP (PC fingerprinting) using LC-MS/MS. A total of 263 shared proteins (present on all three IONP) were identified and represented 60.59% of IONP-PEG associated proteins, for, 64.30% for IONP-bare, and 52.9% for IONP-PVP. The set of non-shared proteins in the respective coronas contributed to the 8.5% of IONP-bare PC, 18.3% of the IONP-PVP PC, and 15.5% for the PC on IONP-PEG (58, 125, and 106 proteins respectively) (Figure 2A). In regard of physicochemical properties of the proteins such as the isoelectric point (pI) and molecular weight (MW), we observed slight differences between the three IONP (Figure 2B and C). For example, IONP-PEG adsorbed more proteins with a pI from 5 to 6, and fewer proteins with a pI from 8 to 9 compared to IONP-bare and IONP-PVP; IONP-PEG do not have a preference for proteins of a particular MW (Figure 2C). IONP-bare adsorbed more proteins with a pI from 6 to 7 and fewer proteins with a pI <5 than to the other two NP. Proteins with MW <20 kDa were more abundant in IONP-bare and proteins from 60 to 100 kDa were less adsorbed. While, IONP-PVP adsorbed more proteins with a pI >9 and high MW but in a lesser extent proteins of lower MW (<20 kDa) when compared to IONP-bare and IONP-PEG.
Figure 2.
Characteristics of recovered in the coronas of the three IONP.
Notes: Venn diagram showing the number of identified proteins and the relative percentage of each group to the total shared (A). Proteins were classified according to their isoelectric point (B) or molecular weight (C) and their relative percentages are shown.
Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone.
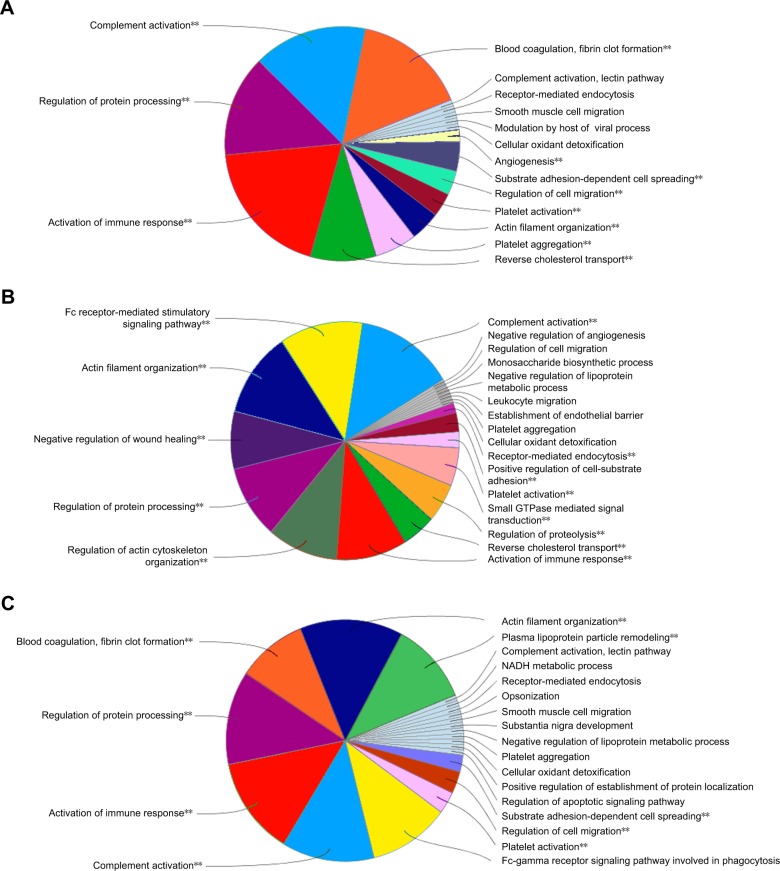
Regarding the identity and function of the adsorbed proteins, the biological processes network of associated proteins on IONP-bare have a role in blood coagulation, fibrin clot formation, angiogenesis, regulation of cell migration, substrate adhesion, and proteins related to the modulation by host of viral process (Figure 3A). On the other hand, IONP-PVP adsorbed proteins related to regulation of proteolysis, small GTPase, receptor-mediated endocytosis, Fc receptor-mediated stimulatory signaling pathway, and negative regulation of wound healing (Figure 3B).
Figure 3.
Biological processes network of associated proteins on IONP.
Notes: Gene ontology was used to group the identified proteins in the corresponding biological process using ClueGo analysis. The label of the most significant term per functional group is shown. IONP-bare (A), IONP-PVP (B), and IONP-PEG (C). **P<0.001 (P-value Corrected with Bonferroni step-down).
Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone.
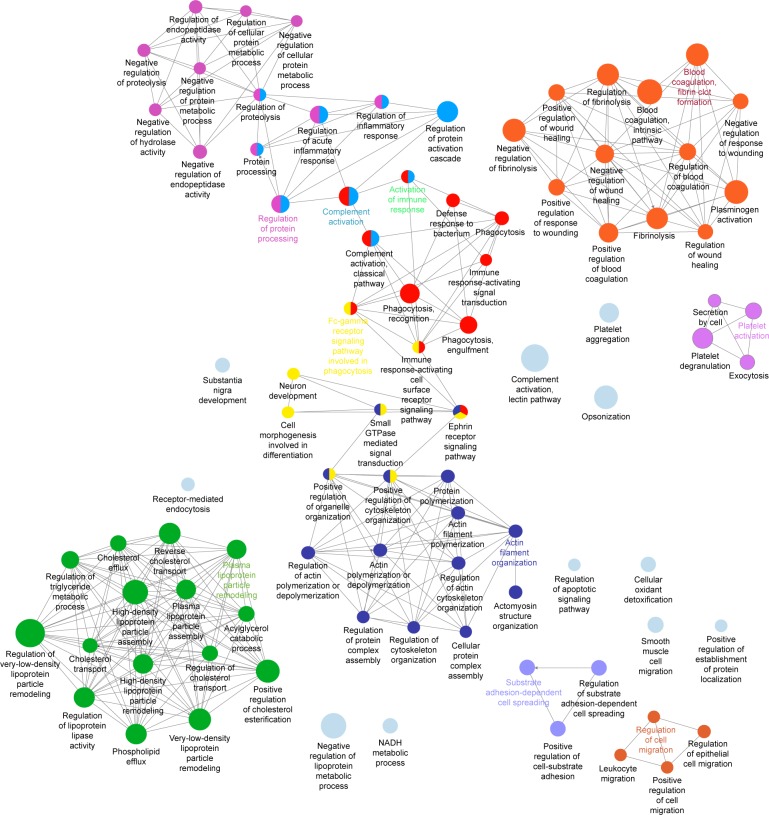
Whereas the corona of IONP-PEG showed that their proteins belong to the activation of the immune response, opsonization, lectin pathway of the complement activation, regulation of protein processing, platelet activation, actin filament organization, and the Fc signaling pathway involved in phagocytosis (Figure 3C). Although the three IONP-coronas share a set of proteins that participate in particular biological processes, there are proteins that were exclusive to each coating (Table S1). For example, IONP-PEG adsorbed proteins involved in plasma lipoprotein particle remodeling (Figure 4).
Figure 4.
Biological processes network of associated proteins on IONP-PEG PC.
Notes: Gene ontology terms are visualized as nodes linked base on their kappa score level (0.5) using ClueGO analysis. The label of the most significant term per functional group is shown in bold. The node size represents the percent of associated genes of proteins per node.
Abbreviations: IONP, iron oxide nanoparticles; PC, protein corona; PEG, polyethyleneglycol.
Complement activation in vitro
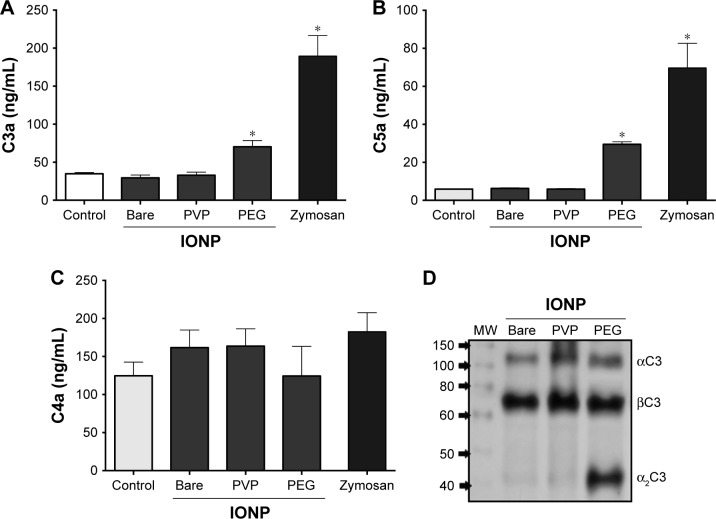
In order to test whether adsorption of complement proteins could translate into an activation of this system, human plasma was exposed to the three different IONP, and the levels of anaphylatoxins were measured. After 30 minutes, samples exposed to IONP-PEG showed a significant 2-fold increment for C3a and 4.8-fold in C5a concentration (Figure 5A and B). However, no evidence of change was observed for C4a (Figure 5C). In contrast, plasma exposed to IONP-PVP and IONP-bare did not induce any significant change in C3a, C4a, or C5a compared to control samples (Figure 5). Figure 5D shows a representative immunoblot of C3a complement component adsorbed in PC of IONP, where the increment for IONP-PEG can be observed. As previously mentioned, complement activation pathways possess common downstream points, the excision of C4 into C4b (which will form the C3 convertase with C2a) and C4a is a common endpoint of the classical and lectin pathways. Consequently, the production of C3a and C5a with the absence of C4a in the human plasma exposed to IONP-PEG hints toward a complement activation that can be predominant through the alternative pathway.
Figure 5.
IONP-PEG activates the complement system and increases the anaphylatoxin concentration in human blood plasma.
Notes: Quantification of complement activation products by ELISA: levels of anaphylatoxin C3a (A), terminal complement complex, C5a (B), and terminal complement complex, C4a (C) in human plasma. Each bar represents the mean ± SEM of three independent experiments. *Statistically significant difference vs control, *P<0.05 ANOVA post hoc Dunnett’s test. Representative immunoblot of C3 complement component adsorbed in protein corona of IONP (D).
Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone; SEM, standard error of the mean.
Complement activation and induction of proinflammatory response in BALB/c mice exposed to IONP
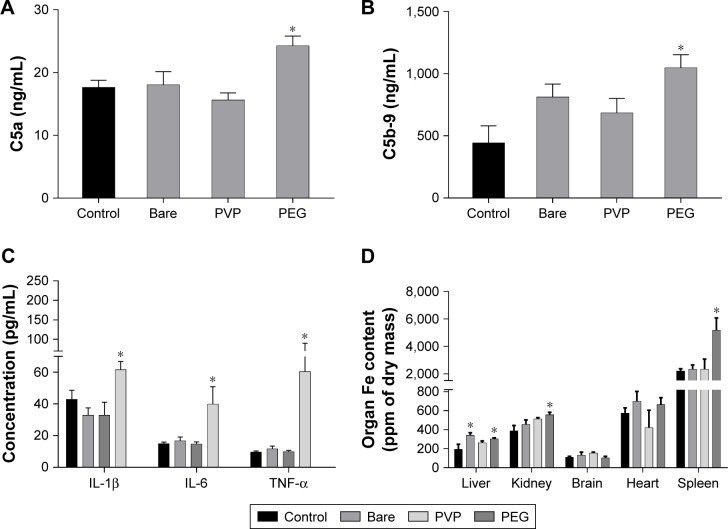
Male BALB/c mice were administered with IONP to get an insight of the capabilities of IONP to activate the complement system and induce a proinflammatory response in vivo. After exposure, the plasma concentration of anaphylatoxin C5a in IONP-bare and IONP-PVP treated animals remained without changes compared to the control group (Figure 6A). In contrast, IONP-PEG induced a 1.3-fold increment of C5a in plasma of exposed mice. Quantification of sC5b-9 showed that IONP-bare and IONP-PVP did not induce a significant increment (P>0.05) in the animals exposed. However, a 2.3-fold increment was observed in the group exposed to IONP-PEG (Figure 6B). The proinflammatory profile of cytokines showed that mice exposed to IONP- bare and IONP-PVP did not exhibit significant increments in IL-1β, TNF-α or IL-6 concentrations vs the control group (Figure 6C). While the group exposed to IONP-PEG showed a 1.60-fold increment of IL-1β, 5.7-fold in TNF-α and 2.6-fold of IL-6 compared to the control group. PIXE measurements revealed that animals exposed to IONP-PEG accumulate higher concentrations of NP in the liver, spleen, and kidney (2.3-fold, 1.57-fold, and 1.43-fold, respectively) compared to control (Figure 6D). IONP-bare induced an increment in the iron content of the liver (1.77-fold) compared to control group. IONP-PVP did not induce a significant change (compared to control) of iron concentration in the selected organs (P>0.05). Given that in such short exposure times IONP dissolution is negligible and the PIXE technique allows the quantification of iron in whole organs,37,38 the retention of initial dose was calculated taking into account the weight of the organs, the concentration of iron in the control, and the IONP dose administered to each animal. These estimations showed that IONP-PEG are retained in larger amounts in the selected organs with a retention of 86% of the initial dose, followed by IONP-bare with 55.9% and IONP-PVP with 33% of the administered dose.
Figure 6.
IONP-PEG activates the complement system and induces a proinflammatory response in mice.
Notes: Quantification of complement activation products by ELISA: levels of anaphylatoxin C5a (A) and terminal complement complex, C5b-9 (B), in plasma of BALB/c mice. Levels of circulating proinflammatory cytokines (C). Biodistribution of IONP in mice exposed (D). Each bar represents the mean ± SEM of n=5 (*P<0.05) ANOVA post hoc Dunnett’s test.
Abbreviations: IONP, iron oxide nanoparticles; PEG, polyethyleneglycol; PVP, polyvinylpyrrolidone; SEM, standard error of the mean.
Discussion
In this study, we investigated whether the interaction of plasma proteins with different coatings on IONP is capable of triggering an adverse effect, namely the proinflammatory response in an in vivo and in vitro model. Despite the fact that PEG grafting is considered as a highly hydrophilic coating that can prevent nonspecific protein adsorption, in the last decade, several reports have shown that PEG and PEGylated nanoformulations can elicit complement activation, which is in agreement with the observed results in this study. For instance, Hamad et al39 demonstrated that PEG molecules of increasing MW were capable of binding complement proteins, enhancing the alternative pathway turnover, and probably to bind directly to L-lectins to further activate the lectin pathway. Moreover, Yang et al40 observed that the grafting of polymeric NP with different PEG sizes reduced protein binding but was not able to prevent effective complement activation and complement-mediated phagocytosis in human and mouse macrophages. Activation of the complement system by PEG coating on IONP could be achieved by direct or indirect mechanisms such as the specific recognition by anti-PEG IgG2 and IgM specific antibodies that could initiate the classical pathway.41 Also, PEGylated nanostructures can interact directly with complement proteins; particularly, if the C3 component is trapped inside the hydrated structure of PEG, the conformational changes of C3 and its spontaneous hydrolysis could be accelerated leading to the assembly of the fluid-phase C3 convertase C3bB.42 Moreover, Szebeni et al43 have proposed that the exposed hydroxyl groups at the end of PEG structures could act as molecular anchors for C3b and thus initiate the alternative pathway.
Recent studies focused on magnetic NP have reported that size, charge, surface, polymer conformation, and molecular structure influences protein adsorption. For example, Hu et al44 reported an effect of the primary size on the composition and abundance of proteins on pristine IONP; at smaller sizes (<30 nm) NP adsorbed fewer proteins from FBS and displayed different composition comparing to IONP of 200 and 400 nm. In contrast, in this study, we did not observe a relation between size and the amount or identity of proteins adsorbed. In regard to the surface charge, Sakulkhu et al45 observed that positively charged IONP coated with polyvinyl alcohol or dextran adsorbed more proteins from FBS rather than ones with a negative charge. In contrast, IONP used in this study had a negative charge and the most abundant proteins in physiological conditions at pH 7.4 are negative; in particular, IONP-PEG adsorbed more proteins with a pI from 5 to 6 and their zeta potential was the highest negative −46.3 mV compared to the other two IONP. Moreover, Hofmann46 proposed that protein positive-charge domains could mediate these interactions, protein–particle, protein–protein interactions as well as protein conformational change and denaturation. These results demonstrated that PC depends on the entire physicochemical characteristics of IONP and suggest that electrostatic force is not the only factor that can modulate protein adsorption on IONP.
The PC profiles were associated with several biological processes; this protein fingerprinting gives an insight into responses that potentially could be triggered by IONP such as blood coagulation, complement activation, regulation of protein processing, lipid metabolism, and cytoskeleton organization. For example, IONP-bare adsorbed ex situ proteins related to blood coagulation and angiogen-esis. It has been reported that IONP adsorbed coagulation factor VII and fibrinogen,45 which can activate the kallikrein system. In this regard, Simberg et al47 demonstrated that amine-modified IONP adsorb plasma kallikrein and high molecular weight kininogen and induce thrombosis and activation of the kallikrein–kinin system in vivo. On the other hand, IONP-PVP adsorbed proteins related to regulation of proteolysis (associated with blood coagulation) and the Fc receptor-mediated stimulatory signaling pathway, which could potentially modify the risk of intravascular coagulation or incidents of vascular thrombosis.48 Moreover, nanomedicines could exacerbate some pathologies by impairment or depletion of plasma proteins. For example, the adsorption of lipoproteins is related to hypercholesterolemia and a higher risk of atherosclerosis. Muller et al49 observed that lipoproteins that are adsorbed to polystyrene NP disintegrate. Also, proteins adsorbed onto redox-active NP can undergo denaturation by oxidation,50 which can induce systemic effects such as thrombosis.51,52 It is noteworthy that some of the unique proteins associated to IONP-PEG corona have a critical role in the complement activation, immune response, and coagulation systems. For example, we identified MBL2, L-ficolin, collectin-liver 1, galectin 3, SKAP2, and protein G6b, PDGF, Annexin V, and collagen alpha chain (Table S1). Particularly, the first three proteins are implicated in complement activation. MBL belongs to the collectin family and is an element in the innate immune system capable of activating the lectin complement pathway.53 Also, MBL binds to late apoptotic cells and necrotic cells facilitating their uptake by macrophages.54 L-ficolin is a pattern recognition molecule that specifically binds to mannan, LPS, 1,3-β-glucans, and lipoteichoic acids.55 Meanwhile, collectin-liver 1 has been recently recognized as a pattern recognition molecule that could interact with carbohydrates through its specific recognition domain and it’s found associated in circulation with other lectins.56 Both MBL2 and L-ficolin are part of the few described activators of the lectin pathway, associated to MASP. The resulting complex activates MASP, which cleaves C2 and C4 to form the C3b convertase.57
Altogether, MBL2, L-ficolin, and collectin-liver 1 evidenced that IONP-PEG induced a selective adsorption of complement recognition molecules, which would be the responsible for the complement system activation. This association of recognition molecules is in agreement with previous observations with polyethyleneoxide polymeric NP,58 soluble PEG,39 PEGylated CNT,59 and iron oxide nanoworms.60 Although the lectin pathway is a strong candidate to explain the route for complement activation, it has been reported that PEG and PEGylated CNT can also increase the C3 turnover leading to an amplification loop in the alternative pathway.39,59 In addition to complement activation proteins and immunoglobulins, we identified proteins implicated in the activation of immune system cells that could explain the proinflammatory responses at other levels. Such is the case of galectin, a protein that activates the inflammatory response in immune cells, promotes the adhesion of neutrophils, and promotes the phagocytosis in macrophages.61 It is clear that after proteomic analysis for fingerprinting PC on nanomedicines, a next logical step is to identify whether the components of the PC may have a causal involvement in physiological disorders, a tool that could be used for both personalized diagnostics and therapeutic treatments.62
Although differences exist between mouse and human immune system, the complement system is highly conserved among these species.63 The main differences in these two species are present in the proteins that modulate the amplification cascade rather than in proteins that participate in the triggering of the proteolytic pathways.64 In agreement with our results, Banda et al60 demonstrated that iron oxide nanoworms coated with dextran induced complement activation both in human and mouse. However, they found that the alternative complement pathway is the predominant pathway in humans, whereas in mice the main activation occurs through the lectin pathway. It has been demonstrated that interspecies differences in protein-binding profiles could exist for some NP; however, adsorption of complement protein appears to be a unifying factor among PCs and the particular differences in the magnitude of the complement activation may be due to the amount of recruited complement triggering proteins.22 The agreement between the observations in mice and humans opens the possibility to include systematic evaluations of complement activation by nanomedicines as an integral part of preclinical studies to use this parameter as a predictor of biocompatibility and bioavailability.
Our biodistribution findings are in agreement with previous reports, wherein IONP have been observed accumulated in the liver and spleen of exposed animals. For example Xue et al65 found that IONP grafted with different PEG MW intravenously administered in mice are rapidly cleared of the circulation (t½ 15–27 minutes) and accumulated predominantly in the liver and spleen. The authors confirmed that in these organs, IONP did not induce visible necrosis or inflammatory infiltration. Moreover, Huang et al66 observed that regardless of the size of PVP coating, IONP are significantly accumulated in the liver and spleen after 1 hour. It is possible that after internalization in the organs, IONP could exert an inflammatory effect through the induction of necrosis, increase of reactive species of oxygen, and lysosomal or mitochondrial damage as previously described in vitro. Due to our experimental approach, it is not possible to exclude some contribution of the cytokines originated in the organs. However, we postulate that given our exposure time (90 minutes), it is probable that the proinflammatory effect was generated as a response to the anaphylatoxins (the soluble mediators of complement activation) generated upon the first contact between IONP-PEG and plasma, which is in agreement with previous results.67 The anaphylatoxin C5a is the most potent proinflammatory mediator released upon complement activation, it has greater potency to induce histamine release compared to C3a, and it acts as strong chemoattract that activates and guides neutrophils, monocytes, and macrophages to the complement activation.68 Additionally, C3a and C5a trigger the proinflammatory response through their corresponding G-protein-coupled receptors (C3aR and C5aR, respectively) causing the release of proinflammatory cytokines such as IL-1β, TNF-α, and IL-6 in monocytes, macrophages, basophils, and neutrophils.11 In addition, interaction with their receptors stimulates oxidative metabolism in neutrophils, the production of ROS, and the release of lysosomal enzymes from various phagocytic cells.
From our results, it can be suggested that in order to predict systemic effects associated to the nanomedicines, it is not enough to know only the identity of proteins that are forming the PC but it is also becoming necessary to develop experimental approaches capable of considering the complex interactions between biomolecules, immune cells, and nanomedicines.
Conclusion
Our results suggest that the elicited biological effects from the interaction with IONP are not only associated with their pristine properties but also to the identity of the protein–IONP complexes. It is highly probable that specific polymeric determinants, projected in the PEGylated structure of IONP-PEG, are recognized as a pathogen associated pattern and promote their recognition by complement proteins. Additionally, we have shown that complement activation measurements in humans and mice are in agreement and could be used as an integral part of preclinical studies to use this parameter as a predictor of biocompatibility and bioavailability. The engineering of nanoparticles, which takes into account the association between complement proteins and nanomedicines, will reduce the immunostimulatory effects through a selective adsorption of plasma proteins and will enable a safer application of IONP in human therapy.
Acknowledgments
The authors would like to thank Dr Jaime Santoyo-Salazar and M.Sc. Roberto Mejía-Olvera, (Physics Department and Nanosciences and Nanotechnology Graduate program in Cinvestav-IPN), for the supply of the IONP-bare. RW was funded by the CONACyT Fronteras project 2015-2/814, the bilateral grant CONACyT-DFG 2016/277850 and PlanTECC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sharma VK, Alipour A, Soran-Erdem Z, Aykut ZG, Demir HV. Highly monodisperse low-magnetization magnetite nanocubes as simultaneous T(1)–T(2) MRI contrast agents. Nanoscale. 2015;7(23):10519–10526. doi: 10.1039/c5nr00752f. [DOI] [PubMed] [Google Scholar]
- 2.Kossatz S, Grandke J, Couleaud P, et al. Efficient treatment of breast cancer xenografts with multifunctionalized iron oxide nanoparticles combining magnetic hyperthermia and anti-cancer drug delivery. Breast Cancer Res. 2015;17(1):1–17. doi: 10.1186/s13058-015-0576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi D, Sadat ME, Dunn AW, Mast DB. Photo-fluorescent and magnetic properties of iron oxide nanoparticles for biomedical applications. Nanoscale. 2015;7(18):8209–8232. doi: 10.1039/c5nr01538c. [DOI] [PubMed] [Google Scholar]
- 4.Anselmo AC, Mitragotri S. A review of clinical translation of inorganic nanoparticles. AAPS J. 2015;17(5):1041–1054. doi: 10.1208/s12248-015-9780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YXJ. Superparamagnetic iron oxide based MRI contrast agents: current status of clinical application. Quant Imaging Med Surg. 2011;1(1):35–40. doi: 10.3978/j.issn.2223-4292.2011.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monopoli MP, Walczyk D, Campbell A, et al. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc. 2011;133(8):2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 7.Kim JA, Salvati A, Åberg C, Dawson KA. Suppression of nanoparticle cytotoxicity approaching in vivo serum concentrations: limitations of in vitro testing for nanosafety. Nanoscale. 2014;6(23):14180–14184. doi: 10.1039/c4nr04970e. [DOI] [PubMed] [Google Scholar]
- 8.Walkey CD, Chan WCW. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 9.Harms C, Datwyler AL, Wiekhorst F, et al. Certain types of iron oxide nanoparticles are not suited to passively target inflammatory cells that infiltrate the brain in response to stroke. J Cereb Blood Flow Metab. 2013;33(5):e1–e9. doi: 10.1038/jcbfm.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inturi S, Wang G, Chen F, et al. Modulatory role of surface coating of superparamagnetic iron oxide Nanoworms in complement opsonization and leukocyte uptake. ACS Nano. 2015;9(11):10758–10768. doi: 10.1021/acsnano.5b05061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moghimi SM, Andersen AJ, Ahmadvand D, Wibroe PP, Andresen TL, Hunter AC. Material properties in complement activation. Adv Drug Deliv Rev. 2011;63(12):1000–1007. doi: 10.1016/j.addr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Szebeni J. Complement activation-related pseudoallergy: a stress reaction in blood triggered by nanomedicines and biologicals. Mol Immunol. 2014;61(2):163–173. doi: 10.1016/j.molimm.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 14.Szebeni J, Baranyi L, Savay S, et al. Role of complement activation in hypersensitivity reactions to doxil and hynic PEG liposomes: experimental and clinical studies. J Liposome Res. 2002;12(1–2):165–172. doi: 10.1081/lpr-120004790. [DOI] [PubMed] [Google Scholar]
- 15.Salvador-Morales C, Flahaut E, Sim E, Sloan J, Green MLH, Sim RB. Complement activation and protein adsorption by carbon nanotubes. Mol Immunol. 2006;43(3):193–201. doi: 10.1016/j.molimm.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Allard-Vannier E, Cohen-Jonathan S, Gautier J, et al. PEGylated magnetic nanocarriers for doxorubicin delivery: a quantitative determination of stealthiness in vitro and in vivo. Eur J Pharm Biopharm. 2012;81(3):498–505. doi: 10.1016/j.ejpb.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Chanan-Khan A, Szebeni J, Savay S, et al. Complement activation following first exposure to pegylated liposomal doxorubicin (Doxil®): possible role in hypersensitivity reactions. Ann Oncol. 2003;14(9):1430–1437. doi: 10.1093/annonc/mdg374. [DOI] [PubMed] [Google Scholar]
- 18.Santoyo Salazar J, Perez L, de Abril O, et al. Magnetic iron oxide nanoparticles in 10−40 nm range: composition in terms of magnetite/maghemite ratio and effect on the magnetic properties. Chem Mater. 2011;23(6):1379–1386. [Google Scholar]
- 19.Mahmoudi M, Abdelmonem AM, Behzadi S, et al. Temperature: the “ignored” factor at the NanoBio interface. ACS Nano. 2013;7(8):6555–6562. doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- 20.Monopoli MP, Pitek AS, Lynch I, Dawson KA. In: Formation and Characterization of the Nanoparticle–Protein Corona BT – Nanomaterial Interfaces in Biology: Methods and Protocols. Bergese P, Hamad-Schifferli K, editors. Totowa, NJ: Humana Press; 2013. pp. 137–155. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Solorio-Rodríguez A, Escamilla-Rivera V, Uribe-Ramírez M, et al. A comparison of the human and mouse protein corona profiles of functionalized SiO2 nanocarriers. Nanoscale. 2017;9(36):13651–13660. doi: 10.1039/c7nr04685e. [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 30.Winkler R. An evolving computational platform for biological mass spectrometry: workflows, statistics and data mining with MASSyPup64. PeerJ. 2015;3(11):e1401. doi: 10.7717/peerj.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vizcaíno JA, Deutsch EW, Wang R, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcaíno JA, Csordas A, del-Toro N, et al. 2016 update of the pride database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinforma. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leiner T, Ho KYJAM, Ho VB, et al. Multicenter phase-II trial of safety and efficacy of NC100150 for steady-state contrast-enhanced peripheral magnetic resonance angiography. Eur Radiol. 2003;13(7):1620–1627. doi: 10.1007/s00330-002-1791-6. [DOI] [PubMed] [Google Scholar]
- 35.Weinstein JS, Varallyay CG, Dosa E, et al. Superparamagnetic iron oxide nanoparticles: diagnostic magnetic resonance imaging and potential therapeutic applications in neurooncology and central nervous system inflammatory pathologies, a review. J Cereb Blood Flow Metab. 2010;30(1):15–35. doi: 10.1038/jcbfm.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Kolk LE, Grillo-López AJ, Baars JW, Hack CE, van Oers MH. Complement activation plays a key role in the side-effects of rituximab treatment. Br J Haematol. 2001;115(4):807–811. doi: 10.1046/j.1365-2141.2001.03166.x. [DOI] [PubMed] [Google Scholar]
- 37.Lozano O, Laloy J, Alpan L, et al. Effects of sic nanoparticles orally administered in a rat model: biodistribution, toxicity and elemental composition changes in feces and organs. Toxicol Appl Pharmacol. 2012;264(2):232–245. doi: 10.1016/j.taap.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Lozano O, Mejia J, Masereel B, Toussaint O, Lison D, Lucas S. Development of a PixE analysis method for the determination of the biopersistence of sic and tic nanoparticles in rat lungs. Nanotoxicology. 2012;6(3):263–271. doi: 10.3109/17435390.2011.572301. [DOI] [PubMed] [Google Scholar]
- 39.Hamad I, Hunter AC, Szebeni J, Moghimi SM, Poly MSM. Poly (ethylene glycol)s generate complement activation products in human serum through increased alternative pathway turnover and a MASP-2-dependent process. Mol Immunol. 2008;46(2):225–232. doi: 10.1016/j.molimm.2008.08.276. [DOI] [PubMed] [Google Scholar]
- 40.Yang A, Liu W, Li Z, Jiang L, Xu H, Yang X. Influence of polyethyleneglycol modification on phagocytic uptake of polymeric nanoparticles mediated by immunoglobulin G and complement activation. J Nanosci Nanotechnol. 2010;10(1):622–628. doi: 10.1166/jnn.2010.1738. [DOI] [PubMed] [Google Scholar]
- 41.Garay RP, El-Gewely R, Armstrong JK, Garratty G, Richette P. Antibodies against polyethylene glycol in healthy subjects and in patients treated with PEG-conjugated agents. Expert Opin Drug Deliv. 2012;9(11):1319–1323. doi: 10.1517/17425247.2012.720969. [DOI] [PubMed] [Google Scholar]
- 42.Moghimi SM, Andersen AJ, Hashemi SH, et al. Complement activation cascade triggered by PEG-PL engineered nanomedicines and carbon nanotubes: the challenges ahead. J Control Release. 2010;146(2):175–181. doi: 10.1016/j.jconrel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Szebeni J, Muggia FM, Alving CR. Complement activation by Cremophor EL as a possible contributor to hypersensitivity to paclitaxel: an in vitro study. J Natl Cancer Inst. 1998;90(4):300–306. doi: 10.1093/jnci/90.4.300. [DOI] [PubMed] [Google Scholar]
- 44.Hu Z, Zhang H, Zhang Y, Wu Ren’an, Zou H. Nanoparticle size matters in the formation of plasma protein coronas on Fe3O4 nanoparticles. Colloids Surf B Biointerfaces. 2014;121:354–361. doi: 10.1016/j.colsurfb.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 45.Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein corona composition of superparamagnetic iron oxide nanoparticles with various physicochemical properties and coatings. Sci Rep. 2015;4(1):5020. doi: 10.1038/srep05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakulkhu U, Mahmoudi M, Maurizi L, et al. Significance of surface charge and shell material of superparamagnetic iron oxide nanoparticle (SPION) based core/shell nanoparticles on the composition of the protein corona. Biomater Sci. 2015;3(2):265–278. doi: 10.1039/c4bm00264d. [DOI] [PubMed] [Google Scholar]
- 47.Simberg D, Zhang W-M, Merkulov S, et al. Contact activation of kallikreinkinin system by superparamagnetic iron oxide nanoparticles in vitro and in vivo. J Control Release. 2009;140(3):301–305. doi: 10.1016/j.jconrel.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemmar A, Al-Salam S, Beegam S, Yuvaraju P, Ali BH. The acute pulmonary and thrombotic effects of cerium oxide nanoparticles after intratracheal instillation in mice. Int J Nanomedicine. 2017;12:2913–2922. doi: 10.2147/IJN.S127180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller J, Prozeller D, Ghazaryan A, et al. Beyond the protein corona – lipids matter for biological response of nanocarriers. Acta Biomater. 2018;71:420–431. doi: 10.1016/j.actbio.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 50.Jayaram DT, Runa S, Kemp ML, Payne CK. Nanoparticle-induced oxidation of corona proteins initiates an oxidative stress response in cells. Nanoscale. 2017;9(22):7595–7601. doi: 10.1039/c6nr09500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat Nanotechnol. 2011;6(1):39–44. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Cui C, Li R, et al. Study on the oxidation of fibrinogen using Fe3O4 magnetic nanoparticles and its influence to the formation of fibrin. J Inorg Biochem. 2018;189:58–68. doi: 10.1016/j.jinorgbio.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Matsushita M, Endo Y, Fujita T. Cutting edge: complement-activating complex of ficolin and mannose-binding lectin-associated serine protease. J Immunol. 2000;164(5):2281–2284. doi: 10.4049/jimmunol.164.5.2281. [DOI] [PubMed] [Google Scholar]
- 54.Petersen SV, Thiel S, Jensenius JC. The mannan-binding lectin pathway of complement activation: biology and disease association. Mol Immunol. 2001;38(2–3):133–149. doi: 10.1016/s0161-5890(01)00038-4. [DOI] [PubMed] [Google Scholar]
- 55.Kilpatrick DC, Chalmers JD. Human L-ficolin (ficolin-2) and its clinical significance. J Biomed Biotechnol. 2012;2012(33):1–10. doi: 10.1155/2012/138797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohtani K, Suzuki Y, Wakamiya N. Biological functions of the novel collectins CL-L1, CL-K1, and CL-P1. J Biomed Biotechnol. 2012;2012(3):1–8. doi: 10.1155/2012/493945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujita T. Evolution of the lectin-complement pathway and its role in innate immunity. Nat Rev Immunol. 2002;2(5):346–353. doi: 10.1038/nri800. [DOI] [PubMed] [Google Scholar]
- 58.Hamad I, Al-Hanbali O, Hunter AC, et al. Distinct polymer architecture mediates switching of complement activation pathways at the nanosphere-serum interface: implications for stealth nanoparticle engineering. ACS Nano. 2010;4(11):6629–6638. doi: 10.1021/nn101990a. [DOI] [PubMed] [Google Scholar]
- 59.Hamad I, Christy Hunter A, Rutt KJ, Liu Z, Dai H, Moein Moghimi S. Complement activation by PEGylated single-walled carbon nanotubes is independent of C1q and alternative pathway turnover. Mol Immunol. 2008;45(14):3797–3803. doi: 10.1016/j.molimm.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banda NK, Mehta G, Chao Y, et al. Mechanisms of complement activation by dextran-coated superparamagnetic iron oxide (SPIO) nanoworms in mouse versus human serum. Part Fibre Toxicol. 2014;11(1):1–10. doi: 10.1186/s12989-014-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu FT, Yang RY, Hsu DK. Galectins in acute and chronic inflammation. Ann N Y Acad Sci. 2012;1253(1):80–91. doi: 10.1111/j.1749-6632.2011.06386.x. [DOI] [PubMed] [Google Scholar]
- 62.Hadjidemetriou M, Kostarelos K. Nanomedicine: evolution of the nanoparticle corona. Nat Nanotechnol. 2017;12(4):288–290. doi: 10.1038/nnano.2017.61. [DOI] [PubMed] [Google Scholar]
- 63.Holers VM, Kinoshita T, Molina H. The evolution of mouse and human complement C3-binding proteins: divergence of form but conservation of function. Immunol Today. 1992;13(6):231–236. doi: 10.1016/0167-5699(92)90160-9. [DOI] [PubMed] [Google Scholar]
- 64.Pouw RB, Vredevoogd DW, Kuijpers TW, Wouters D. Of mice and men: the factor H protein family and complement regulation. Mol Immunol. 2015;67(1):12–20. doi: 10.1016/j.molimm.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Xue W, Liu Y, Zhang N, et al. Effects of core size and PEG coating layer of iron oxide nanoparticles on the distribution and metabolism in mice. Int J Nanomedicine. 2018;13:5719–5731. doi: 10.2147/IJN.S165451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Bu L, Xie J, et al. Effects of nanoparticle size on cellular uptake and liver MRI with polyvinylpyrrolidone-coated iron oxide nanoparticles. ACS Nano. 2010;4(12):7151–7160. doi: 10.1021/nn101643u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolf-Grosse S, Rokstad AM, Ali S, et al. Iron oxide nanoparticles induce cytokine secretion in a complement-dependent manner in a human whole blood model. Int J Nanomedicine. 2017;12:3927–3940. doi: 10.2147/IJN.S136453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bosmann M, Ward PA. Role of C3, C5 and anaphylatoxin receptors in acute lung injury and in sepsis. Adv Exp Med Biol. 2012;946:147–159. doi: 10.1007/978-1-4614-0106-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Escamilla-Rivera V, Uribe-Ramírez M, González-Pozos S, Lozano O, Lucas S, De Vizcaya-Ruiz A. Protein corona acts as a protective shield against Fe3O4-PEG inflammation and ROS-induced toxicity in human macrophages. Toxicol Lett. 2016;240(1):172–184. doi: 10.1016/j.toxlet.2015.10.018. [DOI] [PubMed] [Google Scholar]