Abstract
Cell division cycle 20 (CDC20) is a regulatory molecule and serves critical roles at multiple points of the cell cycle. Recent evidence indicates that CDC20 may serve an oncogenic role in a number of human cancer types. However, the role of CDC20 in primary cutaneous squamous cell carcinoma (cSCC) has not been studied, to the best of our knowledge. The aim of the present study was to investigate whether and how CDC20 is involved in the tumorigenesis of cSCC. The results revealed that CDC20 expression was significantly increased in cSCC tissues and cell lines, and its expression was associated with pathological differentiation. Downregulation of CDC20 inhibited cell proliferation, induced cell cycle arrest, promoted apoptosis and reduced migratory ability through inhibition of the Wnt/β-catenin signaling pathway. Furthermore, all-trans-retinoic acid treatment significantly downregulated CDC20 expression in cSCC. The present results revealed that CDC20 may serve a crucial role in human cSCC, and suggested that CDC20 may be a novel biomarker for the prevention, diagnosis and treatment of cSCC.
Keywords: cell division cycle 20, cutaneous squamous cell carcinoma, β-catenin, all-trans-retinoic acid
Introduction
Cutaneous squamous cell carcinoma (cSCC) is the second most common form of non-melanoma skin cancer, with an annual global incidence exceeding 1 million (1). Although the majority of patients with cSCC have a good prognosis following appropriate therapy, 5-10% of cases have very poor outcomes, which include local recurrence, nodal metastasis and disease-specific mortality (2-4). Given the frequency of its occurrence, cSCC is reported to cause as many annual mortalities as melanoma in the USA (5). However, there are few reliable biomarkers to predict the biological behavior and clinical outcome of cSCC. Therefore, the identification of biomarkers along with a better understanding of the molecular mechanisms of cSCC is urgently required for the development of effective treatments for the disease.
Cell division cycle 20 (CDC20) is a key molecule that serves an important role in the cell cycle, including as a spindle assembly checkpoint and as an activator of the anaphase-promoting complex (APC/C) (6). APC/Ccdc20 serves a crucial role during the metaphase to anaphase transition, in addition to mitotic exit, through the destruction of various critical cell cycle regulators (7). In addition to regulating the cell cycle, evidence has demonstrated that CDC20 serves an important role in human tumorigenesis and cancer progression (8). Abnormal expression of CDC20 may disorder APC/C activation and promote premature anaphase, which frequently results in mitotic abnormalities and aneuploidy in the two daughter cells (8). Overexpression of CDC20 has been observed in a variety of human tumors, including lung cancer, gastric cancer and breast cancer. Its activity is reported to be critical for tumor initiation, maintenance and growth (9-17). However, CDC20 expression and its clinical significance in human cSCC has not been previously studied, to the best of our knowledge.
The present study detected the expression of CDC20 in cSCC patient samples at different stages. Its biological functions were also investigated through a gene knockdown approach in vitro. The present results suggested that CDC20 expression was positively correlated with the progression of cSCC, and that the loss of CDC20 suppressed cell growth and migration, but induced apoptosis and cell cycle arrest in cSCC cells through inhibition of the wnt/β-catenin signaling pathway. In addition, the present findings demonstrated that all-trans-retinoic acid (ATRA) treatment downregulated CDC20 expression in cSCC cells.
Materials and methods
Bioinformatics analyses
The cSCC mRNA expression microarray datasets GSE32628 and GSE45216 were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) (18,19). A total of 39 samples [13 cSCC, 13 actinic keratosis (AK) and 13 normal skin] from 13 patients in GSE32628 were included. The expression values of CDC20 were calculated using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). A total of 40 samples (15 well-differentiated cSCC, 15 moderately/poorly differentiated cSCC and 10 AK) from 40 different patients in GSE45216 were included. The expression values of CDC20 was calculated using GEO2R.
Patient samples
Patient tissue blocks were obtained from the Department of Dermatology at The Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). These tissues were obtained from patients at the hospital between April 2014 and March 2017. Written informed consent for tissue procurement was obtained. Inclusion criteria were as follows: Clinical data and medical history were well-documented; patients were diagnosed with normal skin, SCC in situ, AK or cSCC; and patients had not received any pre-operative treatment. The exclusion criterion was a history of any other cancer. There was a total of 21 paired cSCC tissues and adjacent normal skin tissues, and 144 samples of different pathological differentiation stages (29 well-differentiated cSCC, 25 moderately/poorly differentiated cSCC, 27 SCC in situ, 31 AK and 32 normal skin tissues obtained from cosmetic surgery). The differentiation stages of all samples were determined by two experienced pathologists. Among these patients, there were 52 male and 113 female, their ages ranged from 11 to 92 years, with a median age of 57.
Immunohistochemistry
The paraffin-embedded human tissue were sectioned into 4-µm-thick slices and mounted on slides. Following being de-waxed, rehydrated and blocked with goat serum (reagent A in the SP-9002 kit; OriGene Technologies, Inc., Beijing, China) at room temperature for 1 h, slides were incubated with primary anti-CDC20 antibody (1:100; cat. no. sc-13162; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Subsequently, the sections were incubated with the biotin-labeled goat anti-mouse IgG working solution (reagent B in the SP-9002 kit; OriGene Technologies, Inc.) at room temperature for 30 min. Detection was achieved with diaminobenzidine for 10 sec and counterstaining with hematoxylin for 30 sec at room temperature, and routine dehydration, clearing and mounting were performed. The staining results were evaluated under an optical microscope (magnification, ×100) by two independent pathologists. Each sample was given a score by multiplying the stain intensity grade (0, no staining; 1, low intensity; 2, medium intensity; 3, strong intensity) by the numerical code for the percentage of positive cells (≤5%=0; 6-25%=1; 26-50%=2; 51-75%=3; >75%=4). The average score of five fields was taken as the final score of CDC20 expression for each slide.
Cell lines and cell culture
The human cSCC cell lines A431 and SCL-1 were purchased from the Cell Bank of the Shanghai Institute of Cell Biology (Shanghai, China), and SCC13 was provided as a gift from Dr Yuming Xia (Department of Dermatology, The Second Affiliated Hospital of Xi'an Jiaotong University). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone; GE Healthcare Life Sciencies, Logan, UT, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel). Primary keratinocytes were isolated from the foreskin epidermis of a 23-year old man by Hu et al (20). Keratinocytes were cultured in Keratinocyte Media (ScienCell Research Laboratories, San Diego, CA, USA). The cells were cultured at 37°C in a humidified incubator with 5% CO2.
Transient transfection of CDC20 small interfering (si) RNA and plasmids
The siRNA oligonucleotides were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and their sequences are listed in Table I. A431 and SCL-1 cells were seeded in 6-well/96-well plates and transfected with 100 pmol/10 pmol siRNA, respectively, the following day, according to the recommended procedures for Lipofectamine® 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The CDC20 expression plasmid (HA-CDC20; cat. no. 11594) and the empty control expression plasmid (pCS2P+; cat. no. 17095), which had the same backbone as the CDC20 expression construct, were purchased from Addgene, Inc. (Cambridge, MA, USA). SCC13 cells were seeded in 6-well/96-well plates and transfected with plasmid the following day. Plasmid DNA was diluted in OptiMEM (Gibco; Thermo Fisher Scientific, Inc.) and combined with 1 µg/ml polyethylenimine (PEI) in a 1:3 ratio (DNA:PEI). The PEI/DNA mix was added in a dropwise manner to the cells, and they were incubated for 12 h. The DMEM was refreshed and the cells were harvested or counted after 72 h. Cells treated with the transfection reagent only acted as the mock transfection control.
Table I.
Sequences of the siRNAs used in the present study.
| Name | Sequences |
|---|---|
| si-CDC20-1 | Sense: 5′-GGAUUGGAGUUCUGGGAAUTT-3′ |
| Antisense: 5′-AUUCCCAGAACUCCAAUCCTT-3′ | |
| si-CDC20-2 | Sense: 5′-GCAGAAACGGCUUCGAAAUTT-3′ |
| Antisense: 5′-AUUUCGAAGCCGUUUCUGCTT-3′ | |
| si-CDC20-3 | Sense: 5′-GAUGAGACCCUGAGGCUAUTT-3′ |
| Antisense: 5′-AUAGCCUCAGGGUCUCAUCTT-3′ | |
| Si-ctrl | Sense: 5′-UUCUUCGAACGUGUCACGUTT-3′ |
| Antisense: 5′-ACGUGACACGUUCGGAGAATT-3′ |
si, small interfering; ctrl, control; CDC20, cell division cycle 20.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA samples were isolated using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed to cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). The reaction conditions were 37°C for 15 min and 85°C for 5 sec, followed by rapid cooling to 4°C and storage at -20°C. The CDC20 mRNA levels in each group were detected using SYBR Premix Ex Taq (Takara Bio, Inc.) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions were: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. The gene expression levels were calculated using the 2−ΔΔCq method (21) and each experiment was repeated three times. The primer sequences were as follows: CDC20 (forward, 5′-GACCACTCCTAGCAAACC TGG-3′ and reverse, 5′-GGGCGTCTGGCTGTTTTCA-3′) and GAPDH (forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′).
Western blotting
Total cellular proteins were extracted from the cultured cells using lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 50 mM NaF, 1 mM EDTA and 10 mM β-glycerol-P]. The protein concentration was determined using a Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Subsequently, 40 µg proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Following blocking with non-fat dried milk for 2 h at room temperature, the membranes were incubated overnight with primary antibodies at 4°C and subsequently incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (anti-Rabbit IgG; 1:3,000; cat. no. 7074; Cell Signaling Technology, Inc.; anti-mouse IgG; 1:3,000; cat. no. 7076; Cell Signaling Technology, Inc.) for 2 h at room temperature. The following primary antibodies were used in this study: Anti-CDC20 (1:1,000; cat. no. 14866; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-cyclin A (1:200; cat. no. sc-271682; Santa Cruz Biotechnology, Inc.), anti-cyclin B1 antibody (1:1,000; cat. no. 12231; Cell Signaling Technology, Inc.), anti-apoptosis regulator Bcl-2 (Bcl-2; 1:200; cat. no. sc-492; Santa Cruz Biotechnology, Inc.), anti-apoptosis regulator BAX (Bax; 1:200; cat. no. sc-6236; Santa Cruz Biotechnology, Inc.), anti-β-catenin (1:1,000; cat. no. 8480; Cell Signaling Technology, Inc.), anti-c-Myc proto oncogene protein (c-Myc; 1:1,000; cat. no. 5605; Cell Signaling Technology, Inc.), anti-GAPDH (1:4,000; cat. no. CW0101; CWBIO, Beijing, China) and anti-hemagglutinin (HA; 1:1,000; cat. no. H3663; Sigma-Aldrich; Merck KGaA). Protein bands were visualized using the Immobilon western chemiluminescent HRP substrate (cat. no. WBKLS0500; EMD Millipore, Billerica, MA, USA).
MTT assay
Cells were seeded in 96-well plates (A431, 5×103 cells/well; Scl-1, 3×103 cells/well; and SCC13, 5×103 cells/well) in quintuplicate and transfected with the negative control siRNA, CDC20 siRNA, HA-CDC20 plasmid or empty control plasmid the following day. Relative cell numbers were measured 24, 48 or 72 h subsequently by incubating cells with 20 µl 0.5 mg/ml MTT at 37°C for 4 h. Following complete removal of the medium, 150 µl dimethyl sulfoxide was administrated to dissolve the formazan. The relative absorbance at a wavelength of 490 nm was measured with a microplate reader (model 680; Bio-Rad Laboratories, Inc.). A total of three independent experiments were performed. Meanwhile, cells were seeded in 96-well plates and treated with different concentrations of ATRA (cat. no. R2625; Sigma-Aldrich; Merck KGaA; 1, 10, 20, 40, 60, 80, 100 and 120 µM) the following day. Relative cell numbers were measured 48 h after ATRA treatment using the aforementioned method. A total of three independent experiments were performed.
Cell cycle analysis
The siRNA-transfected cells were harvested 48 h after transfection. For the analysis, cells were harvested, fixed with ice-cold 70% ethanol overnight, and stored overnight at -4°C. The cells were stained with the Cell Cycle Detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), and incubated for 30 min in the dark at room temperature, according to the manufacturer's protocol. The cells were analyzed using flow cytometry (FACSCalibur; BD Biosciences, San Diego, CA, USA). ModFit LT™ V3.3 (Verity Software House, Inc., Topsham, ME, USA) was used to analyze the results. A total of three independent experiments were performed.
Cell apoptosis analysis
The siRNA-transfected cells were harvested 48 h after transfection. For the analysis, the cells were stained with the Annexin V-fluorescein isothiocyanate/propidium iodide (PI) Apoptosis Detection kit (KeyGEN Biotech, Nanjing, China), following the manufacturer's instructions. Stained cells were analyzed by flow cytometry (FACSCalibur; BD Biosciences). FlowJo 10 (FlowJo LLC, Ashland, OR, USA) was used to analyze the results. A total of three independent experiments were performed.
Wound-healing assay
Cells were seeded into six-well plates (A431, 5×105 cells/well; Scl-1, 3×105 cells/well) and transfected with the negative control siRNA or CDC20 siRNA the following day. A total of 48 h after transfection, wounds were created in the cell monolayer using a 200 µl pipette tip. Cell debris was removed by washing once with PBS, followed by the addition of fresh DMEM containing 1% fetal bovine serum. The healing process was followed for the next 72 h.
Immunofluorescent staining
Cells were grown on glass coverslips. Following fixing with 4% paraformaldehyde at room temperature for 30 min, permeabilization with 0.5% Triton X-100 and blocking with 5% bovine serum albumin (Thermo Fisher Scientific., Inc.) for 1 h at room temperature, the cells were incubated with primary antibodies against β-catenin (1:100, cat. no. 8480; Cell Signaling Technology, Inc.) at 4°C overnight. Subsequently, the cells were incubated with the appropriate goat anti-rabbit IgG2 secondary antibody at room temperature for 1 h in the dark. The nuclei were counterstained with DAPI for 10 min at room temperature and imaged with a confocal laser-scanning microscope (magnification, ×200) (Nikon Corporation, Tokyo, Japan).
Statistical analysis
Data are presented as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The Student's t-test was used for comparisons between two groups, and one-way analysis of variance with Dunnett's or Bonferroni's post hoc test was applied for experiments with more than two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
CDC20 is significantly upregulated in cSCC tumor tissues and cSCC cell lines
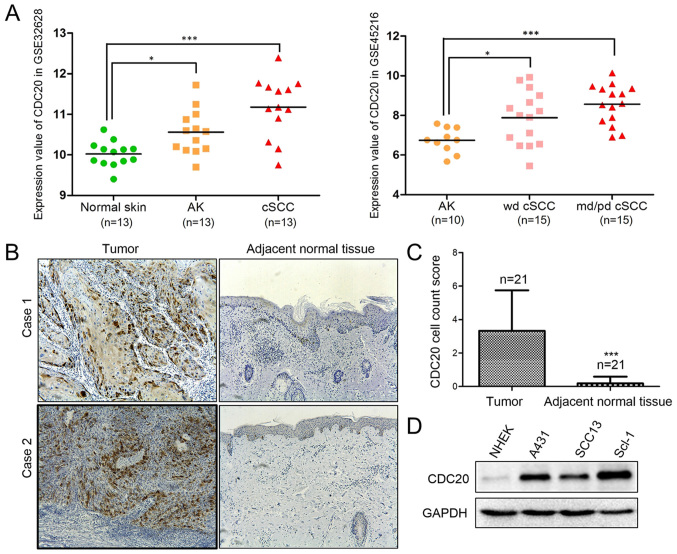
The analysis results from GEO2R for GSE32628 and GSE45216 are presented in Fig. 1A. In GSE32628, the mRNA expression values of CDC20 were 10.02±0.31 in normal skin tissues (n=13), 10.56±0.55 in AK tissues (n=13) and 11.17±0.74 in cSCC tissues (n=13). The CDC20 mRNA expression level was significantly higher in AK and cSCC tumor tissues compared with paired normal skin tissues (Fig. 1A). In GSE45216, the mRNA expression values of CDC20 were 6.74±0.63 in the AK group (n=10), 7.89±1.35 in the well-differentiated cSCC group (n=15) and 8.56±0.99 in the moderately/poorly differentiated cSCC group (n=15). CDC20 expression was significantly increased in well-differentiated cSCC and moderately/poorly differentiated cSCC compared with AK (Fig. 1A).
Figure 1.
Increased expression of CDC20 in cSCC tumors and cell lines. (A) Bioinformatics analysis results for GSE32628 and GSE45216. *P<0.05; ***P<0.001. (B) Immunohistochemical analysis of CDC20 in pairs of tumor (left) and adjacent normal tissue (right) samples from patients with cSCC (magnification, ×100). (C) Semiquantitative analysis of CDC20 staining in 21 paired samples. ***P<0.001 vs. tumor. (D) Western blot analysis of CDC20 protein expression in primary NHEK cells and three cSCC cell lines: A431, SCC13 and SCL-1. CDC20, cell division cycle 20; cSCC, cutaneous squamous cell carcinoma; NHEK, normal human epidermal keratinocytes; AK, actinic keratosis; wd, well-differentiated; md/pd, moderately or poorly differentiated.
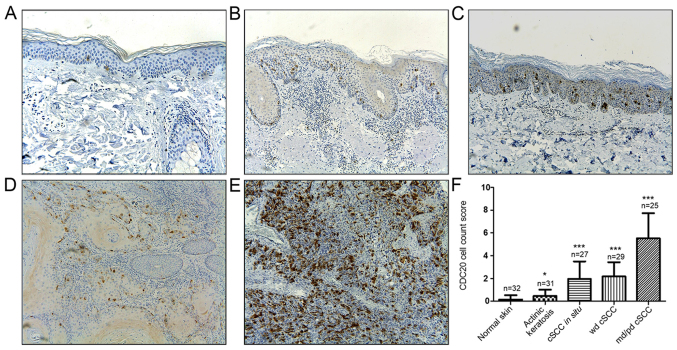
Along with the results of the bioinformatics analysis, the immunohistochemical staining in 21 paired cSCC and corresponding normal tissues confirmed the elevated CDC20 expression in cancerous tissues (Fig. 1B and C). In addition, compared with the primary human epidermal keratinocytes, CDC20 was upregulated in cSCC cell lines A431, SCL-1 and SCC13 at the mRNA (data not shown) and protein level (Fig. 1D). Furthermore, the expression of CDC20 protein was detected in 144 samples from patients with cSCC at different stages, including normal skin tissues (n=32), precancerous actinic keratosis lesions (n=31), cSCC in situ (n=27), well-differentiated cSCC (n=29) and moderately/poorly differentiated cSCC (n=25). Subsequent semiquantitative analysis of the immunohistochemistry results demonstrated that CDC20 expression was significantly higher in cSCC in situ, well-differentiated cSCC, and moderately/poorly differentiated cSCC compared with normal skin, and was associated with disease progression (Fig. 2). The findings indicated that CDC20 may have an oncogenic role in cSCC progression.
Figure 2.
Expression of CDC20 is associated with cSCC progression. (A) Normal skin (n=32) (magnification, ×100). (B) Actinic keratosis (n=31) (magnification, ×100). (C) cSCC in situ (Bowen's disease; n=27) (magnification, ×100). (D) Well-differentiated cSCC (n=29) (magnification, ×100). (E) Moderately or poorly differentiated cSCC (n=25) (magnification, ×100). (F) Semiquantitative analysis of CDC20 expression in the different tissues. CDC20, cell division cycle 20; cSCC, cutaneous squamous cell carcinoma; wd, well-differentiated; md/pd, moderately or poorly differentiated. *P<0.05, ***P<0.001 vs. normal skin.
CDC20 silencing inhibits the proliferation of cSCC cells and leads to cell cycle arrest
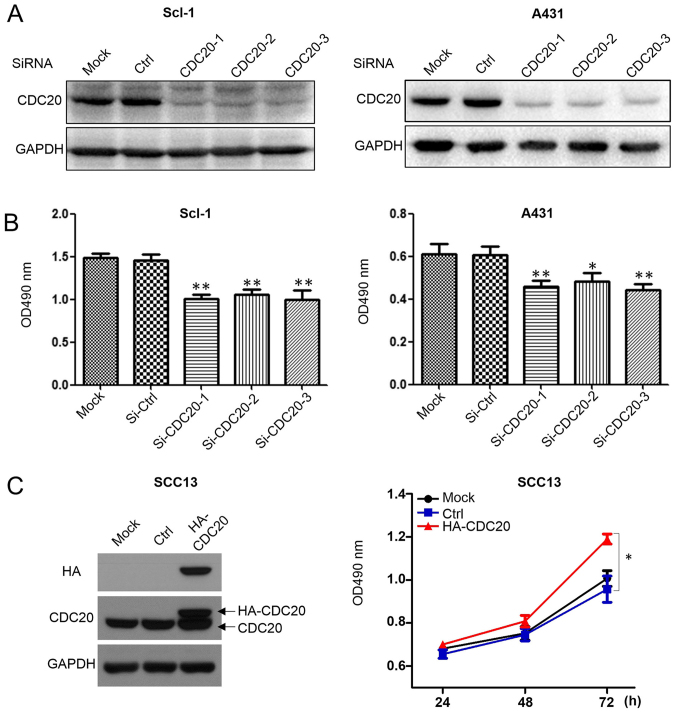
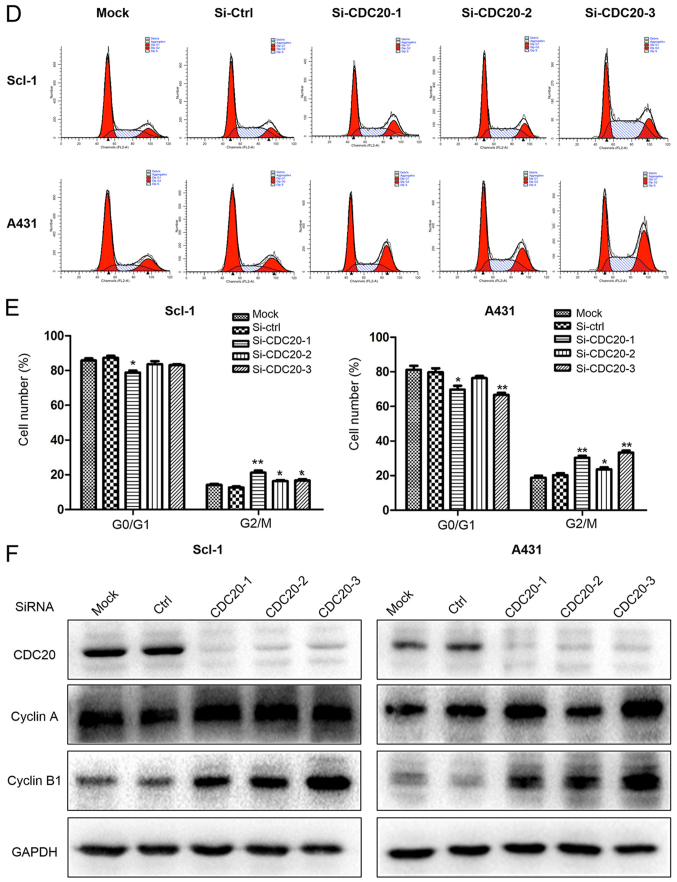
Compared with normal human epidermal keratinocytes, all the cSCC cell lines has increased CDC20 protein expression levels (Fig. 1D); however, among the three cSCC cell lines, SCC13 had the lowest basal level of CDC20. To test the potential oncogenic effects of CDC20, CDC20 was downregulated in Scl-1 and A431 cells, and overexpressed in SCC13. CDC20 silencing suppressed the growth of A431 and Scl-1 cells, as determined by the MTT assay (Fig. 3A and B). Transfection of HA-tagged CDC20 led to an increased total level of CDC20 protein and promoted the growth of SCC13 cells (Fig. 3C). Cell cycle analysis demonstrated that CDC20 depletion led to cells accumulating in the G2/M phase (Fig. 3D and E) and an increased expression level of cyclin B1 and cyclin A (Fig. 3F), suggesting arrest in early mitosis.
Figure 3.
CDC20 silencing inhibits the proliferation of cSCC cells and leads to cell cycle arrest, while overexpression of CDC20 promotes the growth of cSCC cells. (A) Knockdown effect of CDC20 by siRNA in the A431 and SCL-1 cell lines. (B) The number of cells was counted 72 h post-siRNA transfection by MTT assay. (C) Overexpression of CDC20 promoted the growth of SCC13 cells. Control cells were transfected with an empty plasmid which had the same backbone as the CDC20 expression construct. (D) The cell cycle profiles were analyzed and (E) quantified 48 h after siRNA transfection by flow cytometry. (F) Cell cycle regulators were analyzed 48 h after siRNA transfection by western blot analysis. *P<0.05, **P<0.01 vs. respective Ctrl group. CDC20, cell division cycle 20; cSCC, cutaneous squamous cell carcinoma; si, small interfering; Ctrl, control; HA, hemagglutinin; OD, optical density.
CDC20 silencing promotes apoptosis and inhibits migration in cSCC in vitro
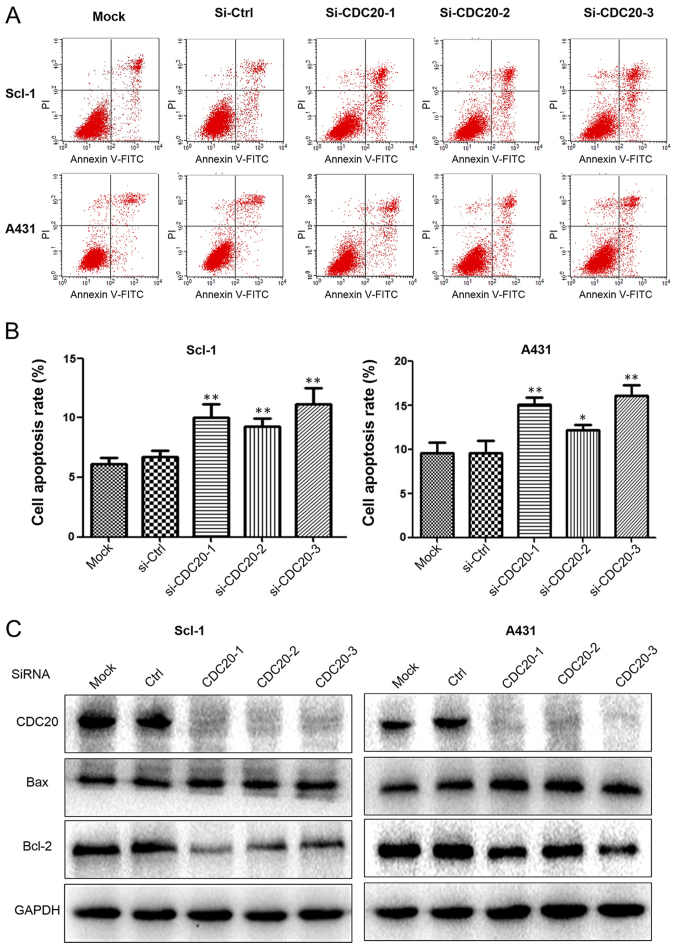
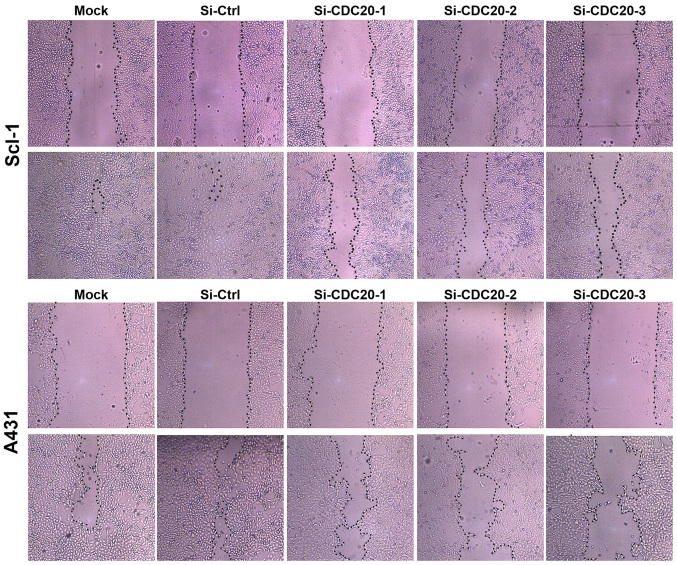
Annexin V/PI staining was performed to detect apoptosis. Non-transfected or control siRNA-transfected cells exhibited a lower apoptosis rate of 11-12% in A431 and 6-7% in SCL-1 cells compared with the increased apoptosis rate of 12-16% in A431 and 9-12% in Scl-1 cells following CDC20 silencing (Fig. 4A and B). The increased apoptosis was associated with increased expression of the apoptosis-promoting gene Bax and decreased expression of the apoptosis-suppressing gene Bcl-2 (Fig. 4C). These results demonstrated that CDC20 was involved in protecting cSCC cells against apoptosis and that depletion of CDC20 induced apoptosis. In addition, to analyze whether CDC20 was able to inhibit the motility of cSCC cells, wound healing assays were performed. The results demonstrated that silencing of CDC20 decreased cell migration in A431 and Scl-1 cells (Fig. 5).
Figure 4.
CDC20 silencing promotes apoptosis in cutaneous squamous cell carcinoma cells. (A) Cell apoptosis was analyzed 48 h after siRNA transfection by flow cytometry. (B) Quantification of the apoptosis rate. (C) Apoptosis indicators were analyzed 48 h after siRNA transfection by western blotting. *P<0.05, **P<0.01 vs. respective Ctrl group. FITC, fluorescein isothiocyanate; PI, propidium iodide; si, small interfering; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator BAX; CDC20, cell division cycle 20; Ctrl, control.
Figure 5.
CDC20 silencing reduces the migratory ability of cutaneous squamous cell carcinoma cells (magnification, ×100). CDC20, cell division cycle 20; Ctrl, control; si, small interfering.
Loss of CDC20 suppresses the Wnt/β-catenin signaling pathway
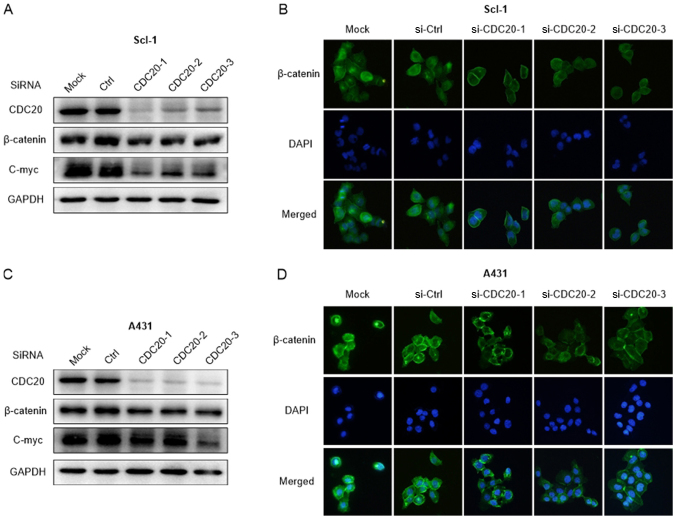
To understand the molecular mechanisms of the oncogenic effects of CDC20, the Wnt/β-catenin signaling pathway, one of the principal signaling pathways involved in the regulation of cell growth, survival and migration, was analyzed. The present results indicated that CDC20 silencing led to a reduced expression level of β-catenin and c-Myc (Fig. 6A and B). In line with this, the immunofluorescence results indicated CDC20 that silencing led to decreased fluorescence and less distribution of β-catenin in the nucleus (Fig. 6C and D), which indicated that the translocation of β-catenin into the nucleus was also inhibited by CDC20 knockdown. These results confirmed that CDC20 contributes to the growth of cSCC cells by activating the Wnt/β-catenin signaling pathway.
Figure 6.
Loss of CDC20 suppresses the Wnt/β-catenin signaling pathway. (A) Knockdown of CDC20 led to a reduced protein expression level of β-catenin and c-Myc in Scl-1 cells. (B) Loss of CDC20 led to a decreased distribution of β-catenin in the nucleus in Scl-1 cells (magnification, ×200). (C) Knockdown of CDC20 led to a reduced protein expression level of β-catenin and c-Myc in A431 cells. (D) Loss of CDC20 led to a decreased distribution of β-catenin in the nucleus in A431 cells (magnification, ×200). CDC20, cell division cycle 20; Ctrl, control; si, small interfering; c-Myc, c-Myc proto oncogene protein.
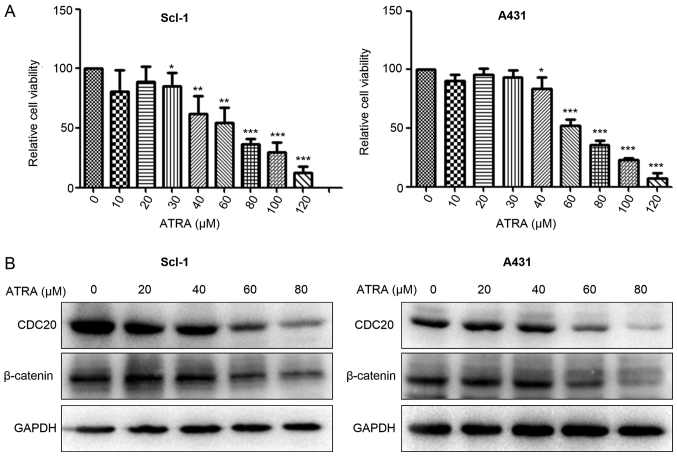
ATRA treatment downregulates CDC20 and β-catenin expression in cSCC cells
To determine the suppressive effects of ATRA on cSCC cells, the MTT assay was conducted. The results demonstrated that ATRA decreased the viability of A431 and SCL-1 cells in a concentration-dependent manner (Fig. 7A). To examine whether the CDC20/β-catenin pathway was involved in ATRA-induced growth inhibition, western blot analysis was used to detect CDC20 and β-catenin expression in cSCC cells following ATRA treatment for 48 h. The results demonstrated that ATRA treatment reduced the CDC20 and β-catenin protein expression in A431 and Scl-1 cells (Fig. 7B).
Figure 7.
ATRA treatment downregulates CDC20 and β-catenin expression in cSCC cells. (A) ATRA decreased the viability of Scl-1 and A431 cells in a concentration-dependent manner. (B) ATRA treatment reduced CDC20 and β-catenin protein expression in A431 and Scl-1 cells. *P<0.05, **P<0.01, ***P<0.001 vs. respective control group (0 µM ATRA). ATRA, all-trans-retinoic acid; CDC20, cell division cycle 20.
Discussion
In recent years, the overexpression of CDC20 has been documented in a wide range of malignancies. In a number of tumor types, its expression has been reported to correlate with a poor clinical outcome, providing evidence for its role in carcinogenesis and tumor progression (10-11,13). However, the role of CDC20 in primary cSCC has not been previously reported, to the best of our knowledge. In the present study, it was demonstrated that CDC20 is overexpressed in cSCC tissues in addition to cSCC cell lines, and its expression is associated with tumor progression. Knockdown of CDC20 suppressed cell growth and migration, and promoted apoptosis and cell cycle arrest in cSCC.
Similar to what has been observed in other tumor types (9), CDC20 expression was low in normal intact skin and was significantly increased in tumor cells in cSCC, suggesting that CDC20 may work as a biomarker for cSCC. Furthermore, the elevated expression of CDC20 in AK and cSCC in situ, and its association with histological classification, provide evidence that the induction of CDC20 expression is an early event in cSCC development and may serve a role in the progression of in situ cSCC to the invasive stage. However, there were no follow-up data for the patients in the present study, thus it was not possible to draw any conclusion about the prognostic value of CDC20.
CDC20 was overexpressed in SCC13 cells since this was the cell line with the lowest basal level of CDC20. When SCC13 was first established in 1981, it was identified that its colony formation efficiency was low and in a nude mouse model it tended to form large differentiating cysts with an organized structure, which is a pathological sign of good differentiation (22,23). Therefore SCC13 was considered to be a well-differentiated cSCC cell line in the present study. The immunohistochemistry results demonstrated that CDC20 was not only elevated in cancerous tissues, but was also associated with tumor differentiation and progression. The low basal protein level of CDC20 may, on the other hand, support the well-differentiated classification of SCC13. Therefore, the overexpression experiment was conducted to see whether CDC20 was able to induce malignancy, and the present results demonstrated that overexpression of CDC20 slightly increased the growth of SCC13 cells.
CDC20 is known to regulate the separation of chromosomes by inducing ubiquitination and the degradation of cell cycle regulators, including cyclin A and cyclin B1 in a D-box dependent manner at the beginning of metaphase, loss of CDC20 results in the accumulation of cyclin B1 and cyclin A (15). While the degradation of cyclin B induces exit from mitosis (telophase and cytokinesis) (16), non-degradable cyclin B1 may arrest human cells in a metaphase-like state (17,24). CDC20 mutation/depletion blocks cell division and stops cell cycle progression toward anaphase and chromosome segregation (25). Consistent with this report, it was observed in the present study that CDC20 knockdown inhibited the proliferation of cSCC cells and led to G2/M cell cycle arrest. CDC20 depletion induces the expression of cyclin A and cyclin B1 (24,26), and exogenous expression of CDC20 slightly increased the growth of SCC13 cells in the present study. Similarly, a recent study confirmed that the overexpression of CDC20 promoted cell growth, inhibited apoptosis, and enhanced cell migration and invasion in osteosarcoma cells (27). In addition, it was reported that CDC20 may control mitotic progression by targeting a number of novel substrates, including ubiquitin interaction motif containing 1, PHD finger protein 8 and BRCA1 associated RING domain 1 (28,29). These findings support the hypothesis that CDC20 serves a crucial role in cell cycle regulation.
In addition to G2/M phase arrest, the present data demonstrated that the inhibition of CDC20 activity led to the induction of apoptosis. CDC20 depletion increased the expression of Bax and reduced the expression of Bcl-2. Similar regulation of apoptosis by CDC20 has been observed in other studies. For example, in multiple tumor-derived cell lines, Bcl-2 like 11 accumulation upon CDC20 knockdown contributes to apoptosis induction and chemoradiation sensitization (30). Another study suggested that CDC20 governs apoptosis by mediating the degradation of the anti-apoptotic protein induced myeloid leukemia cell differentiation protein Mcl-(31). Furthermore, it was demonstrated that CDC20 regulated the migratory ability of cSCC cells. This finding is consistent with the results from a previous study, demonstrating that CDC20 expression was further increased in matched metastatic liver tissues compared with primary colorectal cancer tissues (13).
Wnt signaling through β-catenin has long been recognized to be crucial for skin epidermal development and homeostasis (32). A number of studies have confirmed that it is essential for cSCC initiation and progression, and for maintaining the cancer stem cell niche (33). Based on the primary hypothesis that targeting CDC20 may improve the aggressive properties of cSCC, the present study sought to investigate whether this process was associated with suppression of the canonical Wnt/β-catenin pathway. Wnt/β-catenin is a well-known pathway that is involved in the progression of cSCC. Recently, Zimmerli et al (33) reported that the Wnt/β-catenin pathway controlled the initiation and progression of human papillomavirus-driven cSCC. Although the effect of stimulation of Wnt/β-catenin on the cellular behavior of cSCC was not assessed in the present study, β-catenin and c-Myc, which are classically important components involved in the Wnt/β-catenin signaling pathway, were tested following silencing of CDC20. The present results demonstrated that CDC20 silencing led to a significantly decreased protein expression level of β-catenin and c-Myc, and inhibited the translocation of β-catenin into the nucleus, suggesting that CDC20 contributes to the development of cSCC through the Wnt/β-catenin signaling pathway. However, there were a number of limitations to the signaling pathway analysis. First, the role of other proteins involved in the Wnt/β-catenin pathway, including adenomatous polyposis coli protein, axin and zinc finger protein SNAI1 were not tested in the present study. Second, it was not possible to ascertain whether β-catenin is the substrate of the APC/C-CDC20 complex.
Retinoids have long been considered for use in the treatment and prevention of cancer (34,35). The present finding that ATRA was able to inhibit cSCC cell growth was consistent with the results reported by Zhang et al (36). The fact that ATRA was able to downregulate the expression of CDC20 indicates that CDC20 may work as a potential therapeutic target in cSCC. It was noted that the cell killing effect of ATRA was marked at ~60 µM, a relatively high concentration. However, the majority of retinoids are available for topical use, which means that they may be able to work directly and specifically on skin cancer lesions. Severe side effects of topical retinoids are very rare despite their high concentrations (for example, 0.1% adapalene gel) (37). Thus, for patients with a localized AK or cSCC in situ, topical retinoids may be beneficial. Meanwhile, discovering and validating small molecule inhibitors specifically targeting CDC20 remains critical for future studies. It is noteworthy that a recent study demonstrated that the systemic use of CDC20 siRNA was able to inhibit solid growths of subcutaneously injected melanoma and spontaneous growth of intravenously injected melanoma cells on the mouse lung (38). Therefore, CDC20 is a promising target for the treatment of human cancer.
In conclusion, the present results demonstrated that CDC20 serves a carcinogenic role in human cSCC, and may therefore serve as a novel target for early prevention, diagnosis and treatment.
Acknowledgments
The authors would like to thank Dr Bin Zheng and Dr Yi Lv for technical assistance.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81372912), Funds of Northwest Hospital of Xi'an Jiaotong University [grant no. RC(GG)201602], and the Fundamental Research Funds for the Central Universities (grant no. PY3A0241001016).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZWC and SMG designed and performed the experiments, XYZ and QYL participated in the collection of clinical samples and data analysis, GLH contributed to the isolation of the primary keratinocytes, and CGL interpreted the data and critically revised the manuscript. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The use of human tissues was approved by the Ethics Committee of the Second Affiliated Hospital of Xi'an Jiao Tong University and patient consent was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012;166:1069–1080. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- 2.Stratigos A, Garbe C, Lebbe C, Malvehy J, del Marmol V, Pehamberger H, Peris K, Becker JC, Zalaudek I, Saiag P, et al. European Dermatology Forum (EDF); European Association of Dermato-Oncology (EADO); European Organization for Research and Treatment of Cancer (EORTC): Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline. Eur J Cancer. 2015;51:1989–2007. doi: 10.1016/j.ejca.2015.06.110. [DOI] [PubMed] [Google Scholar]
- 3.Levine DE, Karia PS, Schmults CD. Outcomes of patients with multiple cutaneous squamous cells carcinomas: A 10-year single institution cohort study. JAMA Dermatol. 2015;151:1220–1225. doi: 10.1001/jamadermatol.2015.1702. [DOI] [PubMed] [Google Scholar]
- 4.Schmults CD, Karia PS, Carter JB, Han J, Qureshi AA. Factors predictive of recurrence and death from cutaneous squamous cell carcinoma: A 10-year, single-institution cohort study. JAMA Dermatol. 2013;149:541–547. doi: 10.1001/jamadermatol.2013.2139. [DOI] [PubMed] [Google Scholar]
- 5.Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: Estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68:957–966. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 6.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. J Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 9.Gayyed MF, El-Maqsoud NM, Tawfiek ER, El Gelany SA, Rahman MF. A comprehensive analysis of CDC20 overexpression in common malignant tumors from multiple organs: Its correlation with tumor grade and stage. Tumour Biol. 2016;37:749–762. doi: 10.1007/s13277-015-3808-1. [DOI] [PubMed] [Google Scholar]
- 10.Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol. 2012;106:423–430. doi: 10.1002/jso.23109. [DOI] [PubMed] [Google Scholar]
- 11.Ding ZY, Wu HR, Zhang JM, Huang GR, Ji DD. Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int J Clin Exp Pathol. 2014;7:722–727. [PMC free article] [PubMed] [Google Scholar]
- 12.Karra H, Repo H, Ahonen I, Löyttyniemi E, Pitkänen R, Lintunen M, Kuopio T, Söderström M, Kronqvist P. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer. 2014;110:2905–2913. doi: 10.1038/bjc.2014.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu WJ, Hu KS, Wang DS, Zeng ZL, Zhang DS, Chen DL, Bai L, Xu RH. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med. 2013;11:142. doi: 10.1186/1479-5876-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Gao JZ, Du JL, Huang ZX, Wei LX. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int J Oncol. 2014;45:1547–1555. doi: 10.3892/ijo.2014.2559. [DOI] [PubMed] [Google Scholar]
- 15.Yam CH, Fung TK, Poon RY. Cyclin A in cell cycle control and cancer. Cell Mol Life Sci. 2002;59:1317–1326. doi: 10.1007/s00018-002-8510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rieder CL. Mitosis in vertebrates: The G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 2011;19:291–306. doi: 10.1007/s10577-010-9178-z. [DOI] [PubMed] [Google Scholar]
- 17.Wolf F, Wandke C, Isenberg N, Geley S. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 2006;25:2802–2813. doi: 10.1038/sj.emboj.7601163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hameetman L, Commandeur S, Bavinck JN, Wisgerhof HC, de Gruijl FR, Willemze R, Mullenders L, Tensen CP, Vrieling H. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer. 2013;13:58. doi: 10.1186/1471-2407-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert SR, Mladkova N, Gulati A, Hamoudi R, Purdie K, Cerio R, Leigh I, Proby C, Harwood CA. Key differences identified between actinic keratosis and cutaneous squamous cell carcinoma by transcriptome profiling. Br J Cancer. 2014;110:520–529. doi: 10.1038/bjc.2013.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Liang L, Liu Y, Liu J, Tan X, Xu M, Peng L, Zhai S, Li Q, Chu Z, et al. TWEAK/Fn14 Interaction Confers Aggressive Properties to Cutaneous Squamous Cell Carcinoma. J Invest Dermatol. 2018 Nov 8; Epub ahead of print. [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 23.Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78:237–247. doi: 10.1016/j.jaad.2017.08.059. [DOI] [PubMed] [Google Scholar]
- 24.Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther. 2015;151:141–151. doi: 10.1016/j.pharmthera.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang G, Ma X, Lv G. Cell division cycle 20 promotes cell proliferation and invasion and inhibits apoptosis in osteosarcoma cells. Cell Cycle. 2018;17:43–52. doi: 10.1080/15384101.2017.1387700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim HJ, Dimova NV, Tan MK, Sigoillot FD, King RW, Shi Y. The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol Cell Biol. 2013;33:4166–4180. doi: 10.1128/MCB.00689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, Liu J, Shaik S, Chen G, Deng J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell. 2014;29:377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku AT, Miao Q, Nguyen H. Monitoring Wnt/β-Catenin Signaling in Skin. Methods Mol Biol. 2016;1481:127–140. doi: 10.1007/978-1-4939-6393-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmerli D, Cecconi V, Valenta T, Hausmann G, Cantù C, Restivo G, Hafner J, Basler K, van den Broek M. WNT ligands control initiation and progression of human papillomavirus-driven squamous cell carcinoma. Oncogene. 2018;37:3753–3762. doi: 10.1038/s41388-018-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blomberg M, He SY, Harwood C, Arron ST, Demehri S, Green A. Research gaps in the management and prevention of cutaneous squamous cell carcinoma in organ transplant recipients. Br J Dermatol. 2017;177:1225–1233. doi: 10.1111/bjd.15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: Management of advanced and high-stage tumors. J Am Acad Dermatol. 2018;78:249–261. doi: 10.1016/j.jaad.2017.08.058. [DOI] [PubMed] [Google Scholar]
- 36.Zhang ML, Tao Y, Zhou WQ, Ma PC, Cao YP, He CD, Wei J, Li LJ. All-trans retinoic acid induces cell-cycle arrest in human cutaneous squamous carcinoma cells by inhibiting the mitogen-activated protein kinase-activated protein 1 pathway. Clin Exp Dermatol. 2014;39:354–360. doi: 10.1111/ced.12227. [DOI] [PubMed] [Google Scholar]
- 37.Riahi RR, Bush AE, Cohen PR. Topical Retinoids: Therapeutic Mechanisms in the Treatment of Photodamaged Skin. Am J Clin Dermatol. 2016;17:265–276. doi: 10.1007/s40257-016-0185-5. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee A, Bhattacharyya J, Sagar MV, Chaudhuri A. Liposomally encapsulated CDC20 siRNA inhibits both solid melanoma tumor growth and spontaneous growth of intravenously injected melanoma cells on mouse lung. Drug Deliv Transl Res. 2013;3:224–234. doi: 10.1007/s13346-013-0141-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.