 Target proteins and mechanism of a HeLa-specific cytotoxic compound were investigated by two thermal shift-based label-free proteomics methods.
Target proteins and mechanism of a HeLa-specific cytotoxic compound were investigated by two thermal shift-based label-free proteomics methods.
Abstract
Phenotypic screening can not only identify promising first-in-class drug candidates, but can also reveal potential therapeutic targets or neomorphic functions of known proteins. In this study, we identified target proteins of SB2001, a cytotoxic agent that acts specifically against HeLa human cervical cancer cells. Because SB2001 lacks chemical modification sites, label-free target identification methods including thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis (TS-FITGE) and thermal proteome profiling (TPP) were applied to characterize its mechanism of action. Owing to their differences, the two label-free target identification methods uncovered complementary target candidates. Candidates from both methods were prioritized according to their selective lethality upon the knockdown of those genes in HeLa cells, compared to CaSki cells which were used as a negative control cell line from the human cervix. LTA4H was identified only by TS-FITGE, but not by TPP, because only one isoform was stabilized by SB2001. Furthermore, it was implied that a non-canonical function of LTA4H was involved in the SB2001 activity. MTH1 was identified by both TS-FITGE and TPP, and SB2001 inhibited the function of MTH1 in hydrolyzing oxidized nucleotides. Compared to CaSki cells, HeLa cells displayed downregulated DNA mismatch repair pathways, which made HeLa cells more susceptible to the oxidative stress caused by SB2001, resulting in increased 8-oxoG concentrations, DNA damage, and subsequent cell death.
Introduction
Phenotype-based chemical screening has contributed significantly to the discovery of first-in-class molecular entities with novel mechanisms of action.1 Phenotypic screening is an empirical and discovery-driven approach for identifying new bioactive compounds that modulate a specific cellular outcome of interest, rather than exploring a particular hypothesis-driven molecular target.2 The unbiased nature of phenotypic screening allows the discovery of novel proteins with therapeutic potential, disease-relevant pathways, unrevealed functions of proteins, or polypharmacology with multiple targets.3–7 Therefore, the identification of target proteins that bind to the bioactive compound is a crucial and decisive component of the phenotype-based drug discovery process.8
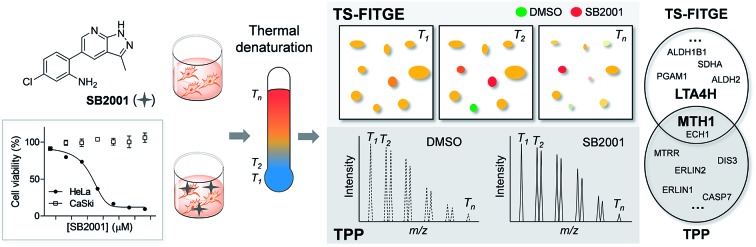
Conventional chemical proteomics methods for affinity-based target identification require functional chemical handles to immobilize the bioactive compound on a solid support.8 It is essential to know the structure–activity relationship (SAR) of bioactive compounds and their synthetic accessibility for the preparation of probe molecules; this has been a major obstacle in identifying the targets of bioactive natural compounds and synthetic compounds without functional handles.9 Therefore, label-free target identification is an important technique for the compounds with tight SARs. Cellular thermal shift assay (CETSA) was the first reported label-free method for verifying the physical engagement of bioactive compounds with target proteins in live cells.10 CETSA is based on the principle that the thermal denaturation curve of a particular protein may shift upon the binding of a bioactive compound. To expand the CETSA principle for an unbiased proteome-wide target identification method, thermal proteome profiling (TPP)11 and thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis (TS-FITGE)12 were developed (Fig. 1). TPP uses quantitative mass spectrometry with isobaric mass reporter-tagged peptides, whereas TS-FITGE utilizes a 2-dimensional (2D) gel electrophoresis with different fluorescence-tagged proteins. Although it has been proven that the thermal stability shift can identify target proteins in live cells, its applications remain limited.11–16
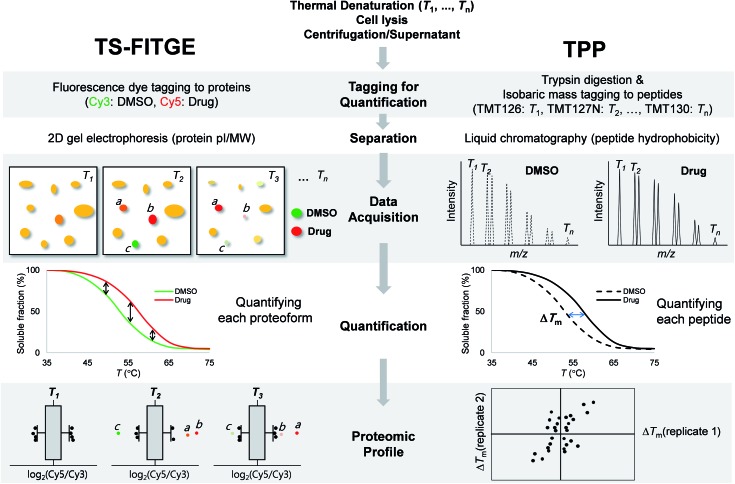
Fig. 1. Workflow and characteristics of TS-FITGE and TPP. In both TS-FITGE and TPP, cells were treated with either DMSO or drug, and heated to various temperatures. After cell lysis, the remaining proteins in the soluble fraction were collected. TS-FITGE: soluble proteins were conjugated with fluorescence dyes (Cy3 for the DMSO-treated group and Cy5 for the drug-treated group) and then pooled, followed by separation on a 2D gel. The Cy5 to Cy3 fluorescence ratio for each proteoform was quantified. The distribution of the ratio was plotted on a box plot to select outliers as hits with significant thermal stability shifts. TPP: soluble proteins were digested with trypsin into peptides, which were conjugated with isobaric mass tags (a different tag was used for each temperature). The resulting peptides were pooled and analyzed by liquid chromatography-tandem mass spectrometry. The reporter ions of each peptide were quantified, and the melting temperatures (Tm) of the corresponding proteins were calculated. Proteins with significant Tm changes between the DMSO- and drug-treated groups were chosen as the target candidates.
In this study, we used phenotypic screening to identify target proteins of a human cervical cancer HeLa cell-specific cytotoxic compound, SB2001 (Fig. 2a). Because any chemical modifications neutralized the function and HeLa cell-selectivity of SB2001, label-free target identification methods such as TPP and TS-FITGE were the only available approaches for target identification. These methods were found to be complementary in terms of the number of total proteins and isoform coverage. Functional experiments to confirm the candidates revealed leukotriene A4 hydrolase (LTA4H) and MutT homolog 1 (MTH1) as target proteins. Protein expression profiling revealed that mismatch repair (MMR) proteins were downregulated in HeLa cells, causing them to be susceptible to oxidative stress. Consequently, HeLa cells could not eliminate the increased concentration of oxidized nucleotides caused by the action of SB2001, which resulted in oxidative DNA damage and apoptosis.
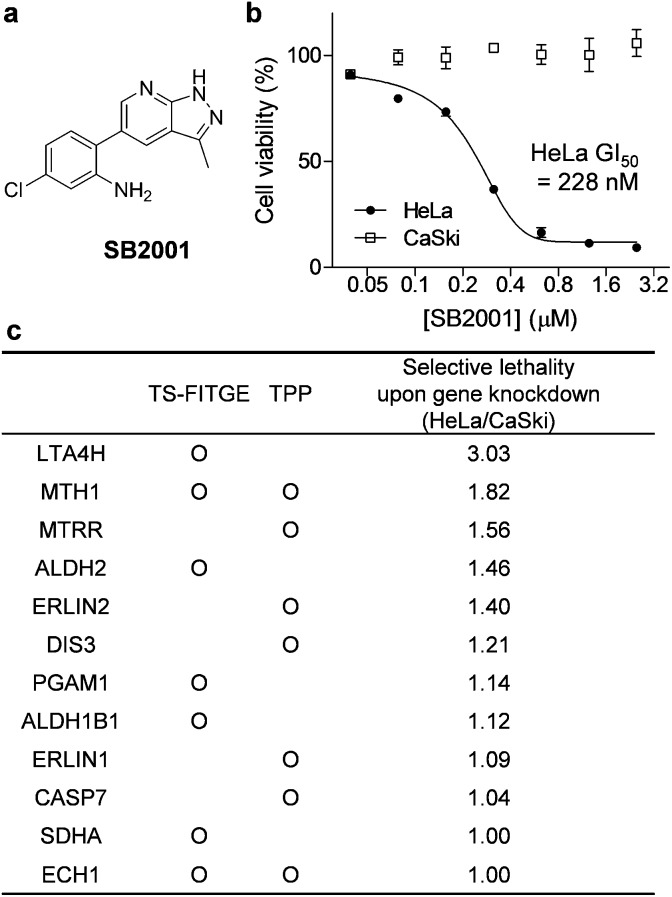
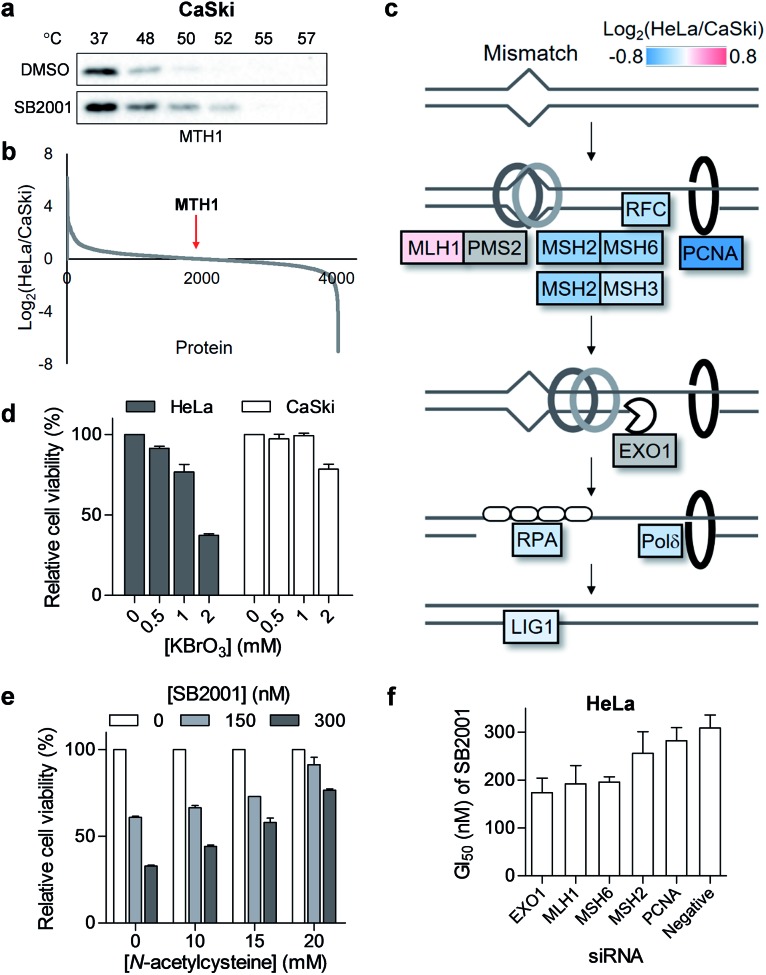
Fig. 2. SB2001, a HeLa cell-specific cytotoxic agent, and the identification of its target candidates. (a) The chemical structure of SB2001. (b) Cell viability after treatment with SB2001 for 2 days in HeLa and CaSki cells. Data are presented as the mean ± SEM (n = 2). (c) Target candidates from TS-FITGE and TPP in HeLa cells. Candidate proteins were prioritized according to the lethality ratio of HeLa cells over CaSki cells after the gene knockdown of each candidate protein.
Results
Discovery of SB2001 as a HeLa-specific cytotoxic agent
Phenotypic screening was conducted using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay to assess the potential anticancer activity of the in-house chemical library containing heterobiaryl pyrazolo[3,4-b]pyridine moieties17 constructed by privileged substructure-based diversity-oriented synthesis (pDOS).18 Interestingly, SB2001 (Fig. 2a) showed cytotoxicity specifically against HeLa cells with a concentration for 50% maximal inhibition of cell growth (GI50) value of 228 nM, but not against CaSki cells and the other eleven cell lines originating from either normal or cancerous tissues (Fig. 2b, cytotoxicity data of SB2001 toward other cell lines are not shown in this manuscript and will be published separately19). Although we attempted to functionalize the active compound for an affinity-based pull-down method, the SAR investigation indicated that minor modifications to the original structure of SB2001 attenuated either its potency or its selectivity.19 Therefore, we decided to identify the target proteins of SB2001 using label-free methods based on the principle of thermal stability shift upon drug engagement.
Label-free target identification using TS-FITGE and TPP
TS-FITGE and TPP share the same basic principle, which is that the binding of a protein to a drug may cause a shift in its thermal denaturation curve. In both TS-FITGE and TPP, the live cells were heat-denatured at various temperature points in the presence or absence of a specific drug. Thereafter, the heat-treated cells were lysed, and the soluble fractions of cell lysates were collected by centrifugation. However, TS-FITGE and TPP differ in several aspects, including the labeling method (fluorescence dyes vs. isobaric mass tags, respectively), the separation of the analytes (2D gel electrophoresis vs. liquid chromatography, respectively), data acquisition (fluorescence scanning vs. tandem mass spectrometry, respectively), quantification (of proteoforms vs. peptides, respectively), and proteomic profiling (Fig. 1). These differences mean that the two label-free techniques can be complementary.
During TS-FITGE, two different fluorescent dyes were added to the soluble protein fraction at each temperature: Cy3 dyes were added to the DMSO-treated group, and Cy5 dyes were added to the drug-treated group. The Cy3- and Cy5-tagged proteomes from the same temperature were then mixed, and the labeled proteins were separated by 2D gel electrophoresis according to the isoelectric point and molecular weight of each proteoform. For inter-gel spot localization and quantification, the proteome of the lowest temperature was tagged with Cy2 and spiked into each gel as an internal standard. Importantly, we used Cy2, Cy3, and Cy5 dyes with almost identical mass and charge to minimize spot separation in 2D gel electrophoresis.12 The ratio of Cy5 to Cy3 fluorescence intensity of each proteoform spot was quantified, and their distributions at each temperature were plotted using a box plot. Outliers in each box plot were considered as differentially stabilized proteins, indicating binding of the target protein to the drug.
In the TPP, on the other hand, the soluble proteins from heat-denatured cells were digested into peptides, followed by the chemical conjugation of 10-plex isobaric mass tags to the peptide at each temperature. The tagged peptides, from ten different temperature points, were pooled and then separated by reverse-phase liquid chromatography according to their hydrophobicity. Using tandem mass spectrometry, each peptide was identified, and its abundance at each temperature was quantified based on its reporter ions. The melting curves of the proteins were inferred from those of their component peptides, and the melting temperatures (Tm) of the proteins in the DMSO- and drug-treated groups were calculated separately. Proteins with significant changes in Tm were considered as target proteins. Although quantitative proteomics using liquid chromatography-tandem mass spectrometry has become a state-of-the-art technology due to its high throughput and high proteome coverage,20 TS-FITGE remains crucial for target identification, in that it can analyze different proteoforms including protein isoforms and various post-translational modifications (PTMs).21
Selection and prioritization of target candidates
As shown in Fig. S1,† the global melting proteomes (meltomes) in the absence or presence of SB2001 were revealed using both TS-FITGE and TPP. Target candidates with a significant thermal stability shift were independently chosen from the results of TS-FITGE and TPP: seven proteins (LTA4H, MTH1, ALDH2, PGAM1, ALDH1B1, SDHA, and ECH1) from TS-FITGE, and seven proteins (MTH1, MTRR, ERLIN2, DIS3, ERLIN1, CASP7, and ECH1) from TPP (Fig. 2c and S2†). Even though the thermal stability shift indicates the target engagement, the intensity of the thermal stability shift does not necessarily correlate with the relevance of the protein's function to its phenotype because each protein is diverse in terms of structure, innate thermal stability, and binding sites. Therefore, we prioritized the candidates not according to the intensity of the thermal shift but based on their functional relevance. As SB2001 caused selective cytotoxicity against HeLa cells, we compared the changes in viability after knocking down each candidate gene using small interfering RNA (siRNA) in HeLa versus CaSki cells (Fig. S3a†). CaSki cells were chosen as a negative control cell line, as both CaSki and HeLa cells originated from human papilloma virus-infected cervical cancer patients22 while SB2001 showed specific cytotoxicity only in HeLa cells but not in CaSki cells (Fig. 2b). As shown in Fig. 2c, we prioritized target candidates according to their selective lethality upon gene knockdown of each candidate protein in HeLa cells over CaSki cells, which phenocopied the HeLa cell-specific cytotoxicity of SB2001, for further functional validation.
Target validation of LTA4H
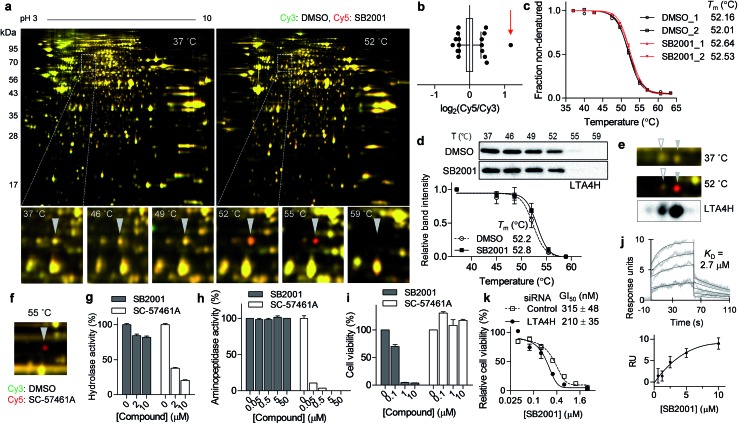
The spot with the highest Cy5/Cy3 ratio in the 55 °C gel (Fig. 3a, b and S1a†) showed temperature-dependent thermal stabilization in the presence of SB2001. In-gel digestion and mass spectrometry analysis revealed this spot to be LTA4H, and the genetic knockdown of LTA4H substantially increased lethality in HeLa cells compared to that in CaSki cells (Fig. 2c). As LTA4H was not selected by TPP, we manually inspected the melting curve of LTA4H from the TPP data. As shown in Fig. 3c, LTA4H was excluded from the candidate list because its Tm shift was relatively small—0.48 and 0.52 °C in the duplicate experiments. CETSA with the LTA4H antibody revealed a marginal shift in its melting curve, consistent with the TPP data (Fig. 3d). However, this was inconsistent with the results of TS-FITGE, where LTA4H showed the most prominent red spot. Through western blot analysis of the 2D gel, we found that LTA4H had more than two isoforms with identical molecular weight (Fig. 3e), as has been previously reported.23,24 Interestingly, only one spot (filled triangle in Fig. 3e) exhibited thermal stabilization by SB2001. Consequently, the single LTA4H isoform that was isolated by TS-FITGE showed significant thermal stabilization, but the overall thermal shift of LTA4H in TPP was marginal because the melting curve included all different LTA4H isoforms owing to the bottom-up analytic nature of shotgun proteomics.21 As the LTA4H antibody detected other LTA4H isoforms as well, CETSA also showed a marginal difference in the melting curve. To confirm that LTA4H was the protein stabilized by SB2001, we conducted TS-FITGE with a known LTA4H inhibitor, SC-57461A.25 As shown in Fig. 3f, SC-57461A clearly stabilized the same LTA4H spot as SB2001 did. On the other hand, SB2024, an inactive analogue of SB2001 (Fig. S4†), did not induce thermal stabilization of LTA4H in TS-FITGE (Fig. S5a and S5b†) and TPP (Fig. S5c†).
Fig. 3. SB2001 stabilizes one isoform of LTA4H and inhibits a non-canonical function of LTA4H. (a) Representative images of TS-FITGE (pH 3–10) in HeLa cells. Images of the Cy3 channel (green, DMSO-treated) and Cy5 channel (red, 10 μM of SB2001-treated) are overlaid. The area in the white box is magnified. (b) Box plot showing the distribution of the Cy5/Cy3 fluorescence ratio of each spot in the 55 °C gel. Center line denotes median, and the upper and lower box limits are the first and third quartiles, respectively. The whiskers indicate the 1st–99th percentiles. The spot denoted by a triangle in (a) is indicated by the red arrow. (c) Melting curves of LTA4H from TPP. (d) CETSA in HeLa cells with the LTA4H antibody (n = 5). (e) LTA4H isoform spots in the 2D gel were detected by anti-LTA4H immunoblotting, and the spots in TS-FITGE are indicated by triangles. (f) TS-FITGE 55 °C gel with SC-57461A, a known LTA4H inhibitor. (g) In vitro hydrolase activity of LTA4H upon treatment with SB2001 or SC-57461A (n = 2). (h) In vitro aminopeptidase activity of LTA4H upon treatment with SB2001 or SC-57461A (n = 2). (i) Viability of HeLa cells upon treatment with SB2001 or SC-57461A. (j) Sensorgrams of surface plasmon resonance (SPR) assay showing the binding kinetics of SB2001 (0.625 to 10 μM) to immobilized LTA4H. The dissociation constant (KD) was calculated as the ratio of rate constants (kd/ka). The figure below shows the steady-state response against various concentrations of SB2001. Representative data from two independent experiments. (k) DMSO-normalized cell viability upon treatment with SB2001 after gene knockdown of LTA4H in HeLa cells (n = 4). Data are presented as the mean ± SEM.
It is known that LTA4H is a bifunctional enzyme that acts as both an epoxide hydrolase and aminopeptidase, and its catalytic sites for those two functions overlap.26 Therefore, we performed in vitro enzymatic assays to investigate whether SB2001 mimics the activity of SC-57461A, which blocks both the hydrolase and peptidase activity of LTA4H.25 However, SB2001 inhibited neither the hydrolase activity nor the aminopeptidase activity of LTA4H (Fig. 3g and h). On the other hand, the viability of HeLa cells was not affected by SC-57461A (Fig. 3i).
In spite of the contradictory activity with the known inhibitor, surface plasmon resonance (SPR) assay confirmed that SB2001 showed one-to-one binding kinetics to LTA4H with a KD value of 2.7 μM while SB2024 hardly showed a binding sensorgram (Fig. 3j and S5d†). Next, we constructed a dose–response curve of SB2001 after the genetic knockdown of LTA4H to validate whether LTA4H was functionally related to the cytotoxic activity of SB2001. Compared to the negative control siRNA, the genetic knockdown of LTA4H significantly sensitized the activity of SB2001 by increasing its potency (Fig. 3k). Taken together, our results showed that the binding of SB2001 to LTA4H is functionally related to its HeLa cell-specific cytotoxicity, but the canonical function of LTA4H was not inhibited by SB2001, suggesting that LTA4H might have a yet undiscovered function. It is worth mentioning that the relatively low KD value in SPR assay compared to the GI50 value of HeLa cells implied that other target proteins might also be involved in the SB2001 activity.
Target validation of MTH1
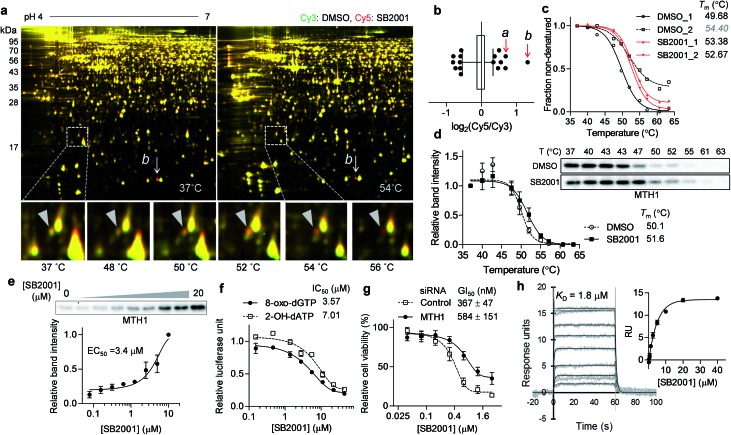
Another promising target candidate in TS-FITGE was found using immobilized pH gradient gel strips spanning pH 4–7 (Fig. S1b†). The red spot “a”, indicated by a triangle in Fig. 4a, had an outlying Cy5/Cy3 ratio in the 52 °C gel (Fig. 4b) and was identified as MTH1 (also known as 7,8-dihydro-8-oxoguanine triphosphatase, NUDT1). The red spot “b”, indicated by an arrow in Fig. 4a, had the highest Cy5/Cy3 ratio (Fig. 4b), but the spot also appeared red in the 37 °C gel; therefore it was thought not to be the target protein stabilized by SB2001. The thermal stabilization of MTH1 was also identified by TPP (Fig. 4c). One melting curve (DMSO_2 in Fig. 4c) had a distinct shape and bottom value from the others; in addition, the comparison between SB2001 and SB2024, a negative control compound (Fig. S2†), supported that SB2001 induced the repetitive thermal stabilization of MTH1 compared to DMSO_1, SB2024_1, and SB2024_2 (Fig. S6a†). Therefore, we excluded the DMSO_2 curve as an outlier shape caused by low coverage of quantified peptides due to the limited running and scanning time for tandem mass analysis. CESTA with the MTH1 antibody confirmed the thermal stabilization of MTH1 upon treatment with SB2001 compared to DMSO and SB2024 (Fig. 4d and S6b†). The isothermal dose–response curve showed a dose-dependent stabilization of MTH1 by SB2001 at 52 °C (Fig. 4e).
Fig. 4. SB2001 stabilizes MTH1 and inhibits its function of hydrolyzing oxidized nucleotides. (a) Representative images of TS-FITGE (pH 4–7) in HeLa cells. Images of the Cy3 channel (green, DMSO-treated) and Cy5 channel (red, 10 μM of SB2001-treated) are overlaid. The area in the white box is magnified. (b) Box plot showing the distribution of the Cy5/Cy3 fluorescence ratios for each spot in the 52 °C gel. Center line denotes median, and the right and left box limits are the first and third quartiles, respectively. The whiskers indicate the 1st–99th percentiles. The a and b spots denoted in (a) are pointed by the red arrows. (c) Melting curves of MTH1 from TPP. (d) CETSA in HeLa cells with the MTH1 antibody (n = 3). (e) Isothermal dose–response stabilization of MTH1 by various concentrations of SB2001 at 52 °C. (f) Inhibition of the in vitro hydrolytic activity of MTH1 by SB2001 using 8-oxo-dGTP or 2-OH-dATP as a substrate. (g) DMSO-normalized cell viability upon treatment with SB2001 after MTH1 knockdown in HeLa cells (n = 4). (h) SPR sensorgrams showing the kinetics of SB2001 (0.16 to 40 μM) to immobilized MTH1. The dissociation constant (KD) was calculated as the ratio of rate constants (kd/ka). The inset shows the steady-state response against various concentrations of SB2001. Representative data from two independent experiments. Data are presented as the mean ± SEM.
MTH1 is a sanitizing enzyme that catalyzes the hydrolysis of oxidized nucleotides to protect DNA from oxidative stress.27,28 SB2001 strongly inhibited the in vitro enzymatic activity of MTH1 toward its substrates, 8-oxo-dGTP and 2-OH-dATP (Fig. 4f), indicating that SB2001 inhibits the canonical function of MTH1, as the other MTH1 inhibitors such as TH287 (ref. 29) and (S)-crizotinib30 do (Fig. S7a–S7c†). In the Lineweaver–Burk plot with various concentrations of 8-oxo-dGTP, KM increased while Vmax remained constant upon treatment of SB2001 (Fig. S8†). These data indicate that SB2001 competes with the substrate to act as a competitive inhibitor such as TH287. The knockdown of MTH1 in HeLa cells also altered the cytotoxic potency of SB2001 by increasing the GI50 value (Fig. 4g), providing further evidence that the biological effect of SB2001 is associated with MTH1. Thus, we performed SPR analysis to biophysically confirm the direct binding of SB2001 to the purified MTH1 protein; the SPR sensorgram clearly showed one-to-one binding kinetics, as well as a concentration-dependent increase in response units (Fig. 4h). The negative control compound SB2024 neither inhibited in vitro enzymatic assay (Fig. S6c†) nor showed the binding events in SPR analysis (Fig. S6d†).
Origin of the selective cytotoxicity of SB2001 toward HeLa cells
Although we revealed that SB2001 inhibits the cellular function of MTH1, the reason for the selective cytotoxicity of SB2001 was still unclear because its function is rather general in many cellular systems. First, we investigated whether the binding of SB2001 to MTH1 would be impaired in CaSki cells. However, the thermal stability of MTH1 was also stabilized upon treatment with SB2001 in CaSki cells (Fig. 5a). Next, we checked whether the expression levels of MTH1 would be significantly higher in HeLa cells to stimulate the differential activity of SB2001. However, through a global protein expression profiling analysis between HeLa and CaSki cells obtained using quantitative proteomics, we found that MTH1 had nearly identical expression patterns in HeLa and CaSki cells (Fig. 5b). We then attempted to identify other differentially expressed proteins between HeLa and CaSki cells. Gene set enrichment analysis indicated that the expression of proteins involved in the DNA mismatch repair (MMR) pathway, including MSH2, MSH3, MSH6, RFC, PCNA, Polδ, and LigI, was consistently downregulated in HeLa cells (Fig. 5c and S9†). The correlation between the downregulated expression of MMR proteins and functional MMR-deficiency was shown in a previous proteomics study with colorectal tumor cells.31 The DNA MMR pathway primarily repairs base mismatches and insertion/deletion loops through recognizing replication errors, excising the errors, resynthesizing the nucleotides, and ligating the gap.32 8-Oxoguanine (8-oxoG), an oxidized form of guanine, can pair with both cytosine and adenine, allowing it to cause G:C to T:A transversions.33 In addition, there has been emerging evidence that MMR also plays important roles in the response to oxidative DNA damage by removing oxidized nucleotides such as 8-oxoG from DNA.34 Thus, it is possible that the downregulation of MMR proteins in HeLa cells makes them more susceptible to oxidative stress. As shown in Fig. 5d, HeLa cells were more vulnerable than CaSki cells to the oxidant KBrO3, supporting this hypothesis. Furthermore, when HeLa cells were co-treated with SB2001 and an antioxidant, N-acetylcysteine, to compensate for the oxidative stress, the cytotoxicity of SB2001 in HeLa cells was significantly neutralized in a dose-dependent manner (Fig. 5e). Therefore, we propose that SB2001 selectively kills HeLa cells through the inhibition of MTH1 and the subsequent increases in cellular oxidative stress and oxidized nucleotides; this can be compensated for by MMR pathways in CaSki cells, but not in HeLa cells. The effect of MMR proteins on the HeLa cell-specific cytotoxicity of SB2001 was also supported by the knockdown of each MMR gene in HeLa cells; silencing of each gene resulted in an increased potency of SB2001 (Fig. 5f and S3b†).
Fig. 5. HeLa cell-specific cytotoxicity of SB2001 originates from the downregulated mismatch repair (MMR) pathway. (a) CETSA in CaSki cells with the MTH1 antibody upon treatment of SB2001 (10 μM). (b) HeLa to CaSki ratio of protein expression levels acquired using quantitative mass spectrometry. MTH1 protein is indicated by a red arrow. (c) Differential expression of the MMR pathway in HeLa cells compared to that in CaSki cells. Undetected proteins (PMS2 and EXO1) are shown in gray color. (d) Viability of HeLa and CaSki cells upon treatment with an oxidant, KBrO3, for 2 d (n = 2). (e) Cytotoxicity of SB2001 in HeLa cells upon co-treatment with an antioxidant, N-acetylcysteine, for 2 d (n = 2). (f) GI50 of SB2001 after the knockdown of MMR genes in HeLa cells (n = 2). Data are presented as the mean ± SEM.
Biological evaluation of the proposed mechanism-of-action of SB2001
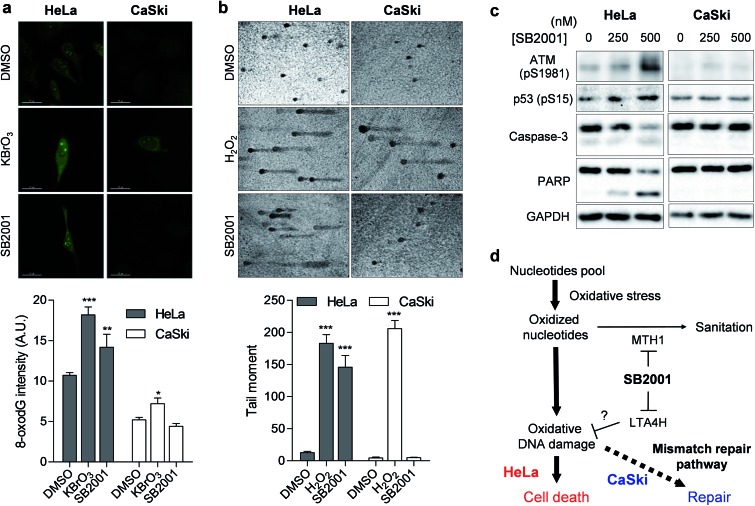
Once MTH1 is inhibited, the concentration of oxidized nucleotides increases, and these oxidized nucleotides are incorporated into DNA.35 When we measured the concentration of 8-oxoG in HeLa and CaSki cells by avidin-Alexa Fluor 488 imaging,36 the basal concentration of 8-oxoG was higher in MMR-downregulated HeLa cells than in CaSki cells (Fig. 6a), which is consistent with previous reports.37 Treatment of the strong oxidant KBrO3 elevated the cellular concentration of 8-oxoG significantly in HeLa cells, but only slightly in CaSki cells, while treatment with SB2001 increased cellular 8-oxoG in HeLa cells exclusively (Fig. 6a). The levels of 8-oxoG were correlated with cell viability upon treatment with SB2001 (Fig. 2b) and KBrO3 (Fig. 5d). This result was consistent with a previous study showing that the increase in 8-oxoG was the key determinant for cytotoxicity by MTH1 inhibition.38 Indeed, the suggested relationship between MTH1 inhibition and MMR was also supported by Bignami et al.,37,39 who clearly showed that the expression of human MTH1 in MMR-defective mouse embryonic fibroblasts reduced the cellular 8-oxoG concentration, mutation rate, and genetic instability.
Fig. 6. SB2001 selectively induces oxidative DNA damage in HeLa cells. (a) Fluorescence imaging of 8-oxoG in live cells. Intensities in the nuclear region were quantified (n > 11). Representative data from three biological replicates. (b) Silver-stained images of the comet assay measuring DNA damage. Tail moments of each comet were quantified (n > 6). Representative data from two biological replicates. (c) Immunoblot of ATM-p53 DNA damage response and apoptosis markers. (d) Schematic model of the selective cytotoxicity of SB2001 toward HeLa cells compared to CaSki cells. Data are presented as the mean ± SEM. The P value was derived from a two-tailed Student's t-test. ***P < 0.001, **P < 0.01, and *P < 0.05.
The increased incorporation of oxidized nucleotides into DNA causes DNA damage, which can be assessed by single-cell gel electrophoresis, also known as a comet assay.40 8-OxoG DNA glycosylase 1 (OGG1) excises 8-oxoG bases in DNA, and the induced strand breaks cause DNA relaxation and migration in the gel electrophoresis, allowing measurement of the extent of lesion-specific DNA damage. The treatment of live cells with hydrogen peroxide for 5 min increased DNA damage and the migration length in both HeLa and CaSki cells (Fig. 6b). However, a 2-day treatment of SB2001 increased DNA damage only in HeLa cells. The comet assay data clearly supported our finding that long-term treatment with SB2001 induces oxidative DNA damage only in HeLa cells but not in CaSki cells—where the oxidative stress could be attenuated by the MMR pathway—compared to the acute treatment of an indiscriminate oxidant, hydrogen peroxide. As DNA damage accumulates, ataxia-telangiectasia mutated (ATM) kinase autophosphorylates and phosphorylates its downstream substrate, p53.41 Immunoblotting of these markers clearly showed that ATM and p53 were selectively activated in HeLa cells upon treatment with SB2001 (Fig. 6c). Consequently, caspase-3 and poly(ADP-ribose) polymerase (PARP) were cleaved only in HeLa cells (Fig. 6c), which corresponded to the observed initial phenotype, the selective apoptosis of HeLa cells by SB2001 (Fig. 2b).
Discussion
Although both TS-FITGE and TPP are based on thermal stability shift induced by drug engagement, they differ in several technical aspects. We utilized both techniques to compensate for the weakness of each of them and to increase the success rate of label-free target identification. TPP, which uses state-of-the-art quantitative mass spectrometry, could identify and quantify thousands of proteins nearly simultaneously. Even though TS-FITGE could visualize only a thousand protein spots in a 2D gel, protein isoforms, including different PTMs, could be separated based on their isoelectric points and molecular weights. LTA4H was a good example of the different characteristics of TPP and TS-FITGE. LTA4H showed only a marginal Tm shift in TPP, but one isoform of LTA4H had a significant thermal shift in TS-FITGE. Therefore, TS-FITGE illustrated its potential to identify relevant target proteins when a compound modulates their specific proteoforms. Moreover, the identification of LTA4H as a target protein demonstrated the merits of phenotypic screening. Phenotypic screening improves the likelihood of finding novel drug targets or new functions of known proteins. Historically, the aminopeptidase activity had been considered as a non-canonical function of LTA4H until the endogenous substrate and physiological relevance of the aminopeptidase function was discovered.42 It is now recognized that both functions play important roles in the regulation of inflammation. Throughout this study, we assumed that LTA4H might have an undiscovered function regulating inflammation-mediated oxidative DNA damage, which is probably mediated by leukotrienes. Indeed, previous studies showed that leukotriene C4 (LTC4), another metabolic product of LTA4, is a major mediator of stress-induced oxidative DNA damage, and antagonizing the LTC4 receptor abolished oxidative DNA damage and cell death.43 Our fluorescence imaging data illustrated that cellular oxidative DNA damage was induced by the knockdown of LTA4H as well as MTH1 (Fig. S10†). It is worth mentioning that the cellular level of 8-oxoG was significantly higher in the case of SB2001 treatment than the knockdown case of either LTA4H or MTH1. The increased HeLa-selective cytotoxicity upon dual knockdown of LTA4H and MTH1, compared to their single knockdown, might support the dual modulation of SB2001 toward LTA4H and MTH1 (Fig. S11a†). We also observed that the conventional LTA4H inhibitor (SC-57461A) in combination with the MTH1 inhibitor (TH287) did not mimic the efficacy of SB2001 (Fig. S11b†), indicating that the canonical LTA4H function is not involved in HeLa cell-specific cytotoxicity. Collectively, through the potential neomorphic function of LTA4H, the dual modulation of LTA4H and MTH1 might exhibit the effects of SB2001 on oxidative DNA damage in HeLa cells.
Synthetic lethality has been exploited to improve the therapeutic index of anticancer therapies by inhibiting one gene in cancer cells where another gene is specifically mutated in the tumor, but normal in healthy cells.44 MMR deficiency has been identified as an important determinant of synthetic lethality.45–47 The mechanism of SB2001 seemed analogous to the synthetic lethality in that target inhibition by SB2001 was only lethal in HeLa cells where MMR proteins were consistently downregulated. Actually, SB2001 showed mild cytotoxicity in CaSki cells after the knockdown of MSH2, MSH6, PMS2, or EXO1 (Fig. S12 and S3c†). We assume that SB2001 did not exhibit its full efficacy because not just single gene but a group of MMR genes are upregulated in CaSki cells. Taken together, these results suggest that as the cellular level of 8-oxoG increased upon treatment with SB2001, the oxidative stress exceeded the capacity limit of MMR in HeLa cells, resulting in apoptosis, rather than DNA repair. We inferred that the compound could be suitable to treat MMR-downregulated HeLa cells specifically, while not damaging other tissues with an undamaged MMR system. Indeed, SB2001 exhibited selective in vivo efficacy in the mice xenografted with MMR-downregulated HeLa cells but not in C33A, another cervical cancer cell.19
Interestingly, SB2001 exhibited differential behavior compared to the typical MTH1 inhibitors, (S)-crizotinib29 and TH287 (ref. 30) (Fig. S7a†). Although these molecules inhibited the in vitro MTH1 activity (Fig. S7b†), (S)-crizotinib and TH287 did not potently and selectively kill HeLa cells (Fig. S7d and S7e†). Additionally, while SB2001 decreased MSH2 and MSH6 expression levels exclusively in HeLa cells, (S)-crizotinib and TH287 did not differentially downregulated either MSH2 or MSH6 between HeLa and CaSki cells (Fig. S7f†). Therefore, SB2001 has a different mechanism from the typical MTH1 inhibitors via dual inhibition of LTA4H and MTH1 along with downregulated MMR in cellular systems.
Conclusions
In conclusion, we identified target proteins and the mechanism of action of SB2001, a HeLa cell-specific cytotoxic agent discovered in a cell viability assay. Both TS-FITGE and TPP were used for target identification in a label-free manner because SB2001 has a tight SAR without any available modification sites. LTA4H was identified only by TS-FITGE, as only one of its isoforms showed significant thermal stabilization. Knockdown of LTA4H phenocopied the selective cytotoxicity of SB2001; functional validation suggested that a non-canonical function of LTA4H might be involved in the SB2001 activity. Both TS-FITGE and TPP identified MTH1 as a target protein candidate, and the function of MTH1, the hydrolysis of oxidized nucleotides, was inhibited by SB2001. Protein expression profiling revealed that proteins in the MMR pathway were downregulated in HeLa cells, causing them to become more vulnerable to oxidative stress and subsequent DNA damage. As a result, SB2001 was selectively cytotoxic to HeLa cells due to the increased levels of 8-oxoG, which led to DNA damage and apoptosis (Fig. 6d). This study demonstrates the merits of phenotypic screening followed by label-free target identification, as a potential neomorphic function of LTA4H related to oxidative DNA damage was implied, and polypharmacological application related to oxidative DNA damage involving LTA4H and MTH1 was suggested.
Data availability statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009903.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the Creative Research Initiative Grant (2014R1A3A2030423) and Bio & Medical Technology Development Program (2012M3A9C4048780) through the National Research Foundation of Korea (NRF) funded by the Korean Government (Ministry of Science & ICT). We appreciate Dr Sanghee Lee and Dr Heejun Kim, former members of our laboratory, for the early stage discovery of SB2001 and its chemical synthesis, respectively. We also thank Dr Jin Young Kim at the Korea Basic Science Institute for providing access to the Orbitrap Fusion Lumos mass spectrometer.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc05465g
References
- Swinney D. C., Anthony J. Nat. Rev. Drug Discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Wagner B. K. Expert Opin. Drug Discovery. 2016;11:121–125. doi: 10.1517/17460441.2016.1122589. [DOI] [PubMed] [Google Scholar]
- Schenone M., Dancik V., Wagner B. K., Clemons P. A. Nat. Chem. Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J. G., Vincent F., Lee J. A., Eder J., Prunotto M. Nat. Rev. Drug Discovery. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., Handa H. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- de Waal L., Lewis T. A., Rees M. G., Tsherniak A., Wu X., Choi P. S., Gechijian L., Hartigan C., Faloon P. W., Hickey M. J., Tolliday N., Carr S. A., Clemons P. A., Munoz B., Wagner B. K., Shamji A. F., Koehler A. N., Schenone M., Burgin A. B., Schreiber S. L., Greulich H., Meyerson M. Nat. Chem. Biol. 2016;12:102–108. doi: 10.1038/nchembio.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior M., Chiruta C., Currais A., Goldberg J., Ramsey J., Dargusch R., Maher P. A., Schubert D. ACS Chem. Neurosci. 2014;5:503–513. doi: 10.1021/cn500051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S., Pries V., Hedberg C., Waldmann H. Angew. Chem., Int. Ed. Engl. 2013;52:2744–2792. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- Laraia L., Waldmann H. Drug Discovery Today: Technol. 2017;23:75–82. doi: 10.1016/j.ddtec.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E. A., Dan C., Sreekumar L., Cao Y., Nordlund P. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Savitski M. M., Reinhard F. B., Franken H., Werner T., Savitski M. F., Eberhard D., Martinez Molina D., Jafari R., Dovega R. B., Klaeger S., Kuster B., Nordlund P., Bantscheff M., Drewes G. Science. 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- Park H., Ha J., Koo J. Y., Park J., Park S. B. Chem. Sci. 2017;8:1127–1133. doi: 10.1039/c6sc03238a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard F. B. M., Eberhard D., Werner T., Franken H., Childs D., Doce C., Savitski M. F., Huber W., Bantscheff M., Savitski M. M., Drewes G. Nat. Methods. 2015;12:1129–1131. doi: 10.1038/nmeth.3652. [DOI] [PubMed] [Google Scholar]
- Huber K. V. M., Olek K. M., Muller A. C., Tan C. S. H., Bennett K. L., Colinge J., Superti-Furga G. Nat. Methods. 2015;12:1055–1057. doi: 10.1038/nmeth.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher I., Werner T., Doce C., Zaal E. A., Togel I., Khan C. A., Rueger A., Muelbaier M., Salzer E., Berkers C. R., Fitzpatrick P. F., Bantscheff M., Savitski M. M. Nat. Chem. Biol. 2016;12:908–910. doi: 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Liao P.-J., Lee K. H., Wong J., Shang S. C., Minami N., Sampetrean O., Saya H., Lingyun D., Prabhu N., Diam G. K., Sobota R., Larsson A., Nordlund P., McCormick F., Ghosh S., Epstein D. M., Dymock B. W., Lee S. H. Nat. Commun. 2017;8:2200. doi: 10.1038/s41467-017-02287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Park S. B. Org. Lett. 2009;11:5214–5217. doi: 10.1021/ol902147u. [DOI] [PubMed] [Google Scholar]
- Kim J., Kim H., Park S. B. J. Am. Chem. Soc. 2014;136:14629–14638. doi: 10.1021/ja508343a. [DOI] [PubMed] [Google Scholar]
- Lee S., Cho W. and Park S. B., manuscript in preparation.
- Yates J. R. J. Am. Chem. Soc. 2013;135:1629–1640. doi: 10.1021/ja3094313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R., Agar J. N., Amster I. J., Baker M. S., Bertozzi C. R., Boja E. S., Costello C. E., Cravatt B. F., Fenselau C., Garcia B. A., Ge Y., Gunawardena J., Hendrickson R. C., Hergenrother P. J., Huber C. G., Ivanov A. R., Jensen O. N., Jewett M. C., Kelleher N. L., Kiessling L. L., Krogan N. J., Larsen M. R., Loo J. A., Ogorzalek Loo R. R., Lundberg E., MacCoss M. J., Mallick P., Mootha V. K., Mrksich M., Muir T. W., Patrie S. M., Pesavento J. J., Pitteri S. J., Rodriguez H., Saghatelian A., Sandoval W., Schlüter H., Sechi S., Slavoff S. A., Smith L. M., Snyder M. P., Thomas P. M., Uhlén M., Van Eyk J. E., Vidal M., Walt D. R., White F. M., Williams E. R., Wohlschlager T., Wysocki V. H., Yates N. A., Young N. L., Zhang B. Nat. Chem. Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Yim E.-K., Kim C.-J., Namkoong S.-E., Um S.-J., Park J.-S. Gynecol. Oncol. 2005;98:45–53. doi: 10.1016/j.ygyno.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Odlander B., Claesson H.-E., Bergman T., Rådmark O., Jörnvall H., Haeggström J. Z. Arch. Biochem. Biophys. 1991;287:167–174. doi: 10.1016/0003-9861(91)90402-5. [DOI] [PubMed] [Google Scholar]
- Bito H., Ohishi N., Miki I., Minami M., Tanabe T., Shimizu T., Seyama Y. J. Biochem. 1989;105:261–264. doi: 10.1093/oxfordjournals.jbchem.a122650. [DOI] [PubMed] [Google Scholar]
- Askonas L. J., Kachur J. F., Villani-Price D., Liang C.-D. D., Russell M. A., Smith W. G. J. Pharmacol. Exp. Ther. 2002;300:577–582. doi: 10.1124/jpet.300.2.577. [DOI] [PubMed] [Google Scholar]
- Numao S., Hasler F., Laguerre C., Srinivas H., Wack N., Jäger P., Schmid A., Osmont A., Röthlisberger P., Houguenade J., Bergsdorf C., Dawson J., Carte N., Hofmann A., Markert C., Hardaker L., Billich A., Wolf R. M., Penno C. A., Bollbuck B., Miltz W., Röhn T. A. Sci. Rep. 2017;7:13591. doi: 10.1038/s41598-017-13490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y., Kasai H. J. Biol. Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- Huber K. V. M., Salah E., Radic B., Gridling M., Elkins J. M., Stukalov A., Jemth A. S., Gokturk C., Sanjiv K., Stromberg K., Pham T., Berglund U. W., Colinge J., Bennett K. L., Loizou J. I., Helleday T., Knapp S., Superti-Furga G. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad H., Koolmeister T., Jemth A.-S., Eshtad S., Jacques S. A., Strom C. E., Svensson L. M., Schultz N., Lundback T., Einarsdottir B. O., Saleh A., Gokturk C., Baranczewski P., Svensson R., Berntsson R. P. A., Gustafsson R., Stromberg K., Sanjiv K., Jacques-Cordonnier M.-C., Desroses M., Gustavsson A.-L., Olofsson R., Johansson F., Homan E. J., Loseva O., Brautigam L., Johansson L., Hoglund A., Hagenkort A., Pham T., Altun M., Gaugaz F. Z., Vikingsson S., Evers B., Henriksson M., Vallin K. S. A., Wallner O. A., Hammarstrom L. G. J., Wiita E., Almlof I., Kalderen C., Axelsson H., Djureinovic T., Puigvert J. C., Haggblad M., Jeppsson F., Martens U., Lundin C., Lundgren B., Granelli I., Jensen A. J., Artursson P., Nilsson J. A., Stenmark P., Scobie M., Berglund U. W., Helleday T. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- Halvey P. J., Wang X., Wang J., Bhat A. A., Dhawan P., Li M., Zhang B., Liebler D. C., Slebos R. J. C. Cancer Res. 2014;74:387–397. doi: 10.1158/0008-5472.CAN-13-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.-M. Cell Res. 2007;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Bridge G., Rashid S., Martin S. Cancers. 2014;6:1597–1614. doi: 10.3390/cancers6031597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Struthers L., Patel R., Clark J., Thomas S. Anal. Biochem. 1998;255:20–31. doi: 10.1006/abio.1997.2354. [DOI] [PubMed] [Google Scholar]
- Colussi C., Parlanti E., Degan P., Aquilina G., Barnes D., Macpherson P., Karran P., Crescenzi M., Dogliotti E., Bignami M. Curr. Biol. 2002;12:912–918. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- Warpman Berglund U., Sanjiv K., Gad H., Kalderén C., Koolmeister T., Pham T., Gokturk C., Jafari R., Maddalo G., Seashore-Ludlow B., Chernobrovkin A., Manoilov A., Pateras I. S., Rasti A., Jemth A. S., Almlöf I., Loseva O., Visnes T., Einarsdottir B. O., Gaugaz F. Z., Saleh A., Platzack B., Wallner O. A., Vallin K. S. A., Henriksson M., Wakchaure P., Borhade S., Herr P., Kallberg Y., Baranczewski P., Homan E. J., Wiita E., Nagpal V., Meijer T., Schipper N., Rudd S. G., Bräutigam L., Lindqvist A., Filppula A., Lee T. C., Artursson P., Nilsson J. A., Gorgoulis V. G., Lehtiö J., Zubarev R. A., Scobie M., Helleday T. Ann. Oncol. 2016;27:2275–2283. doi: 10.1093/annonc/mdw429. [DOI] [PubMed] [Google Scholar]
- Russo M. T., Blasi M. F., Chiera F., Fortini P., Degan P., Macpherson P., Furuichi M., Nakabeppu Y., Karran P., Aquilina G., Bignami M. Mol. Cell. Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive P. L., Banath J. P. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- Maréchal A., Zou L. Cold Spring Harbor Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelgrove R. J., Jackson P. L., Hardison M. T., Noerager B. D., Kinloch A., Gaggar A., Shastry S., Rowe S. M., Shim Y. M., Hussell T., Blalock J. E. Science. 2010;330:90–94. doi: 10.1126/science.1190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvash E., Har-Tal M., Barak S., Meir O., Rubinstein M. Nat. Commun. 2015;6:10112. doi: 10.1038/ncomms10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin Jr W. G. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Martin S. A., McCarthy A., Barber L. J., Burgess D. J., Parry S., Lord C. J., Ashworth A. EMBO Mol. Med. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish M., Martin S. A., Elliott R., Cunningham D., Lord C. J., Ashworth A. Br. J. Cancer. 2013;108:983–992. doi: 10.1038/bjc.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. A., Hewish M., Sims D., Lord C. J., Ashworth A. Cancer Res. 2011;71:1836–1848. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009903.