Abstract
Emerging evidence indicates that mesenchymal stem cells (MSCs) serve an indispensable role in the tumor microenvironment. However, whether MSCs participate in the development of oral carcinogenesis remains unclear. The present study isolated MSCs from clinical tissues and investigated the differences of MSCs derived from normal oral mucosa (N-MSC), oral leukoplakia with dysplasia (LK-MSC) and oral carcinoma (Ca-MSC). The results revealed that the LK-MSCs exhibited reduced proliferation and migration, compared with the N-MSCs and Ca-MSCs. Furthermore, it was demonstrated that the exosomes secreted by LK-MSCs have significant roles in promoting proliferation, migration and invasion in vitro, which was similar to the Ca-MSC-derived exosomes. The promoting effect was also demonstrated in a 3D coculture model. When the secretion of exosomes was blocked, the promoting effect of LK-MSCs was reversed. Based on a microarray analysis of MSC-derived exosomes, microRNA-8485 (miR-8485) was identified to be ectopically expressed. The exosomal miR-8485 was capable of promoting the proliferation, migration and invasion of tumor cells. Therefore, the present study highlights the significance of MSC-derived exosomes and exosomal miR-8485 in premalignant lesions and carcinogenesis. Intervention with the secretion of MSC-derived-exosomes may be an innovative strategy to retard the carcinogenesis.
Keywords: oral premalignant lesion, carcinogenesis, mesenchymal stem cell, exosome, microRNA-8485
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common malignancy types globally, with an annual incidence of 354,864 cases according to a paper published in 2018 (1). The majority of OSCC cases in the oral mucosa evolve from oral premalignant lesions (2). Although erythroplakia has an increased malignant transformation rate, oral leukoplakia is more common in the clinic (3). Oral leukoplakia is one of the most common types of precancerous lesion (4), but its pathogenesis and carcinogenesis have not yet been thoroughly elucidated. Generally, the clinical treatment of oral leukoplakia includes photodynamic therapy, microwave therapy and surgical resection (5). However, oral leukoplakia is prone to recur and transform into carcinoma in situ, which notably affects the quality of life of patients in a negative way (6).
Malignant transformation is not only attributable to the cancer cells, as the complex biological interactions between the tumor and the stroma-tumor microenvironment (TME) also contribute to this transformation (7,8). The TME involves different types of cells, including mesenchymal stem cells (MSCs), fibroblasts, immune cells and endothelial cells (9). During carcinogenesis, MSCs sustain tumor cell growth and downregulate antitumor effector lymphocytes (10). MSCs also inhibit the antitumor immune response through carcinoma-associated fibroblasts or bone marrow stromal cells (11). However, the nature of the association between MSCs and tumor cells is controversial (12). In mouse xeno-graft models, tumor growth could be inhibited following the injection of MSCs (13,14). MSC may inhibit tumor growth by increasing inflammatory infiltration, inhibiting angiogenesis and suppressing tumor-associated signaling pathways (15). MSCs derived from oral mucosa have a distinct neural crest origin and possess superior immunomodulation, exhibiting a number of unique stem cell-like properties, including enhanced proliferative capacity, compared with MSCs derived from bone marrow and other postnatal tissues (16,17). Therefore, MSCs derived from oral tissues may have different properties. In oral leukoplakia, whether local MSCs participate in the process of carcinogenesis has not yet been thoroughly elucidated.
Cell-to-cell interactions are direct and indirect, and the exchange of extracellular vesicles has profound effects (18). Exosomes are vesicles secreted by the majority of cells and are rich in protein, mRNA, microRNA (miRNA) and lipids that mediate cell-to-cell signaling (19-21). The contents in an exosome vary according to cell type and environment (22). Recent studies demonstrated that exosomes encapsulating microRNAs are delivered to regulate recipient cells through serum (23,24) or saliva (25). Thus, exosomes are promising prognostic biomarkers and are targets for the treatment of various types of disease.
In the present study, it was proposed that exosomes secreted by MSCs are influenced by abnormal microenvironments and MSCs derived from oral leukoplakia with dysplasia (LK-MSC) may be the pivotal factor in the process of oral carcinogenesis. Additionally, the exosomal miR-8485 was also investigated and the tumor promotion function was identified.
Materials and methods
Clinical samples
Oral mucosal tissues were obtained from clinical patients in the Peking University School of Stomatology (Beijing, China) from September 2016. to September 2018. The patients were suffering from mucosal cysts or the third molar extraction (n=16), oral dysplasia (n=17) and oral carcinoma (n=15). The inclusion criteria were as follows: For oral normal mucosal tissues, biopsy results indicated that there was no inflammation; for dysplasia tissues, the clinical diagnosis was oral leukoplakia or erythema and the biopsy results were mild to moderate hyperplasia; and for carcinoma tissues, the clinical diagnosis was carcinoma and biopsy results were early infiltration or carcinoma in situ. All patients provided informed consent. The patient information is presented in Table I. The present study was approved by the Ethics Committee of the Peking University School of Stomatology (Beijing, China).
Table I.
Clinical characteristic of the patients for reverse transcription-quantitative polymerase chain reaction.
Cell culture
Human MSCs were isolated and identified according to a previous study (26). Briefly, the primary tissues were treated by dispase (2 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in sterile PBS and the mesenchyme part was separated and cut into 2 mm3, and then cultured in a 60-mm dish in MSC medium (Sciencell Research Laboratories, Inc., San Diego, CA, USA) for 5-7 days at 37°C. The MSCs were obtained from three donors for each type and the patient information is presented in Table II. Cells at passages 3-5 were used for the subsequent experiments. The DOK cell line (oral hyperplasia cell line) was purchased from the Cell Laboratory of Central South University (Changsha, China) and the SCC15 cell line (oral carcinoma cell line) was provided by the Department of Pathology, Peking University School of Stomatology (Beijing, China). DOK cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and SCC15 cells were cultured in Dulbecco's modified Eagle's medium: F12 (DMEM:F12; Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured at 37°C in a humidified 5% CO2 atmosphere with 10% fetal bovine serum (FBS).
Table II.
Clinical characteristic of the patients for primary cell culture.
| Patient | Sex | Age, years |
|---|---|---|
| N-MSC-1 | Female | 31 |
| N-MSC-2 | Female | 35 |
| N-MSC-3 | Male | 42 |
| LK-MSC-1 | Female | 32 |
| LK-MSC-2 | Male | 40 |
| LK-MSC-3 | Female | 54 |
| Ca-MSC-1 | Male | 29 |
| Ca-MSC-2 | Female | 48 |
| Ca-MSC-3 | Male | 55 |
MSC, mesenchymal stem cell; N-MSC, MSCs derived from normal oral mucosa; LK-MSC, MSCs derived from oral premalignant lesion; Ca-MSC, MSCs derived from oral carcinoma.
Osteogenic and adipogenic differentiation
The MSCs were characterized by differentiation into osteogenic and adipogenic lineages according to the protocols of our previous study (26). For osteogenic differentiation, 10 nM dexamethasone, 10 mM β-glycerophosphate, 0.1 mM L-ascorbic acid-2-phosphate and 2 mM glutamine (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were supplemented in α-minimum Eagle's medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) for cell culture at 37°C in a humidified 5% CO2 atmosphere. For adipogenic differentiation, 1 µM dexamethasone, 0.5 mM 3-isobutyl-ethylxanthine, 10 mg/ml insulin, 60 mM indomethacin and 2 mM glutamine were supplemented in α-MEM. Alizarin Red S and Oil Red O staining assays were performed respectively for 30 min at room temperature, to detect osteogenic and adipogenic differentiation. The staining was imaged with a light microscope (Olympus IX51; Olympus Corporation, Tokyo, Japan) at ×10 magnification.
Cell proliferation assay
An IncuCyte® live-cell imager (Essen Bioscience, Ann Arbor, MI, USA) was utilized for the proliferation assay. In brief, cells (MSC, 1×103; SCC15, 5×103; and DOK, 3×103) were seeded in a 96-well-plate with 100 µl α-MEM (MSCs), DMEM:F12 (SCC15) or RPMI-1640 (DOK cells), medium containing 10% FBS/well and incubated at 37°C in a humidified 5% CO2 atmosphere for 2-5 days. Fresh medium was applied every day. The plate was scanned, and phase-contrast images were captured at ×10 magnification with a light microscope (Nikon Corporation, Tokyo, Japan). The ratios of cell growth confluence were analyzed.
Wound healing assay
The cells (MSC, 1×104; SCC15, 3×104; DOK, 3×104) were seeded in a 96-well-plate with 100 µl α-MEM, DMEM:F12 or RPMI-1640 medium containing 10% FBS at 37°C and then when a confluence of 90% was reached, a uniform wound ~800 µm in width was inflicted by a woundmaker (Essen Bioscience). To remove all cellular debris, the cells were washed twice with PBS. Subsequently, the plates were cultured in serum-free medium for 6 h (SCC15 cells, DMEM:F12 medium) or 24 h (MSC, α-MEM medium; and DOK cells, RPMI-1640 medium) at 37°C. The closure of the wounds was evaluated using a light microscope (Nikon Corporation) at ×10 magnification and the relative mobility width was calculated.
Cell invasion assay
A cell invasion assay was conducted using a Transwell chamber (8 µm, 24-well insert; Corning Inc., Corning, NY, USA). The Transwell membranes were coated with Matrigel (cat. no. 356234; BD Biosciences; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) prior to the seeding of SCC15 or DOK cell suspensions (1×106 cells/ml) with medium (SCC15 cells, DMEM:F12 medium; and DOK cells, RPMI-1640 medium) without FBS in the upper chamber. After culturing for 12 h, the cells invading into the lower chambers, which contained 10% FBS and RPMI-1640 medium for DOK cells and DMEM:F12 for SCC15 cells, were fixed with methanol at room temperature for 20 min, stained with 0.1% crystal violet at room temperature for 5 min and observed in ≥6 fields using a light microscope (Olympus BX51; Olympus Corporation) at ×20 magnification in order to count the cells.
Exosome isolation, characterization and uptake
Exosomes were collected from the supernatants of the MSCs cultured for 48 h in α-MEM containing 10% FBS and centrifuged for 16 h at 100,000 × g at 4°C. The exosomes were then collected by density gradient differential centrifugation (27). Briefly, the supernatants were centrifuged at 300 × g for 10 min, 2,000 × g for 20 min and 10,000 × g for 30 min all at 4°C to remove the cell debris and large vehicles. Subsequently, the supernatants were passed through centrifugal filters at 5,000 × g for 30 min at 4°C. The concentrated supernatants were centrifuged at 100,000 × g for 70 min at 4°C in a 30% sucrose/D2O solution, and were then washed and purified with PBS by centrifugation at 5,000 × g for 30 min repeated 3 times at 4°C in centrifugal filters. The shape of exosomes was observed with an electron microscope (JEOL, Ltd., Tokyo, Japan) at ×350,000 magnification. The concentration of the total exosome proteins was quantified using a Bicinchoninic Acid (BCA; Thermo Fisher Scientific, Inc.) protein assay. Exosomes were labeled by PKH26 (Sigma-Aldrich; Merck KGaA), according to the manufacturer's protocols. Briefly, 100 µg exosomes suspended in PBS were added into 1 ml Dilute C, and then 4 µl PKH26 was added into Dilute C and mixed for 5 min at room temperature. Subsequently, 2 ml 0.5% bovine serum albumin (Huaxingbio Biotechnology, Beijing, China) was added to bind excess dye. The labeled exosome suspensions were then centrifuged at 100,000 × g for 70 min at 4°C and resuspended with 100 µl PBS. The labeled exosomes were incubated with SCC15 cells for 24 h at 37°C. Actin-Tracker Green [F-actin-fluorescein isothiocyanate (FITC); Beyotime Institute of Biotechnology, Haimen, China] was used to label the cytoskeleton at room temperature for 20 min and DAPI was used to label the cellular nuclei at room temperature for 5 min, respectively. The uptake images were captured with a LSM 5 Exciter confocal laser scanning microscope (Carl Zeiss AG, Oberkochen, Germany) at ×100 magnification. In total, 100 µg/ml exosomes were predicted to be the best concentration and were therefore applied in the subsequent experiments.
Western blotting
The proteins was extracted by radioimmunoprecipitation assay lysis buffer (cat. no. R0020; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The protein concentration was determined by BCA method. A total of 30 µg proteins were loaded and separated on 10% SDS-PAGE. The proteins were transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking with 5% evaporated milk at room temperature for 1 h, the membrane was incubated with the mouse antibodies cluster of CD63 (1:1,000; cat. no. ab59479; Abcam, Cambridge, MA, USA) and CD9 (1:1,000; cat. no. ab92726; Abcam) and the rabbit antibody p53 (1:1,000; cat. no. 2527; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight. The mouse antibody GAPDH (1:5,000; cat. no. HX1828; Huaxingbio Biotechnology) was used as reference protein at 4°C overnight. Goat FITC-conjugated anti-mouse IgG (1:10,000; cat. no. HX2032; Huaxingbio Biotechnology) or goat anti-rabbit IgG (1:5,000; cat. no. CW0103; CWbio, Beijing, China) was used as the secondary antibody at room temperature for 30 min. The results were detected with a chemiluminescence reagent (CWbio).
3D coculture model
A total of 5×105 MSCs were embedded in 1 ml rat tail collagen type-I (Beijing Solarbio Science & Technology Co., Ltd.). After the collagen had solidified, the gel was overlaid with 5×105 SCC15 cells and then cultured at 37°C, 24 h later, the gel was lifted at the cell-air interface with a Transwell chamber (8 µm, 24-well insert; Corning Inc.) and cultured for 5 days at 37°C with the stimulation of transforming growth factor (TGF)-β1 (5 ng/ml; Proteintech Group, Inc., Chicago, IL, USA). Meanwhile, GW4869 (10 mM; cat. no. HY-19363/CS-6865; MedChemExpress, Monmouth Junction, NJ, USA), an inhibitor of exosomal secretion, was added to the coculture models. Subsequently, the cocultures were fixed with 4% paraformaldehyde for 20 min at room temperature for the following staining procedures.
Immunohistochemical staining and haematoxylin and eosin (H&E) staining
The tissues were fixed by 4% paraformaldehyde at 4°C for 24 h and the paraffin-embedded sections (5 µm) were then dewaxed in xylene and dehydrated in ethanol (100, 95, 90, 80 and 70%). For immunohistochemical staining, the tissue sections were placed into sodium citrate buffer (OriGene Technologies, Inc., Beijing, China) and heated to 120°C for 1 min for antigen retrieval. Subsequently, 3% hydrogen peroxide was applied as blocking reagent at room temperature for 10 min. CK-Pan (1:1,000; cat. no. 4545; Cell Signaling Technology, Inc.) and vimentin (1:1,000; cat. no. ab92547; Abcam) primary antibodies were applied to the tissue sections overnight at 4°C. The endogenous peroxidase activity was blocked by 3% hydrogen peroxide pretreatment. The sections were then incubated with peroxidase-conjugated mouse anti-goat secondary antibody (ready to use; cat. no. PV-9000; OriGene Technologies, Inc.) for 30 min at room temperature, and DAB staining at room temperature for 30 sec was used for staining. For H&E staining, the tissue sections were stained followed by standard protocols (28) after dewaxing in xylene and dehydrated in ethanol (100, 95, 90, 80 and 70%). The results were observed with a light microscope (Olympus BX51; Olympus Corporation) at ×20 magnification.
Reverse transcription-quantitative polymerase chain reaction (PCR) and microarray analysis
Total RNA was extracted by TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) from the 6-well plates and tissue RNA was obtained, according to standard protocols (29). An All-in-One™ First-Strand cDNA Synthesis kit (cat. no. QP008; GeneCopoeia, Inc., Rockville, MD, USA) was used to reverse transcribe the miRNAs. Subsequently, quantitative PCR was performed and the expression of miR-8485 (cat. no. HmiRQP4656; GeneCopoeia, Inc.) was normalized to the expression of U6 [Sangon Biotech Co., Ltd., Shanghai, China; sequence (5′-3′), forward primer: CTCGCTTCGGCAGCACA; and reverse primer: AACGCTT CACGAATTTGCGT]. The experiment was performed with an Applied Biosystems 7500 instrument. A 20 µl reaction system was used with 10 µl 2X SYBR® Green (Roche Applied Science, Rotkreuz, Switzerland), 1 µl primer, 1 µl 2X Universal Adapter (cat. no. QP029; GeneCopoeia, Inc.), 2 µl cDNA and 2 µl H2O. Reactions were incubated at 95°C for 10 min; 95°C for 15 sec; 60°C for 30 sec; 72°C for 20 sec for 40 cycles; 95°C for 15 sec; 60°C for 1 min; 95°C for 15 sec; 60°C for 15 sec. The results were analyzed using the 2-∆∆Cq method (30). Microarray analysis was performed by Sangon Biotech Co., Ltd., according to standard Agilent protocols (31-33).
miRNA transfection
DOK or SCC15 cells were pretreated with 50 nM miR-8485 mimics (cat. no. miR1180323091501) or negative control (NC-mimics; cat. no. miR1N0000001), as well as 100 nM miR-8485 inhibitor (cat. no. miR2180709052553) or the corresponding negative control (NC-inhibitor; cat. no. miR2N0000001) (Guangzhou RiboBio Co., Ltd., Guangzhou, China) for 48 h at 37°C. Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) was used to transfer the mimics or inhibitors into the cells, according to the manufacturer's instructions.
Statistical analysis
All data conforming to a normal distribution is expressed as the mean ± standard deviation. For all other data, the median ± standard deviation is expressed. A Student's t-test, least significant difference test of one-way analysis of variance or the Mann-Whitney U test was applied to evaluate differences among groups using SPSS software, version 17.0 (SPSS, Inc., Chicago, IL, USA). All in vitro data was repeated at least 3 independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification and comparison of MSCs derived from normal oral mucosa (N-MSCs), LK-MSCs and oral carcinoma (Ca-MSCs)
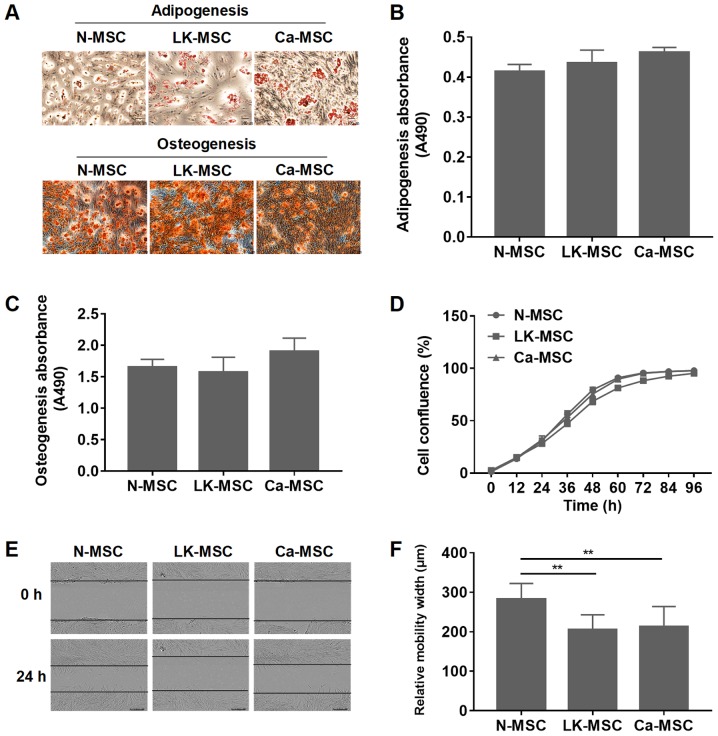
N-MSCs, LK-MSCs and Ca-MSCs were successfully generated according to the methods of our previous study (26). For osteogenic and adipogenic differentiation, all of the MSCs formed mineralized nodules and oil droplets, and there was no statistically significant difference among the three groups (P>0.05; Fig. 1A-C). However, the proliferation rate of the LK-MSCs was reduced at 24-72 h, compared with N-MSCs and Ca-MSCs (Fig. 1D). The wound healing assay demonstrated that compared with the N-MSCs, the migration rates of the LK-MSCs and Ca-MSCs were significantly reduced; however, there was no significant difference between those of the LK-MSCs and Ca-MSCs (P>0.05; Fig. 1E and F).
Figure 1.
Characteristics of the N-MSCs, LK-MSCs and Ca-MSCs. (A) Adipogenic and osteogenic differentiation were assessed. Representative images with a magnification of ×10 are depicted. (B) Quantitative analysis of adipogenic differentiation was calculated using the absorbance value at 490 nm. (C) Quantitative analysis of osteogenic differentiation was calculated using the absorbance value at 490 nm. (D) MSCs were cultured and the cell confluence was analyzed with an InCucyte monitoring microscope. (E and F) The relative migration width was determined by a wound healing assay. Representative images with a magnification of ×10 are depicted. **P<0.01. MSC, mesenchymal stem cell; N-MSC, MSCs derived from normal oral mucosa; LK-MSC, MSCs derived from oral leukoplakia with dysplasia; Ca-MSC, MSCs derived from oral carcinoma.
Characterization of MSC-derived exosomes
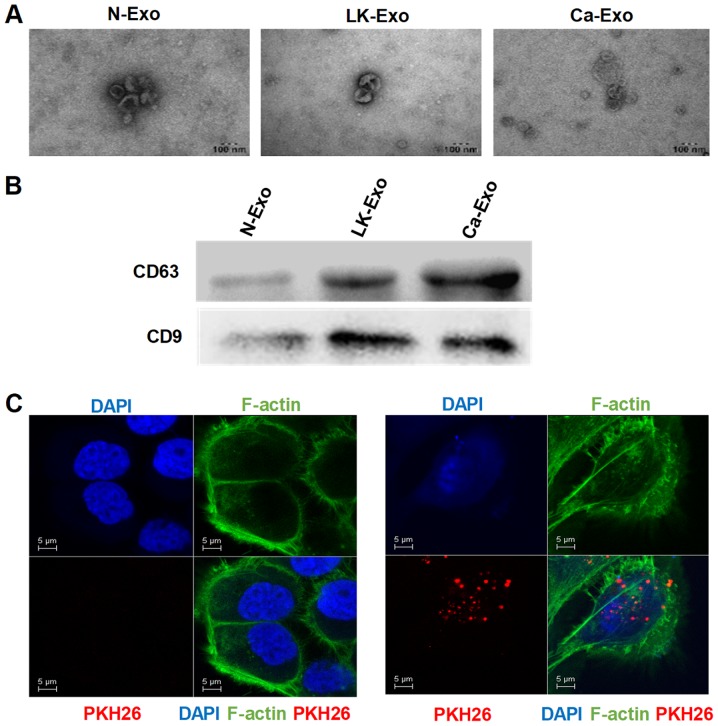
In oral premalignant lesions, the interaction between epithelial cells and MSCs is probably via paracrine signaling with cytokines or extracellular vesicles. Exosomes are small membrane vesicles (diameter, 30-200 nm) that are constitutively released via fusion with the cell membrane (22). In order to investigate whether exosomes participate in the interaction between MSCs and epithelial cells, exosomes from MSCs were isolated and characterized in the present study. The electron microscopy results revealed that the exosomes had a cup-shaped morphology with diameters of <100 nm (Fig. 2A). Additionally, CD63 and CD9 were enriched among the exosome proteins (Fig. 2B). To confirm the uptake of exosomes, the fluorescent dye PKH26 was applied to label the exosomes. The PKH26-labeled exosomes were localized in the cytoplasm of the SCC15 cells (Fig. 2C), indicating that exosomes can be internalized by tumor cells.
Figure 2.
Identification of exosomes. (A) Exosomes were isolated from MSCs and observed using transmission electron microscopy. (B) The western blotting revealed that the CD63 and CD9 proteins were expressed in the exosomes. (C) Confocal microscopy was used to identify the uptake of PKH26-labeled exosomes, which were secreted by N-MSC and internalized in the cytoplasm. N-exo, exosomes secreted by MSCs derived from normal oral mucosa; LK-exo, exosomes secreted by MSCs derived from oral leukoplakia with dysplasia; Ca-exo, exosomes secreted by MSCs derived from oral carcinoma; MSCs, mesenchymal stem cells; CD63, cluster of differentiation 63.
LK-MSC- and Ca-MSC-derived exosomes enhance the proliferation, migration and invasion abilities of epithelial cells
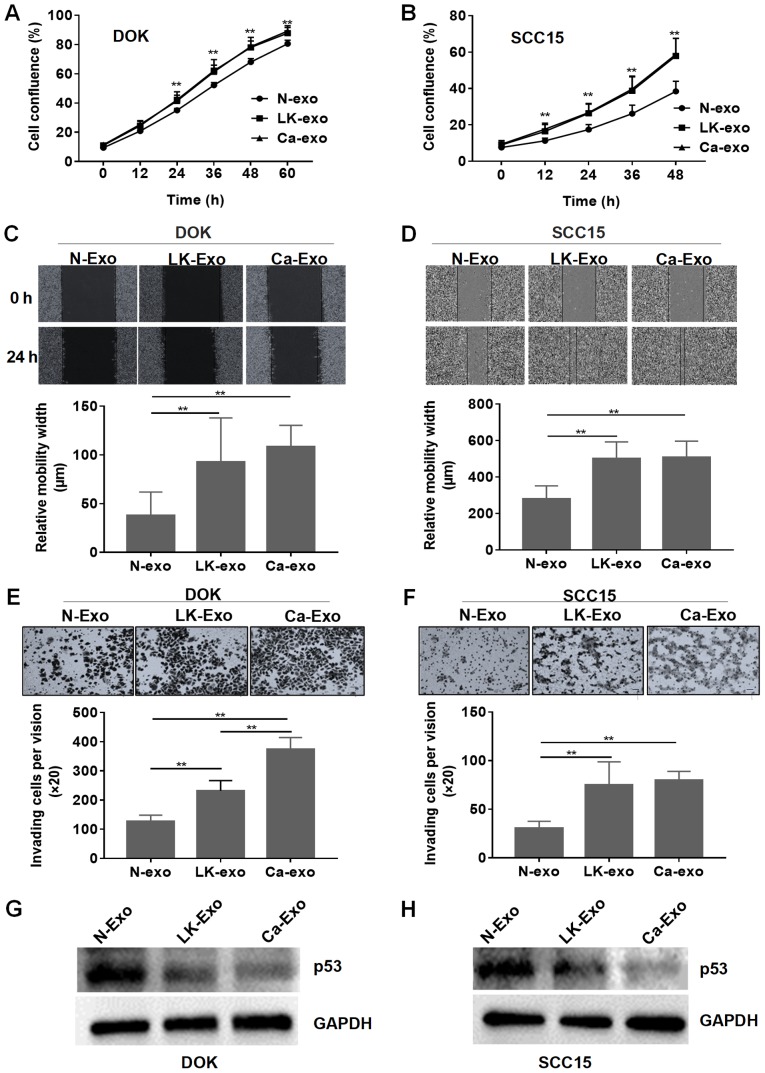
To identify the function of the exosomes, SCC15 and DOK cells were treated with them separately. The cell proliferation assay demonstrated that the exosomes from the LK-MSCs (LK-exo) and Ca-MSCs (Ca-exo) accelerated the proliferation of DOK and SCC15 cells, compared with the exosomes derived from the N-MSCs (N-exo) (P<0.01; Fig. 3A and B). However, there was no significant difference between the LK-MSC and Ca-MSC groups (P>0.05).
Figure 3.
Effects of MSC-derived exosomes on epithelial cells. (A) The effect of the exosomes on proliferation was determined by the confluence of DOK cells. (B) The effect of the exosomes on proliferation was determined by the confluence of SCC15 cells. (C) The relative migration width of DOK cells was determined by a wound healing assay. Representative images with a magnification of ×10 are depicted. (D) The relative migration width of SCC15 cells was determined by a wound healing assay. Representative images with a magnification of ×10 are depicted. (E) The invasion ability of DOK cells treated with exosomes was assessed by a Matrigel cell invasion assay. Representative images with a magnification of ×20 are depicted. (F) The invasion ability of SCC15 cells treated with exosomes was assessed by a Matrigel cell invasion assay. Representative images with a magnification of ×20 are depicted. (G) The expression of p53 was assessed by western blotting for DOK cells. (H) The expression of p53 was assessed by western blotting for SCC15 cells. **P<0.01, compared with N-exo. N-exo, exosomes secreted by MSCs derived from normal oral mucosa; LK-exo, exosomes secreted by MSCs derived from oral leukoplakia with dysplasia; Ca-exo, exosomes secreted by MSCs derived from oral carcinoma; MSCs, mesenchymal stem cells.
Subsequently, the role of the exosomes secreted by MSCs in inducing migration and invasion was investigated. The wound healing assay demonstrated that the relative migration widths of the SCC15 and DOK cells pretreated with LK-exo and Ca-exo were significantly increased, compared with the N-exo-treated group (P<0.01; Fig. 3C and D). The Transwell cell invasion experiment revealed that the LK-exo and Ca-exo groups had an increased number of invading cells, compared with the N-exo group (Fig. 3E and F). The p53 tumor suppressor gene is widely reported to be implicated in oral carcinogenesis (34-36). The western blot assay in the present study demonstrated that the LK-exo- and Ca-exo-pretreated SCC15 and DOK cells exhibited reduced expression of p53, compared with the N-exo-pretreated group (Fig. 3G and H). These results implied that exosomes secreted by LK-MSCs have similar functions to those secreted by Ca-MSCs, which indicates that exosomes secreted by residual LK-MSCs may be a cause of the recurrence of oral leukoplakia and may accelerate the process of malignant transformation, which will be further identified in animal models.
Interaction between MSCs and tumor cells in a 3D coculture model
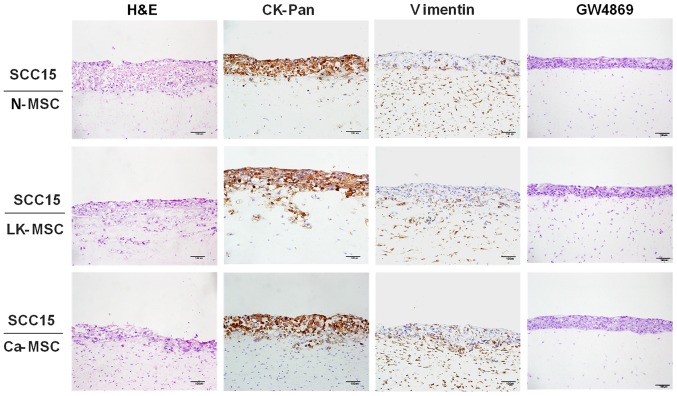
In order to fully understand the interaction between MSCs and epithelial cells, a TGF-β1-3D-coculture model was prepared to mimic in vivo interactions (37). Immunohistochemical staining of CK-Pan and vimentin was conducted to clarify the boundaries between the MSCs and SCC15 cells. Notably, the LK-MSCs and Ca-MSCs, particularly LK-MSCs, were more susceptible to TGF-β1 stimulation, compared with N-MSCs, promoting SCC15 cell invasion via collagen hydrolysis and fracture, with CK-Pan expressed in a deeper part of the MSCs; however, the integrity of the basement membrane was maintained in the N-MSC group. Furthermore, the enhancing nature of LK-MSC- and Ca-MSC-derived exosomes was blocked by the exosomal secretion inhibitor GW4869, as they exhibited similar H&E staining to the N-MSC group (Fig. 4).
Figure 4.
SCC15 cells cocultured with MSCs in a 3D coculture model. MSCs and SCC15 cells were cocultured in a 3D coculture model for 5 days. The cocultures were assessed with H&E and immunohistochemical staining. Representative images with a magnification of ×20 are depicted. MSCs, mesenchymal stem cells. MSC, mesenchymal stem cell; N-MSC, MSCs derived from normal oral mucosa; LK-MSC, MSCs derived from oral premalignant lesion; Ca-MSC, MSCs derived from oral carcinoma; H&E, haematoxylin and eosin.
Exosomes derived from LK-MSCs and Ca-MSCs contain increased miR-8485, compared with N-MSCs
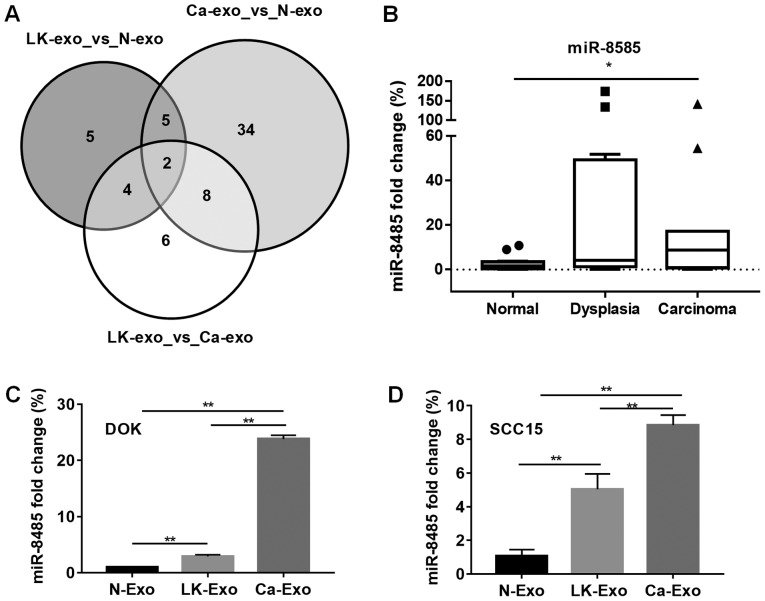
To investigate whether miRNAs contained in exosomes from different MSCs function differently, microarray analysis was performed. The results demonstrated that there were 16 differentially-expressed miRNAs between the N-exo and LK-exo groups, 49 between the N-exo and Ca-exo groups, and 20 between the LK-exo and Ca-exo groups. Among these miRNAs, the expression levels of miR-4433a and miR-8485 were different between all three groups (Fig. 5A). Therefore, miR-8485, which may participate in manipulating tumor cells, was selected for the subsequent experiments. The clinical tissues and cells were treated with the exosomes. The expression levels of miR-8485 were elevated in the oral carcinoma tissues (P<0.05) and oral dysplasia tissues, compared with the normal mucosa tissues (Fig. 5B). Furthermore, the cells treated with LK-exo and Ca-exo expressed significantly increased levels of miR-8485, compared with the N-exo group (P<0.01; Fig. 5C and D).
Figure 5.
Expression of exosomal miR-8485 was gradually elevated. (A) The miRNA from the N-exo, LK-exo and Ca-exo groups was analyzed. Among the miRNAs measured with fold change >2, the expression levels of miR-4433a and miR-8485 were both changed. (B) The expression of miR-8485 in the clinical tissues was assessed by reverse transcription-quantitative polymerase chain reaction. The box represents the interquartile range (25-75th percentile) and the line within this box is the median value. The bottom and top bars of the whisker plot indicate the 10th and 90th percentiles, respectively. (C) DOK cells treated with LK-exo and Ca-exo exhibited elevated expression of miR-8485. (D) SCC15 cells treated with LK-exo and Ca-exo exhibited elevated expression of miR-8485. *P<0.05 and **P<0.01. miR, microRNA; N-exo, exosomes from mesenchymal stem cells derived from normal oral mucosa; LK-exo, exosomes from mesenchymal stem cells derived from oral premalignant lesion; Ca-exo, exosomes from mesenchymal stem cells derived from oral carcinoma.
miR-8485 promotes the proliferation, migration and invasion abilities of epithelial cells
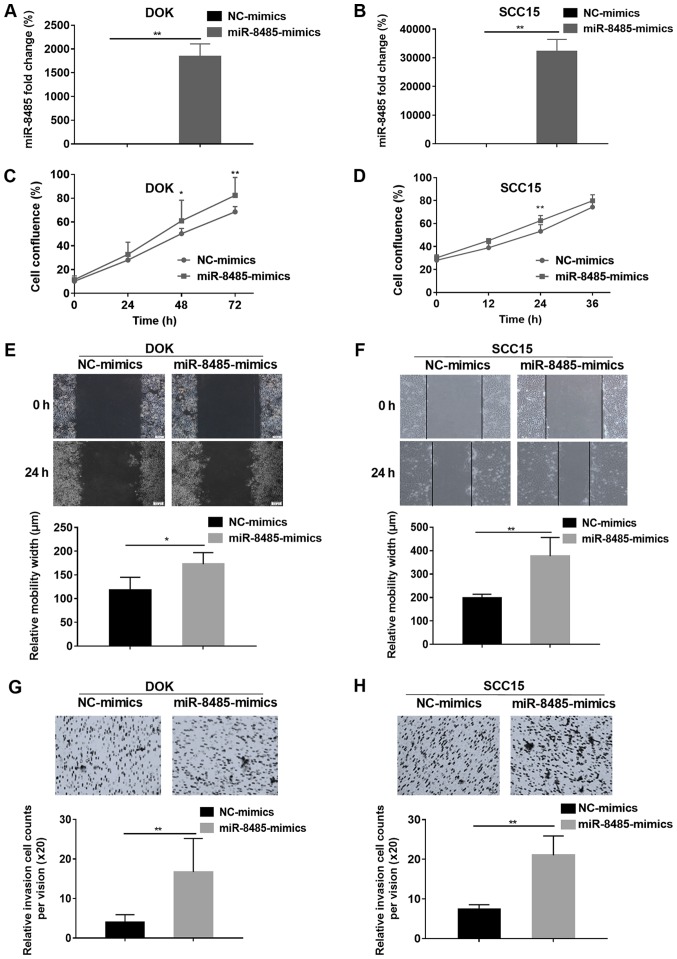
Based on the aforementioned results, miR-8485 mimics were transfected into DOK and SCC15 cells and the transfection efficiency was assessed by a reverse transcription-quantitative PCR assay (Fig. 6A and B). Compared with the NC-mimics group, the miR-8485 mimics caused rapid growth of the DOK (48 and 72 h) and SCC15 cells (24 h) (P<0.01; Fig. 6C and D). Overexpression of miR-8485 promoted migration of the DOK cells (P<0.05) and SCC15 cells (P<0.01) (Fig. 6E and F). Furthermore, the cell invasion assay demonstrated that the miR-8485 mimics increased the invasive ability of the DOK and SCC15 cells (P<0.01; Fig. 6G and H).
Figure 6.
Overexpression of miR-8485 promotes the migration and invasion of DOK and SCC15 cells. (A) The miR-8485 mimics and mimics control (NC-mimics) were transfected into DOK cells, and the efficiency was assessed by reverse transcription-quantitative polymerase chain reaction. (B) The miR-8485 mimics and mimics control (NC-mimics) were transfected into SCC15 cells, and the efficiency was assessed by reverse transcription-quantitative polymerase chain reaction. (C) The effect of the miR-8485 mimics on proliferation was analyzed by assessing cell confluence of DOK cells. (D) The effect of the miR-8485 mimics on proliferation was analyzed by assessing cell confluence of SCC15 cells. (E) The relative migration width of the DOK cells following overexpression of miR-8485 was determined with a wound healing assay. Representative images with a magnification of ×10 are depicted. (F) The relative migration width of the SCC15 cells following overexpression of miR-8485 was determined with a wound healing assay. Representative images with a magnification of ×10 are depicted. (G) The Matrigel invasion assay indicated the invasion ability of the DOK cells. Representative images with a magnification of ×20 are depicted. (H) The Matrigel invasion assay indicated the invasion ability of the SCC15 cells. Representative images with a magnification of ×20 are depicted. *P<0.05 and **P<0.01, compared with NC groups. NC, negative control; miR, microRNA.
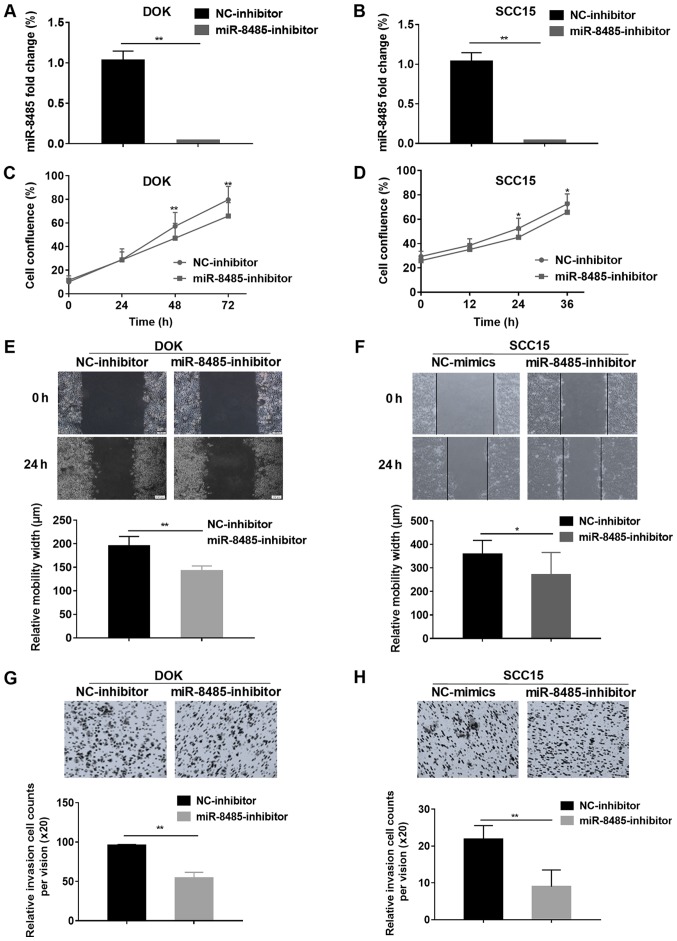
To further confirm the effects of miR-8485, DOK and SCC15 cells were transfected with a miR-8485 inhibitor (Fig. 7A and B). As expected, the DOK and cells transfected with the miR-8485 inhibitor exhibited marked inhibition of proliferation at 48 and 72 h, and the SCC15 cells at 24 and 36 h (Fig. 7C and D). Additionally, the miR-8485 inhibitor inhibited the migrative and invasive ability of the DOK and SCC15 cells in the wound healing assay and Matrigel cell invasion assay (P<0.01; Fig. 7E-H).
Figure 7.
miR-8485 inhibitor suppresses the migration and invasion of DOK and SCC15 cells. (A) miR-8485 expression was detected by reverse transcription-quantitative polymerase chain reaction after miR-8485 inhibitor was transfected into the DOK cells. (B) miR-8485 expression was detected by reverse transcription-quantitative polymerase chain reaction after miR-8485 inhibitor was transfected into the SCC15 cells. (C) The effect of the miR-8485 inhibitor on proliferation was analyzed by the cell confluence of DOK cells. (D) The effect of the miR-8485 inhibitor on proliferation was analyzed by the cell confluence of SCC15 cells. (E) The wound healing assay revealed that compared with the NC-inhibitor, the miR-8485 inhibitor suppressed the migration of the DOK cells. Representative images with a magnification of ×10 are depicted. (F) The wound healing assay revealed that compared with the NC-inhibitor, the miR-8485 inhibitor suppressed the migration of the SCC15 cells. Representative images with a magnification of ×10 are depicted. (G) The Matrigel invasion assay indicated that compared with the NC-inhibitor, the miR-8485 inhibitor suppressed the invasion ability of the DOK cells. Representative images with a magnification of ×20 are depicted. (H) The Matrigel invasion assay indicated that compared with the NC-inhibitor, the miR-8485 inhibitor suppressed the invasion ability of the SCC15 cells. Representative images with a magnification of ×20 are depicted. *P<0.05 and **P<0.01, compared with NC groups. NC, negative control; miR, microRNA.
Discussion
Oral leukoplakia is one of the common oral potentially malignant disorders worldwide, and the malignant transformation rate of oral leukoplakia varies between 0.13-34% (38). To date, studies on the etiology of oral leukoplakia carcinogenesis have primarily focused on local irritation factors, including abuse of tobacco and alcohol, infection factors, such as human papillomavirus (39), and epithelial cell factors, including oxidative stress injury on DNA (40). However, recent studies demonstrated that microenvironmental factors serve a critically important role in tumor development (9,41). MSCs are a cellular component of the TME, and the interaction between MSCs and tumor cells is bidirectional (12). In normal tissues, MSCs maintain normal structure and function, as well as organizational homeostasis (42). However, during the process of malignant transformation, MSCs are vulnerable and acquire the abnormal phenotypes of tumor cells, thereby sustaining cancer cell growth and tumor progression (43). In the present study, MSCs derived from normal mucosa, oral leukoplakia and oral carcinoma in situ tissues were separated. Compared with the N-MSCs, the LK-MSCs and Ca-MSCs exhibited a decreased migration capacity. The functions of MSCs are notably diverse and depend on the tissue-specific origins and the special microenvironment in which MSCs are embedded (44). During the process of carcinogenesis, MSC heterogeneity is characterized by altered proliferative capacity and aging properties, which may also include epigenetic changes (45). The result indicates that there is a process of functional transformation in MSCs during the TME maturing.
In the 3D coculture models, which were affected by oral epithelial dysplasia or carcinoma in situ, the LK-MSCs and Ca-MSCs exhibited clear compatibility with the tumor cells. Furthermore, the LK-MSCs were more vulnerable and sensitive to the TGF-β1 stimulation, thus promoting the migration and invasion capacity of the tumor cells via the hydrolysis and rupture of collagen type I. Therefore, MSCs are affected by epithelial cells, and thus MSCs could regulate epithelial cells by negative feedback.
As previously described, the exosome is capable of mediating intercellular communication between cells (46). Exosomes secreted by MSCs orchestrate various autocrine and paracrine functions, including receptor-binding, direct fusion or endocytosis by target cells (47), thereby transforming the biological behavior of epithelial cells (48,49). The exosome contents of MSCs cocultured with tumor cells differs from normal MSCs (50). Blocking the secretion of exosomes using the exosomal inhibitor GW4869 reverses the development of diseases caused by MSCs (51,52). Additionally, with regards to histocompatibility and reproducibility, exosomes have favorable application prospects in cell-free treatments (53). According to the present study, exosomes derived from MSCs from normal mucosa or that suffering from oral leukoplakia with dysplasia and carcinoma exhibit dissimilar effects. The exosomes isolated from LK-MSCs exhibited similar effects to the Ca-MSCs, promoting proliferation and migration, and reducing the expression of p53 in SCC15 and DOK cells in vitro. Additionally, the application of GW4869 reversed the promoting function of the LK-MSC- and Ca-MSC-derived exosomes.
MicroRNAs are preferentially encapsulated by exosomes (54). Exosomal microRNAs have various effects on tumor biological behaviors, including proliferation, migration and invasion (55), apoptosis and chemoresistance (8) and epigenetic modification of TME (56). Considering the importance of exosomal microRNAs, the N-exo, LK-exo and Ca-exo groups were subjected to microarray analysis in the present study. miR-8485, a rarely reported gene that may be associated with tumor development, was selected. In order to identify the function of miR-8485 in the cells, transfection of mimics and inhibitors was utilized. miR-8485 promoted the proliferation, migration and invasion of the tumor cells in vitro, indicating that miR-8485 is a novel microRNA associated with malignant transformation.
In summary, the present study clarifies the function of MSCs associated with both dysplastic oral keratinocyte cells and tumor cells. LK-MSCs share similar effects with Ca-MSCs, and are more susceptible to the surrounding environment. LK-MSCs may be involved in the recurrence of oral leukoplakia and malignant transformation by secreting exosomes. Additionally, the observation that the exosomes contained miR-8485 demonstrates that miR-8485 acts as an oncogene in oral carcinogenesis and therefore is a potential novel biomarker for clinical treatment. The present study emphasizes the importance of MSCs derived from premalignant lesions and the exosomes they secrete. As it is difficult to convert tumor cells to normal cells, intervention with MSC-derived exosomes from premalignant lesions may be an excellent choice to reduce the malignant transformation rate during clinical therapy.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81771071 and 81772873), Beijing Natural Science Foundation (grant nos. 7172240 and 7182181), Nonprofit Industry Research Specific Fund of National Health and Family Planning Commission of China (grant no. 201502018) and Foundation of Capital Health Development (grant no. 2014-2-4102).
Availability of data and materials
All data used or analyzed in this study are included in this article.
Authors' contributions
WL performed the majority of the experiments and wrote the manuscript. HL and YW made notable contributions to the design, data interpretation and the manuscript revision. YH, ZZ, XJ and XW were involved in the validation of data and responsible for the statistic analysis. JJ, QW and XG helped with the collection of tissue samples and ZC, ML and GW involved in the table drafting of the manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Experiments using tissue samples from human subjects were approved by the Ethics Committee of the School of Stomatology, Peking University. (Beijing, China). All patients provided written informed consent for the whole study.
Patient consent for publication
All participants provided written informed consent for the publication of the study.
Competing interest
The authors declare that they have no competing interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bouquot JE, Whitaker SB. Oral leukoplakia - rationale for diagnosis and prognosis of its clinical subtypes or 'phases'. Quintessence Int. 1994;25:133–140. [PubMed] [Google Scholar]
- 3.Kao SY, Mao L, Jian XC, Rajan G, Yu GY. Expert consensus on the detection and screening of oral cancer and precancer. Chin J Dent Res. 2015;18:79–83. [PubMed] [Google Scholar]
- 4.van der Waal I, Schepman KP, van der Meij EH, Smeele LE. Oral leukoplakia: A clinicopathological review. Oral Oncol. 1997;33:291–301. doi: 10.1016/S1368-8375(97)00002-X. [DOI] [PubMed] [Google Scholar]
- 5.Vohra F, Al-Kheraif AA, Qadri T, Hassan MI, Ahmed A, Warnakulasuriya S, Javed F. Efficacy of photodynamic therapy in the management of oral premalignant lesions. A systematic review. Photodiagn Photodyn Ther. 2015;12:150–159. doi: 10.1016/j.pdpdt.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Bewley AF, Farwell DG. Oral leukoplakia and oral cavity squamous cell carcinoma. Clin Dermatol. 2017;35:461–467. doi: 10.1016/j.clindermatol.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15:409–425. doi: 10.1038/nrc3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015;368:7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 10.Poggi A, Musso A, Dapino I, Zocchi MR. Mechanisms of tumor escape from immune system: Role of mesenchymal stromal cells. Immunol Lett. 2014;159:55–72. doi: 10.1016/j.imlet.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Poggi A, Giuliani M. Mesenchymal stromal cells can regulate the immune response in the tumor microenvironment. Vaccines (Basel) 2016;4:4. doi: 10.3390/vaccines4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klopp AH, Gupta A, Spaeth E, Andreeff M, Marini F., III Concise review: Dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells. 2011;29:11–19. doi: 10.1002/stem.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khakoo AY, Pati S, Anderson SA, Reid W, Elshal MF, Rovira II, Nguyen AT, Malide D, Combs CA, Hall G, et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi's sarcoma. J Exp Med. 2006;203:1235–1247. doi: 10.1084/jem.20051921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Cao F, De A, Cao Y, Contag C, Gambhir SS, Wu JC, Chen X. Trafficking mesenchymal stem cell engraftment and differentiation in tumor-bearing mice by bioluminescence imaging. Stem Cells. 2009;27:1548–1558. doi: 10.1002/stem.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee KJ, Lee JI, Eom YW. Mesenchymal Stem Cell-Mediated Effects of Tumor Support or Suppression. Int J Mol Sci. 2015;16:30015–30033. doi: 10.3390/ijms161226215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boink MA, van den Broek LJ, Roffel S, Nazmi K, Bolscher JG, Gefen A, Veerman EC, Gibbs S. Different wound healing properties of dermis, adipose, and gingiva mesenchymal stromal cells. Wound Repair Regen. 2016;24:100–109. doi: 10.1111/wrr.12380. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, Chen C, Akiyama K, Chai Y, Le AD, Wang Z, Shi S. Gingivae contain neural-crest- and mesoderm-derived mesen-chymal stem cells. J Dent Res. 2013;92:825–832. doi: 10.1177/0022034513497961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melzer C, von der Ohe J, Hass R. Concise Review: Crosstalk of mesenchymal stroma/stem-like cells with cancer cells Provides therapeutic potential. Stem Cells. 2018;36:951–968. doi: 10.1002/stem.2829. [DOI] [PubMed] [Google Scholar]
- 19.Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 2018;502:515–521. doi: 10.1016/j.bbrc.2018.05.208. [DOI] [PubMed] [Google Scholar]
- 20.Kim JE, Eom JS, Kim WY, Jo EJ, Mok J, Lee K, Kim KU, Park HK, Lee MK, Kim MH. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thorac Cancer. 2018;9:911–915. doi: 10.1111/1759-7714.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong L, Bao Q, Hu C, Wang J, Zhou Q, Wei L, Tong L, Zhang W, Shen Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem Biophys Res Commun. 2018;500:170–176. doi: 10.1016/j.bbrc.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 23.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tengda L, Shuping L, Mingli G, Jie G, Yun L, Weiwei Z, Anmei D. Serum exosomal microRNAs as potent circulating biomarkers for melanoma. Melanoma Res. 2018;28:295–303. doi: 10.1097/CMR.0000000000000450. [DOI] [PubMed] [Google Scholar]
- 25.Machida T, Tomofuji T, Ekuni D, Maruyama T, Yoneda T, Kawabata Y, Mizuno H, Miyai H, Kunitomo M, Morita M. MicroRNAs in salivary exosome as potential biomarkers of aging. Int J Mol Sci. 2015;16:21294–21309. doi: 10.3390/ijms160921294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X, Zhang Z, Han Y, Song J, Xu X, Jin J, Su S, Mu D, Liu X, Xu S, et al. Mesenchymal stem cells derived from normal gingival tissue inhibit the proliferation of oral cancer cells in vitro and in vivo. Int J Oncol. 2016;49:2011–2022. doi: 10.3892/ijo.2016.3715. [DOI] [PubMed] [Google Scholar]
- 27.Yang WW, Yang LQ, Zhao F, Chen CW, Xu LH, Fu J, Li SL, Ge XY. Epiregulin promotes lung metastasis of salivary adenoid cystic carcinoma. Theranostics. 2017;7:3700–3714. doi: 10.7150/thno.19712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4986. pdb prot4986. [DOI] [PubMed] [Google Scholar]
- 29.Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5439. pdb prot5439. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Killeen K. The fundamental aspects and applications of Agilent HPLC-Chip. J Sep Sci. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.López-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinevici N, O'sullivan J. Oral cancer: Deregulated molecular events and their use as biomarkers. Oral Oncol. 2016;61:12–18. doi: 10.1016/j.oraloncology.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Verma R, Singh A, Jaiswal R, Chandra A, Verma R, Tak J. Association of Ki-67 antigen and p53 protein at invasive tumor front of oral squamous cell carcinoma. Indian J Pathol Microbiol. 2014;57:553–557. doi: 10.4103/0377-4929.142660. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Li J, Wang L, Tian Z, Zhang P, Xu Q, Zhang C, Wei F, Chen W. Prognostic significance of p21, p27 and survivin protein expression in patients with oral squamous cell carcinoma. Oncol Lett. 2013;6:381–386. doi: 10.3892/ol.2013.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tkach M, Théry C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 38.Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: A systematic review of observational studies. J Oral Pathol Med. 2016;45:155–166. doi: 10.1111/jop.12339. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Zhao Y. Human papillomavirus infection in oral potentially malignant disorders and cancer. Arch Oral Biol. 2017;83:334–339. doi: 10.1016/j.archoralbio.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Singla S, Singla G, Zaheer S, Rawat DS, Mandal AK. Expression of p53, epidermal growth factor receptor, c-erbB2 in oral leukoplakias and oral squamous cell carcinomas. J Cancer Res Ther. 2018;14:388–393. doi: 10.4103/0973-1482.191027. [DOI] [PubMed] [Google Scholar]
- 41.Yuan Y, Jiang YC, Sun CK, Chen QM. Role of the tumor microenvironment in tumor progression and the clinical applications (Review) Oncol Rep. 2016;35:2499–2515. doi: 10.3892/or.2016.4660. [DOI] [PubMed] [Google Scholar]
- 42.Hass R. Retrodifferentiation - a mechanism for cellular regeneration? Biol Chem. 2009;390:409–416. doi: 10.1515/BC.2009.050. [DOI] [PubMed] [Google Scholar]
- 43.Cortini M, Massa A, Avnet S, Bonuccelli G, Baldini N. Tumor-activated mesenchymal stromal cells promote osteosarcoma stemness and migratory potential via IL-6 secretion. PLoS One. 2016;11:e0166500. doi: 10.1371/journal.pone.0166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melzer C, Yang Y, Hass R. Interaction of MSC with tumor cells. Cell Commun Signal. 2016;14:20. doi: 10.1186/s12964-016-0143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majore I, Moretti P, Hass R, Kasper C. Identification of subpopulations in mesenchymal stem cell-like cultures from human umbilical cord. Cell Commun Signal. 2009;7:6. doi: 10.1186/1478-811X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kourembanas S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 47.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 48.Sharma A. Role of stem cell derived exosomes in tumor biology. Int J Cancer. 2018;142:1086–1092. doi: 10.1002/ijc.31089. [DOI] [PubMed] [Google Scholar]
- 49.Shi S, Zhang Q, Xia Y, You B, Shan Y, Bao L, Li L, You Y, Gu Z. Mesenchymal stem cell-derived exosomes facilitate nasopharyngeal carcinoma progression. Am J Cancer Res. 2016;6:459–472. [PMC free article] [PubMed] [Google Scholar]
- 50.Schepers K, Campbell TB, Passegué E. Normal and leukemic stem cell niches: Insights and therapeutic opportunities. Cell Stem Cell. 2015;16:254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770–1778. doi: 10.1038/onc.2016.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Yao K, Huuskes BM, Shen HH, Zhuang J, Godson C, Brennan EP, Wilkinson-Berka JL, Wise AF, Ricardo SD. Mesenchymal stem cells deliver exogenous microRNA-let7c via exosomes to attenuate renal fibrosis. Mol Ther. 2016;24:1290–1301. doi: 10.1038/mt.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phinney DG, Pittenger MF. Concise Review: MSC-derived exosomes for cell-free therapy. Stem Cells. 2017;35:851–858. doi: 10.1002/stem.2575. [DOI] [PubMed] [Google Scholar]
- 54.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Zhao C, Shi H, Zhang B, Zhang L, Zhang X, Wang S, Wu X, Yang T, Huang F, et al. Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: Novel biomarkers and a mechanism for gastric cancer. Br J Cancer. 2014;110:1199–1210. doi: 10.1038/bjc.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gajos-Michniewicz A, Duechler M, Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347:29–37. doi: 10.1016/j.canlet.2014.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used or analyzed in this study are included in this article.