Abstract
Introduction
Twin-to-twin transfusion syndrome (TTTS) is a potentially lethal condition that affects pregnancies in which twins share a single placenta. The definitive treatment for TTTS is fetoscopic laser photocoagulation, a procedure in which placental blood vessels are selectively cauterized. Challenges in this procedure include difficulty in quickly identifying placental blood vessels due to the many artifacts in the endoscopic video that the surgeon uses for navigation. We propose using deep-learned segmentations of blood vessels to create masks that can be recombined with the original fetoscopic video frame in such a way that the location of placental blood vessels is discernable at a glance.
Methods
In a process approved by an institutional review board, intraoperative videos were acquired from ten fetoscopic laser photocoagulation surgeries performed at Yale New Haven Hospital. A total of 345 video frames were selected from these videos at regularly spaced time intervals. The video frames were segmented once by an expert human rater (a clinician) and once by a novice, but trained human rater (an undergraduate student). The segmentations were used to train a fully convolutional neural network of 25 layers.
Results
The neural network was able to produce segmentations with a high similarity to ground truth segmentations produced by an expert human rater (sensitivity=92.15%±10.69%) and produced segmentations that were significantly more accurate than those produced by a novice human rater (sensitivity=56.87%±21.64%; p < 0.01).
Conclusion
A convolutional neural network can be trained to segment placental blood vessels with near-human accuracy and can exceed the accuracy of novice human raters. Recombining these segmentations with the original fetoscopic video frames can produced enhanced frames in which blood vessels are easily detectable. This has significant implications for aiding fetoscopic surgeons—especially trainees who are not yet at an expert level.
Keywords: Segmentation, Vessels, Deep learning, Convolutional neural network, Fetoscopy, Twin-to-twin transfusion syndrome
Introduction
Monochorionic pregnancies—pregnancies involving twins that share a single placenta—account for 0.3% of deliveries worldwide [1]. At current birth rates, this amounts to several hundred thousand such pregnancies per year.
Twin-to-twin transfusion syndrome (TTTS), which is estimated to affect 10–15% of all monochorionic pregnancies [2], is a disease that arises from the formation of abnormal vascular connections within placental circulation that disproportionately redirect blood from one fetus to the other. This unequal distribution of blood can have serious consequences for both twins, including cardiac dysfunction in the twin that serves as a net blood recipient, injury to the central nervous system in the twin that serves as a net donor, and death in either twin [3].
There are several options for managing TTTS but only one definitive treatment: fetoscopic laser photocoagulation surgery [4]. In this procedure, a surgeon inspects placental blood vessels with a specialized endoscope known as a fetoscope. Any problematic vascular connections that are found are cauterized with a laser. This procedure is illustrated in Fig. 1.
Fig. 1.
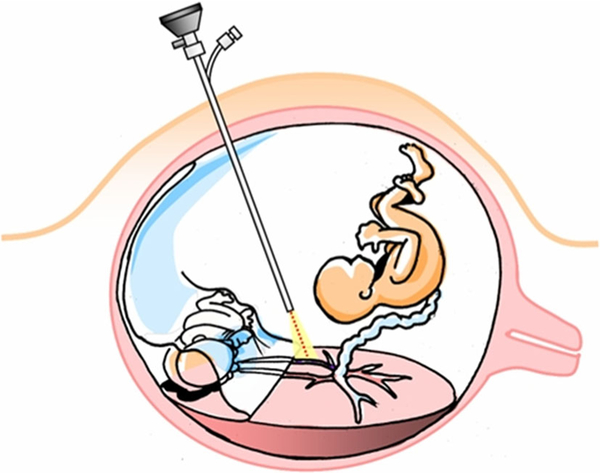
A diagram of fetoscopic laser photocoagulation surgery for twin-to-twin transfusion syndrome by Luks [5]. Pictured are twin fetuses, each within their own amniotic sac. The fetuses have a single shared placenta, with problematic vascular connections that allow a net flow of blood from the donor fetus (lower left) to the recipient fetus (upper right). An endoscope (center) is used to inspect the placental vasculature and find problematic connections. When such connections are found, they are cauterized with a laser (bottom)
The challenges of fetoscopic laser photocoagulation surgery are well described in the literature [6–8]. There is no imaging modality by which problematic blood vessels can be visualized preoperatively. The surgeon must therefore identify them intraoperatively, but the limited field of view of the fetoscope impairs his or her ability to remain oriented during the procedure. Additionally, the poor depth of field of the fetoscope, the turbidity of the amniotic fluid, and intermittent changes in illumination caused by the activation of the cautery laser all impair the surgeon’s ability to quickly and accurately identify blood vessels within the fetoscopic image (Fig. 2).
Fig. 2.
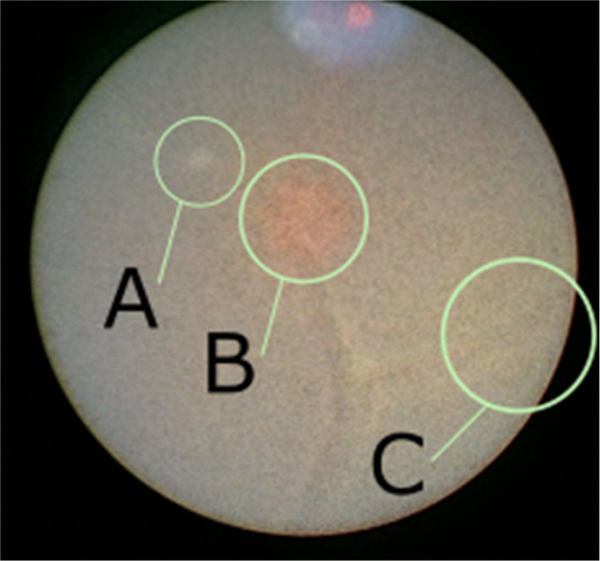
An intraoperative video frame from an endoscope that demonstrates the effects that can obscure the view of placental vasculature during fetoscopic surgery: a floating debris within the amniotic fluid, b illumination effects—here caused by the targeting light for the laser, and c poor contrast near the periphery of the field of view
While it might initially appear that only abnormal vascular formations are of interest to the surgeon, as these are the structures that the surgeon seeks to cauterize, normal blood vessels are also important. The limited field of view of the fetoscope only allows the surgeon to view a small fraction of the placental surface at any given point in time. The surgeon must therefore rely on visual landmarks to find abnormal vascular formations. In particular, the surgeon relies on the fact that abnormal vascular formations tend to be found at the smallest branches of normal blood vessels [9]. Vascular formations can therefore be found by tracking a normal blood vessel as it divides into smaller and smaller branches.
One can conceive of four manners in which the identification of placental blood vessels could be made easier:
Physically enhancing the fetoscopic image by improving the optical characteristics of the fetoscope.
Physically improving environmental visibility factors.
Devising alternative methods of visualizing the placenta.
Digitally enhancing the fetoscopic image.
The first approach is largely impracticable. The nature of fetoscopic surgery, which involves highly constrained spaces, requires that fetoscopes are smaller in diameter than standard endoscopes. Modern “ultra-thin” fetoscopes can have diameters as small as 1.0 mm [10]. These small diameters place a physical maximum constraint on image quality [9].
The second approach, like the first, is typically not feasible in practice. While there are methods by which environmental visibility factors can be manipulated—the turbidity of amniotic fluid, for example, can be reduced by amniotic fluid exchange, a procedure in which a portion of the amniotic fluid is removed and replaced with a saline solution [11]—the risks associated with manipulating the intrauterine environment mid-pregnancy likely outweigh any benefit that would be provided to the surgeon during a fetoscopic surgery.
Most existing work on improved vascular visualization in fetoscopic surgery has focused on the third approach. In particular, much of the existing work has focused on using software to combine fetoscopic video frames in real time to create a mosaic of the placental surface that the surgeon can use as a map [8, 12, 13]. This approach is attractive for multiple reasons: First, the additional context provided by a map of the placental surface can not only help with the identification of placental blood vessels, but also help to address the issue of limited field of view that impedes the surgeon’s ability to remain oriented. Second, a placental map can be used intraoperatively or postoperatively as a check to ensure that all regions of the placental surface have been inspected.
While there has been progress in the real-time construction of placental maps from fetoscopic video frames, this technology is still not ready for clinical use. In particular, there has been limited success in finding methods of describing visual features within video frames in such a way that they can be matched across frames to compute a homography for pasting a frame into the mosaic [12, 13]. Current methods work well for short runs of fetoscopic video but struggle with longer video segments because they cannot reliably recover features between frames [13] and because they can completely fail to find features in poor lighting conditions [12].
This work focuses on the fourth strategy, which is to digitally postprocess fetoscopic video in such a way that blood vessels are more easily identifiable.
There are existing studies that describe methods for segmenting placental blood vessels. Almoussa et al. [14], Park et al. [15], and Chang et al. [16] describe methods for segmenting blood vessels within images of ex vivo placentas. The segmentation of ex vivo placentas is useful for applications in pathology, such as the postpartum diagnosis of placental diseases by analyzing the structure of the placental vascular network. If segmentations are to be used to enhance intraoperative fetoscopic video, however, then the segmentation algorithm must be able to process in vivo placental images.
While the vascular anatomy of an ex vivo placenta is identical to that of an in vivo placenta, the appearance is dramatically different, and this has significant implications for image analysis. Gaisser et al. [13], for example, simulated ex vivo and in vivo settings using a placental phantom and found that the performance of various feature detection algorithms could fall dramatically in the translation to in vivo. A feature detector could detect as many as 73% fewer features in images acquired in vivo settings as opposed to images acquired in ex vivo settings.
Recently, there have been efforts to analyze in vivo placental images. Perera Bel [17] describes a method for segmenting blood vessels from Doppler-flow ultrasound. This method can locate large blood vessels such as the umbilical arteries and vein, which are often several dozen centimeters long. Finding these large vessels is important as it informs the surgeon’s choice of insertion point for the fetoscope. However, once the surgeon has inserted the fetoscope into the uterine environment, he or she is interested in much smaller blood vessels that are on the order of a few millimeters in length. These blood vessels are too small to be reliably resolved on ultrasound. Gaisser et al. [13] made an important step by demonstrating that it is possible to use a region-based convolutional neural network (R-CNN) to detect stable center points within placental blood vessels. This deep-learned approach far outperformed standard feature detection algorithms when applied to in vivo images in which the amniotic fluid had a yellow coloration, but the performance significantly degraded when the amniotic fluid had a green coloration, suggesting that the algorithm was not robust to variations that might be seen in the intrauterine environment.
In this work, we present the design of a fully convolutional neural network (FCNN) that can detect placental blood vessels within fetoscopic video. We demonstrate that the FCNN is able to identify blood vessels with human-level accuracy across a varied dataset consisting of fetoscopic video from ten different patients. We further show that the FCNN can be used to highlight blood vessels in fetoscopic video, producing images in which blood vessels are more easily identifiable and potentially providing a benefit to the surgeon.
Materials and methods
Image acquisition
In a process approved by an institutional review board, intraoperative videos were obtained from ten fetoscopic laser coagulation procedures performed at Yale New Haven Hospital. All videos were recorded using a Storz miniature 11540AA endoscope with incorporated fiber optic light transmission and had a resolution of 1920×1080 pixels with RGB color channels. A total of 544,975 video frames were collected in total, accounting for approximately 5 h of video. These video frames were downscaled, and the far left and right regions of the frames were then cropped to create square images of a resolution of 256×256 pixels.
Neural network architecture
The design of our FCNN builds upon the “U-Net” architecture described by Ronneberger et al. [18]. This architecture consists of a convolutional layer that is divided into contracting and expanding segments. The contracting segment has a structure similar to that of the typical convolutional network for image classification: Convolutional layers that apply learned filters to the image data are interspersed with max pooling layers that simultaneously reduce the dimensionality of the image and increase the receptive field of downstream convolutional layers. The expanding segment is essentially the contracting segment in reverse: This segment consists of upconvolutional layers, which are convolutional layers followed by image upscaling.
Max pooling operations are a critical part of convolutional neural network, as their dimensionality reduction allows for successive convolutional layers to learn increasingly high-level features within the image. However, the max pooling layers also reduce the resolution of the image, making it difficult to produce a pixel-perfect segmentation. The key contribution of the U-Net architecture is to add bridges that connect convolutional layers within the contracting segment to convolutional layers within the expanding segment. These bridges transfer image details that may have been lost during max pooling operations in the contracting segment, thus allowing detailed information to be incorporated during segmentation, while still allowing for the learning of high-level image features that may be useful during segmentation.
The U-Net architecture is divided into “convolutional units,” which consist of one or more convolutional layers followed by a max pooling layer (within the contracting segment) or an upscaling operation (within the expanding segment). This architecture is described in Fig. 3. Our implementation of the U-Net differs from the original architecture as described by Ronneberger et al. in that it contains three contracting convolutional units and three expanding convolutional units, with each unit containing eight convolutional layers (as opposed to the original U-Net architecture, which has two convolutional layers per unit). The FCNN consists of a total of twenty-five layers.
Fig. 3.
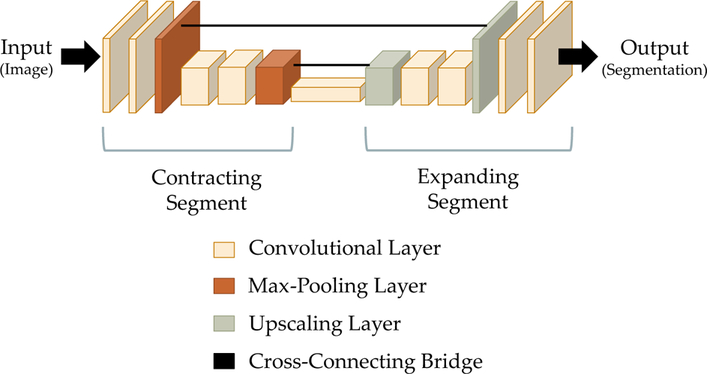
A schematic diagram of the U-Net architecture, a fully convolutional neural network that is often used for the semantic segmentation of biomedical images [18]. The architecture consists of a contracting segment (left), in which convolutional layers are followed by max pooling layers that downscale the image, followed by a expanding segment (right), in which convolutional layers are intermixed with upscaling layers that increase the size of the image. Cross-connecting bridges (center) connect corresponding layers in the contracting and expanding segments. These bridges preserve detailed information that would otherwise be lost during the max pooling operations. The height and width of each block in the block diagram reflect the height and width of the image as it is encoded at that point. The thickness reflects the number of feature channels
Training
A total of 345 frames were selected from the ten fetoscopic videos at regularly spaced intervals. The blood vessels within these video frames were manually labeled twice: once by an expert rater (a maternal–fetal medicine fellow), and once by a trained, but novice, rater (an undergraduate student). To increase the number of samples available to train the neural network and to make the training process computationally tractable, we used patch-based training with a mini-batch of size 64. In each mini-batch, we randomly selected overlapping 128×128 pixel patches from the entire set of images with replacement. These patches were augmented with vertical and horizontal reflections. Due to the class imbalance between the vessel and non-vessel segments of the training data, we used a class-weighted cross-entropy loss [19]. Given a vector x of per-pixel predictions, a corresponding vector y of per-pixel ground truth labels, and a positive weight w by which correctly labeling vessels is valued over correctly labeling background, the class-weighted cross-entropy loss of the nth element of x is defined as follows:
In this equation, S represents the sigmoid function .
The FCNN was trained by stochastic gradient descent with a positive weight of 3000 and an initial learning rate of 10−4 that was adjusted as needed by adaptive moment estimation. Training was stopped after 20,000 iterations, as the loss over the training dataset had shown signs of convergence by this point.
Prediction
The neural network generates predictions for novel images in a patch-based manner. Input video frames are divided into nine regularly spaced 128×128 pixel patches with an isotropic stride of 64 pixels. The neural network is evaluated once on each patch to produce nine 128×128 patch segmentations. These patch segmentations are then pieced together into a complete segmentation of the original 256×256 image: At any pixel p in the reconstructed image where multiple patches overlap, the value of the reconstructed pixel p is calculated as a weighted sum of the corresponding patch pixels {p1, p2, p3,..., pn} where each patch pixel’s weight is inversely proportional to its distance from the center of its patch:
| (1) |
The weighted sum images are thresholded to produce a binary segmentation.
Quantification
The segmentations generated by the FCNN were compared to the ground truth segmentations provided by the expert human rater. Sensitivities and specificities were calculated.
As accuracy can be sensitive to large imbalances in the ratio of positive class values to negative class values, we instead evaluate the generated segmentations using the Dice coefficient [20], which is less sensitive to such imbalances. The Dice coefficient was originally formulated in terms of set intersections and unions, but can be equivalently defined in terms of true positives (TP), false positives (FP), and false negatives (FN):
| (2) |
A perfectly accurate test will produce only true positives and will not produce any false negatives or false positives and will thus have a Dice coefficient of one. The worst possible test will produce many false negatives and false positives but no true positives and will accordingly have a Dice coefficient of zero. Geometrically, the Dice coefficient can be interpreted as a measurement of the degree of spatial overlap between two regions. Under this interpretation, two segmentations in which the positive (white) portions overlap exactly will have a Dice coefficient of one. Two segmentations in which there is no overlap will have a Dice coefficient of zero. Two segmentations that have a partial overlap will have an intermediate Dice coefficient.
To evaluate the ability of the FCNN to generalize to new patients, we use a leave-one-out strategy: One patient is selected from the pool of ten patients, and all images obtained from this patient are excluded from the pool of training images sent to the FCNN. Once trained, the FCNN is evaluated on the images from the excluded patient. As the FCNN did not see images from this patient during the training phase, its performance on this patient is indicative of its ability to generalize to new patients. This process is repeated for all ten patients such that every patient serves as the excluded patient exactly once.
Implementation of the Frangi filter
We compare our deep-learned segmentation method to the Frangi vesselness filter [21], considered to be the de facto standard for vessel enhancement. While the Frangi vesselness filter is a widely used tool, it often needs fine-tuning to the particular application. When comparing a novel algorithm to the Frangi vesselness filter, it is therefore important to specify exactly how the Frangi filter is used.
We model our usage of the Frangi filter upon the strategy described by Srivastava et al. [22]. Briefly, we preprocess each image with Gaussian blurring to remove local irregularities within the image, followed by histogram equalization to increase the image contrast. The Frangi filter is applied to compute a “vesselness” score at each pixel, and the vesselness is thresholded to create a binarized segmentation.
Unlike Srivastava et al., we do not use a modification of the original Frangi filter; Srivastava et al. used a modified algorithm because they wished to detect only lesions to blood vessels and not the blood vessels themselves. As we are interested in the blood vessels themselves, we do not adopt this modification.
The output of the Frangi filter is often postprocessed with some form of connectivity analysis to eliminate “island” blood vessels. However, this requires prior knowledge of the geometry of the blood vessels. Jiang et al., for example, enforced the constraint that vessels must follow a tree-like branching pattern [23]. Given the limited field of view of the fetoscope, the branching points of blood vessels within a given video frame could be outside of the field of view, making it impossible for us enforce a similar constraint.
The Frangi filter has three tunable parameters: the scale range, β1, and β2. These values define the sizes of the blood vessels that the algorithm detects, the algorithm’s sensitivity to nonlinear vessels, and the algorithm’s sensitivity to noise, respectively. The ideal values of these parameters depend on the specific application. We selected values for these parameters empirically by evaluating a number of different values on a subset of the available data and selecting those that gave the best average Dice coefficient.
Selection of ideal thresholds
As described in “Prediction” section, our patch-based segmentation method produces grayscale images rather than binary segmentations. We must apply an arbitrary threshold to binarize the output images. This is also true of the Frangi vesselness filter, which produces a vesselness score at every pixel rather than a binary vessel or non-vessel designation.
An ideal threshold for the FCNN-based segmentations was determined by evaluating the FCNN on the validation dataset and thresholding the grayscale images that it produced at every possible threshold value. At each threshold value, the binarized segmentations were compared to the ground truth segmentations from the validation dataset. The threshold that yielded binarized segmentations that had the highest Dice overlap with the ground truth segmentations was defined as the ideal threshold. Given that we use grayscale images with a depth of 8-bits, this threshold must be an integer between 0 and 255. The ideal threshold was determined to be 230.
For the Frangi filter, the choice of threshold affects the optimal alpha and beta parameters as described in “Implementation of the Frangi filter” section. It is therefore impossible to determine optimal alpha and beta parameters without fixing the threshold to an arbitrary value or to determine an optimal threshold without fixing the alpha and beta parameters to arbitrary values. We chose to fix the threshold at 230 and to optimize alpha and beta accordingly.
Results
All 345 fetoscopic video frames for which ground truth segmentations were available were resegmented in three ways: once by a novice (but trained) human rater, once with the Frangi vesselness filter, and once with the U-Net. The grayscale images produced by the Frangi vesselness filter and the U-Net were binarized into segmentations by applying an ideal threshold as described in “Selection of ideal thresholds” section. For each generated segmentation, a sensitivity, specificity, and Dice coefficient was calculated relative to a ground truth segmentation provided by an expert human rater. The average sensitivities, specificities, and Dice coefficients and the associated standard deviations are reported in Table 1.
Table 1.
Accuracy of segmentations produced by a novice human rater, the Frangi vesselness filter, and our trained FCNN model relative to ground truth segmentations provided by an expert rater
| Sensitivity | Specificity | Dice Coeff. | |
|---|---|---|---|
| Novice | 56.87%±21.64% | 99.16%±1.34% | 0.42±0.34 |
| Frangi | 23.32%±17.68% | 95.53%±2.91% | 0.19±0.19 |
| FCNN | 92.15%±10.69% | 94.12%±3.23% | 0.55±0.22 |
The deep-learned approach to vessel segmentation, the FCNN, far exceeds the novice human rater and the Frangi filter in terms of sensitivity and Dice coefficient. While both the novice human rater and the Frangi filter are able to segment blood vessels with a higher specificity than the FCNN, this is likely an artifact of the class imbalance in the dataset.
Most images in the dataset have many more pixels that are part of the background than pixels that are within blood vessels. We would therefore expect segmentation strategies that are heavily biased toward classifying pixels as background to yield better Dice coefficients. This seems to be the case for the Frangi filter, which was optimized to maximize the Dice coefficient and therefore has a high specificity and a low sensitivity. The novice human rater has a similarly poor sensitivity relative to the expert human rater. Indeed, there were many instances across the dataset where the novice human rater did not identify any blood vessel within the video frame, whereas the expert human rater identified several. The FCNN is able to identify many of the same blood vessels that the novice human rater missed, leading to its higher sensitivity score (Fig. 4).
Fig. 4.
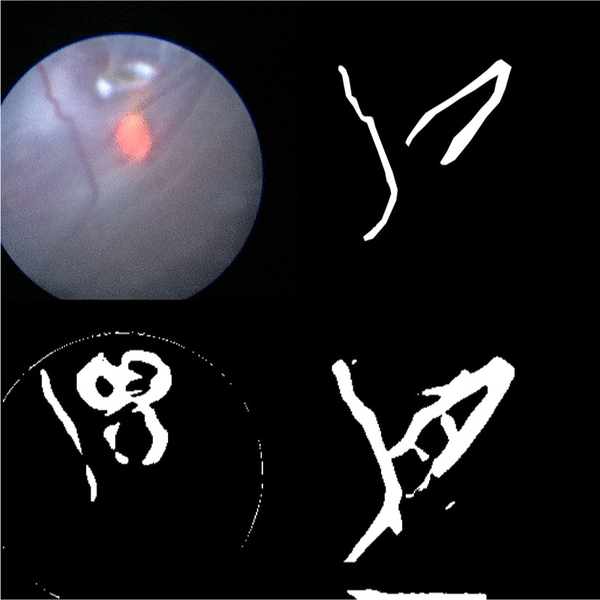
The results of running various segmentation algorithms on a particularly difficult fetoscopic video frame that was not included in the training data. Top left: the input fetoscopic video frame. Top right: the ground truth segmentation. Bottom left: the segmentation generated by thresholding the Frangi filtered image. The Frangi filter mistakenly identifies the glare at the top of the image and the guide light at the center of the image as blood vessels. Bottom right: the segmentation generated by the FCNN. Note the close correspondence to the image created by the human rater
Generalizability
The ability of the neural network to generalize to novel patients—that is, patients whose images were not included in the training set—is critical if this technology is to be applied to clinical practice, as training images for a particular patient will not be available preoperatively. The sensitivity, specificity, and Dice coefficients listed for the FCNN in Table 1 were calculated exclusively on patients that were not included in the training set using a leave-one-out strategy as described in “Quantification” section. The high accuracy of the FCNN on novel patients demonstrates that it has good generalizability.
Robustness to threshold variations
As described in “Selection of ideal thresholds” section, our patch-based method produces a grayscale image that must be binarized with an arbitrary threshold to produce a segmentation. To determine the robustness of the segmentation algorithm to variations in this threshold, we binarized the grayscale images produced by the neural network at varying thresholds and calculated the sensitivities and specificities of the segmentations at each threshold value. The results are summarized by the receiver operating characteristic in Fig. 5, which shows that the segmentations produced by the FCNN are robust across a wide variety of thresholds. The area under the curve (AUC) is 0.987.
Fig. 5.
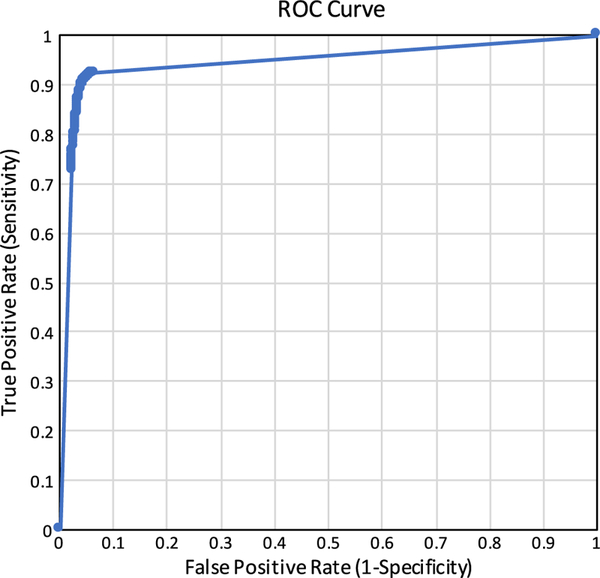
The receiver operating characteristic for binarizing the segmentations produced by the FCNN at different decision thresholds. All possible thresholds (integers between 0 and 255, inclusive) were evaluated and included in this plot. The area under the curve (AUC) is 0.987
Video frame enhancement
Segmentations produced by the FCNN were recombined with the original input fetoscopic video frames in several ways to produce enhanced images, as might be presented to the surgeon if this technology were to be used intraoperatively. The enhanced images are summarized in Fig. 6.
Fig. 6.
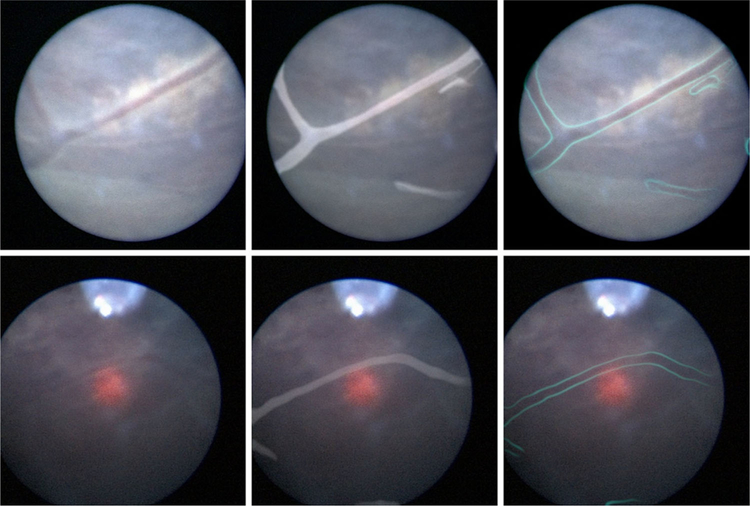
Top left: An unenhanced fetoscopic video frame with easily visible blood vessels. This is the same frame that is shown in Fig. 4. Bottom left: An unenhanced video frame with a difficult to discern blood vessel. By combining the segmentation provided by the FCNN with the original fetoscopic video frame, it is possible to enhance the original frame in various ways, such as highlighting the blood vessels themselves (middle) or their edges (right). We envision a system in which surgeons can toggle between the enhanced video and unaltered video at will depending on which modality bests suits their needs at the given moment
Discussion
Accurately identifying blood vessels is arguably the most important task in fetoscopic laser photocoagulation surgery. The surgeon’s ultimate goal is to find and cauterize abnormal vascular formations, and the surgeon’s main tool for finding these abnormal formations is to track normal blood vessels. However, the surgeon’s ability to visualize blood vessels is impaired by the poor quality of video acquired by standard fetoscopes and by environmental factors such as the turbidity of amniotic fluid. There is therefore good reason to believe that a system for digitally enhancing the visibility of placental vasculature within fetoscopic video would aid the surgeon’s performance during laser photocoagulation.
Traditional techniques for the computerized enhancement of blood vessels such as Frangi vesselness filtering and matched filtering do not translate well to fetoscopic video. This work demonstrates that deep-learned vessel segmentation is quantifiably better than non-deep-learned methods and novice humans at identifying blood vessels in in vivo fetoscopic images. We believe that the enhancement of fetoscopic video using deep-learned segmentations has the potential to be useful to surgeons in the near future: The ground truth segmentations required for training the network can be prepared in a matter of a few days, no new hardware is required, and the software can be easily integrated with existing surgical computer towers. Furthermore, this method is noninvasive and carries no risk to the patient or fetus.
There are limitations to the approach outlined in this paper: The FCNN operates on still video frames and is therefore ignorant of the temporal aspect of fetoscopic video. Temporal information could be useful in eliminating false positive vessels that “flicker” into existence in one video frame only to be eliminated one or two frames later frames later. Future work could leverage temporal information to give a more accurate result. While we demonstrated that the FCNN is able to accurately detect and enhance blood vessels in patients that it has not yet encountered, we have not yet demonstrated that the FCNN is able to generalize to new fetoscopes, as all data in this study were collected using fetoscopes of the same brand and model. We plan to investigate whether a single FCNN can be used to process images from multiple different fetoscope models, or whether separate FCNNs must be trained.
The utility of automatic blood vessel segmentation is not limited to highlighting blood vessels within fetoscopic video; blood vessel segmentations could also be used as features for computing frame-to-frame homographies for the creation of a map of the placental surface. Most existing work on constructing panoramic maps of the placenta has relied on general-purpose feature descriptors such as SIFT, SURF, and ORB [8, 10, 11]. Such feature descriptors are attractive initial options for mosaicking algorithms, as they are easy to describe mathematically and do not require application-specific training datasets. They have poor repeatability when applied to fetoscopic video frames, however, which makes it difficult to consistently extract enough frame-to-frame correspondences to estimate the motion of the fetoscope [13]. Deep-learned vessel segmentations have the potential to be invariant to many of the visual distractors present in fetoscopic video frames and therefore have the potential to serve as more robust and more repeatable features for panorama construction. We plan to investigate the utility of incorporating our placental vessel detection technique into the placental map construction process.
Acknowledgments
Funding This work was supported by the National Institutes of Health Grant Number T35DK104689 (NIDDK Medical Student Research Fellowship).
Footnotes
Compliance with ethical standards
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11548–018-1886–4) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent Informed consent was obtained from all individual participants included in the study.
References
- 1.Cordero L, Franco A, Joy D, O’Shaughnessy W (2005) Monochorionic diamniotic infants without twin-to-twin transfusion syndrome. J Perinatol 25:753–758. 10.1038/sj.jp.7211405 [DOI] [PubMed] [Google Scholar]
- 2.Bahtiyar O, Emery P, Dashe S, Wilkins-Haug E, Johnson A, Paek W, Moon-Grady J, Skupski W, O’Brien M, Harman R, Simpson L (2015) The North American Fetal Therapy Network consensus statement: prenatal surveillance of uncomplicated monochorionic gestations. Obstet Gynecol 125:118–123. 10.1097/AOG.0000000000000599 [DOI] [PubMed] [Google Scholar]
- 3.Faye-Petersen M, Crombleholme M (2008) Twin-to-twin transfusion syndrome. NeoReviews 9:370–379 [Google Scholar]
- 4.Emery P, Bahtiyar O, Moise J (2015) The North American Fetal Therapy Network consensus statement: management of complicated monochorionic gestations. Obstet Gynecol 126:575–584. 10.1097/AOG.0000000000000994 [DOI] [PubMed] [Google Scholar]
- 5.Luks F (2009) Schematic illustration of endoscopic fetal surgery for twin-to-twin trans-fusion syndrome
- 6.Pratt R, Deprest J, Vercauteren T, Ourselin S, David L (2015) Computer-assisted surgical planning and intraoperative guidance in fetal surgery: a systematic review. Prenat Diagn 35:1159–1166. 10.1002/pd.4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller R, Novotny J, Laidlaw H, Luks F, Merck D, Collins S (2016) Virtually visualizing vessels: a study of the annotation of placental vasculature from MRI in large-scale virtual reality for surgical planning Brown University, Providence [Google Scholar]
- 8.Tella-Amo M, Daga P, Chadebecq F, Thompson S, Shakir I, Dwyer G, Wimalasundera R, Deprest J, Stoyanov D, Vercauteren T, Ourselin S (2016) A combined EM and visual tracking probabilistic model for robust mosaicking: application to fetoscopy. In: Proceedings of IEEE CVPR workshops, vol 31, pp 84–92. 10.1515/10.1109/cvprw.2016.72 [DOI] [Google Scholar]
- 9.Graves E, Harrison R, Padilla E (2017) Minimally invasive fetal surgery. Clin Perinatol 44:729–751. 10.1016/j.clp.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Tchirikov M, Oshovskyy V, Steetskamp J, Falkert A, Huber G, Entezami M (2011) Neonatal outcome using ultrathin fetoscope for laser coagulation in twin-to-twin-transfusion syndrome. J Perinat Med 10.1515/jpm.2011.091 [DOI] [PubMed]
- 11.Olguner M, Akgür M, Özdemir T, Aktug˘ T, Özer E (2000) Amniotic fluid exchange for the prevention of neural tissue damage in myelomeningocele: an alternative minimally invasive method to open in utero surgery. Pediatr Neurosurg 33:252–256. 10.1159/000055964 [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Wang J, Ando T, Kubota A, Yamashita H, Sakuma I, Chiba T, Kobayashi E (2016) Towards scene adaptive image correspondence for placental vasculature mosaic in computer assisted fetoscopic procedures. Int J Med Robot Comput Assist Surg 12:375–386. 10.1002/rcs.1700 [DOI] [PubMed] [Google Scholar]
- 13.Gaisser F, Peeters S, Lenseigne B, Jonker P, Oepkes D (2018) Stable image registration for in vivo fetoscopic panorama reconstruction. J Imaging 4:24 10.3390/jimaging4010024 [DOI] [Google Scholar]
- 14.Almoussa N, Dutra B, Lampe B, Getreuer P, Wittman T, Salafia C, Vese L (2011) Automated vasculature extraction from placenta images. In: Medical imaging 2011: image processing, vol 7962 International Society for Optics and Photonics [Google Scholar]
- 15.Park M, Yampolsky M, Shlakhter O, VanHorn S, Dygulska B, Kiryankova N, Salafia C (2013) Vessel enhancement with multi-scale and curvilinear filter matching for placenta images. Placenta 34:A12 [Google Scholar]
- 16.Chang JM, Huynh N, Vazquez M, Salafia C. (2013) Vessel enhancement with multiscale and curvilinear filter matching for placenta images. In: 2013 20th international conference on systems, signals and image processing (IWSSIP), pp 125–128 [Google Scholar]
- 17.Perera Bel E (2017) Ultrasound segmentation for vascular network reconstruction in twin-to-twin transfusion syndrome. M.S. Thesis, Pompeu Fabra University, Barcelona, Spain: https://repositori.upf.edu/handle/10230/33180. Accessed 13 Nov 2018 [Google Scholar]
- 18.Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. MICCAI 18:234–241. 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 19.Panchapagesan S, Sun M, Khare A, Matsoukas S, Mandal A, Hoffmeister B, Vitaladevuni S (2016) Multi-task learning and weighted cross-entropy for DNN-based keyword spotting. In: Inter-speech, pp. 760–764. 10.21437/Interspeech.2016-1485 [DOI] [Google Scholar]
- 20.Dice R (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302. 10.2307/1932409 [DOI] [Google Scholar]
- 21.Frangi F, Niessen J, Vincken L, Viergever A (1998) Multiscale vessel enhancement filtering. MICCAI 1496:130–137 [Google Scholar]
- 22.Srivastava R, Wong K, Duan L, Liu J, Wong TY (2015) Red lesion detection in retinal fundus images using Frangi-based filters. IEEE EMBC 37:5663–5666. 10.1109/EMBC.2015.7319677 [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, Zhuang W, Sinusas J, Staib H, Papademetris X (2011) Vessel connectivity using Murray’s hypothesis. MICCAI 14:528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]


