Abstract
Nanoparticles improve drug efficacy by delivering drugs to sites of disease. To effectively deliver a drug in vivo, a nanoparticle must overcome physical and physiological hurdles that are not present in cell culture, yet in vitro screens are used to predict nanoparticle delivery in vivo. An ideal nanoparticle discovery pipeline would enable scientists to study thousands of nanoparticles in vivo. Here, we discuss technologies that enable high throughput in vivo screens, focusing on DNA barcoded nanoparticles.
RNA therapies are limited by inefficient delivery to target tissues
Advances in genomics have armed scientists with lists of genes that cause diseases. This has brought about a significant change in the way drugs are designed and discovered. Researchers were previously limited to targeting broad cellular phenotypes; for example, cisplatin intercalates into double stranded DNA, causing toxicity in any cell undergoing cell division. Although many of these drugs successfully treated disease, they also caused severe side effects driven by drug activity in ‘off-target’ cells. Researchers now often use small molecules that target specific mutations. However, only 15% of the protein coding genome – and a much smaller percentage of the non-coding genome – is ‘druggable’ using small molecules [1]. This has led scientists to develop technologies to target all genes.
RNA therapies have emerged as a promising solution to the problem of ‘undruggable’ targets [2]. RNAs can specifically turn any gene in the genome on or off, regardless of its eventual protein structure. Gene silencing can be mediated by RNA interference (RNAi), a well characterized mechanism where base pairing between small interfering RNA (siRNA) and target mRNA catalyzes RISC-mediated mRNA degradation. Gene silencing can also be mediated by DNA nucleases, including those derived from zinc fingers (ZFNs) [3], transcription factor-like effectors (TALENs) [4], or clustered regularly interspaced short palindromic repeats and their associated proteins (CRISPR-Cas) systems [5,6]. Similarly, mRNA-based therapies can transiently upregulate genes [7]. To date siRNA therapies have shown the most promise in patients. In one example, a degenerative disease that was uniformly fatal has been halted (and in some cases, reversed) in Phase III clinical trials [8,9]. More generally, over 1,200 patients have been treated with siRNA; some for as long as 36 months. With a few exceptions, primarily limited to antisense nucleotides (which are distinct from siRNA) [10], siRNA therapies have been well tolerated and efficacious. Advances in siRNA biochemistry [11] and improved delivery vehicles [12] raise the exciting prospect that once yearly injections to treat genetic disease are within reach.
Although siRNA is currently the most clinically advanced RNA therapy, DNA nucleases and mRNA therapies are up and coming. They have shown efficacy in mice [13,14] and nonhuman primates [15], and are poised to treat disease in humans [16–18]. The results described above have one limitation: most clinical trials have involved RNAs locally injected into muscle (e.g., vaccines), eye, or lymph nodes, administered to cells ex vivo, or systemically delivered to hepatocytes. Systemically delivering therapeutic RNA outside the liver remains a substantial unsolved problem [19] that limits the development of gene therapies targeting other organs. RNA therapies will require delivery because naked RNAs are quickly degraded by nucleases, and their large molecular weight and highly negative phosphodiester backbone prevents them from crossing the anionic cell membrane [20].
Thousands of nanoparticles are screened in vitro; this may not predict in vivo delivery
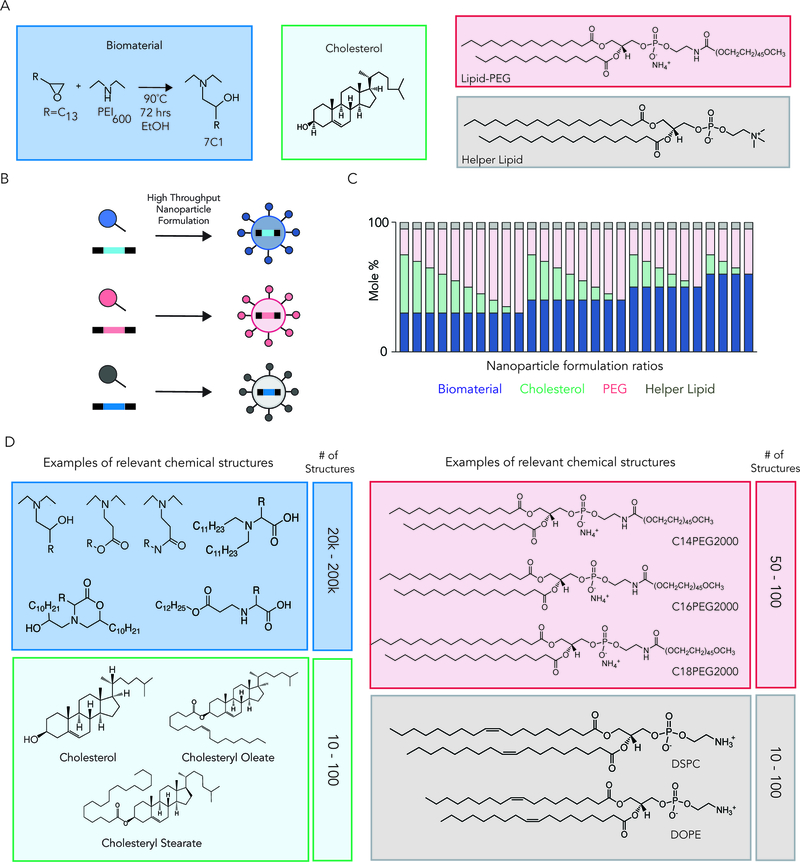
RNA delivery vehicles are designed to protect the nucleic acid and transport it to the target cell. Although many drug delivery vehicles have been used to deliver RNA, we will focus on nanoparticles, which have generated promising clinical data [8]. Here we define a nanoparticle as a structure with all 3 dimensions less than 1,000 nm. Scientists have made steady advances in nanoparticle design. Nanoparticles can be made with variable size [21], ionizability [22], hydrophilicity [23], shape [24], and with varying degrees of active targeting ligands [25]. Large, chemically diverse libraries have been synthesized using simple synthetic routes including, but not limited to, Michael addition [26], epoxides [27], peptide [28], and thiol chemistry [29]. Importantly, advances in nanoparticle formulation – defined here as the process of ‘loading’ the nucleic acid into the nanoparticle – have also been reported. High throughput microfluidics has been shown to reliably make small, consistent batches of nanoparticles that are stable for weeks [30,31]. It is still difficult to cover the entire nanoparticle chemical space; formulating nanoparticles using available chemistries, it is feasible to formulate between 100 million and 200 billion chemically distinct nanoparticles (Fig. 1).
Figure 1.
(A) Lipid nanoparticle libraries can be generated with a variety of chemical compounds. LNPs are formulated by combining different biomaterials with cholesterols, lipid-PEG compounds, and helper lipids. (B) High-throughput nanoparticle formulation allows for the rapid production of large, diverse libraries. (C) Nanoparticle libraries can be generated by varying the molar ratio of biomaterials, cholesterols, PEG, and helper lipids. (D) Between 1 × 108 and 2 × 1011 chemically distinct nanoparticles can be made by combining these compounds. Notably, these numbers do not include variations in the molar ratio of the compounds (e.g., Figure 1C).
After nanoparticles are formulated, they are screened in vitro. More specifically, scientists (a) synthesize thousands of different nanoparticles before (b) evaluating whether they deliver RNA in easily expandable cell lines (e.g., HeLa). Scientists have also used primary cells in place of immortalized cells [32]. Based on in vitro results, a very small number of nanoparticles are (c) tested in vivo. This process is only an efficient use of time and resources if in vitro nanoparticle delivery predicts in vivo delivery. To test this, we compared how the same 300 nanoparticles delivered nucleic acids in vitro and in vivo in multiple cell types; we found no correlation [33] (Fig 2A). The discrepancy between in vitro and in vivo delivery is not necessarily surprising. If a nanoparticle is systemically administered, it must travel through the blood stream and enter the target tissue. Several physical factors dictate where the nanoparticles go. For example, it is difficult for nanoparticles to access the brain, which is physically cordoned off by the blood brain barrier. Nanoparticles can also be physically disassembled by the glomerular membrane in the kidney [34]. As a counterexample, nanoparticle often target hepatocytes since nanoparticles can exit the blood via nanoscale pores in sinusoidal blood vessels and basement membrane [35]. Physiological factors also influence nanoparticle delivery. For example, serum proteins can bind to nanoparticles in the blood [36,37], changing nanoparticle interactions with the immune system and target cells [38]. Interestingly, systemic nanoparticle delivery can also change with local disease states; in one example, scientists found that delivery to systemic organs was affected by the presence of a primary tumor [39]. Even if the nanoparticle reaches its target cell, it must enter the cytoplasm, often by escaping an endosome. Endosomal escape is inefficient; a LNP that delivers siRNA to hepatocytes very efficiently (50% target gene silencing after a 0.01 mg / kg injection) still had >95% of its siRNA sequestered within endosomes [40]. The nanoparticle must also evade liver, kidney, and splenic clearance, as well as avoid initiating an immune response. The majority of these obstacles are not recapitulated in vitro. Some processes are required for in vitro delivery and in vivo delivery (e.g., endocytosis and endosomal escape). However, these processes are carefully regulated [41] by gene expression that is likely to change with microenvironmental cues. Increasing evidence suggests that gene expression in cultured cells may not reliably predict gene expression in primary cells or in vivo [42].
Figure 2.
DNA barcodes have analyzed nanoparticle delivery in vivo. (A) Chemically diverse LNPs are tagged with unique DNA barcodes. LNPs were pooled together and administered both in vitro and in vivo. For in vitro studies, delivery at condition 1 often correlates with delivery at condition 2. However, in vitro delivery does not correlate to in vivo delivery. These results were consistent across many cell types, and across different experiments, for >300 different LNPs [33]. (B) Liposomes carrying different chemotherapies were formulated with unique barcodes. After administering the liposomes in vivo, tissues were removed and cells were stained with propidium iodine (PI), which distinguishes dead cells from live cells. Barcodes enriched in dead cells corresponded to successful chemotherapy agents.
Nanoparticle barcoding enables simultaneous analysis of >100 nanoparticles in vivo
The lines of evidence described above suggest screening nanoparticles directly in vivo would be useful. However, the expensive nature of in vivo experiments has limited the field to testing a few in vivo. This is a universal problem in nanomedicine, and it has driven groups to design systems that facilitate high throughput in vivo nanoparticle screens. In all cases, these systems utilize ‘multiplexed’ signals, which are signals that can be quantified without interfering with one another. In the simplest case, nanoparticle 1, with chemical structure 1, is ‘barcoded’ (i.e., ‘tagged’) with signal 1; nanoparticle N, with chemical structure N, is barcoded with signal N. The nanoparticles are co-administered, and later, signals 1 and N are quantified simultaneously. In one example, scientists isotopically-barcoded silver nanoparticles with different functional peptides to evaluate potential targeting ligands [43]. Another group has used quantum dots to barcode polymeric nanoparticles; in vivo screens were performed to understand how nanoparticle surface modifications altered delivery through the blood brain barrier [44].
One multiplexed signal that has been used by groups (including ours) is DNA [33,45,46]. DNA is an excellent molecule for multiplexing. The number of different barcodes scales exponentially faster than any other signaling molecule; 4N different barcodes can be generated with a DNA sequence N nucleotides long. An 8 nucleotide barcode can create 65,536 different sequences. Additionally, DNA sequence readouts are easy to analyze. The cost of DNA sequencing has decreased more rapidly than Moore’s Law; it is now possible to generate 400 million DNA reads for $2,000 using a machine that fits on a desktop. Because DNA sequencing has become an increasingly more common tool in several fields [47], easy to use software packages are readily available to help analyze and interpret the data.
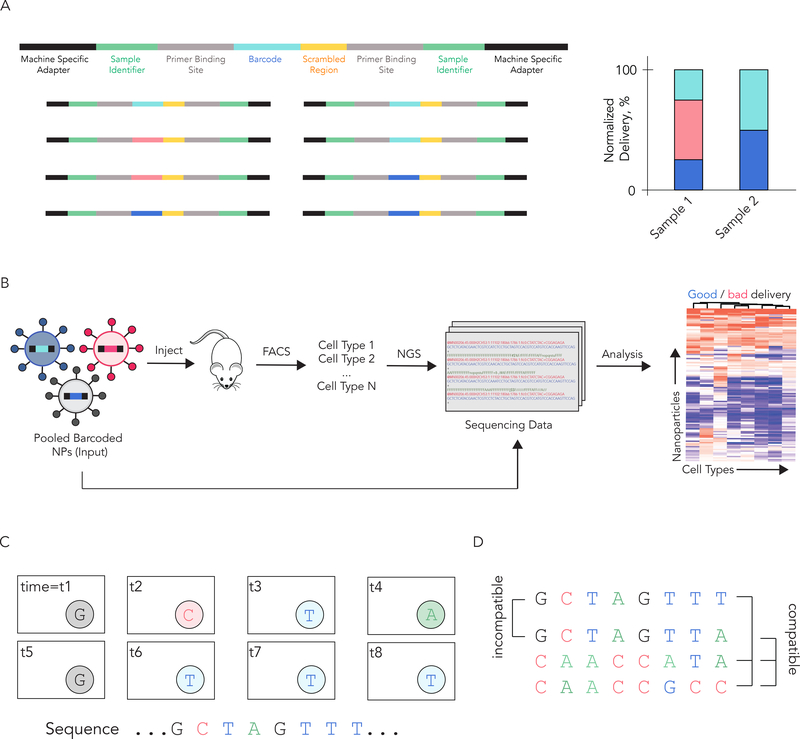
Two distinct nanoparticle DNA barcoding systems have been reported to date [33,45,46]. In one example, DNA was formulated into liposomes alongside different chemotherapies [45] (Fig. 2B). By varying the primers or altering the barcode length, different sequences were detected using gel electrophoresis or real time PCR. The authors tested several drugs at once in vivo and concluded that the DNA barcodes found in dead cells corresponded to nanoparticles containing effective chemotherapies. This method may be used in the future to test hundreds of different cancer drugs in vivo. We developed a separate DNA barcoding system that utilizes next generation sequencing. Our barcodes contain (i) universal primer sites, (ii) a 7 nucleotide region with fully randomized sequences (to monitor for biased PCR amplification), and (iii) an 8 nucleotide barcode region in the center [46] (Fig 3A). Initially, we demonstrated that this system predicted siRNA delivery in vivo and generated DNA sequencing outputs that were linear with respect to the administered DNA [46]. Later, we demonstrated that this system, which we named Joint Rapid DNA Analysis of Nanoparticles (JORDAN), could simultaneously analyze over 150 nanoparticles simultaneously in vivo [33] (Fig 3B).
Figure 3.
DNA barcodes can be rationally designed to track hundreds of nanoparticles in vivo using next generation sequencing. (A) JORDAN barcodes contain universal primer sites, a 7 nucleotide randomized region, and an 8 nucleotide barcode region. This barcode design allows us to multiplex hundreds of different barcodes. The normalized delivery for every barcoded LNP is determined; this is analogous to ‘counts per million’ in RNAseq studies. (B) JORDAN uses DNA barcodes and next generation sequencing to analyze the biodistribution of thousands of particles in vivo. Next generation sequencing is an effective way to read DNA barcodes. (C) Solid phase next generation sequencing reads each nucleotide of the sequencing using fluorescent nucleotides. Understanding how NGS generates data is important to understanding barcode design; NGS is reviewed extensively in reference 47. (D) For example, it is helpful if the sequence of a barcode differs from all other barcodes at 3 of the 8 positions.
Although there are different ways to design DNA barcodes, specific traits help increase the robustness of the data. Most importantly, universal primer sites – which are primer sites that do not change - confer an important advantage, maximizing the chance all barcodes are amplified in an unbiased way. This seems counterintuitive, but the vast majority of a DNA barcode should be identical; the barcode region of the DNA should be small. Adding chemical modifications including phosphorothioates to the 5’ and 3’ termini increase barcode stability. The barcode regions should also be designed to work together on solid phase next generation sequencing (NGS) platforms like Illumina. Since solid phase NGS relies on fluorophores (Fig. 3C), each individual barcode must have a ‘base distance’ of at least 3; in other words, each barcode must be different from all other barcodes at 3 of the 8 positions (Fig. 3D). We have designed >200 barcodes with base distances of 3 or more [33]. It is also critical to sequence the DNA ‘input’ administered to the animals. This allows us to normalize the data and compare different cell types to each other within an experiment. Finally, like all big data systems, nanoparticle DNA barcoding experiments should include proper controls in each experiment. Two examples include a liposome loaded with caffeine (instead of a chemotherapeutic) [45] and a naked barcode [33]; in both cases, the negative controls performed poorly relative to the experimental conditions, as expected. A detailed protocol describing barcode design and a nanoparticle bioinformatics pipeline is available on DahlmanLab.org.
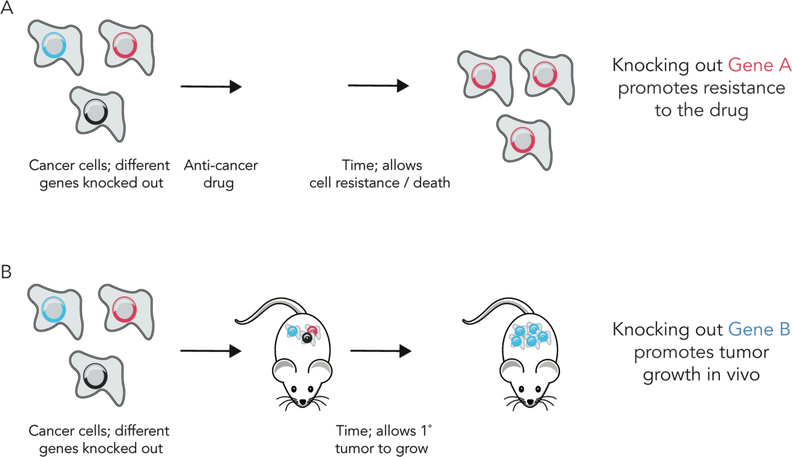
DNA barcodes enable nanotechnologists to track how hundreds of distinct nanoparticles deliver drugs in vivo for the first time; this enables nanoparticle studies that were not feasible using traditional methods. As an example, we compared how dozens of different nanoparticles delivered DNA to 8 different cell sub-types in the spleen. Using unbiased Euclidean clustering – a common bioinformatics technique that analyzes large datasets – we found that specific types of immune cells tended to be targeted by the same nanoparticles [33]. DNA barcodes have enabled large scale experiments in fields as varied as oncology, developmental biology, and viral delivery. As an example, by using the single guide RNA (sgRNA) sequence as a ‘barcode’ to denote a gene that was knocked out, scientists performed whole genome screens to identify genes and non-coding regions that regulate cellular response to cancer and immunotherapies [48–51]. More specifically, a system named Genome-Scale CRISPR-Cas9 Knockout Screening (GeCKO) helped determine which genes promote resistance to anti-cancer drugs [50]. GeCKO was used to targeted 18,080 in the same experiment; multiple sgRNAs were designed per gene. The pool of sgRNAs were administered to human cancer cell lines that also expressed Cas9; as a result, each cell – on average – had no gene knocked out, or 1 gene knocked out. After adding an anti-cancer drug, cells were allowed to grow; enriched sgRNA sequences in live cells corresponded to genes that increased drug resistance (Fig 4A). The same approach has been used to study metastasis in vivo. In this case, a pool of tumor cells were edited using GeCKO and administered in the hind limb of a mouse. After waiting a period of time, the authors isolated metastatic tumor cells from different organs, and thereby identified genes that promoted metastasis [52]. Similar approaches have been used to identify genes that suppress primary tumor growth in the liver and brain (Fig. 4B) [53,54]. Scientists also designed a combinatorial DNA barcode system named ‘PolyLox’ that identified stem and progenitor cells that led to the development of the immune system [55]. Finally, there are reports of ‘capsid shuffling’ and other combinatorial cloning strategies; these use sequences that code for amino acids as barcodes that denote which amino acids successfully enabled Adeno-associated viral (AAV) vectors that facilitate cellular entry and DNA delivery [56–58]. Given that DNA is a highly efficient method to store and access information, it is likely that many other biology and engineering fields will use DNA barcodes in the future.
Figure 4.
DNA barcodes are regularly used by biologists to perform whole genome studies. (A) In cancer cells, each gene in the genome of Cas9 expressing cells was knocked out individually using sgRNAs. The sgRNA served as the barcode that denoted which gene was silenced. After knocking every protein coding gene, an anti-cancer drug is added to the cells and time is allotted to facilitate drug resistance. Enriched sgRNA sequences in the live cells correspond to genes that affect cancer drug resistance. In this example, knocking down Gene A promotes drug resistance. (B) The same whole genome studies have identified genes that – when knocked down – promote primary tumor growth.
Outlook
RNA therapies have tremendous clinical potential but will require on-target drug delivery to reach the clinic. siRNA drugs in the liver are a great example; several patient populations are already benefiting from treatments that silence genes in hepatocytes. However, without new delivery vehicles, the clinical impact will be limited to local injection, and to the liver [19]. To target new cell types, it is important to test as many nanoparticles as possible. Traditional screening methods test delivery vehicles in vitro; however, in vitro particle screens may inaccurately predict delivery in vivo. In conjunction with rapid nanoparticle synthesis, highthroughput in vivo nanoparticle screening methods will allow scientists to track more particles simultaneously than ever before. Over time, this may allow scientists to better understand how nanoparticles behave in the body. We envision a time where enough nanoparticles have been tested in vivo that nanoparticles which target a given cell type can be rationally designed. We find it likely these high throughput approaches will help accelerate the development of new genetic therapies.
Acknowledgments.
The authors thank Nirav N. Shaw for his helpful input.
Funding.
M.L., C.D.S. and J.E.D. were funded by Georgia Tech startup funds (awarded to J.E.D.). C.D.S. was funded by the NIH-sponsored Research Training Program in Immunoengineering (T32EB021962). This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research was funded by the Cystic Fibrosis Research Foundation (DAHLMA15XX0, awarded to J.E.D.), the Parkinson’s Disease Foundation (PDF-JFA-1860, awarded to J.E.D.), and the Bayer Hemophilia Awards Program (AGE DTD, awarded to J.E.D.).
Footnotes
Competing interests.
There are no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dixon SJ, Stockwell BR: Identifying druggable disease-modifying gene products. Current opinion in chemical biology (2009) 13(5–6):549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaczmarek JC, Kowalski PS, Anderson DG: Advances in the delivery of rna therapeutics: From concept to clinical reality. Genome medicine (2017) 9(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD: Genome editing with engineered zinc finger nucleases. Nature reviews Genetics (2010) 11(9):636–646. [DOI] [PubMed] [Google Scholar]
- 4.Joung JK, Sander JD: Talens: A widely applicable technology for targeted genome editing. Nature reviews Molecular cell biology (2013) 14(1):49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna JA, Charpentier E: Genome editing. The new frontier of genome engineering with crispr-cas9. Science (2014) 346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- 6.Hsu PD, Lander ES, Zhang F: Development and applications of crispr-cas9 for genome engineering. Cell (2014) 157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin U, Kariko K, Tureci O: Mrna-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov (2014) 13(10):759–780. [DOI] [PubMed] [Google Scholar]
- 8. **.Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M et al. : Safety and efficacy of rnai therapy for transthyretin amyloidosis. N Engl J Med (2013) 369(9):819–829. An early demonstration that lipid nanoparticles can safely deliver siRNAs that treat disease in human patients. These data were later confirmed in Phase III trials, which are reviewed in Reference 9. [DOI] [PubMed] [Google Scholar]
- 9.Rizk M, Tuzmen S: Update on the clinical utility of an rna interference-based treatment: Focus on patisiran. Pharmacogenomics and personalized medicine (2017) 10(267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooke ST, Baker BF, Kwoh TJ, Cheng W, Schulz DJ, Xia S, Salgado N, Bui HH, Hart CE, Burel SA, Younis HS et al. : Integrated safety assessment of 2’-o-methoxyethyl chimeric antisense oligonucleotides in nonhuman primates and healthy human volunteers. Mol Ther (2016) 24(10):1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. *.Foster DJ, Brown CR, Shaikh S, Trapp C, Schlegel MK, Qian K, Sehgal A, Rajeev KG, Jadhav V, Manoharan M, Kuchimanchi S et al. : Advanced sirna designs further improve in vivo performance of galnac-sirna conjugates. Mol Ther (2018) 26(3):708–717. This paper describes the rational improvement of siRNA sequences. Using biochemistry, the authors design siRNAs that confer long-lasting silencing in vivo. It is possible that these sequence modifications will enable once-yearly injections of siRNA to treat disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sehgal A, Barros S, Ivanciu L, Cooley B, Qin J, Racie T, Hettinger J, Carioto M, Jiang Y, Brodsky J, Prabhala H et al. : An rnai therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat Med (2015) 21(5):492–497. [DOI] [PubMed] [Google Scholar]
- 13.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ: Nonviral crispr/cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of cas9 mrna and sgrna. Angew Chem Int Ed Engl (2017) 56(4):1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, Wang Q, Guo Y, Dong Y, Tan X: A nonviral crispr/cas9 delivery system for therapeutically targeting hbv DNA and pcsk9 in vivo. Cell research (2017) 27(3):440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, Schlake T: Sequence-engineered mrna without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther (2015) 23(9):1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, Laska ME, Smith M, Almarsson O, Thompson J, Ribeiro AM et al. : Preclinical and clinical demonstration of immunogenicity by mrna vaccines against h10n8 and h7n9 influenza viruses. Mol Ther (2017) 25(6):1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC et al. : Gene editing of ccr5 in autologous cd4 t cells of persons infected with hiv. N Engl J Med (2014) 370(10):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, Bukur V, Tadmor AD, Luxemburger U, Schrors B, Omokoko T et al. : Personalized rna mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature (2017) 547(7662):222–226. [DOI] [PubMed] [Google Scholar]
- 19.Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J: Going beyond the liver: Progress and challenges of targeted delivery of sirna therapeutics. J Control Release (2015) 203(1–15. [DOI] [PubMed] [Google Scholar]
- 20.Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, Dowdy CF, Presente A et al. : Efficient delivery of rnai prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol (2014) 32(12):1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang W, Kim BY, Rutka JT, Chan WC: Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol (2008) 3(3):145–150. [DOI] [PubMed] [Google Scholar]
- 22.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM et al. : Rational design of cationic lipids for sirna delivery. Nat Biotechnol (2010) 28(2):172–176. [DOI] [PubMed] [Google Scholar]
- 23.Stefanick JF, Ashley JD, Bilgicer B: Enhanced cellular uptake of peptide-targeted nanoparticles through increased peptide hydrophilicity and optimized ethylene glycol peptide-linker length. ACS nano (2013) 7(9):8115–8127. [DOI] [PubMed] [Google Scholar]
- 24.Champion JA, Katare YK, Mitragotri S: Making polymeric micro- and nanoparticles of complex shapes. Proceedings of the National Academy of Sciences of the United States of America (2007) 104(29):11901–11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedmi R, Veiga N, Ramishetti S, Goldsmith M, Rosenblum D, Dammes N, HazanHalevy I, Nahary L, Leviatan-Ben-Arye S, Harlev M, Behlke M et al. : A modular platform for targeted rnai therapeutics. Nat Nanotechnol (2018) 13(3):214–219. [DOI] [PubMed] [Google Scholar]
- 26.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A et al. : A combinatorial library of lipid-like materials for delivery of rnai therapeutics. Nat Biotechnol (2008) 26(5):561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M et al. : Lipid-like materials for lowdose, in vivo gene silencing. Proceedings of the National Academy of Sciences of the United States of America (2010) 107(5):1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA et al. : Lipopeptide nanoparticles for potent and selective sirna delivery in rodents and nonhuman primates. Proceedings of the National Academy of Sciences of the United States of America (2014) 111(11):39553960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, Siegwart DJ: Modular degradable dendrimers enable small rnas to extend survival in an aggressive liver cancer model. Proceedings of the National Academy of Sciences of the United States of America (2016) 113(3):520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung AK, Tam YY, Chen S, Hafez IM, Cullis PR: Microfluidic mixing: A general method for encapsulating macromolecules in lipid nanoparticle systems. J Phys Chem B (2015). [DOI] [PubMed] [Google Scholar]
- 31. *.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG: Rapid discovery of potent sirna-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc (2012) 134(16):6948–6951. These papers describe practical microfluidic devices that enable scientists to formulate many nanoparticles in the same day. Without these devices, high throughput in vitro screens and in vivo screens would be much more challenging. Reference 30 is also an important paper that describes microfluidic systems for nanoparticle formulation. [DOI] [PubMed] [Google Scholar]
- 32.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D et al. : Degradable lipid nanoparticles with predictable in vivo sirna delivery activity. Nature communications (2014) 5(4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. **.Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV et al. : A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett (2018) 18(3):2148–2157. First demonstration that >100 nanoparticles can be analyzed simultaneously in vivo. This paper also directly compared in vitro to in vivo delivery, and found no correlation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuckerman JE, Choi CHJ, Han H, Davis ME: Polycation-sirna nanoparticles can disassemble at the kidney glomerular basement membrane. Proceedings of the National Academy of Sciences of the United States of America (2012) 109(8):31373142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsoi KM, MacParland SA, Ma XZ, Spetzler VN, Echeverri J, Ouyang B, Fadel SM, Sykes EA, Goldaracena N, Kaths JM, Conneely JB et al. : Mechanism of hardnanomaterial clearance by the liver. Nat Mater (2016) 15(11):1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS et al. : Targeted delivery of rnai therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther (2010) 18(7):1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertrand N, Grenier P, Mahmoudi M, Lima EM, Appel EA, Dormont F, Lim JM, Karnik R, Langer R, Farokhzad OC: Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nature communications (2017) 8(1):777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monopoli MP, Aberg C, Salvati A, Dawson KA: Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol (2012) 7(12):779–786. [DOI] [PubMed] [Google Scholar]
- 39.Rohner NA, Thomas SN: Melanoma growth effects on molecular clearance from tumors and biodistribution into systemic tissues versus draining lymph nodes. J Control Release (2016) 223(99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H et al. : Image-based analysis of lipid nanoparticle-mediated sirna delivery, intracellular trafficking and endosomal escape. Nat Biotechnol (2013) 31(7):638–646. [DOI] [PubMed] [Google Scholar]
- 41.Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP: Endocytosis and signaling: Cell logistics shape the eukaryotic cell plan. Physiological reviews (2012) 92(1):273–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA, 3rd et al. : Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron (2016) 89(1):37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willmore AM, Simon-Gracia L, Toome K, Paiste P, Kotamraju VR, Molder T, Sugahara KN, Ruoslahti E, Braun GB, Teesalu T: Targeted silver nanoparticles for ratiometric cell phenotyping. Nanoscale (2016) 8(17):9096–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medina DX, Householder KT, Ceton R, Kovalik T, Heffernan JM, Shankar RV, Bowser RP, Wechsler-Reya RJ, Sirianni RW: Optical barcoding of plga for multispectral analysis of nanoparticle fate in vivo. J Control Release (2017) 253(172–182. [DOI] [PubMed] [Google Scholar]
- 45. **.Yaari Z, da Silva D, Zinger A, Goldman E, Kajal A, Tshuva R, Barak E, Dahan N, Hershkovitz D, Goldfeder M, Roitman JS et al. : Theranostic barcoded nanoparticles for personalized cancer medicine. Nature communications (2016) 7(13325 First nanoparticle DNA barcoding study published. This – along with reference 46 – demonstrate that DNA barcoding can be used to track many nanoparticles simultaneously in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, Langer R, Anderson DG, Wang ET: Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proceedings of the National Academy of Sciences of the United States of America (2017) 114(8):2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. *.Metzker ML: Sequencing technologies - the next generation. Nature reviews Genetics (2010) 11(1):31–46. Important review that outlines advances in next generation sequencing. [DOI] [PubMed] [Google Scholar]
- 48.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, Merchant AS et al. : Identification of essential genes for cancer immunotherapy. Nature (2017) 548(7669):537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY et al. : Genome-scale activation screen identifies a lncrna locus regulating a gene neighbourhood. Nature (2017) 548(7667):343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. **.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F: Genome-scale crispr-cas9 knockout screening in human cells. Science (2014) 343(6166):84–87. This paper describes GeCKO, a system now used throughout biology to simultaneously study how every gene in the genome affects a phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O et al. : Genome-scale transcriptional activation by an engineered crispr-cas9 complex. Nature (2015) 517(7536):583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H et al. : Genome-wide crispr screen in a mouse model of tumor growth and metastasis. Cell (2015) 160(6):1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, Renauer P et al. : Aav-mediated direct in vivo crispr screen identifies functional suppressors in glioblastoma. Nat Neurosci (2017) 20(10):1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G, Chow RD, Ye L, Guzman CD, Dai X, Dong MB, Zhang F, Sharp PA, Platt RJ, Chen S: Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using aav-crispr-mediated direct in vivo screening. Science advances (2018) 4(2):eaao5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei W, Feyerabend TB, Rossler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, Chen W et al. : Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature (2017) 548(7668):456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, Gradinaru V: Cre-dependent selection yields aav variants for widespread gene transfer to the adult brain. Nat Biotechnol (2016) 34(2):204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA: Selection and evaluation of clinically relevant aav variants in a xenograft liver model. Nature (2014) 506(7488):382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mingozzi F, High KA: Therapeutic in vivo gene transfer for genetic disease using aav: Progress and challenges. Nature reviews Genetics (2011) 12(5):341–355. [DOI] [PubMed] [Google Scholar]