Abstract
Objective:
In this systematic review, we aim to synthesize the literature on the use of natural language processing (NLP) and text mining as they apply to symptom extraction and processing in electronic patient-authored text (ePAT).
Materials and Methods:
A comprehensive literature search of 1,964 articles from PubMed and EMBASE was narrowed to 21 eligible articles. Data related to purpose, text source, number of users and/or posts, evaluation metrics, and quality indicators were recorded.
Results:
Pain (n=18) and fatigue and sleep disturbance (n=18) were the most frequently evaluated symptom clinical content categories. Studies accessed ePAT from sources such as Twitter and online community forums or patient portals focused on diseases, including diabetes, cancer, and depression. Fifteen studies used NLP as a primary methodology. Studies reported evaluation metrics including the precision, recall, and F-measure for symptom-specific research questions.
Discussion:
NLP and text mining have been used to extract and analyze patient-authored symptom data in a wide variety of online communities. Though there are computational challenges with accessing ePAT, the depth of information provided directly from patients offers new horizons for precision medicine, characterization of sub-clinical symptoms, and the creation of personal health libraries as outlined by the National Library of Medicine.
Conclusion:
Future research should consider the needs of patients expressed through ePAT and its relevance to symptom science. Understanding the role that ePAT plays in health communication and real-time assessment of symptoms, through the use of NLP and text mining, is critical to a patient-centered health system.
Keywords: Natural language processing, signs and symptoms, electronic patient-authored text, review
BACKGROUND AND SIGNIFICANCE
According to the most recent study from the Pew Research Center, a leading nonpartisan data-driven research organization, up to 80% of Internet users, or about 93 million distinct individuals in the United States, use the Internet to get information about a health condition1. Of that percentage, almost 8 million user posts are about specific symptoms, adverse effects from medication, and/or general healthcare experiences1. The amount of symptom-related information directly from patients outside of the electronic health record (EHR) is unprecedented and not routinely integrated with the formal healthcare system. These texts, referred to as electronic patient-authored text (ePAT), are proliferating at a time of increased healthcare costs, a lack of access to care, and higher rates of technological literacy among patients2. While we know this information exists, it is stored in diverse text formats (e.g., tweets, comments, blogs) across hundreds of host websites (e.g., Twitter, WebMD, Reddit). In addition, medical terms are often embedded with other patient narratives which make further research on symptoms more challenging without advanced computational tools.
One algorithmic toolbox for identifying and managing text of interest is natural language processing (NLP) and more broadly, text mining. Text mining comprises a range of techniques used for characterizing and transforming text. Within text mining, NLP is a collection of syntactic and/or semantic rule- or statistical-based processing algorithms that can be used to parse, segment, extract, or analyze text data3. What differentiates these approaches is the use of text analysis; text mining uses the words themselves as a unit of analysis (e.g., frequencies, the presence or absence of specific words of interest), while NLP methods utilize the underlying metadata including content and phrase patterns. Both NLP and text mining have been used in an array of health-related online domains such as mental health4, oncology5, and infectious disease6.
ePAT is a critical component of understanding symptoms and patient experiences associated with their symptom status. Symptoms are the self-reported perceptions and experiences of a disease or condition7. Symptoms include physical experiences, such as pain and disturbed sleep, or emotional experiences, such as anxiety or forgetfulness7. Most patients experience several concurrent symptoms contributing to their current health status. To illustrate the diverse benefits of sharing symptom information online, Wicks et al. conducted a study which included contacting patients who were participating in an epilepsy forum on PatientsLikeMe.com, a website developed for patients to share their experiences with the intention to improve their outcomes. Over half of patients in this study reported perceived benefits such as finding another patient experiencing the same symptoms and learning more about relevant symptoms8.
While some patients seek Web 2.0 (a term used to describe communication rich, dynamic, user-generated wed-based communities9) applications for sharing medical experiences, others use it to characterize undiagnosed symptoms. Identifying sub-clinical symptoms from ePAT can help to further refine diagnostic criteria leading to better and more efficient care. An analysis of Crowdmed (crowdmed.com), an online crowdsourcing diagnostic platform where users post their signs and symptoms in the hopes of diagnostic resolution, found that patients interacting on Crowdmed spent a median of 50 hours researching their illnesses online and had symptoms for a median of 2.6 years before finding a diagnosis10. Another study on the same platform found that users had fewer medical visits (P=.01) and lower medical costs after resolution (P=.03)11. These studies show that symptom-focused ePAT can have significant implications for experience sharing, time between symptom onset and diagnosis, and healthcare-related expenses.
Despite the widespread availability of symptom-related ePAT, symptom information is routinely assessed only in the context of the EHR and formal review by clinical experts. This practice has clear limitations because symptom information documented in the EHR is often not well captured and may not be self-reported directly by the patient. Further, the use of symptom-based ePAT on a population-level without access to specific patient characteristics or confounding variables can introduce errors in future studies or clinical decision-making. In addition, patients usually experience symptoms for a measurable time-period before seeking care. However, the increased use of host websites to communicate symptom information has created an opportunity for NLP and text mining to leverage the potential of ePAT to advance symptom science and management.
OBJECTIVE
The purpose of the present study is to systematically review the literature on the use of NLP and text mining to process and/or analyze symptom information from ePAT. In particular, we report the following aspects of the included studies: 1) study characteristics including the purpose and data source, 2) relevant symptom information, 3) NLP or text mining use, evaluation, and performance, and 4) indicators of quality. We further synthesize and discuss the current trends across studies, values and challenges to how ePAT could influence clinical symptoms and care and persistent gaps related to ePAT.
METHODS
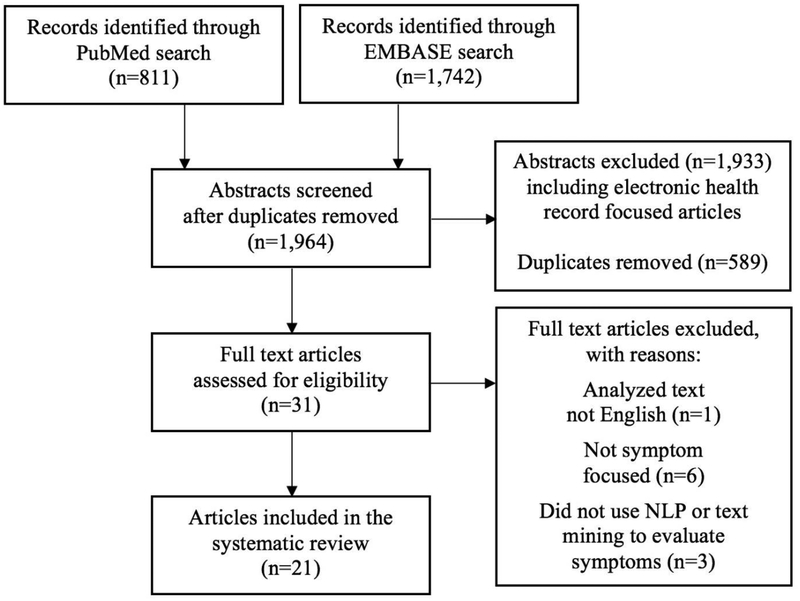
We searched PubMed and EMBASE in February 2018 with the original goal of identifying all relevant abstracts related to NLP, text mining, and symptoms from all types of free-text. A corpus distinction between EHRs and ePAT became apparent during the review process. Therefore, we decided to conduct two corpus-specific systematic reviews – the first focused on EHRs12 and the second on ePAT. We defined a symptom as a subjective indication of disease (e.g., anxiety, depressed mood, fatigue, disturbed sleep, impaired cognition, and nausea). Table 1 shows the search terms, derived from the Medical Subject Headings vocabulary (MeSH; US National Library of Medicine) for the database queries, that were used. Inclusion of terms for specific symptoms was guided by presence of the symptom in National Institute of Nursing Research common data element (CDE) measures which focus on patient-reported outcomes. Consequently, the term nurs* was included to increase the depth of the search and potentially capture additional symptom science concepts (e.g., patient-reported outcomes) that may not be formally labeled as “signs and symptoms.” The searches, which were limited to English language with no publication date restriction, resulted in 811 records from PubMed and 1,742 records from EMBASE; 589 articles were duplicates and removed from further assessment. Our review utilized the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) recommendations including assessment of research content as well as a detailed report of the number of records identified through the search, number of studies that were included and excluded in the review, and the reasons for exclusion. See Figure 1 for an illustration of our PRISMA workflow.
Table 1.
Search terms used to retrieve records
| Database | Search Terms |
|---|---|
| PubMed | (natural language processing [mh] OR natural language processing [tw] OR NLP [tw] OR text mining [tw]) AND (signs and symptoms [mh] OR symptom* [tw] OR nursing [mh] OR nurs* [tw] OR pain [mh] OR pain [tw] OR anxiety [mh] OR anxi* [tw] OR cognition [mh] OR cognit* [tw] OR cognitive function [tw] OR attention [tw] OR memory [tw] OR executive function [tw] OR sleep [mh] OR dyssomnias [mh] OR sleep* [tw] OR fatigue [mh] OR fatigue [tw] OR depression [mh] OR depress* [tw] OR affect [mh] OR affective symptoms [mh] OR affect* [tw] OR mood [tw] OR well being [tw] OR well-being [tw] OR nausea [mh] OR nausea [tw]) AND english [la] |
| EMBASE | (‘natural language processing’/exp OR ‘natural language processing’:ab,ti,kw OR ‘nlp’:ab,ti,kw OR ‘text mining’/exp OR ‘text mining’:ab,ti,kw) AND (‘symptom’/exp OR ‘symptomatology’/exp OR ‘symptom*’:ab,ti,kw OR ‘nursing’/exp OR ‘nurs*’:ab,ti,kw OR ‘pain’/exp OR ‘pain’:ab,ti,kw OR ‘anxiety’/exp OR ‘anxi*’:ab,ti,kw OR ‘cognition’/exp OR ‘cognit*’:ab,ti,kw OR ‘cognitive function’:ab,ti,kw OR ‘sleep’/exp OR ‘sleep disorder’/exp OR ‘sleep*’:ab,ti,kw OR ‘fatigue’/exp OR ‘fatigue’:ab,ti,kw OR ‘depression’/exp OR ‘depress*’:ab,ti,kw OR ‘mood disorder’/exp OR ‘mood’:ab,ti,kw OR ‘affect*’:ab,ti,kw OR ‘wellbeing’/exp OR ‘well being’:ab,ti,kw OR ‘well-being’:ab,ti,kw OR ‘nausea’/exp OR ‘nausea’:ab,ti,kw) AND [english]/lim |
Figure 1.
PRISMA flow diagram of included articles.
The articles that resulted from the search were imported into Covidence (covidence.org), a web-based tool designed to facilitate screening and data extraction for systematic reviews. To be eligible for inclusion in the review, the primary requirement was that the article needed to focus on the description, evaluation, or use of an NLP or text mining algorithm/pipeline to process and/or analyze patient symptoms. Review articles, non-English language articles, and articles without full text available were excluded. Two authors (TAK, CD) independently reviewed each title and abstract with discussion to provide consensus. After the initial screening, 31 articles were included in the full text review. The same authors narrowed the final included articles for this review to 21. There were several reasons for exclusion including: analyzed text not English (n=1), not focused on symptoms (n=6), and did not use NLP or text mining to evaluate symptoms (n=3).
Data related to study purpose, corpus, patient-authors, symptoms, methodology, and outcomes were manually extracted (CD, TAK) from the 21 articles included in the review (Table 2). Trends related to extracted elements were evaluated over time. In addition, while a formal quality assessment was not conducted (as relevant reporting standards have not been established for NLP and text mining articles), we report indicators of quality based on elements included in previous NLP-focused systematic reviews13–15.
Table 2.
Study characteristics
| Author< | Purpose | Data Source | Text Type | Number of Docum ents | Number of Users |
|---|---|---|---|---|---|
| Brennan & Aron | To evaluate the application of MetaMap for detecting the presence of terms found in the UMLS within the electronic messages of patients | Internet-based home care post-discharge support service intervention | Emails sent by patients to a clinical nurse | 241 electronic messages | Not reported |
| Portier et al, 201333 | To examine whether sentiment change is influenced by the main topic of the initiating post | Online peer support community for cancer patients (ACS’s Cancer Survivors Network) forums for breast and colorectal cancer | Discussion posts | 29,384 threaded discussions | Not reported |
| Freifeld et al, 201427 | To evaluate the level of concordance between Twitter posts mentioning adverse event-like reactions and spontaneous reports received by a regulatory agency | Tweets with mentions of 23 drugs and 4 vaccines and resemblance to adverse events | 4,401 tweets | Not reported | |
| Gupta et al, 201419 | To extract symptoms and conditions as well as drugs and treatments from patient-authored text by learning lexico-syntactic patterns from data annotated with seed dictionaries | Online community forum (MedHelp.org) for asthma, ENT, adult type II diabetes, acne, and breast cancer | Sentences | 680,071 sentences | Not reported |
| Park & Ryu, 201425 | To evaluate the possibility of using text-mining to identify clinical distinctions and patient concerns in online memoirs posted by patients with fibromyalgia | Online social networking community (Experienceproject.com) forum with title “I Have Fibromyalgi a” | Patient narratives | 399 narratives | Not reported |
| Janies et al, 201528 | To create a web-based workflow application that uses chief complaints from Twitter as a syndromic surveillance tool and correlates outbreak signals to pathogens known to circulate a geographic area | Tweets | >1,000,000 tweets | Not reported | |
| Jimeno-Yepes et al, 201520 | To develop an annotated data set from Twitter feeds that can be used to train and evaluate methods to recognize mentions of diseases, symptoms, and pharmacologic substances in social media | Tweets with 2 out of 3 entity types – diseases symptoms, or pharma cological substances | 1,300 tweets | Not reported | |
| Karmen et al, 201521 | To develop a method that detects symptoms of depression in free text from social media sources | Online public mental health message board (Psycho-Babble) “Grief” forum | User posts within a 20–200 word interval | 1,304 posts | Not reported |
| Liu & Chen, 201523 | To develop a research framework for patient reported adverse drug event extraction | 3 online patient forums for diabetes (ADA online community, Diabetes Forum, and Diabetes Forums) and 1 for heart disease (MedHelp.org) | Patient discussi on posts | 1,072,474 posts | Not reported |
| Nikfarjam et al, 201524 | To design a machine learning-based approach to extract mentions of adverse drug reactions from social media text | Twitter and a health related social network (DailyStrength) | Tweets and user posts for 81 widely used drugs | 1,784 tweets and 6,279 post from DailyStrength | Not reported |
| Tighe et al, 201532 | To examine the type, context, and dissemination of pain-related tweets from 50 cities around the world | Tweets with mention s of pain | 47,958 tweets | Not reported | |
| Eshleman & Singh, 201618 | To described a framework based on graph-theoretic modeling of drug-effect relationships drawn from various data sources | Tweets with mentions of 200 commonly prescribed drugs | 157,735 tweets | 7,981 | |
| Lee & Donovan, 201635 | To understand the symptom experiences and strategies that were associated with fatigue management among women with ovarian cancer | Web-based ovarian cancer symptom management intervention | Patient respons es to prompts | Not reported | 165 |
| Marshall et al, 201629 | To compare symptom cluster patterns derived from messages on a breast cancer forum with those from a symptom checklist completed by breast cancer survivors participating in a research study | Online community forum (MedHelp.or g) for breast cancer | Messag e posts | 50,426 posts | 12,991 |
| Topaz et al, 201630 | To compare electronic health record data and social media data to clinician-reported adverse drug reactions and patients’ concerns regarding aspirin and atorvastatin | Treato Ltd (treato.com) database of health-related websites, forums, blogs, and Treato discussion platform | List of potential adverse drug reactions for aspirin and atorvast atin | 42,594 potential adverse drug reactions | Not reported |
| Sunkureddi et al, 201634 | To describe patient experiences reported online to better understand the day-to-day disease burden of ankylosing spondylitis | 52 online sources, including social networking sites, patient- physician Q&A sites, and ankylosing spondylitis forums | Patient narratives | 34,780 narratives | 3,449 |
| Cocos et al, 201717 | To develop a scalable, deep-learning approach for adverse drug reaction detection in social media data | Tweets for 81 widely used drugs and 44 ADHD drugs | 844 tweets | Not reported | |
| Cronin et al, 201736 | To develop automated patient portal message classifiers for communication type (i.e., informational, logistic, social, medical, and other) | Patient portal (My Health at Vanderbilt) | Portal messages | 3,253 messages | 3,116 |
| Lamy et al, 201722 | To examine synthetic cannabinoid receptor agonist-related effects and their variations through a longitudinal content analysis of web-forum data | 3 drug focused web forums (Bluelight.or g and 2 anonymized forums) | User posts related to synthetic cannabinoid receptor agonists | 19,052 posts | 2,543 |
| Lu et al, 201731 | To develop a content analysis method for stakeholder analysis, topic analysis, and sentiment analysis health care social media | Online community forum (MedHelp.org) for lung cancer, diabetes, and breast cancer | Messag e posts | 138,161 posts | 39,606 |
| Patel et al, 201726 | To detect and quantify glucocorticoid-related adverse events using a computerized system for automated suspected adverse drug reaction detection from narrative text in Twitter | Tweets with mentions of | 15,730 | Not reported | |
| prednis one or prednis olone | tweets |
Note.
Studies have been arranged in chronological order to assess trends over time; ACS=American Cancer Society; ADA=American Diabetes Association; ADHD=attention deficit hyperactivity disorder; ENT=ear, nose, and throat; UMLS=Unified Medical Language System
RESULTS
Twenty-one articles were included in the review. Years of publication ranged from 2003 to 2017 with over 95% (n=20) of articles published in the last 10 years.
Study characteristics
The purposes of the 21 articles were diverse and focused on identification, detection, extraction, and/or description of symptom terms, including adverse drug events (n=11)16–26; comparison of ePAT to another data source (n=4), including a regulatory agency27, molecular database28, research study29, or EHR30; content and/or sentiment analysis (n=3)31–33; description of the patient experience (n=2)34,35; and development of a classifier (n=1)36. Table 2 organizes each study purpose as well as data source, text type, number of documents, and number of users chronologically to assess overall trends. Over a third of the included studies (n=8) used Twitter as a main data source17,18,20,24,26–28,32. Ten studies used ePAT from disease-specific online communities including, but not limited to, breast cancer19,29,31,33, diabetes19,23,31, and mental health conditions21. The number of documents used for analysis ranged from 241 electronic messages16 to greater than 1 million tweets28. The text type of these documents varied between studies but was typically comprised of short length text documents including tweets17,18,20,24,26–28,32, message posts16,36, forum posts19,21–23, and patient responses to prompts35. Seven studies reported the number of users18,22,29,31,34–36. The number of users varied greatly, ranging from 165 to 39,606. Interestingly, all studies that reported the numbers of users were published in 2016 or later.
Relevant symptom information
Symptoms named in the methods, results, discussion, or supplementary sections of the review articles were manually extracted and grouped into clinically meaningful categories by the authorship team (TAK, CD). Symptom terms extracted included symptom-related words and phrases used by patients to described their current state of health or symptom concepts from or mapped to a medical vocabulary. Symptom clinical content categories included: pain (e.g., sore, ache), fatigue and sleep disturbance (e.g., insomnia, exhaustion), digestive system and abdomen (e.g., abdominal cramp, anorexia, nausea), affective mood (e.g., depressed mood, altered mood, anger), perception (e.g., dizziness, paresthesia), affective anxiety (e.g., restlessness, worry), circulatory and respiratory system (e.g., chest heaviness, dyspnea), musculoskeletal system (e.g., arthralgia, weakness), cognitive function (e.g., forgetful, dazed), skin and subcutaneous tissue (e.g., irritation, pruritus), genitourinary system (e.g., dysuria, sex drive), and general (e.g., chills). Table 3 lists symptoms mentioned in the included studies grouped by their clinical content category. Symptom terms represent both traditional medical vocabulary (e.g., insomnia, dyspnea, pruritus, paresthesia) and colloquial words/phrases (e.g., floaty head, jittery, head spinning, sluggishness).
Table 3.
Symptoms included in articles by clinical content category
| Clinical content category | Representative symptom concepts |
|---|---|
| Pain | ache, pain, sore, tenderness; arm pain; back pain; breast pain; ear pain, ear ache; eye irritation; flank pain; general aches, general pain, body discomfort; hand pain; headache, head pain, head discomfort; kidney pain; knee pain; leg pain; neck-skull ache, neck-skull pain; rib pain; shoulder pain; sore throat; tooth ache |
| Fatigue and sleep disturbance | abnormal dreams, nightmares, vivid dreams; asthenia; disturbed sleep, insomnia; drowsiness; energy, lassitude, listlessness; exhaustion; fatigue; lethargic; sluggishness; somnolence; tiredness; weariness |
| Digestive system and abdomen | abdominal pain; abdominal cramp; anorexia, changes in appetite, hunger; bloating; constipation; difficulty swallowing; dry mouth; dyspepsia, heartburn, indigestion; epigastric pain; gastrointestinal upset, upset stomach; nausea; stomach ache, stomach pain; thirst |
| Affective mood | affect lability, emotional instability, emotional stories, emotional support; aggressiveness, snappy; agitated, irritable mood; altered mood, mood changes, mood swing; anger, frustration; apathy, loss of interest, indifference; bliss, elation, euphoria, happiness, joy, pleasure, warm fuzziness; calmness, relaxed; depressed mood, depression; down feelings, feeling low, melancholy, miserable, sadness; feeling empty; fussiness; hopelessness; self-confidence; stress; suicidal ideation |
| Perception | blackout; blind, blurred vision, diplopia, loss of vision, visual disturbances, visual distortions; burning, formication, neuropathy, numbness, pins and needles, paresthesia, sensation, tingling; dizziness, lightheadedness, head rush, head spinning, wooziness; |
| Affective anxiety | antsy, restlessness; anxiety; concern about the future; dread; fear; nervousness; panic, panic attacks; paranoia; worry |
| Circulatory and respiratory system | angina, chest pain, chest heaviness, chest tightness; breathless, difficulty breathing, dyspnea, short of breath; congestion, stuffy; heart flutter, heart racing, palpitation; hot flash; lung discomfort, lung hurt, lung irritation, lung pain; sinus pressure |
| Musculoskeletal system | arthralgia, joint pain, joint tenderness; clonus, dyskinesia, musclespasms; cramping legs, muscle cramp; debility, loss of strength, weakness; muscle aches, muscle pain, musculoskeletal discomfort, musculoskeletal pain, myalgia; stiffness |
| Cognitive function | amnesia, forgetful, memory impairment; brain fog; clean head, cloudy head, empty head, floaty head, lethargic head; cognitive difficulties, cognitive impairment; confusion; craving; daydreaming, difficulty concentrating, easily distracted, focus, short attention span; dazed; difficulty thinking, trouble thinking; dissociation; psychomotor agitation, psychomotor impairment; ear distortion, deafness, tinnitus; feeling high; feeling jittery; hallucinations, hearing things; tightness |
| Skin and subcutaneous tissue | irritation; itchiness, pruritus; skin discomfort |
| Genitourinary system | dysuria; sex drive, sexual inhibition, sexual urge; sexual dysfunction; urgent urination; vaginal dryness |
| General | chills; malaise |
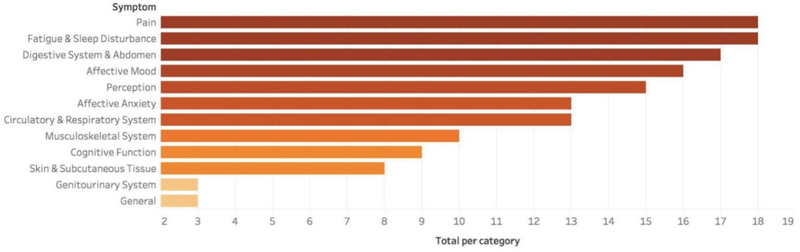
The number of studies with mentions of one or more symptoms in the corresponding clinical categories is visualized in Figure 2. Pain and fatigue and sleep disturbance were the most frequently analyzed categories (n=18 per category), followed by digestive system and abdomen (n=17), affective mood (n=16), and perception (n=15). In addition, 15 studies did not specifically name all of the symptoms used in the analysis and/or referred to standardized vocabularies (e.g., Unified Medical Language System [UMLS]).
Figure 2.
Number of studies grouped by symptom category.
Evaluation and performance of algorithms
Of the studies reviewed, 14 studies used NLP as the primary methodology16,17,19–24,26–28,30,34,36, six studies used text mining exclusively25,29,31–33,35, and one study used a combination of both approaches18. Table 4 summarizes the approach (NLP or text mining implementation), primary evaluation metric, and whether or not a comparison was performed for the analysis. The evaluation metric listed in the table reflects the relevant statistics related to the extraction or understanding of symptoms, not any secondary outcomes within the study.
Table 4.
Evaluation and performance metrics
| Author< | NLP/Text Mining | Primary Evaluation Metric | Comparative Evaluation |
|---|---|---|---|
| Brennan & Aronson, 200316 | NLP | Number of matched terms to vocabularies; reported matched frequencies higher for nursing complemented vocabularies | Compared several models on nursing, MeSH, and SNOMED terms |
| Portier et al, 201333 | Text mining | Descriptive results including sentiment analysis | None reported |
| Freifeld et al, 201427 | NLP | Automated, dictionary-based symptom classification had 72% recall and 86% precision | Results of annotation were compared to FDA Adverse Event Reporting System data |
| Gupta et al, 201419 | NLP | Extracts symptoms and conditions with a F-measure of 66–76% | Compared performance of two other programs, the OBA and the MetaMap annotator, with baseline and default parameters |
| Park & Ryu, 201425 | Text mining | Descriptive results including symptoms and clinical distinctions | None reported |
| Janies et al, 201528 | NLP | No reported algorithm evaluation metrics | None reported |
| Jimeno-Yepes et al, 201520 | NLP | Highest performing model (Micromed+Meta) had precision, recall, and F-measure as 72%, 60%, and 66%, respectively | Compared across exact and partials matches for five models |
| Karmen et al, 201521 | NLP | Average precision of 84% and an average F-measure of 79% | Compared algorithm results to independent expert ratings |
| Liu & Chen, 201523 | NLP | Average F-measure of 90% for drug entity extraction and average F- measure of 80% for medical event extraction | Compared several methods across patient-authored forums |
| Nikfarjam et al, 201524 | NLP | 86%, 78%, and 82% for precision, recall, and F-measure, respectively | Comparison between several methods including SVM, ADRMine, MetaMap, and a lexicon-based method |
| Tighe et al, 201532 | Text mining | Descriptive results including the average degree centrality of the reduced pain tweet corpus graph was 60.7 | Compared sentiment for relevant terms to objective terms |
| Eshleman & Singh, 201618 | NLP & text mining | Precision exceeding 85% and F- measure over 81% | Compared sentiment analysis to graph topology with co-occurring symptoms |
| Lee & Donovan, 201635 | Text mining | Descriptive results for symptom findings | None reported |
| Marshall et al, 201629 | Text mining | Descriptive results including cooccurrence and clustering for symptom findings | None reported |
| Topaz et al, 201630 | NLP | Descriptive results including symptom extraction | None reported |
| Sunkureddi et al, 201634 | NLP | Descriptive results including frequency ranking of reactions and patients’ concerns | None reported |
| Cocos et al, 201717 | NLP | Approximate match F-measure for RNN of 75% for ADR identification | Compared the BLSTM-RNN ADR classification, a baseline lexicon system, and a condition random-field model |
| Cronin et al, 201736 | NLP | Logistic regression for medical communications with AUC of 0.899 | Compared naive bayes, logistic regression, and random forest across different types of patient portal messages |
| Lamy et al, 201722 | NLP | No reported algorithm evaluation metrics | None reported |
| Lu et al, 201731 | Text mining | Descriptive results including sentiment scores, clustering of groups, and Jaccard similarities | None reported |
| Patel et al, 201726 | NLP | No reported algorithm evaluation metrics | Compared method between two datasets |
Note.
Studies have been arranged in chronological order to assess trends over time; ADR=adverse drug reaction; ADRMine (a machine learning-based concept extraction system that uses Conditional Random Fields); AUC=area under the curve; BLST=Binarized Long Short-Term Memory Network; FDA=Federal Drug Administration; F-measure=also known as F1 score or F-score in the published literature; MeSH=Medical Subject Headings; NLP=Natural Language Processing; OBA=open biomedical annotator; MetaMap (tool for recognizing Unified Medical Language System [UMLS] concepts in text); RNN=recurrent neural network; SVM=support vector machine
In reviewing the studies that utilized text mining, descriptive results included symptom characterization in the online community of interest (n=5)25,29,30,34,35; similarity scores, such as the Jaccard index, for clustering of symptoms (n=3)29,31,32; and sentiment analysis (n=2)31,33. Descriptive statistics in the text mining articles included symptom-specific analyses to understand patient reported co-occurrence of symptoms. Only one study that used text mining, Tighe et al., had a sentiment analysis comparison for non-relevant symptom terms32. The majority of studies, more than 70% as mentioned above, utilized NLP approaches for their main analytic method. The primary reported evaluation metrics were precision, recall, and F-measure. Other reported metrics depended on the task but included an area under the curve (AUC) score36 and an indicator score26. The indicator score, used in Patel et al., quantified the probability that the tweet contained an adverse drug reaction26. Across all studies, no specific trends in approach, evaluation, and performance were identified over time.
Indicators of quality across studies
Indicators of quality are summarized and compared across studies by year of publication in Table 5. Specifically, clarity of the study purpose statement, adequate description of the study approach, and presence of information related to the data source/text type, number of documents, number of users, evaluation metrics, and comparative evaluation. All studies have at least three of the seven quality indicators. Thirteen studies have at least six quality indicators16,17,19–21,23,24,26,27,29,31,32,34, with only two studies18,36 addressing all seven.
Table 5.
Indicators of quality across articles
| Author< | Clearly defined purpose% | Approach Adequately described@ | Data source/text type described* | Number of docum ents specifi ed* | Number of users specified* | Evaluation metric s reported*! |
Inclusion of comparative evaluation*# |
|---|---|---|---|---|---|---|---|
| Brennan & Aronson, 200316 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Portier et al, 201333 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Freifeld et al, 201427 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Gupta et al, 201419 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Park & Ryu, 201425 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Janies et al, 201528 | ✓ | ✓ | ✓ | ✓ | |||
| Jimeno-Yepes et al, 201520 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Karmen et al, 201521 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Liu & Chen, 201523 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Nikfarjam et al, 201524 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Tighe et al, 201532 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Eshleman & Singh, 201618 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Lee & Donovan, 201635 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Marshall et al, 201629 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Topaz et al, 201630 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Sunkureddi et al, 201634 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Cocos et al, 201717 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Cronin et al, 201736 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Lamy et al, 20 1 722 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Lu et al, 201731 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Patel et al, 201726 | ✓ | ✓ | ✓ | ✓ | ✓ |
Note.
Studies included in this table have been arranged in chronological order to assess trend over time;
A checkmark denotes reviewer judgement of clear statement of the study purpose;
A checkmark denotes reviewer judgement of adequate description of the study approach;
A checkmark denotes the presence of information in the article;
Evaluation metrics include descriptive results, accuracy, area under the curve, sensitivity, specificity, recall, or precision;
Comparison includes another algorithm or dataset
DISCUSSION
In this systematic review, over 1,900 abstracts were narrowed to 21 full-text articles on the identification and analysis of symptom-related ePAT in online communities. More than 50% of articles focused on the identification, detection, extraction, and/or description of symptom terms. Symptoms were studied in a variety of both general (e.g., Twitter, patient portals) and disease specific (e.g., mental health, diabetes, cancer, heart disease, asthma, fibromyalgia, ankylosing spondylitis) online communities.
A broad range of symptom content categories were evaluated by studies, including pain, fatigue and sleep disturbance, digestive system and abdomen, mood, perception, anxiety, circulatory and respiratory system, musculoskeletal system, cognitive function, skin and subcutaneous tissue, genitourinary system, and general. Inclusion of terms from the pain and fatigue and sleep disturbance categories tied as the most frequent clinical content category evaluated in studies in our review. Sleep, in particular, is important to note because research conducted by the American Academy of Family Practice has identified symptoms related to insomnia, tiredness, and daytime sleepiness as under-reported symptoms yet crucial to more significant diagnoses such as sleep apnea and narcolepsy37. On the other hand, pain was reported by almost 80 million patients seen in the healthcare system between 1999–2002 according to a National Center for Health Statistics Report38. Yet, the majority of physicians (60%) feel uncomfortable with managing pain in patients due to the subjectivity in measurement and fear of misidentifying patients at risk of misuse39. This incongruity may play a factor in the proper resolution of pain and patients seeking support online.
Upon further review of symptom-related information, we noted that all but one of the articles35 evaluated symptoms from at least two symptom clinical concept categories. Moreover, while no single article mentioned symptoms from all twelve of the symptom categories, five articles evaluated symptoms in ten or more categories17,18,22,26. Given that patients rarely experience a single symptom in isolation, studying two or more concurrent symptoms that may share underlying mechanisms and outcomes is especially advantageous40. In fact, a component of symptom science as outlined in National Institutes of Health Symptom Science Model (NIH-SSM), is the identification of symptoms or clusters of symptoms41. Two of the studies represented in this review specifically aimed to identify clusters of symptoms to find relationships and sub-types amongst symptoms29,31. This research is substantial to addressing the complexity of symptoms as they relate to each other and/or subtypes of the symptoms as patient’s experience them.
The results of this review also highlight important findings related to the differences between symptoms reported in formal settings (e.g., EHRs, databases) and informal communities, such as those represented in Web 2.0. It is of interest to note that the purpose of four of the studies included in the review was the comparison of ePAT to another, more formal data source27–30. Specifically, these findings include (1) the widespread use of real-time social networking sites (i.e., Twitter) and short online documentation as the data source for ePAT and (2) the colloquial nature of symptoms and the diverse nature of language online.
Our review revealed that Twitter was the online community most frequently used to access and study symptom-related ePAT. The eight studies that used Twitter largely focused on pharmacological vigilance and assessment of public health outbreaks. Posts made by users on Twitter are termed “tweets” and are limited to 140, and more recently 280, English characters. In contrast to other textual sources of patient symptom information, such as clinical notes within EHRs (which are typically much longer in length and can be limited by reporting lags), the short length of tweets in combination with real-time, interactive posting of online communities, has the potential to facilitate immediate, scalable access for pharmaceutical companies and governmental agencies, like the Centers for Disease Control or Federal Drug Administration (FDA), to monitor changes in symptoms. This access could have significant implications for examining the recent opioid crisis and patient management of pain. Of note, over 85% of studies included in our review included mentions of terms from the pain clinical content category, which supports the potential for future monitoring and research for this purpose.
While Web 2.0 (particularly social media platforms aimed at user sharing) have great potential for monitoring patient symptoms and understanding patient reported symptoms, there continues to be a gap in being able to leverage text mining and NLP to study heterogeneity of symptoms and experiences. Several studies that were ultimately excluded from this review used NLP to identify terms related to disease conditions yet manually reviewed symptom information. For example, Curtis et al. used NLP to analyze social media data for posts mentioning inflammatory arthritis. In their study, all symptom information was manually extracted and then analyzed using disproportionality methods42. These methods are used to identify statistical associations between products, in this case medications, and specific events according to the FDA Adverse Event Reporting System. Treato, the NLP algorithm used for this study, includes resolution of patient-specific symptom terms but it was not implemented for FDA reporting. Martinez et al., also using the Treato system, performed manual coding of symptoms for qualitative analysis43. These two studies call into question the true efficacy of Treato to extract relevant symptoms.
One reason for issues regarding automation versus manual extraction of symptoms is that the medical community continues to lack gold-standard formal and lay or colloquial symptom lexicons. Our categorization of symptom terms into clinical content categories clearly illustrates the broad assortment of words that patients use to describe their current health state with many of the words being colloquial in nature. Informal symptom-related terms such as brain fog, jittery, and sluggishness exude clear dysfunction but are not traditional medical terms (e.g., dyspnea, congestion, amnesia) captured in EHRs or medical lexicons. In addition to informal terms, a 2015 study of emojis, the symbols often tagged on social media posts, found that sentiments of Twitter posts that have emojis are significantly different to those without44. Understanding how patients report their symptoms online, and the rationale for doing so, is an open area of research which the studies included in this review begin to address.
Several studies have attempted to build a lexicon of terms that help to annotate social media and web communities with medical terms. Alvaro et al. created a corpus of 1,000 tweets and 1,000 PubMed sentences with semantically correct annotations for a set of drugs, diseases, and corresponding symptoms45. Similarly, Mowery et al. developed a comprehensive coding scheme for annotating Twitter data with symptoms (e.g., depressed mood, agitation) from the Diagnostic and Statistical Manual of Mental Disorders, Edition 546. A study by Keselman et al. attempted to find consumer health concepts that were unable to be mapped in the UMLS lexicon47. The study affirmed 64 unmapped concepts for which 17 were labeled as lay terms47. Fascinatingly, only one study in our review mapped terminology to UMLS codes16. Following up on the Keselman et al. study, Zeng-Treitler and her team created an automated computer assisted update (CAU) system to identify new lay terms that could be included in the consumer health vocabulary (CHV)48. CHVs are patient-used terms and phrases that uniquely describe the patient experience and are distinctly separate from medical terminology48. The CAU system found 88,994 unique terms by web scraping over 300 PatientsLikeMe.com webpages. These corpora are critical to lay the groundwork for future research in this area as mappings to more standard vocabularies allow for the consistency and comparability across studies needed to advance the science.
We would like to acknowledge one additional computational challenge of using ePAT to study symptoms – accessing ePAT data. Accessing ePAT data in Web 2.0 from a host website using “web scraping” software is required prior to processing symptom information with techniques like NLP and text mining. Web scraping, or crawling, is a term that identifies the procedures (i.e., fetching the website, identifying the data of interest, advancing pages and links, and organizing the output) needed to pull free text from the World Wide Web. Twitter is a social media exception because daily releases of tweets are made available for research purposes via NodeXL49 (Microsoft open source software) or directly through Twitter’s application programming interfaces (API). Retrospective releases of tweets are also available for a fee. Nevertheless, ePAT in the majority of web communities is organized in non-direct ways that require additional scraping. It is clear that simple methods, such as copy and pasting text through human intervention, are not scalable and should be automated. This automation is significant and likely requires customization for each web platform. Some browsers such as Internet Explorer and Google Chrome offer add-on tools that aid in web scraping but further quality assurance and validation is needed to confirm that data is not systematically missed. As one example from the articles in our review, Liu & Chen developed their own automated web scraping program to extract specific fields in the patient discussion forum23. The relevant extracted data included the post unique identifier, the URL, a topic title, the user ID of the poster, the post date, and the post content23. In contrast, EHR data is often sourced from clinical data repositories that make pulling data fairly straightforward (notwithstanding missing data). Likely due to the computational and programming expense of web scraping for relevant symptom data, a higher proportion (28.6%, n=21) of studies represented in this review specifically employed text mining approaches (e.g., word frequencies and sentiment analysis) as the primary methodology rather than NLP compared to studies in our recent EHR focused systematic review (7.4%, n=27)12. Regardless of the specific strategy, both NLP and text mining of ePAT help to provide a pulse on the conversation of symptoms in the Web 2.0 space. Notwithstanding the potential impact of ePAT, it is also important to consider the ethical implications of gathering this data type and the lack of consent within the public sphere.
Future applications of NLP and text mining in Web 2.0 integrate well with the goals of the National Library of Medicine’s interest in funding data science research regarding personal health libraries (PHL). PHLs are central informatics approaches that help a patient gather, manage, and organize their health information (e.g., symptom information, family history, medication records, genomic information, diagnostic images, physical activity) from multiple data sources (e.g., online communities or patient portals, electronic self-management applications, and electronic health records)50. PHLs could help to increase health literacy, better inform patient-centered care, give patients the tools for improved symptom management, and facilitate quality data collection in large scale patient-consented studies. Quality data collection must include privacy considerations that aim to protect patient health information (e.g., creating requisite firewalls, respecting user pseudonyms, and monitoring for unintended use).
While this review provides a general scope for the use of ePAT for addressing symptom science, there are limitations of this review. First, we only queried two databases for our review. It is possible that additional relevant articles not indexed in EMBASE or PubMed were omitted; however, we feel that these databases were the most relevant for our research question due to their content focus on biomedical literature. To make the review clinically meaningful and practically manageable (as there are hundreds of unique symptoms/symptom terms), we used both general (e.g., signs and symptoms, nursing) as well as specific symptom search terms (e.g., pain, cognition). Our selection of specific symptoms search terms was guided by evaluation of a symptom using a National Institute of Nursing Research CDE measure. Exclusion of additional specific symptom search terms (e.g., chills) may have caused us to inadvertently miss relevant articles and may have influenced which symptoms, and consequently symptoms categories, that we found to be most frequently evaluated. However, inclusion of search terms for symptoms from CDEs provides best-practice guidelines for identifying literature addressing symptom science51. Additionally, the original search was implemented to address all EHR and ePAT-related articles but the dichotomy between the goals of these papers lead to the creation of separate analyses. We report on 27 studies focusing on EHR data in a separate systematic review12.
CONCLUSION
In this systematic review, we synthesized data from 21 articles on the use of NLP and/or text mining to extract and analyze symptom information from ePAT in online communities. Considering the prevalence of active Internet users sharing their symptom experiences and/or seeking symptom information online, the current focus of the field is on extraction of relevant medical terms and appropriate classification of symptoms. Research on symptoms in the Web 2.0 space would advance more rapidly with further development and access to applicable lexicons across clinical areas. Future research should consider the reliability of outcomes related to the size of the text corpus, role of health disparities and a lack of diverse representation in the corpus, and relevance of ePAT to symptom science. Understanding the role that ePAT plays in health communication, through the use of NLP and text mining techniques, is critical to characterization of sub-clinical symptoms and improved symptom self-management.
HIGHLIGHTS.
We know that electronic patient-authored text (ePAT) is a critical component of understanding symptoms and experiences in a patient-centered health system.
Understanding the role that ePAT plays in health communication, through the use of natural language processing (NLP) and text mining techniques, is critical to characterization of sub-clinical symptoms and improved symptom self-management.
This review synthesizes the literature on the use of NLP and text mining as they apply to symptom extraction and processing in ePAT.
Future applications of NLP and text mining in integrate well with the goals of the National Institutes of Health’s interest in funding data science research regarding personal health libraries and symptom science.
SUMMARY POINTS.
We know that electronic patient-authored text (ePAT) is a critical component of understanding symptoms and experiences in a patient-centered health system.
Understanding the role that ePAT plays in health communication, through the use of natural language processing (NLP) and text mining techniques, is critical to characterization of sub-clinical symptoms and improved symptom self-management.
This review synthesizes the literature on the use of NLP and text mining as they apply to symptom extraction and processing in ePAT.
Future applications of NLP and text mining in integrate well with the goals of the National Institutes of Health’s interest in funding data science research regarding personal health libraries and symptom science.
FUNDING
This work is supported by the Reducing Health Disparities Through Informatics training grant (T32NR007969), the Precision in Symptom Self-Management (PriSSM) Center (P30NR016587) and Advancing Chronic Condition Symptom Cluster Science Through Use of Electronic Health Records and Data Science Techniques (K99NR017651). CD is also supported by a National Research Service Award (F31NR017821).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
We have no conflicts of interest to disclose.
REFERENCES
- 1.Fox S, Duggan M. Health Online 2013 Pew Research Center Internet & American Life Project; 2013:1–55. [Google Scholar]
- 2.MacLean DL, Heer J. Identifying medical terms in patient-authored text: a crowdsourcing-based approach. J. Am. Med. Inform. Assoc 2013;20(6):1120–1127. doi: 10.1136/amiajnl-2012-001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J. Am. Med. Inform. Assoc 2011;18(5):544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo RA, Milne DN, Hussain MS, Christensen H. Natural language processing in mental health applications using non-clinical texts. Nat. Lang. Eng 2017:1–37. doi: 10.1017/S1351324916000383. [DOI] [Google Scholar]
- 5.Yim W-W, Yetisgen M, Harris WP, Kwan SW. Natural language processing in oncology: A review. JAMA Oncol 2016;2(6):797–804. doi: 10.1001/jamaoncol.2016.0213. [DOI] [PubMed] [Google Scholar]
- 6.Paul MJ, Sarker A, Brownstein JS, et al. Social media mining for public health monitoring and surveillance. Pac Symp Biocomput 2016;21:468–479. doi: 10.1142/9789814749411_0043. [DOI] [Google Scholar]
- 7.Dodd M, Janson S, Facione N, et al. Advancing the science of symptom management. J. Adv. Nurs 2001;33(5):668–676. doi: 10.1046/j.1365-2648.2001.01697.x. [DOI] [PubMed] [Google Scholar]
- 8.Wicks P, Keininger DL, Massagli MP, et al. Perceived benefits of sharing health data between people with epilepsy on an online platform. Epilepsy Behav 2012;23(1):16–23. doi: 10.1016/j.yebeh.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Lupiáñez-Villanueva F, Mayer MA, Torrent J. Opportunities and challenges of Web 2.0 within the health care systems: an empirical exploration. Inform. Health Soc. Care 2009;34(3):117–126. doi: 10.1080/17538150903102265. [DOI] [PubMed] [Google Scholar]
- 10.Meyer AND, Longhurst CA, Singh H. Crowdsourcing diagnosis for patients with undiagnosed illnesses: an evaluation of crowdmed. J. Med. Internet Res 2016;18(1):e12. doi: 10.2196/jmir.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juusola JL, Quisel TR, Foschini L, Ladapo JA. The impact of an online crowdsourcing diagnostic tool on health care utilization: A case study using a novel approach to retrospective claims analysis. J. Med. Internet Res 2016;18(6):e127. doi: 10.2196/jmir.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koleck T, Dreisbach C, Bourne P, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J. Am. Med. Inform. Assoc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pons E, Braun LMM, Hunink MGM, Kors JA. Natural language processing in radiology: A systematic review. Radiology 2016;279(2):329–343. doi: 10.1148/radiol.16142770. [DOI] [PubMed] [Google Scholar]
- 14.Canan C, Polinski JM, Alexander GC, Kowal MK, Brennan TA, Shrank WH. Automatable algorithms to identify nonmedical opioid use using electronic data: a systematic review. J. Am. Med. Inform. Assoc 2017;24(6):1204–1210. doi: 10.1093/jamia/ocx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra R, Bian J, Fiszman M, et al. Text summarization in the biomedical domain: a systematic review of recent research. J. Biomed. Inform 2014;52:457–467. doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan PF, Aronson AR. Towards linking patients and clinical information: detecting UMLS concepts in e-mail. J. Biomed. Inform 2003;36(4–5):334–341. doi: 10.1016/j.jbi.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Cocos A, Fiks AG, Masino AJ. Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J. Am. Med. Inform. Assoc 2017;24(4):813–821. doi: 10.1093/jamia/ocw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eshleman R, Singh R. Leveraging graph topology and semantic context for pharmacovigilance through twitter-streams. BMC Bioinformatics 2016;17(Suppl 13):335. doi: 10.1186/s12859-016-1220-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S, MacLean DL, Heer J, Manning CD. Induced lexico-syntactic patterns improve information extraction from online medical forums. J. Am. Med. Inform. Assoc 2014;21(5):902–909. doi: 10.1136/amiajnl-2014-002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jimeno-Yepes A, MacKinlay A, Han B, Chen Q. Identifying diseases, drugs, and symptoms in twitter. Stud. Health Technol. Inform 2015;216:643–647. [PubMed] [Google Scholar]
- 21.Karmen C, Hsiung RC, Wetter T. Screening Internet forum participants for depression symptoms by assembling and enhancing multiple NLP methods. Comput Methods Programs Biomed 2015;120(1):27–36. doi: 10.1016/j.cmpb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Lamy FR, Daniulaityte R, Nahhas RW, et al. Increases in synthetic cannabinoids-related harms: Results from a longitudinal web-based content analysis. Int. J. Drug Policy 2017;44:121–129. doi: 10.1016/j.drugpo.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Chen H. A research framework for pharmacovigilance in health social media: Identification and evaluation of patient adverse drug event reports. J. Biomed. Inform 2015;58:268–279. doi: 10.1016/j.jbi.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J. Am. Med. Inform. Assoc 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J, Ryu YU. Online discourse on fibromyalgia: text-mining to identify clinical distinction and patient concerns. Med. Sci. Monit 2014;20:1858–1864. doi: 10.12659/MSM.890793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel R, Belousov M, Jani M, et al. Frequent discussion of insomnia and weight gain with glucocorticoid therapy: an analysis of Twitter posts. npj Digital Med 2017;1(1):7. doi: 10.1038/s41746-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freifeld CC, Brownstein JS, Menone CM, et al. Digital drug safety surveillance: monitoring pharmaceutical products in twitter. Drug Saf 2014;37(5):343–350. doi: 10.1007/s40264-014-0155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janies D, Wittera Z, Gibsona C, Krafta T, Senturkb I, Çatalyürekb Ü. Syndromic Surveillance of Infectious Diseases meets Molecular Epidemiology in a Workflow and Phylogeographic Application. Studies in health technology and informatics 2015;216:766–70. [PubMed] [Google Scholar]
- 29.Marshall SA, Yang CC, Ping Q, Zhao M, Avis NE, Ip EH. Symptom clusters in women with breast cancer: an analysis of data from social media and a research study. Qual. Life Res 2016;25(3):547–557. doi: 10.1007/s11136-015-1156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topaz M, Lai K, Dhopeshwarkar N, et al. Clinicians’ reports in electronic health records versus patients’ concerns in social media: A pilot study of adverse drug reactions of aspirin and atorvastatin. Drug Saf 2016;39(3):241–250. doi: 10.1007/s40264-015-0381-x. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Wu Y, Liu J, Li J, Zhang P. Understanding health care social media use from different stakeholder perspectives: A content analysis of an online health community. J. Med. Internet Res 2017;19(4):e109. doi: 10.2196/jmir.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tighe PJ, Goldsmith RC, Gravenstein M, Bernard HR, Fillingim RB. The painful tweet: text, sentiment, and community structure analyses of tweets pertaining to pain. J. Med. Internet Res 2015;17(4):e84. doi: 10.2196/jmir.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portier K, Greer GE, Rokach L, et al. Understanding topics and sentiment in an online cancer survivor community. J. Natl. Cancer Inst. Monogr 2013;2013(47):195–198. doi: 10.1093/jncimonographs/lgt025. [DOI] [PubMed] [Google Scholar]
- 34.Sunkureddi P, Gibson D, Doogan S, Heid J, Benosman S, Park Y. Using self-reported patient experiences to understand patient burden: Learnings from digital patient communities in ankylosing spondylitis. Arthritis and Rheumatology 2016;68:1785–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, Donovan H. Application of Text Mining in Cancer Symptom Management. Studies in health technology and informatics 2016;225:930–1. [PubMed] [Google Scholar]
- 36.Cronin RM, Fabbri D, Denny JC, Rosenbloom ST, Jackson GP. A comparison of rule-based and machine learning approaches for classifying patient portal messages. Int. J. Med. Inform 2017;105:110–120. doi: 10.1016/j.ijmedinf.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramar K, Olson EJ. Management of common sleep disorders. Am. Fam. Physician 2013;88(4):231–238. [PubMed] [Google Scholar]
- 38.National Center for Health Statistics. Health, United States, 2006 With Chartbook on Trends in the Health of Americans. Hyattsville, MD; 2006. [PubMed] [Google Scholar]
- 39.Pearson AC, Moman RN, Moeschler SM, Eldrige JS, Hooten WM. Provider confidence in opioid prescribing and chronic pain management: results of the Opioid Therapy Provider Survey. J. Pain Res 2017;10:1395–1400. doi: 10.2147/JPR.S136478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miaskowski C, Barsevick A, Berger A, et al. Advancing symptom science through symptom cluster research: expert panel proceedings and recommendations. J. Natl. Cancer Inst 2017;109(4). doi: 10.1093/jnci/djw253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cashion AK, Gill J, Hawes R, Henderson WA, Saligan L. National Institutes of Health Symptom Science Model sheds light on patient symptoms. Nurs Outlook 2016;64(5):499–506. doi: 10.1016/j.outlook.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis JR, Chen L, Higginbotham P, et al. Social media for arthritis-related comparative effectiveness and safety research and the impact of direct-to-consumer advertising. Arthritis Res. Ther 2017;19(1):48. doi: 10.1186/s13075-017-1251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez B, Dailey F, Almario CV, et al. Patient Understanding of the Risks and Benefits of Biologic Therapies in Inflammatory Bowel Disease: Insights from a Large-scale Analysis of Social Media Platforms. Inflamm. Bowel Dis 2017;23(7):1057–1064. doi: 10.1097/MIB.0000000000001110. [DOI] [PubMed] [Google Scholar]
- 44.Kralj Novak P, Smailović J, Sluban B, Mozetič I. Sentiment of Emojis. PLoS One 2015;10(12):e0144296. doi: 10.1371/journal.pone.0144296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvaro N, Miyao Y, Collier N. Twimed: twitter and pubmed comparable corpus of drugs, diseases, symptoms, and their relations. JMIR Public Health Surveill 2017;3(2):e24. doi: 10.2196/publichealth.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mowery D, Smith H, Cheney T, et al. Understanding Depressive Symptoms and Psychosocial Stressors on Twitter: A Corpus-Based Study. J. Med. Internet Res 2017;19(2):e48. doi: 10.2196/jmir.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keselman A, Smith CA, Divita G, et al. Consumer health concepts that do not map to the UMLS: where do they fit? J. Am. Med. Inform. Assoc 2008;15(4):496–505. doi: 10.1197/jamia.M2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doing-Harris KM, Zeng-Treitler Q. Computer-assisted update of a consumer health vocabulary through mining of social network data. J. Med. Internet Res 2011;13(2):e37. doi: 10.2196/jmir.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon S, Elhadad N, Bakken S. A practical approach for content mining of Tweets. Am. J. Prev. Med 2013;45(1):122–129. doi: 10.1016/j.amepre.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Institutes of Health. PAR-17–159: Data Science Research: Personal Health Libraries for Consumers and Patients (R01) 2017. Available at: https://grants.nih.gov/grants/guide/pa-files/par-17-159.html. Accessed August 16, 2018.
- 51.Redeker NS, Anderson R, Bakken S, et al. Advancing symptom science through use of common data elements. J Nurs Scholarsh 2015;47(5):379–388. doi: 10.1111/jnu.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]