Abstract
Drug resistance is a major cause of cancer-associated mortality. Epirubicin-based chemotherapy initially benefits patients with metastatic or advanced gastric cancer; however, tumor recurrence can occur following several courses of treatment. Mitochondrial ribosomal protein L33 (MRPL33)-long (L) and MRPL33-short (S), isoforms of MRPL33 that arise from AS, have been reported to regulate cell growth and apoptosis in cancer; however, few studies have evaluated the roles of MRPL33-L and MRPL33-S in gastric cancer. In the present study, MRPL33-L was demonstrated to be significantly more abundant in gastric tumor tissues than the MRPL33-S isoform. MRPL33-S promoted chemosensitivity to epirubicin in gastric cancer as demonstrated by a chemoresponse assay; chemosensitivity was suppressed in response to MRPL33-L. Gene microarray analysis was performed to investigate the underlying mechanisms. Bioinformatic analysis revealed that overexpression of MRPL33-L and MRPL33-S served critical roles in transcription, signal transduction and apoptosis. In particular, the phosphoinositide 3-kinase (PI3K)/AKT serine/threonine kinase (AKT) signaling pathway was markedly regulated. A total of 36 target genes, including PIK3 regulatory subunit α, AKT2, cAMP response element-binding protein (CREB) 1, forkhead box 3, glycogen synthase kinase 3β and mammalian target of rapamycin, which are involved in the PI3K/AKT signaling pathway, were selected for further investigation via protein-protein interaction network and Kyoto Encyclopedia of Genes and Genomes pathway analyses. Furthermore, western blot analysis indicated that MRPL33-S promoted the chemoresponse to epirubicin by deactivating PI3K/AKT/CREB signaling and inducing apoptosis, while MRPL33-L had the opposite effects. In conclusion, the results of the present study revealed that isoforms S and L of MRPL33, which arise from alternative splicing, exhibited opposing roles in the chemoresponse to epirubicin in gastric cancer via the PI3K/AKT signaling pathway. These findings may contribute to the development of potential therapeutic strategies for the resensitization of patients with gastric cancer to epirubicin treatment.
Keywords: gastric cancer, alternative splicing, mitochondrial ribosomal protein L33-long/short, chemoresponse, epirubicin, phosphoinositide 3-kinase, AKT serine/threonine kinase
Introduction
Gastric cancer, one of the most common types of malignant cancer worldwide, is often diagnosed at advanced stages and is associated with poor prognosis (1). Chemotherapy remains one of the most important therapeutic strategies for patients with gastric cancer of advanced stages. Initially, the efficacy of chemotherapy is high; however, chemoresistance tends to be acquired during therapy. At present, epirubicin-based chemotherapy is recommended as the first-line treatment with notable survival benefits for patients with metastatic or advanced gastric cancer (2,3). Although patient outcome has improved, tumor recurrence following several courses of epirubicin-based chemotherapy is frequently observed (4,5). Epirubicin chemoresistance accounts for the failures in clinical treatment; however, the molecular mechanism underlying this resistance in patients with gastric cancer is poorly understood.
Alternative splicing (AS) is a complex process that involves the post-transcriptional regulation of pre-RNA processing via exon inclusion/skipping, resulting in alterations in a protein domain rather than variations in the genome. Notably, AS occurs in cancer and serves a role in the resistance to cancer therapy (6-9). The modulation of AS using inhibitors of the spliceosome (10) or oligonucleotides directed at specific genes (11) may be promising strategies to alleviate drug resistance; however, these approaches have only been approved in the treatment of several diseases in the absence of cancer (12,13). Thus, it is important to identify and characterize more AS events associated with the regulation of the chemoresponse in cancer therapy.
Mammalian mitochondrial ribosomes, which comprise a small 28S subunit and a large 39S subunit, are required for protein synthesis in the mitochondria (14). In addition to the regulation of cellular respiration, another role of mitochondrial ribosomes has been reported in the control of apoptosis and autophagy via mitochondrial dysregulation in cancer (14,15). Mitochondrial ribosomal protein L33 (MRPL33), composed of four exons, is one of the 50 genes that encode the large subunit of the mitochondrial ribosome. There are two different transcript variants of MRPL33, MRPL33-L (NM_004891.3) and MRPL33-S (NM_145330.2), which arise from the regulation of AS on exon 3 (16). MRPL33-L and MRPL33-S exhibit opposing effects on the growth and apoptosis of cancer cells (16); however, whether the two MRPL33 isoforms exert differing effects on the chemoresponse to cancer therapy is unknown. Further investigation into the exact functions and mechanisms of the MRPL33 transcript variants may aid the development of effective and personalized treatment strategies to resensitize gastric cancer patients to chemotherapy.
The present study demonstrated that MRPL33-S could promote the sensitivity of gastric cancer cells to epirubicin; however, the splice variant MRPL33-L suppressed this effect. Gene microarray analysis revealed that overexpression of MRPL33-L and MRPL33-S affected transcription, the regulation of transcription, signal transduction and apoptosis. In particular, the phosphoinositide 3-kinase (PI3K)/AKT serine/threonine kinase (AKT) signaling pathway, which is involved in the survival, cell cycle progression, metabolism and proliferation of cells, was markedly regulated. Furthermore, the PI3K/AKT/cAMP response element-binding protein (CREB) axis in apoptosis was involved in the effects of the MRPL33 isoforms, which may underlie epirubicin chemoresistance in gastric cancer.
Materials and methods
Tumor specimens and cell lines
Gastric cancer tissues were obtained from 10 patients in the Tumor Center of Changhai Hospital affiliated to the Second Military Medical University (Shanghai, China). The average age of these patients was 60 years old, and the specific information of every patient is as follows: patient 1, 64 years, female, recruitment date November 30, 2017; patient 2, 36 years, female, recruitment date, November 24, 2017; patient 3, 66 years, male, recruitment date November 24, 2017; patient 4, 46 years, male, recruitment date November 24, 2017; patient 5, 66 years, female, recruitment date November 23, 2017; patient 6, 66 years, male, recruitment date November 24, 2017; patient 7, 75 years, male, recruitment date November 23, 2017; patient 8, 57 years, female, recruitment date November 24, 2017; patient 9, 66 years, male, recruitment date November 24, 2017; and patient 10, 58 years, female, recruitment date November 21, 2017. Fresh samples of normal and tumor tissues were collected from the patients upon obtaining written informed consent. The present study was approved by the Internal Review and Ethics Boards of Changhai Hospital. The gastric cancer cell lines AGS and MGC-803 were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
RNA isolation, reverse transcription-polymerase chain reaction (RT-PCR), vector construction and transfection
Total RNA was extracted from tissues and cultured cells using NucleoSpin RNA (Machery-Nagel GmbH, Düren, Germany), and served as the template for the synthesis of cDNA using 5X All-In-One RT MasterMix (with AccuRT Genomic DNA Removal kit; Applied Biological Materials, Inc., Richmond, Canada), according to the manufacturer's protocols. PCR of MRPL33-L and MRPL33-S isoforms was performed with Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs, Inc., Ipswich, MA, USA), using the following primers: sense, 5′-GTTCCTCTCCGCGGTCTTCTT-3′ and antisense, 5′-GGAGCGTATTTTTTTCTTTTCCAC-3′. Actin β (ACTB) served as internal reference with the following primers: sense, 5′-CCATCATGAAGTGTGACG-3′ and antisense, 5′-GCCGATCCACACGGAGTA-3′. The thermocycling conditions for PCR comprised of an initial activation step at 98°C for 30 sec, followed by 35 cycles of 10 sec at 98°C, 30 sec at 55°C, 30 sec at 72°C, followed by a final extension at 72°C for 10 min. After PCR amplification, 5 µl of products were mixed with 1 µl of 6X Gel Loading Dye Purple (New England Biolabs, Inc., Ipswich, MA, USA) and loaded on a 1% agarose gel stained with Goldview (Yeasen Biotech Co., Ltd., Shanghai, China). Products were electrophoresed for 30 min under 120 V with 0.5X TAE liquid as the electrophoretic liquid. Bands were imaged using the ChemiDOC XRS system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The gene expression levels were normalized to the levels of ACTB using ImageJ software (version 1.51r; National Institutes of Health, Bethesda, MD, USA).
Considering the similarity of nucleotide sequences of MRPL33-L and MRPL33-S, overexpression of MRPL33-L and MRPL33-S were performed instead of knockdown. The expression plasmids were constructed by cloning the coding sequence of MRPL33-L (NM_004891.3) and MRPL33-S (NM_145330.2) into pLenti-GIII-CMV-GFP-2A-Puro vector (Applied Biological Materials, Inc.). Subsequently, recombinant vectors (plenti-MRPL33-L, plenti-MRPL33-S and plenti-vector), and packaging plasmids PSPAX2 and PMD2.G were transfected into 293T cells (American Type Culture Collection; Manassas, VA, USA) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) to produce recombinant lentivirus, in accordance with the manufacturer's protocols. At 48 h post-transfection, overexpression-stable cells were selected using 4 µg/ml puromycin.
Gene microarray and bioinformatic analysis
Total RNA from the stable AGS cell lines (plenti-MRPL33-L, plenti-MRPL33-S and plenti-vector) underwent preparation, quality control, RT, labeling, hybridization, image acquisition and analysis, performed by Oebiotech (17) (Shanghai, China). Briefly, total RNA was reverse transcribed into double-stranded cDNA, and the cDNA was labeled with biotin, followed by hybridization with the GeneChip Prime View™ Human Gene Expression Array (18) (Affymetrix; Thermo Fisher Scientific, Inc.). Raw data was obtained from array images via analysis with Affymetrix GeneChip Command Console (version 4.0, Affymetrix; Thermo Fisher Scientific, Inc.). Subsequently, raw data were normalized with a robust multi-array average algorithm and analyzed using GeneSpring software (19) (version 13.1; Agilent Technologies, Inc., Santa Clara, CA, USA). Differentially expressed genes (DEGs) were then identified via the fold-change in expression, as well as the P-value calculated from a t-test. The threshold set for upregulated and downregulated genes was: fold-change >2.0 and P<0.05. Hierarchical clustering was then performed to demonstrate the expression profile of the DEGs among samples using MultiExperiment Viewer (20) (MeV, version 4.9.0, http://mev.tm4.org/). Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG, https://www.genome.jp/kegg/) pathway analyses of the identified DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) database. Finally, protein-protein interaction (PPI) networks were constructed using the Search Tool for the Retrieval of Interacting Genes (http://string.embl.de/) database and visualized using Cytoscape software (21) (version 3.4.0, https://cytoscape.org/).
Chemoresponse assay
Cells were seeded in 384-well plates at 150 cell/well in RPMI-1640 medium, supplemented with 10% fetal bovine serum at 37°C with 5% CO2 and cultured overnight. For chemoresponse assay to epirubicin, serial concentrations of epirubicin (0.003, 0.03, 0.3, 3 and 30 µM) were added to the cells for 72 h. For rescue assay, 50 µM LY294002 (PI3K inhibitor; Cell Signaling Technology, Inc., Danvers, MA, USA) was added to plenti-MRPL33-L-transfected cells, and 50 µM 1,3-Dicaffeoylquinic acid (PI3K activator; MedChemExpress, Monmouth Junction, NJ, USA) was added to plenti-MRPL33-L transfected cells. After incubation for 1 h, 0.3 µM epirubicin was added to the cells for 72 h. Cell numbers were calculated following staining with NucBlueTM Live ReadyProbes Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and imaged using an IN Cell Analyzer 2000 (GE Healthcare, Chicago, IL, USA). The cell viability rate was calculated as: (cell number of experiment/cell number of control) × 100%.
Western blotting
Total protein was extracted with radioimmunoprecipitation assay lysis buffer (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and then quantified using a Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 10 µg protein was denatured and separated via 10% SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membranes (EMD Millipore, Bedford, MA, USA), followed by blocking with 5% bovine serum albumin for 1 h at room temperature. Next, the membranes were incubated with primary antibodies (1:1,000 dilution; all from Cell Signaling Technology, Inc., Danvers, MA, USA) against GAPDH (cat. no. 5174S), AKT (cat. no. 4685S), phosphorylated (p)-AKT (cat. no. 4060S), CREB (cat. no. 9197S), p-CREB (cat. no. 9198S), myeloid cell leukemia 1 (Mcl-1; cat. no. 94296S), B-cell lymphoma 2 (Bcl-2; cat. no. 3498S) overnight at 4°C. The membranes were then incubated with secondary antibody (anti-rabbit horseradish peroxidase-conjugated; cat. no. 7074V; 1:2,000; Cell Signaling Technology, Inc.) for 1 h at 37°C. Finally, the membranes were imaged using the ChemiDOC XRS system (Bio-Rad Laboratories, Inc.) following detection with an enhanced chemiluminescence kit (Beijing Solarbio Science & Technology Co., Ltd.). The protein expression levels were normalized to the levels of GAPDH using ImageJ software (version 1.51r; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
SPSS statistical software (version 22.0; IBM Corp., Armonk, NY, USA) was used for statistical analysis. One-way analysis of variance followed by Tukey's multiple comparison post hoc test was performed for multiple group comparisons. The Student's t-test was used when only two groups were compared. Data were presented as the mean ± standard deviation from three independent biological replicates. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of MRPL33-L and MRPL33-S in gastric cancer
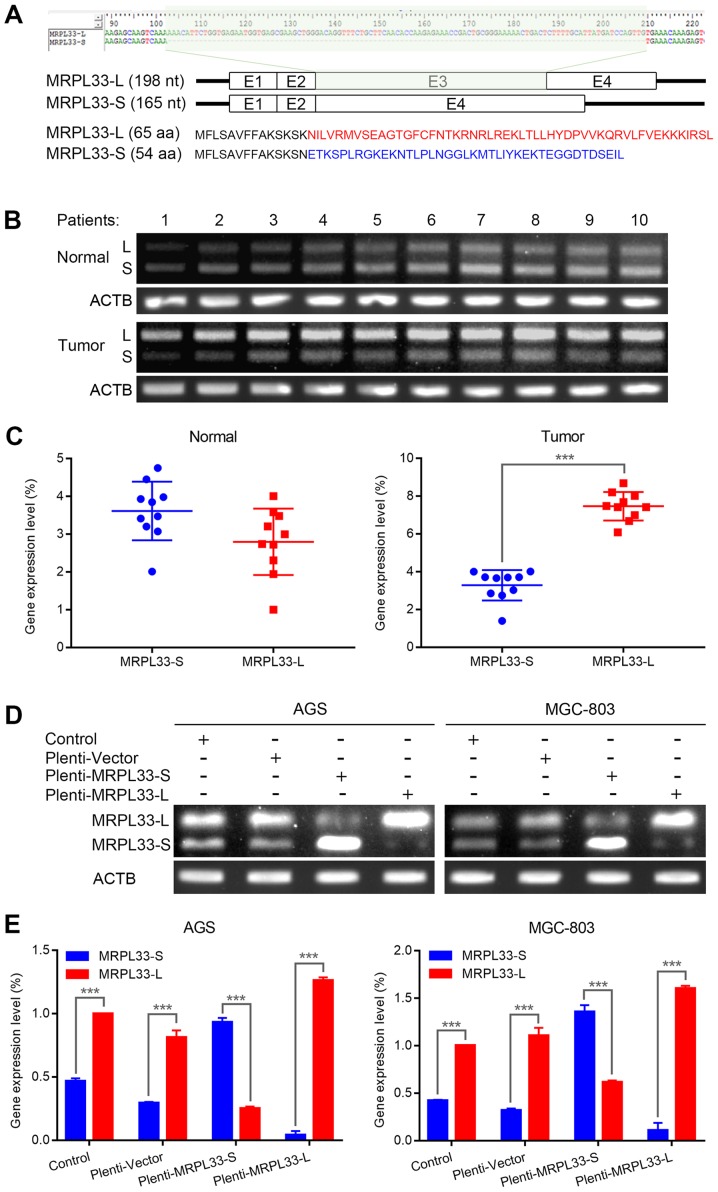
Human MRPL33 mRNA exists in long (L) and short (S) variants due to alternative splicing (Fig. 1A, top). As the exclusion of exon 3 causes a frame-shift mutation, the two isoforms notably differ in their C-terminal amino acid sequences (Fig. 1A, bottom). To determine the expression levels of MRPL33-L and MRPL33-S in clinical gastric cancer specimens, 10 paired samples of tumor and matched adjacent non-tumor tissues from patients with gastric cancer were analyzed. The results revealed that MRPL33-L was markedly more abundant than the MRPL33-S isoform in gastric tumor tissues (Fig. 1B and C). Furthermore, the expression of MRPL33-L was upregulated compared with MRPL33-S in the gastric cancer cell lines, AGS and MGC-803 (Fig. 1D and E). In order to investigate their isoform-specific functional roles, MRPL33-L and MRPL33-S were overexpressed in these two cell lines (Fig. 1D and 1E).
Figure 1.
Expression of MRPL33-L and MRPL33-S in gastric cancer. (A) Schematic diagram of the splice variants of MRPL33, including or lacking alternative exon 3 (MRPL33-L and MRPL33-S; top). Different amino acid sequences are presented in red font for of MRPL33-L and in blue font for MRPL33-S (bottom). (B) Agarose gel electrophoresis photograph and (C) corresponding scatter diagrams of expression levels of MRPL33-L and MRPL33-S in 10 paired clinical specimens of tumor and matched adjacent normal tissues from patients with gastric cancer. (D) Agarose gel electrophoresis photograph and (E) corresponding histograms of expression levels of MRPL33-L and MRPL33-S in AGS and MGC-803 cells. Three independent biological replicates were performed and data were presented as the mean ± standard deviation. ***P<0.001 with comparisons shown by brackets. MRPL33, mitochondrial ribosomal protein L33; L, long variant; S, short variant; ACTB, actin β.
Upregulation of MRPL33-S promotes the chemoresponse of gastric cancer cells to epirubicin, whereas the chemoresponse is suppressed by MRPL33-L
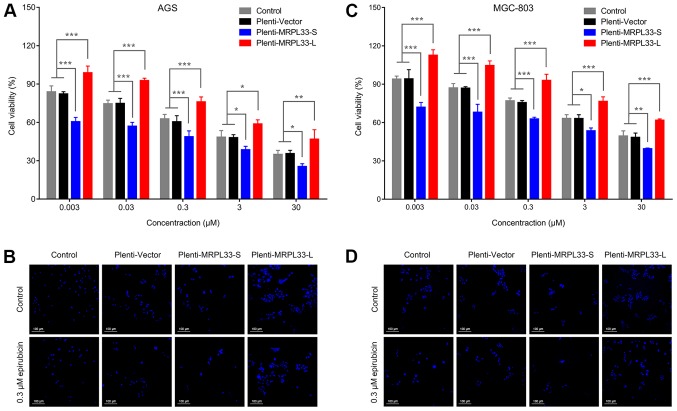
To investigate the regulatory effects of alternative splice variants MRPL33-S and MRPL33-L on the chemoresponse to epirubicin in gastric cancer, a chemoresponse assay was performed using MRPL33-S- and MRPL33-L-overexpressing cells (plenti-MRPL33-S and plenti-MRPL33-L cells). The cells were treated with serial concentrations of epirubicin. Analysis of the AGS cells transfected with plenti-MRPL33-S revealed that cell viability was suppressed compared with control and plenti-vector-transfected cells (Fig. 2A and B). Conversely, plenti-MRPL33-L-transfected cells demonstrated increased cell viability compared with the control and plenti-vector-transfected cells (Fig. 2A and B). These effects were similarly observed in MGC-803 cells (Fig. 2C and D). Collectively, upregulation of MRPL33-S could promote the chemoresponse of gastric cancer cells to epirubicin, while this was suppressed following MRPL33-L upregulation.
Figure 2.
Chemoresponse to epirubicin is regulated by MRPL33-L and MRPL33-S in gastric cancer. (A) Histogram of the chemoresponse of AGS cell groups (AGS control, plenti-vector, plenti-MRPL33-L and plenti-MRPL33-S), which were treated with epirubicin (0.003, 0.03, 0.3, 3 and 30 µM) for 72 h. (B) Fluorescent staining of nuclei in AGS cell groups treated with 0.3 µM epirubicin for 72 h. (C) Histogram of the chemoresponse of MGC-803 cell groups (MGC-803 control, plenti-vector, plenti-MRPL33-L and plenti-MRPL33-S), which were treated with epirubicin (0.003, 0.03, 0.3, 3 and 30 µM) for 72 h. (D) Fluorescent staining of nuclei in MGC-803 cell groups treated with 0.3 µM epirubicin for 72 h. Three independent biological replicates were performed and data were presented as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 with comparisons shown by brackets. MRPL33, mitochondrial ribosomal protein L33; L, long variant; S, short variant.
Effects of MRPL33-L and MRPL33-S overexpression in gastric cancer
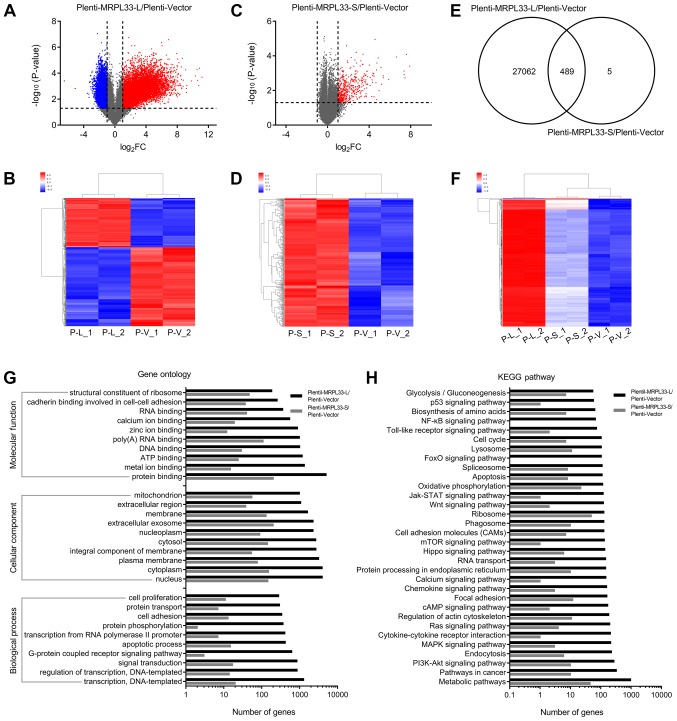
To further investigate the mechanism underlying the regulatory effects of MRPL33-L and MRPL33-S, gene microarray analysis was performed using RNA extracted from plenti-MRPL33-L, plenti-MRPL33-S and plenti-vector-transfected AGS cells. There were 27,551 DEGs, including 10,594 upregulated genes and 16,957 downregulated genes, in plenti-MRPL33-L-transfected cells compared with plenti-vector-transfected cells (Fig. 3A and B). In addition, there were 494 DEGs which were upregulated in plenti-MRPL33-S-transfected cells compared with plenti-vector-transfected cells (Fig. 3C and D). A total of 489 DEGs were detected in plenti-MRPL33-L in common with the plenti-MRPL33-S-transfected cells (Fig. 3E and F). GO analysis ('biological process', 'cellular component' and 'molecular function') was used to classify the functions of the DEGs. In the 'biological process' category, 'transcription', 'regulation of transcription', 'signal transduction' and 'apoptotic process' were predominant (Fig. 3G). In the 'cellular component' category, 'nucleus', 'cytoplasm', 'plasma membrane' and 'integral component of membrane' were predominant (Fig. 3G). In the 'molecular function' category, 'protein binding', 'metal ion binding', 'ATP binding' and 'DNA binding' were predominant (Fig. 3G). Additionally, KEGG pathway analysis based on the DAVID database was performed to determine the potential biological roles of the DEGs. 'Metabolic pathway', 'pathway in cancer', and 'PI3K/AKT signaling pathway' were the most represented pathways of the DEGs (Fig. 3H).
Figure 3.
Effects of MRPL33-L and MRPL33-S overexpression in AGS gastric cancer cells. (A) Volcano plot and (B) heatmap indicating upregu-lated and downregulated genes in plenti-MRPL33-L-transfected cells. (C) Volcano plot and (D) heatmap indicating upregulated and downregulated genes in plenti-MRPL33-S-transfected cells. (E) Venn diagram and (F) heatmap showing the number of overlapping DEGs in plenti-MRPL33-L and plenti-MRPL33-S-transfected cells. (G) Gene Ontology analysis of DEGs in plenti-MRPL33-L and plenti-MRPL33-S-transfected cells. (H) KEGG pathway analysis of DEGs in plenti-MRPL33-L and plenti-MRPL33-S-transfected cells. MRPL33, mitochondrial ribosomal protein L33; L, long variant; S, short variant; DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
PI3K/AKT signaling pathway is regulated by MRPL33-L and MRPL33-S in gastric cancer
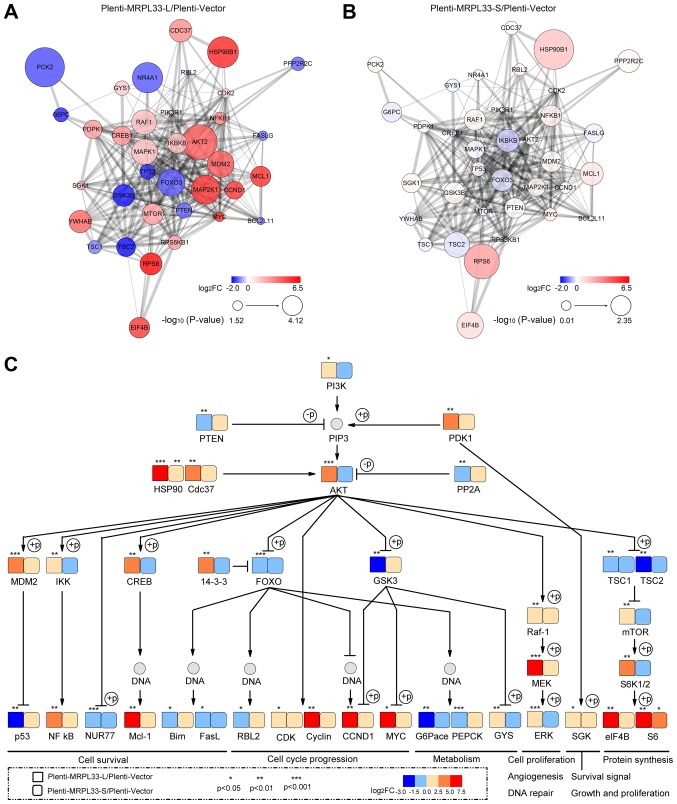
To determine whether the PI3K/AKT signaling pathway is affected by MRPL33-L and MRPL33-S, 36 target genes were selected, including PIK3 regulatory subunit α (PIK3R1), AKT2, CREB1, forkhead box 3 (FOXO3), glycogen synthase kinase 3β (GSK3B) and mammalian target of rapamycin (mTOR). In the plenti-MRPL33-L-transfected cells, 23 target genes were upregulated [including PIK3R1, AKT2, mouse double minute 2 homolog (MDM2), inhibitor of nuclear factor-κB (NF-κB) kinase subunit β (IKBKB), CREB1 and mTOR] and 13 target genes were downregulated [including phosphatase and tensin homolog 1 (PTEN), FOXO3, GSK3B and tumor protein 53 (TP53)] with a fold-change >2.0 and P<0.05, compared with plenti-vector-transfected cells. By contrast, in plenti-MRPL33-S-transfected cells, there were no significantly differences in the target genes based on the criteria of fold-change >2.0 and P<0.05 compared with the plenti-vector-transfected cells. A PPI network was constructed based on these target genes of plenti-MRPL33-L (Fig. 4A) and plenti-MRPL33-S-transfected cells (Fig. 4B) compared with plenti-vector-transfected cells, respectively.
Figure 4.
Analysis of the PI3K/AKT signaling pathway based on the KEGG database. (A) PPI network analysis of 36 target genes involved in the PI3K/AKT signaling pathway in plenti-MRPL33-L-transfected cells and in (B) plenti-MRPL33-S-transfected cells. (C) Map of the PI3K/AKT signaling pathway with the 36 target genes based on the KEGG database. Red and blue represent the upregulation and downregulation of genes, respectively. The size of circle in (A) and (B) indicates significance based on P-value. The square and rounded square in (C) represent genes in plenti-MRPL33-L- and plenti-MRPL33-S-trans-fected cells, respectively. *P<0.05, **P<0.01 and ***P<0.001. PI3K, phosphoinositide 3-kinase; AKT, AKT serine/threonine kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; MRPL33, mitochondrial ribosomal protein L33; L, long variant; S, short variant.
In addition, a map of the PI3K/AKT signaling pathway was generated with these target genes based on the KEGG database (Fig. 4C). 'Cell survival', 'cell cycle progression', 'metabolism', 'cell proliferation', 'angiogenesis', 'DNA repair', 'survival signal', 'growth and proliferation', and 'protein synthesis' were summarized as the predominant biological processes observed. Genes that were associated with 'cell survival' were involved in the 'p53 signaling pathway' (MDM2 and TP53), 'NF-κB signaling pathway' (IKBKB and NF-κB subunit 1), 'apoptosis' (nerve growth factor IB, CREB1, MCL1) and 'FOXO signaling pathway' (FOXO3, Bcl-2 like protein 11 and Fas ligand). Genes associated with 'cell cycle progression' were involved in the 'FoxO signaling pathway' (FOXO3 and retinoblastoma-like protein 2), as well as cyclin-dependent kinase 2, cyclin D1, GSK3B and MYC. As for 'metabolism', the genes involved in the 'FOXO signaling pathway' (FOXO3, glucose-6-phosphatase and phosphoenolpyruvate carboxykinase 2), GSK3B and glycogen synthase 1 were associated with 'glycolysis/gluconeogenesis'. RAF1, mitogen-activated protein (MAPK) kinase kinase 1 and MAPK1 genes which are associated with the MAPK signaling pathway, serve roles in 'cell proliferation', 'angiogenesis' and 'DNA repair'. Serum and glucocorticoid-regulated kinase 1 was involved in 'survival signal', 'growth' and 'proliferation'. Tuberous sclerosis (TSC)1, TSC2, mTOR, ribosomal protein S6 (RPS6) kinase β1, eukaryotic translation initiation factor 4B, RPS6, which are involved in the 'mTOR signaling pathway', may regulate 'protein synthesis'.
Chemoresponse to epirubicin is dependent on the PI3K/AKT/CREB/apoptosis axis, which is regulated by MRPL33-L and MRPL33-S in gastric cancer
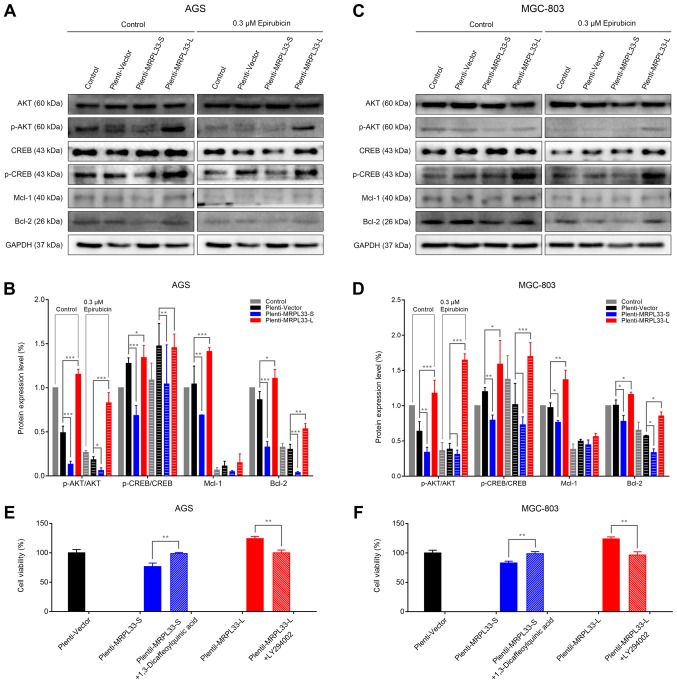
To investigate whether the chemoresponse of epirubicin, which is regulated by MRPL33-L and MRPL33-S, is dependent on the PI3K/AKT signaling pathway in gastric cancer, the expression levels of proteins that are associated with the PI3K/AKT/CREB/apoptosis axis were examined by western blotting. In the AGS cell groups, the results revealed that the protein expression levels of p-CREB were positively associated with p-AKT (Fig. 5A and B). The expression levels of p-AKT and p-CREB decreased in plenti-MRPL33-S-transfected cells, but were increased in plenti-MRPL33-L-transfected cells. The expression levels of the antiapoptotic proteins Mcl-1 and Bcl-2, both downstream targets of CREB, were consistent with the protein expression levels of p-CREB (Fig. 5A and B). Following treatment with 0.3 µM epirubicin, the protein expression levels of p-AKT, CREB, p-CREB, Mcl-1 and Bcl-2 were decreased in AGS cells compared with the control (Fig. 5A and B). In addition, similar results for the expression levels of AKT, p-AKT, CREB, p-CREB, Mcl-1 and Bcl-2 were observed in the MGC-803 cell groups (Fig. 5C and D).
Figure 5.
Chemoresponse to epirubicin is dependent on the PI3K/AKT/CREB/apoptosis axis, which is regulated by MRPL33-L and MRPL33-S in gastric cancer cells. (A) Western blot analysis and (B) corresponding histogram of ratio of p-AKT/AKT, ratio of p-CREB/CREB, Mcl-1 and Bcl-2 expression levels in the AGS cell groups (control, plenti-vector-plentil-MRPL33-S and plenti-MRPL33-L-transfected), with or without 0.3 µM epirubicin. (C) Western blot analysis and (D) corresponding histogram of ratio of p-AKT/AKT, ratio of p-CREB/CREB, Mcl-1 and Bcl-2 expression levels in the MGC-803 cell groups (control, plenti-vector, plentil-MRPL33-S and plenti-MRPL33-L), with or without 0.3 µM epirubicin. (E) Histograms of chemoresponse in AGS cell groups and (F) MGC-803 cell groups treated with 0.3 µM epirubicin for 72 h. Three independent biological replicates were performed and data were presented as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 with comparisons shown by brackets. PI3K, phosphoinositide 3-kinase; AKT, AKT serine/threonine kinase; CREB, cAMP response element-binding protein; MRPL33, mitochondrial ribosomal protein L33; L, long variant; S, short variant; p-, phosphorylated; Mcl-1, myeloid cell leukemia 1; Bcl-2, B-cell lymphoma 2.
To further confirm the key role of PI3K/AKT signaling in chemoresponse to epirubicin, the kinase activity of PI3K was activated by a PI3K activator (1,3-Dicaffeoylquinic acid) in plenti-MRPL33-S-transfected cells, and suppressed by a PI3K inhibitor (LY294002) in plenti-MRPL33-L-transfected cells. By use of chemoresponse assays, cell viability was increased in plenti-MRPL33-S-transfected cells following treatment with 1,3-Dicaffeoylquinic acid and 0.3 µM epirubicin (Fig. 5E and F). Conversely, cell viability was decreased in plenti-MRPL33-L-transfected cells following treatment with LY294002 and 0.3 µM epirubicin (Fig. 5E and F). These results suggested that MRPL33-S-upregulation or MRPL33-L-downregulation may serve as targeting strategies to promote the chemoresponse of cells to epirubicin, by suppressing PI3K/AKT/CREB signaling and inducing apoptosis in gastric cancer cells.
Discussion
Due to abnormal post-transcriptional regulation of pre-RNA by the spliceosome, alternative splice variants are frequently differentially expressed in tumors. For example, the alternative splice variants of doublecortin like kinase 1 (DCLK1; isoforms 2 and 4) exhibit notable upregulation compared with normal tissue and the transcript expression of canonical DCLK1 isoform 1 in renal cell carcinoma (22). The inclusion of exon 3 of MRPL33 was significantly increased in 60 human clinical colorectal cancer tissues compared with paired noncancerous samples (16). In the present study, the abundance of the alternative splice variant of MRPL33, MRPL33-L, was markedly increased compared with MRPL33-S in clinical gastric cancer specimens and cell lines. Additionally, MRPL33-S could promote the chemosensitivity to epirubicin in gastric cancer cells; however, this was suppressed by MRPL33-L overexpression, indicating that the regulation of MRPL33-S and MRPL33-L may serve a role in chemoresistance. Recent studies have demonstrated that the modulation of AS may be applied for the treatment of certain types of cancer. Serine and arginine-rich splicing factor 6 (SRSF6) mediates the progression of colorectal cancer by regulating AS; indacaterol was identified as an inhibitor of SRSF6, which suppressed tumorigenicity (23). A splice-switching oligonucleotide was reported to alter breast cancer 1 (BRCA1) pre-mRNA splicing by inducing BRCA1 exon 11 skipping, and simultaneously reducing the expression of BRCA1-FL and BRCA1-11Q; thus, the effects of poly (ADP-ribose) polymerase inhibitor on the proliferation and survival of BRCA1 wild-type cells were enhanced (24). In addition, a review concluded that several therapeutic approaches, including conventional small molecule compounds, oligonucleotide and RNAi-based gene therapies, and the modulation of AS, inhibited the development and/or altered the course of diseases (25). Therefore, therapeutic strategies involving the upregulation of MRPL33-S or downregulation of MRPL33-L may be developed for epirubicin-based chemotherapy in gastric cancer.
Mitochondrial ribosomes, which are involved in mitochondrial protein synthesis, could mediate numerous important cellular processes, including proliferation, apoptosis and the cell cycle, via the dysregulation of mitochondrial function in various types of cancer (26-29). Research has indicated that exon 3 of MRPL33 involves several members of serine/arginine-rich proteins and heterogeneous nuclear ribonucleoprotein, which leads to the MRPL33 isoforms S and L with opposite roles in tumorigenesis (16). In addition, knockdown of MRPL33-L results in decreased expression levels of 16S rRNA; dysfunctional mitochondria lead to excessive ROS production, increased apoptosis and reduced ATP generation (16). In the present study, overexpression of MRPL33-S and MRPL33-L served critical roles in signal transduction, apoptosis and cell proliferation, which may be the consequence of mitochondrial dysfunction. In particular, the PI3K/AKT signaling pathway is notably regulated in gastric cancer. AKT has been reported to rapidly accumulate in mitochondria following PI3K activation (30). In addition, previous studies have revealed that p-AKT is a crucial modulator of cellular processes, including cell survival (31,32), apoptosis (33,34), the cell cycle (35,36) and glycolysis (37,38) in various types of cancer. The findings of the present study suggested that the pathways downstream of AKT, including the p53 signaling pathway, NF-κB signaling pathway, apoptosis, FOXO signaling pathway, the cell cycle, glycolysis/gluconeogenesis, MAPK signaling pathway and the mTOR signaling pathway, were regulated by MRPL33-L and MRPL33-S; however, further investigation into the biological effects of MRPL33-L and MRPL33-S in gastric cancer is required.
The downstream signaling of the AKT/CREB/apoptosis axis was analyzed to demonstrate the chemoresponse to epirubicin, which was regulated by MRPL33-L and MRPL33-S in gastric cancer. It has been reported that CREB was associated with chemoresistance in cancer. For example, p-AKT and its downstream effector CREB were detected in temozolomide-resistant glioblastoma, indicating that the associated signaling pathways are dysregulated and may be involved in the development of chemoresistance (39). Intercellular adhesion molecule-3 promoted paclitaxel and vincristine resistance via activation of the AKT/CREB signaling pathway and the inhibition of apoptosis in non-small cell lung cancer (40). Mcl-1 and Bcl-2 are downregulated via inhibition of CREB activity, which critically affects the sensitivity of renal carcinoma cells to dicoumarol (41). The findings of the present study revealed that MRPL33-S promoted the chemoresponse to epirubicin by suppressing the PI3K/AKT/CREB signaling pathway and inducing apoptosis. By contrast, MRPL33-L suppressed the chemoresponse to epirubicin by activating the PI3K/AKT/CREB signaling pathway and suppressing apoptosis in gastric cancer. Thus, targeting the PI3K/AKT/CREB-apoptosis axis may be a potential therapeutic strategy to induce the chemosensitivity of cancer cells to treatment. In combination with epirubicin, this therapeutic approach may be valuable in the treatment of gastric cancer.
Acknowledgments
Not applicable.
Abbreviations
- AS
alternative splicing
- MRPL33
mitochondrial ribosomal protein L33
- PCR
polymerase chain reaction
- DEGs
differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- PPI
protein-protein interaction
Funding
The present study was supported by the Changhai Hospital Youth (grant no. CH201719).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL, DF, CG and XZ designed the experiments. JL, YZ and JX performed the experiments and analyzed the results. CG and MW performed the bioinformatic analysis and JL wrote the manuscript. All authors have reviewed and approved the final version of the manuscript.
Ethics approval and consent to participate
Protocols involving the use of clinical samples were approved by the Internal Review and Ethics Boards of Changhai Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Catenacci DVT, Tebbutt NC, Davidenko I, Murad AM, Al-Batran SE, Ilson DH, Tjulandin S, Gotovkin E, Karaszewska B, Bondarenko I, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1467–1482. doi: 10.1016/S1470-2045(17)30566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iveson T, Donehower RC, Davidenko I, Tjulandin S, Deptala A, Harrison M, Nirni S, Lakshmaiah K, Thomas A, Jiang Y, et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: An open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet Oncol. 2014;15:1007–1018. doi: 10.1016/S1470-2045(14)70023-3. [DOI] [PubMed] [Google Scholar]
- 4.Peng L, Yang W, Zhang Z, Liu H, Hua Y. Clinical features and prognosis analysis of 21 gastric cancer patients with pathological complete response after neoadjuvant chemotherapy. Zhonghua Wei Chang Wai Ke Za Zhi. 2017;20:1168–1173. In Chinese. [PubMed] [Google Scholar]
- 5.Polkowska-Pruszyńska B, Rawicz-Pruszyński K, Ciseł B, Sitarz R, Polkowska G, Krupski W, Polkowski WP. Liver metastases from gastric carcinoma: A Case report and review of the literature. Curr Probl Cancer. 2017;41:222–230. doi: 10.1016/j.currproblcancer.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 7.Siegfried Z, Karni R. The role of alternative splicing in cancer drug resistance. Curr Opin Genet Dev. 2018;48:16–21. doi: 10.1016/j.gde.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Wojtuszkiewicz A, Assaraf YG, Maas MJ, Kaspers GJ, Jansen G, Cloos J. Pre-mRNA splicing in cancer: The relevance in oncogenesis, treatment and drug resistance. Expert Opin Drug Metab Toxicol. 2015;11:673–689. doi: 10.1517/17425255.2015.993316. [DOI] [PubMed] [Google Scholar]
- 9.Dehm SM. mRNA splicing variants: Exploiting modularity to outwit cancer therapy. Cancer Res. 2013;73:5309–5314. doi: 10.1158/0008-5472.CAN-13-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 11.Kole R, Krainer AR, Altman S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godfrey C, Desviat LR, Smedsrød B, Piétri-Rouxel F, Denti MA, Disterer P, Lorain S, Nogales-Gadea G, Sardone V, Anwar R, et al. Delivery is key: Lessons learnt from developing splice-switching antisense therapies. EMBO Mol Med. 2017;9:545–557. doi: 10.15252/emmm.201607199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopisetty G, Thangarajan R. Mammalian mitochondrial ribosomal small subunit (MRPS) genes: A putative role in human disease. Gene. 2016;589:27–35. doi: 10.1016/j.gene.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Kim HJ, Maiti P, Barrientos A. Mitochondrial Ribosomes in Cancer. Semin Cancer Biol. 2017;47:67–81. doi: 10.1016/j.semcancer.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Luo C, Luo Y, Chen L, Liu Y, Wang Y, Han J, Zhang Y, Wei N, Xie Z, et al. MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene. 2018;37:86–94. doi: 10.1038/onc.2017.314. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Sun H, Zhu D, Dong X, Liu F, Liang X, Chen C, Shao B, Wang M, Wang Y, et al. TRA2A Promoted Paclitaxel Resistance and Tumor Progression in Triple-Negative Breast Cancers via Regulating Alternative Splicing. Mol Cancer Ther. 2017;16:1377–1388. doi: 10.1158/1535-7163.MCT-17-0026. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Wang J, Yu J, Zhao Y, Dong Y, Fan Y, Li N, Zhang Y, Wang Y. Knockdown of POLE2 expression suppresses lung adenocarcinoma cell malignant phenotypes in vitro. Oncol Rep. 2018;40:2477–2486. doi: 10.3892/or.2018.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499–2512. doi: 10.3892/or.2018.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun S, Wang Y, Wu Y, Gao Y, Li Q, Abdulrahman AA, Liu XF, Ji GQ, Gao J, Li L, et al. Identification of COL1A1 as an invasion related gene in malignant astrocytoma. Int J Oncol. 2018;53:2542–2554. doi: 10.3892/ijo.2018.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge Y, Weygant N, Qu D, May R, Berry WL, Yao J, Chandrakesan P, Zheng W, Zhao L, Zhao KL, et al. Alternative splice variants of DCLK1 mark cancer stem cells, promote self-renewal and drug-resistance, and can be targeted to inhibit tumorigenesis in kidney cancer. Int J Cancer. 2018;143:1162–1175. doi: 10.1002/ijc.31400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, Wu Y, Han F, Zhang L, Yu T, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68:118–129. doi: 10.1136/gutjnl-2017-314983. [DOI] [PubMed] [Google Scholar]
- 24.Smith LD, Leme de Calais F, Raponi M, Mellone M, Buratti E, Blaydes JP, Baralle D. Novel splice-switching oligonucleotide promotes BRCA1 aberrant splicing and susceptibility to PARP inhibitor action. Int J Cancer. 2017;140:1564–1570. doi: 10.1002/ijc.30574. [DOI] [PubMed] [Google Scholar]
- 25.Le KQ, Prabhakar BS, Hong WJ, Li LC. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol Sin. 2015;36:1212–1218. doi: 10.1038/aps.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson GR, Wardell SE, Cakir M, Yip C, Ahn YR, Ali M, Yllanes AP, Chao CA, McDonnell DP, Wood KC. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat Commun. 2018;9:1677. doi: 10.1038/s41467-018-04033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen A, Tiosano D, Guran T, Baris HN, Bayram Y, Mory A, Shapiro-Kulnane L, Hodges CA, Akdemir ZC, Turan S, et al. Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Hum Mol Genet. 2018;27:1913–1926. doi: 10.1093/hmg/ddy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter U, Lahtinen T, Marttinen P, Myöhänen M, Greco D, Cannino G, Jacobs HT, Lietzén N, Nyman TA, Battersby BJ. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Hao C, Duan H, Li H, Wang H, Liu Y, Fan Y, Zhang C. Knockdown of MRPL42 suppresses glioma cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Rep. 2018;38:BSR20171456. doi: 10.1042/BSR20171456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivanco I, Chen ZC, Tanos B, Oldrini B, Hsieh WY, Yannuzzi N, Campos C, Mellinghoff IK. A kinase-independent function of AKT promotes cancer cell survival. eLife. 2014;3:e03751. doi: 10.7554/eLife.03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal E, Brattain MG, Chowdhury S. Cell survival and metastasis regulation by Akt signaling in colorectal cancer. Cell Signal. 2013;25:1711–1719. doi: 10.1016/j.cellsig.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nath S, Mandal C, Chatterjee U, Mandal C. Association of cytosolic sialidase Neu2 with plasma membrane enhances Fas-mediated apoptosis by impairing PI3K-Akt/mTOR-mediated pathway in pancreatic cancer cells. Cell Death Dis. 2018;9:210. doi: 10.1038/s41419-017-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suyama K, Yao J, Liang H, Benard O, Loudig OD, Amgalan D, McKimpson WM, Phillips GR, Segall J, Wang Y, et al. An Akt3 Splice Variant Lacking the Serine 472 Phosphorylation Site Promotes Apoptosis and Suppresses Mammary Tumorigenesis. Cancer Res. 2018;78:103–114. doi: 10.1158/0008-5472.CAN-15-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Zhao X, Mo Z, Huang W, Yan H, Ling Z, Ye Y. Formononetin promotes cell cycle arrest via downregulation of Akt/Cyclin D1/CDK4 in human prostate cancer cells. Cell Physiol Biochem. 2014;34:1351–1358. doi: 10.1159/000366342. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Xu W, Li B, Zhang K, Wu Y, Xu H, Wang J, Zhang J, Fan R, Wei J. Curcumin Promotes Cell Cycle Arrest and Inhibits Survival of Human Renal Cancer Cells by Negative Modulation of the PI3K/AKT Signaling Pathway. Cell Biochem Biophys. 2015;73:681–686. doi: 10.1007/s12013-015-0694-5. [DOI] [PubMed] [Google Scholar]
- 37.Wei J, Wu J, Xu W, Nie H, Zhou R, Wang R, Liu Y, Tang G, Wu J. Salvianolic acid B inhibits glycolysis in oral squamous cell carcinoma via targeting PI3K/AKT/HIF-1α signaling pathway. Cell Death Dis. 2018;9:599. doi: 10.1038/s41419-018-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avanzato D, Pupo E, Ducano N, Isella C, Bertalot G, Luise C, Pece S, Bruna A, Rueda OM, Caldas C, et al. High USP6NL levels in breast cancer sustain chronic AKT phosphorylation and GLUT1 stability fueling aerobic glycolysis. Cancer Res. 2018;78:3432–3444. doi: 10.1158/0008-5472.CAN-17-3018. [DOI] [PubMed] [Google Scholar]
- 39.Haas B, Klinger V, Keksel C, Bonigut V, Kiefer D, Caspers J, Walther J, Wos-Maganga M, Weickhardt S, Röhn G, et al. Inhibition of the PI3K but not the MEK/ERK pathway sensitizes human glioma cells to alkylating drugs. Cancer Cell Int. 2018;18:69. doi: 10.1186/s12935-018-0565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn KC, Choi JY, Kim JS, Hwang SG, Kim WJ, Park JK, Um HD. ICAM-3 endows anticancer drug resistance against microtubule-damaging agents via activation of the ICAM-3-AKT/ERK-CREB-2 pathway and blockage of apoptosis. Biochem Biophys Res Commun. 2013;441:507–513. doi: 10.1016/j.bbrc.2013.10.096. [DOI] [PubMed] [Google Scholar]
- 41.Park EJ, Min KJ, Choi KS, Kwon TK. Dicoumarol sensitizes renal cell carcinoma Caki cells to TRAIL-induced apoptosis through down-regulation of Bcl-2, Mcl-1 and c-FLIP in a NQO1-independent manner. Exp Cell Res. 2014;323:144–154. doi: 10.1016/j.yexcr.2014.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.