Abstract
We report a novel method named LITPOMS (ligand titration, fast photochemical oxidation of proteins and mass spectrometry) to characterize protein-ligand binding stoichiometry, binding sites, and site-specific binding constants. The system used to test the method is melittin – calmodulin, in which the peptide melittin binds to calcium-bound calmodulin. Global-level measurements reveal the binding stoichiometry of 1:1 whereas peptide-level data coupled with fitting reveal the binding sites and the site-specific binding affinity. Moreover, we extended the analysis to the residue level and identified six critical binding residues. The results show that melittin binds to the N-terminal, central linker, and C-terminal regions of holo-calmodulin with an affinity of 4.6 nM, in agreement with results of previous studies. LITPOMS, for the first time, brings high residue-level resolution to affinity measurements, providing simultaneously qualitative and quantitative understanding of protein-ligand binding. The approach can be expanded to other binding systems without tagging the protein to give high spatial resolution.
Keywords: LITPOMS, Fast Photochemical Oxidation of Proteins (FPOP), Ligand Titration, Binding Affinity, Melittin, Calmodulin
Graphical Abstract
Protein-ligand interactions are the basis for numerous biological processes for which quantitative understanding is vital [1, 2]. To quantify binding, investigators have used circular dichroism [2], fluorescence and fluorescence polarization [3, 4], nuclear magnetic resonance [5], surface plasma resonance [6, 7] and isothermal titration calorimetry [8]. The macroscopic binding affinity, however, does not permit complete understanding of a protein-ligand system. The requirements for specific fluorescence labeling [3, 4], special sample preparation [5, 6] and large sample amounts [5] also limit these approaches.
Among the mass spectrometry (MS)-based approaches for quantitative protein-ligand interactions, one class, termed direct methods, utilize the spectrometer to measure concentrations at equilibrium [9–12]. Despite their convenience, there is always a question of whether the measured gas-phase concentrations represent those in solution. To avoid this, indirect methods using hydrogen/deuterium exchange (HDX) can also be used. Stability of unpurified proteins from rates of HDX (SUPREX) [13] yields a protein-ligand binding affinity by measuring the stability of proteins as bound and unbound.
Another method, protein-ligand interactions by MS, titration and HDX (PLIMSTEX) [14, 15], gives binding affinity in a titration-based experiment [16–19]. Titrating the protein at high concentration (e.g., 100 times Kd) yields binding stoichiometry, whereas titration at ~ Kd gives the binding affinity without any modifications (e.g., tagging by a fluorophore) [14, 15]. Indirect methods also characterize binding-induced conformational changes at a regional level represented by peptides from digestion [20–22]; an example is apolipoprotein E3 and a small-molecule drug candidate where location and affinity was determined [19]. HDX is reversible and occurs at sec-to-min timescale [22], which may be competitive with off-rates of protein-ligand equilibrium, giving convergence in HDX for bound and unbound at long times. Further, D2O dilution decreases protein concentration, making tight binging challenging. Back-exchange and D scrambling during fragmentation challenge the extension beyond the peptide level.
Fast photochemical oxidation of proteins (FPOP) labels proteins with hydroxyl radicals generated by hydrogen peroxide photolysis [23–29]. Hydroxyl radicals irreversibly label solvent-accessible areas [23, 24] in sub-milliseconds, faster than changes in protein conformation [24, 26, 27]. This ensures that FPOP labeling will not distort or compete with the binding equilibrium. Because the labeling is irreversible, experiments can be executed off-line, where concentrations are no longer determined by the MS detection limit. Moreover, there are no back-exchange and scrambling, facilitating detection at the amino-acid level [26, 28, 29].
Here, we report a novel FPOP-based ligand-titration method, protein-ligand interaction by Ligand Titration, Fast Photochemical Oxidation of Proteins and Mass Spectrometry (LITPOMS). This new approach allows measurement of binding stoichiometry and site-specific binding constants for any protein-ligand system that experience a change in solvent accessible area upon binding. Such measurements also access the equilibrium composition indirectly and importantly provide the affinity in the liquid phase. LITPOMS overcomes disadvantages of PLIMSTEX, as stated above, providing another significant strategy to assess binding stoichiometries, affinities, and dynamics.
Results and Discussion
To demonstrate LITPOMS, we chose calmodulin-melittin as a model. Melittin (Mel) binds to calcium-bound calmodulin (holo-CaM) at a ratio of 1:1 with a Kd of 3 nM [30–33]. Crosslinking suggests that melittin binds at the N-terminus, C-terminus, and the central linker region of the holo-CaM [31, 32]. In the current study, we equilibrated holo-CaM with melittin overnight in Tris buffer (pH = 7.4). A series of aliquots were prepared at [Mel]:[Holo-CaM] of 0–3 to fulfill the titration process. Hydrogen peroxide and L-histidine were added followed by a pulsed 248 nm laser irradiation to footprint the complex. The sample was introduced by a syringe pump into a capillary tube with a transparent window. L-histidine works as a hydroxyl radical scavenger to control the lifetime of the radicals. The footprinted sample was collected at the end of the capillary in a solution of L-methionine and catalase to quench any residual oxidants. Global-level responses were by direct measurement of the intact complex with a Bruker MaXis 4G Q-TOF mass spectrometer. For peptide-level responses, the complex was digested with trypsin/Lys-C mix [29] and the peptides characterized with a Thermo Scientific Q Executive Plus Orbitrap instrument. Two series of samples with different holo-CaM concentrations (i.e., 10 μM and 200 nM) were prepared for global- and peptide-level studies. Detailed descriptions of the experimental setup and data processing is in supporting information.
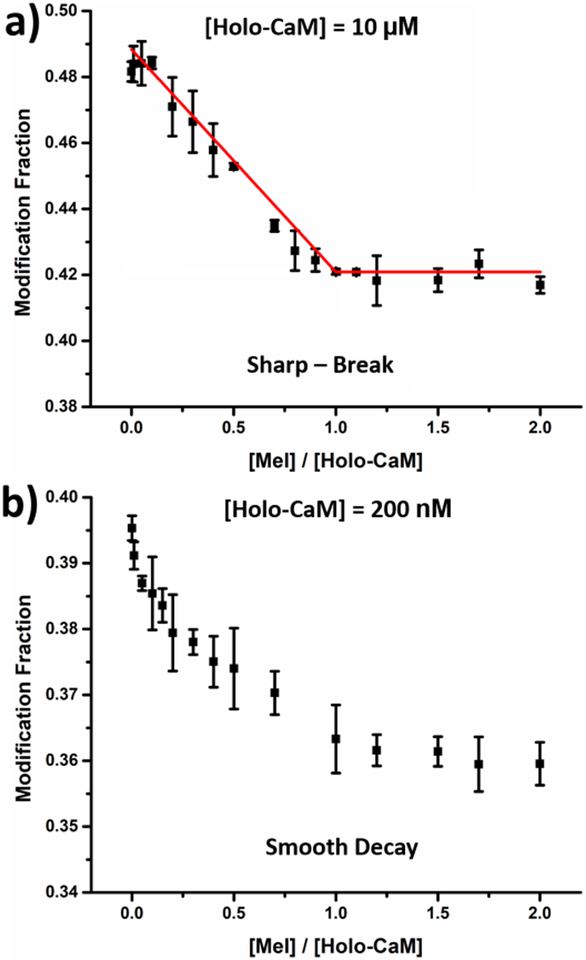
Given that the shape of the titration curve is sensitive to protein concentration, experiments were carried out at the two protein concentrations (Figure 1). A more compact conformation forms as holo-CaM binds with melittin, leading to a lower overall solvent accessibility and a decrease in the macroscopic modification fraction. At [holo-CaM] = 10 μM, 3000-fold > Kd of the system, a sharp-break curve is obtained (Figure 1a) with a break at [Mel]/[Holo-CaM] of 1.0, indicating the binding stoichiometry is 1:1. For a low-concentration titration ([holo-CaM] = 200 nM), the fraction modified decays smoothly as [Mel] is increased (Figure 1b). This promising tendency encouraged us to digest the protein and obtain spatial resolution for binding and affinity.
Figure 1.
Modification fraction measured at global level as a function of melittin concentration with initial protein concentration of (a) 10μM and (b) 200 nM. Error bars are standard deviations from two independent runs whereas the data points are the average of the two runs.
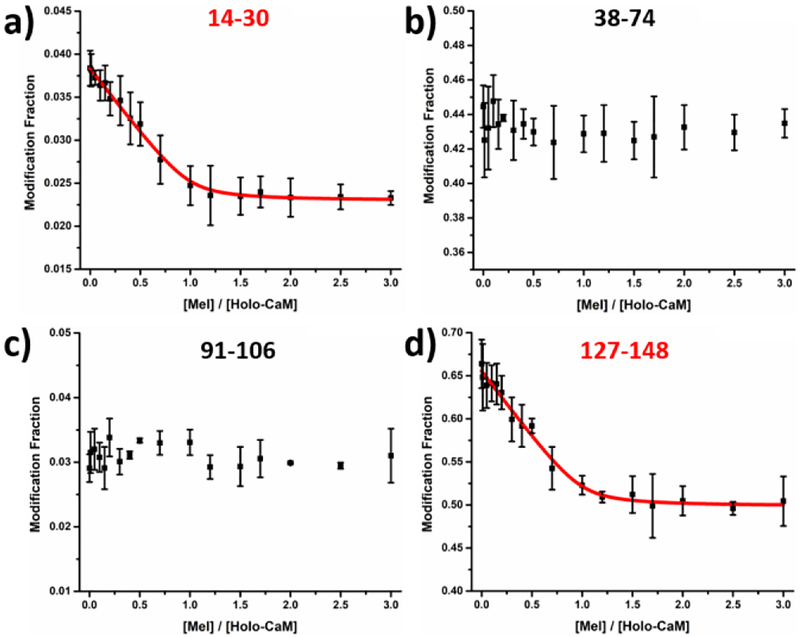
Taking advantage of the irreversible labeling of the protein through FPOP, we combined titration with FPOP-based bottom-up proteomics to report the modification change as a function of ligand concentration. To model the titration process and obtain the binding affinity, we carried out peptide-level experiments at a protein concentration of 200 nM, enabling a plot of modification fraction as a function of ligand concentration for each individual peptide (Figure 2 and Figure S5 in SI). Note that sequence coverage for calmodulin is 99%. Two different behaviors can be distinguished (Figure 2 and Figure S5 in SI), namely, increased protection for binding regions and no change in protection for non-binding regions. Fractional modifications for peptides 1–13, 31–37, 38–74, 76–90 and 91–106 remain relatively stable as the titration proceeds, indicating that the conformations for these regions remain unchanged upon binding. Thus, these regions are unlikely binding sites and serve as controls.
Figure 2.
Modification fraction as a function of melittin:holo-calmodulin for selected peptides. Red solid lines in (a) and (d) represents the fitting result with the algorithm described previously [14, 15]. Error bars are standard deviations from two independent runs whereas the data points represent the average of two runs. Response curves for the remaining
On the contrary, regions 14–30, 107–126 and 127–148 experience increased protection as reported by decreases in fractional modification as the titration proceeds. The increase of protection becomes constant at the later titration stages (i.e., when [Mel]/[Holo-CaM] ≥ 2), indicating complete formation of the complex. These regions are likely to contain residues that are involved in binding. Moreover, the decrease in modification, whether caused by a conformation change or direct binding, provides quantitative information of the binding interaction (Kd). Because melittin binds with holo-CaM at 1:1 (Figure 1a), the three curves were modeled together to afford a single binding affinity (modeling algorithm described previously [14, 15]). In brief, both holo-CaM and the melittin/holo-CaM complex have distinct modification fraction. A search utilizes both of the fractions and the overall equilibrium binding Kd to find the best fit of the experimental data in the plot. Because all the data were measured at equilibrium, modeling is simplified, allowing extraction of a dissociation constant of 4.6 nM (R2 > 99%), in good agreement with a reported affinity of 3 nM [30]. Although each sample was measured in duplicate, statistical certainty of the data increases through curve fitting and resampling. A total of 128 resampling trials were executed based on a previously developed strategy [15], and the estimated standard deviation is 2.7 nM.
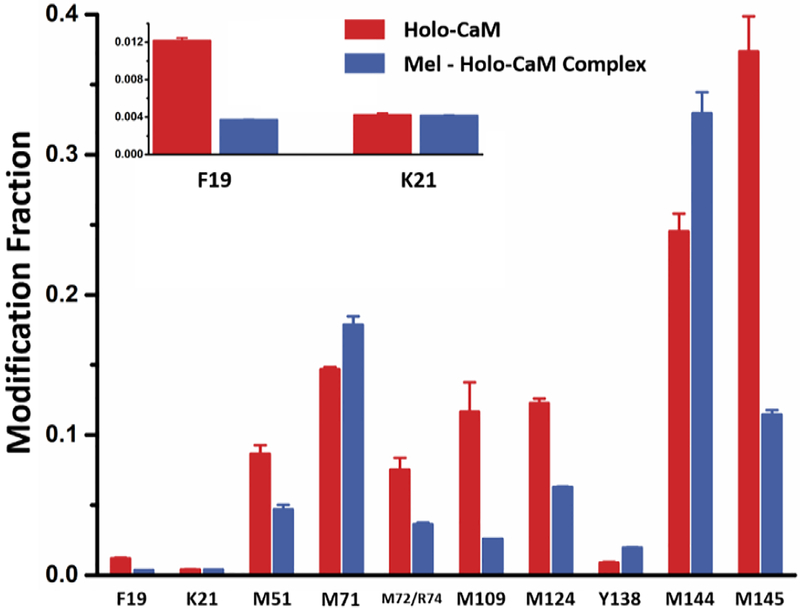
Accepting that FPOP footprints in sub-msecs, faster than binding-induced conformational changes [24, 26, 27], we propose FPOP “snapshots” the real-time equilibrium composition. These features provide a solid foundation for making LITPOMS reliable and informative. The irreversible modifications preserve structural information upon sample workup and MS/MS fragmentation, allowing extension to the residue level [26, 28, 29] (Figure 3) at the three binding regions. For the full protein, 10 amino-acid residues can be successfully resolved (Figure 3). Among them, K21 does not show any significant change upon binding, indicating it is not involved. Residues F19, M109, M124 and M145 show decreases in modification fraction upon binding, indicating that these residues likely bind with melittin, thus experiencing decreases in solvent accessibility owing to spatial hindrance from the melittin ligand. Residues Y138 and M144, however, become more exposed upon binding, suggesting a binding-induced conformational change.
Figure 3.
Modification fraction as a function of melittin:holo-calmodulin for selected amino-acid residues. Red bars show modification fraction for holo-calmodulin itself whereas blue bars represent melittin-bound holo-calmodulin where [Mel]/[Holo-CaM]
Another promising observation involves the two adjacent residues M144 and M145, where M144 becomes exposed whereas M145 becomes protected upon binding. The observations not only provide deeper understanding of melittin/holocalmodulin binding but also demonstrate the promising spatial resolution for LITPOMS.
Out of the previously identified binding regions, three different residues M51, M71 and M72/R74 were successfully identified. Note that the spectral quality is currently not sufficient to distinguish M72 and R74, and the result is reported as M72/R74. M71 becomes more exposed through binding-induced conformational changes given that the modification fraction increases upon binding. Further, M51 and M72/R74 are both involved in binding owing to a decrease in modification fraction. Given that peptide 38–74 does not exhibit a noticeable change upon binding (Figure 2b), this once again reveals that LITPOMS brings more detailed understanding than does PLIMSTEX.
In summary, not only the N-terminal (peptide 14–30) and C-terminal region (peptide 107–126 and peptide 127–148), but also central linker region (residue M51, M72/R74) of holo-CaM are involved in binding with melittin. We identified six critical binding residues and three conformational-change residues and determine a Kd = 4.6 nM. These findings agree with those in a previous crosslinking study [32]. The coupling of FPOP with titration and MS permits characterization of protein-ligand binding stoichiometry, binding sites (peptide and amino-acid levels), and site-specific binding constants, even for a tight binding system like melittin:holo-CaM.
In conclusion, the high-concentration mode for determining stoichiometry and the low-concentration mode for locating binding sites and determining site-specific affinities make LITPOMS promising for characterizing protein-ligand binding for high picomole quantities of protein. Moreover, the irreversible labeling allows rigorous post-labeling digestion without erasing any labeling information (e.g., from back exchange). LITPOMS is also readily compatible with various buffers, pH, salts, lipid-based media (e.g., nano and pico disks), and binding affinities (ranging from nM to μM), making it generally applicable even to membrane proteins. The approach should not be affected by high off-rates for weaker binding systems as for HDX, but that remains to be established. Importantly, residue-level analysis becomes possible owing to the irreversible labeling that will be maintained for MS/MS, and more complete residue locations will be enabled by complementary footprinters (e.g., CF3∙ [34], carbenes [35]). Although we demonstrated the applicability to tight binding (nmolar) of 1:1 binding system, we plan to extend the method to other, more complex binding systems with stoichiometries greater than 1:1.
Supplementary Material
Acknowledgement
This work is financially supported by National Institute of Health NIGMS Grant 5P41GM103422 and 1S10OD016298-01A1 (to M.L.G.). Authors are grateful to Dr. Jagat Adhikari for helpful discussions and to Protein Metrics for software support.
Footnotes
Supporting Information Available Detailed experimental conditions, FPOP labeling optimization, an example of both peptide and residue level data processing as well as remaining LITPOMS response curves at peptide level are in supporting information.
References
- 1.Schellman JA: Macromolecular Binding Biopolymers 14, 999–1018 (1975) [Google Scholar]
- 2.Willams MA; Daviter T Protein-Ligand interactions, Methods and Applications, 2nd ed.; Humana Press: New York, 2013. [Google Scholar]
- 3.Rossi AM; Taylor CW: Analysis of protein-ligand interactions by fluorescence polarization Nat. Protoc 6, 365–387 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y; Marriott G: Analysis of protein interactions using fluorescence technologies Curr. Opin. Chem. Biol 7, 635–640 (2003) [DOI] [PubMed] [Google Scholar]
- 5.Meyer B; Peters T: NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors Angew. Chem. Int. Ed 42, 864–890 (2003) [DOI] [PubMed] [Google Scholar]
- 6.Johnsson B; Lofas S; Lindquist G: Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors Anal.Biochem 198, 268–277 (1991) [DOI] [PubMed] [Google Scholar]
- 7.Willets KA; Van Duyne RP: Localized surface plasmon resonance spectroscopy and sensing Annu.Rev. Phys. Chem 58, 267–297 (2007) [DOI] [PubMed] [Google Scholar]
- 8.Wiseman T; Williston S; Brandts JF; Lin L -N.: Rapid measurement of binding constants and heats of binding using a new titration calorimeter Anal.Biochem 179, 131–137 (1989) [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen TJD; Roepstorff P; Heck AJR: Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogues and vancomycin group antibiotics by electrospray ionization mass spectrometry Anal. Chem 70, 4427–4432 (1998) [Google Scholar]
- 10.Wang W; Kitova EN; Klassen JS: Influence of solution and gas phase processes on protein-carbohydrate binding affinities determined by nanoelectrospray fourier transform ion cyclotron resonance mass spectrometry Anal. Chem 75, 4945–4955 (2003) [DOI] [PubMed] [Google Scholar]
- 11.Daneshfar R; Kitova EN; Klassen JS: Determination of protein-ligand association thermochemistry using variable-temperature nanoelectrospray mass spectrometry J. Am. Chem. Soc 126, 4786–4787 (2004) [DOI] [PubMed] [Google Scholar]
- 12.Gulbakan B; Barylyuk K; Schneider P; Pillong M; Schneider G; Zenobi R: Native electrospray ionization mass spectrometry reveals multiple facets of aptamer-ligand interactions: from mechanism to binding constants J. Am. Chem. Soc 140, 7486–7497 (2018) [DOI] [PubMed] [Google Scholar]
- 13.Powell KD; Ghaemmaghami S; Wang MZ; Ma L; Oas TG; Fitzgerald MC: A general mass spectrometry-bases assay for quantitation of protein-ligand binding interactions in solution J. Am. Chem. Soc 124, 10256–10257 (2002) [DOI] [PubMed] [Google Scholar]
- 14.Zhu MM; Remple DL; Du Z; Gross ML: Quantification of protein-ligand interactions by mass spectrometry, titration and H/D exchange: PLIMSTEX J. Am. Chem. Soc 125, 5252–5253 (2003) [DOI] [PubMed] [Google Scholar]
- 15.Zhu MM; Remple DL; Gross ML: Modeling data from titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants J. Am. Soc. Mass Spectrom 15, 388–397 (2004) [DOI] [PubMed] [Google Scholar]
- 16.Sperry JB; Shi X; Rempel DL; Nishimura Y; Akashi S; Gross ML: A mass spectrometric approach to the study of DNA-binding proteins: interaction of Human TRF2 with telomeric DNA Biochemistry 47, 1797–1807 (2008) [DOI] [PubMed] [Google Scholar]
- 17.Sperry JB; Huang RY; Zhu MM; Rempel DL; Gross ML: Hydrophobic peptides affect binding of calmodulin and Ca2+ as explored by H/D amide exchange and mass spectrometry Int. J. Mass Spectrom 302, 85–92 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang RY-C; Rempel DL; Gross ML: HD exchange and PLIMSTEX determine the affinities and order of binding Ca2+ with Troponin C Biochemistry 50, 5426–5435 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H; Rempel DL; Giblin D; Frieden C; Gross ML: Peptide-level interactions between proteins and small-molecule drug candidates by two hydrogen-deuterium exchange MS-based methods: the example of apolipoprotein E3 Anal. Chem 89, 10687–10695 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aebersold R; Goodlett DR: Mass Spectrometry in Proteomics Chem. Rev 101, 269–296 (2001) [DOI] [PubMed] [Google Scholar]
- 21.Aebersold R; Mann M: Mass spectrometry-based proteomics Nature 422, 198–207 (2003) [DOI] [PubMed] [Google Scholar]
- 22.Weis DD Hydrogen Exchange Mass Spectrometry of Proteins, Fundamentals, Methods, and Applications; Wiley: Chichester, 2016. [Google Scholar]
- 23.Xu G; Chance MR: Hydroxyl radical-mediated modification of proteins as probes for structural proteomics Chem.Rev 107, 3514–3543 (2007) [DOI] [PubMed] [Google Scholar]
- 24.Hambly DM; Gross ML: Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale J. Am. Soc. Mass Spectrom 16, 2057–2063 (2005) [DOI] [PubMed] [Google Scholar]
- 25.Li KS; Shi L; Gross ML: Mass spectrometry-based fast photochemical oxidation of proteins (FPOP) for higher order structure characterization Acc. Chem. Res 51, 736–744 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J; Rempel DL; Gau BC; Gross ML: Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level J. Am. Chem. Soc 134, 18724–18731 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gau BC; Sharp JS; Rempel DL; Gross ML: Fast photochemical oxidation of proteins footprints faster than protein unfolding Anal. Chem 81, 6563–6571 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B; Cheng M; Rempel D; Gross ML: Implementing fast photochemical oxidation of proteins (FPOP) as a footprinting approach to solve diverse problems in structural biology Methods 144, 94–103 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H; Gau BC; Jones LM; Vidavsky I; Gross ML: Fast photochemical oxidation of protein for comparing structures of protein-ligand complexes: the calmodulin-peptide model system Anal. Chem 83, 311–318 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comte M; Maulet Y; Cox JA: Ca2+-dependent high-affinity complex formation between calmodulin and melittin Biochem. J 209, 269–272 (1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scaloni A; Miraglia N; Orru S; Amodeo P; Motta A; Marino G; Pucci P: Topology of the calmodulin-melittin complex J. Mol. Biol 277, 945–958 (1998) [DOI] [PubMed] [Google Scholar]
- 32.Schulz DM; Ihling C; Clore GM; Sinz A: Mapping the topology and determination of a low-resolution three-dimensional structure of the calmodulin-melittin complex by chemical crosslinking and high-resolution FTICRMS: direct demonstration of multiple binding modes Biochemistry 43, 4703–4715 (2004) [DOI] [PubMed] [Google Scholar]
- 33.Wong JWH; Maleknia SD; Downard KM: Hydroxyl radical probe of the calmodulin-melittin complex interface by electrospray ionization mass spectrometry J. Am. Soc. Mass. Spectrom 16, 225–233 (2005) [DOI] [PubMed] [Google Scholar]
- 34.Cheng M; Zhang B; Cui W; Gross ML: Liser-initiated trifluoromethylation of peptides and proteins: application to mass-spectrometry-based protein footprinting Angew. Chem. Int. Ed 56, 14007–14010 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B; Rempel DL; Gross ML: Protein footprinting by carbenes on a fast photochemical oxidation of proteins (FPOP) platform J. Am. Soc. Mass Spectrom 27, 552–555 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.