Abstract
The disease of freshwater sponges was first discovered in 2011, when pink samples were found in the Central Basin of Lake Baikal. Subsequently, the visible signs of the disease have changed, and now sponges appear with various symptoms of damage to the body, such as discoloration, tissue necrosis, the formation of brown patches and dirty-purple biofilms on some branches. These signs of the disease are accompanied by the mass death of sponges. We identified differences in microbiomes by sequencing 16S rRNA genes and found changes in the consortium of microorganisms of freshwater Baikal sponges. We found that the observed imbalance in the studied microbial communities of diseased sponges is caused by several different conditionally pathogenic microorganisms that increase their negative effect by acting together and in concert, which leads to the death of photosynthetic microalgae and sponges. Sponges are an important component of coastal communities, and the massive loss of sponges can obviously affect the structure of benthic communities and the purity of water.
Introduction
Sponges (phylum Porifera) are sessile benthic metazoans and with most belonging to class Demospongiae [1,2]. Sponges are unique filter feeder organisms and serve as efficiently filtering organic particles and plankton from the water column. This ecological function increases water purity and maintains water quality. Most types of sponges are marine; freshwater sponges are much less diverse. All freshwater sponges belong to the suborder Spongillina, consisting of 47 genera with many endemic species [3–5]. Freshwater sponges inhabiting the photic zone habitation are colored in green tones due to symbionts, unicellular green algae or Cyanobacteria [6–10]. Sponges also contain prokaryotic symbionts, which are either an object of nutrition or participate in associative symbiosis with sponges, avoiding the influence of the immune system [11–16]. Thus, a various consortium of bacteria, archaea, algae, fungi, protozoa and viruses and their associative community is structurally defined as a sponge holobiome [17,18].
These microorganisms are able to promote the growth and development of the macro organism, due to the production of regulatory signaling molecules, antibiotics, active metabolites or nutritional components [16,19–22]. Recently, such relationships have recently been actively studied in both marine [23–25] and freshwater sponges [26–30], especially as the response of microbiomes or holobiomes to natural stress (for example, a rise in temperature), and in the context of intrapopulation and biogeographical stability and variability [31–35]. Violation of symbiotic relationships between macro- and microorganisms often leads to diseases and death of sponges, and in the case of the most massive lesions, up to 95% of hosts are affected by the disease [36–38].
An increasing number of reports about sponge diseases and their mass mortality has been published [39–49]. These events can significantly affect the ecosystem as a whole, at least in some cases [36–38,50–54]. Despite the fact that infectious agents are often considered the main factors that cause the mass death of hydrobionts [38–40,48,55,56], only in two cases listed below it has it been possible to isolate and prove the pathogenicity of a certain type of microorganism for marine sponges. In the first case, the strain identified as Alphaproteobacteria NW4327 was isolated in 1998 from necrotic tissues of the Rhopaloeides odorabile sponge, which caused all symptoms of sponge disease in laboratory experiments [55]. Later, the strain was identified as Pseudoalteromonas agarivorans, its study showed that it is able to produce collagenase, which destroys the sponge cytoskeleton [57,58]. In the second case, sponge infestation by a consortium of microorganisms in the disease described as a 'sponge necrosis syndrome' affected 30–36% of the sponge Calypsonian (Euplacella) population in the Maldives [48]. Bacterium Rhodobacteraceae and fungus Rhabdocline were identified in this consortium and only a combination of these microorganisms caused characteristic signs of disease in the sponges in laboratory experiments, which allowed describing this consortium as an etiological agent.
Numerous hypotheses about the assumed role of microorganisms in the development of marine sponge diseases have been put forward in various studies [41,52,59–64]. Researchers noted significant shifts in the structure of microbial associations, including a decrease in the relative abundance of Proteobacteria and an increase in Bacteroidetes, Firmicutes and Deltaproteobacteria, individual phylotypes of which were found only in infected sponges [41,65,66]. Diseases of marine sponges and corals have been observed for a long time throughout the world, while at the same time there have been no cases of freshwater sponge diseases [36].
The disease of freshwater sponges was first discovered in 2011, when samples of pink color were found in the central basin of Lake Baikal. In subsequent years, the external signs of the disease have changed and from 2013 to the present time, there are sponges with various symptoms of damage to the body, such as discoloration, tissue necrosis, the formation of brown plaque and dirty-violet bacterial covers on separate branches (S1 Video). These signs of the disease are observed especially clearly in the branchy sponge Lubomirskia baicalensis, while the cortical and globular sponges have other external signs of the disease. The number of sponges has decreased significantly and now diseased sponges are found throughout Lake Baikal. Disease and mortality of sponges is accompanied by multiple changes in the littoral ecosystem of Lake Baikal [67–72].
The aim of our study is to analyze the changes in the composition of microbiomes in patients and “healthy” freshwater sponges L. baicalensis collected in the three basins of Lake Baikal in 2015 compared with samples collected in 2010 and 2011. Changes in microbiomes were analyzed by using an approach based on the analysis of sequencing data of the 16S rRNA gene fragments.
Materials and methods
Ethics statement
We confirm that the field studies did not involve endangered or protected species. For the described field studies in the water area of Lake Baikal, special permits were not required. Ethical restrictions do not apply to sponges and no permits were required to collect sponge samples.
Sample collection and treatment
Fresh samples of sponges L. baicalensis were collected by scuba diving during field trips conducted in 2010, 2011 and 2015 from the southern, central and northern Baikal basins (S1 Fig). The samples were frozen at -20°C immediately after lifting and transported to the laboratory in the refrigerator for subsequent DNA isolation and sequencing analysis. As a control, we used a sample of a healthy sponge (Sp2010healthy) collected in 2010 before the onset of the disease, as well as two samples of 2011, sponge (Sp2011green) without visible symptoms and sick pink sponge (Sp2011pink).
DNA extraction, PCR amplification, and sequencing
DNA was extracted from the triplicate samples of frozen sponge tissue (0.1–0.2 g) after bead beating using the QIAamp DNA micro Kit (Qiagen Ltd., Crawley, UK) or TRIzol LS reagent (Invitrogen, Ambion, USA) according to the manufacturer’s protocols. Total DNA was suspended in 18μl of RNase free water and stored at—60°C waiting for further analysis. DNA samples of 2015 were transferred to Irkutsk Research Anti-Plague Institute of Siberia and the Far East (Russia) for pyrosequencing on a 454 GS Junior System and samples of 2010 and 2011 were transferred to the RTL Genomics, Lubbock, TX (USA) for high-throughput sequencing on an Illumina platform. Primer sets 357wF/785R and 515yF/926R [73] were used to amplify the variable regions 3–4 and 4–6 of the 16S rRNA gene, using Illumina MiSeq 250 bp chemistry. The universal bacterial primers 518F and 1064R [74] were used to amplify the V4–V6 hypervariable region of the bacterial 16S rRNA gene using the 454 GS Junior sequencing System and with GS FLX Titanium series reagents.
All three sets of primers correspond to the 16S rRNA genes of prokaryotes and chloroplasts, therefore gene fragments of chloroplasts can be amplified and sequenced together with prokaryotes in one experiment. The raw sequencing reads are available under BioProjects ID: PRJNA369024 (454 GS platform) and PRJNA503292 (Illumina MiSeq platform).
OTU picking
The sequencing reads obtained by the 454 GS technology were preprocessed using Mothur package [75] following conventional setup for filtering unreliable and short oligonucleotides; trim.seq function was applied to raw data files, with parameters ' maxambig = 0, maxhomop = 8, flip = T, bdiffs = 1, pdiffs = 2, qwindowaverage = 35, qwindowsize = 50'). For sequencing reads obtained using Illumina technology, the pipeline of RTL Genomics was used to obtain clean data files. Samples of sponges listed in Table 1 were used in this study.
Table 1. Summary of samples used in the survey.
| Disease state/ Sample ID |
Collection place, near | Group ID | Read archives/ columns in the dataset |
|---|---|---|---|
| Illumina technology (6 in total): | |||
| Healthy before disease | Bolshie Koty village (southern basin) | Sp2010healthy | primers 357, 515 |
| Healthy in appearance | Hoboy Cape (central basin) | Sp2011green | primers 357, 515 |
| Diseased | Hoboy Cape (central basin) | Sp2011pink | primers 357, 515 (*) |
| 454 GS technology (23 in total, collected in 2015 year): | |||
| Sponges healthy in appearance: | |||
| L1,L2,L3,L7,L8 | Lystvyanka (southern basin) | Sp2015green | replicates 1–5 |
| OV1,OV3,OV4,OV6 | Olkhonskiye Vorota (central basin) | replicates 1–4 | |
| T1,T4,T6 | Turali Cape (northern basin) | replicates 1–3 | |
| Diseased sponges: | |||
| L4,L5,L6 | Lystvyanka (southern basin) | Sp2015diseased | replicates 1–3 |
| OV2,OV5,OV7,OV8 | Olkhonskiye Vorota (central basin) | replicates 1–4 | |
| T2,T3,T5,T7 | Turali Cape (northern) | replicates 1–4 | |
In the aggregate processing of data files obtained using two different sequencing technologies, an open-reference OTU picking implemented in the QIIME package [76,77] was used. A reference database within the QIIME platform was the database gg_13_5 of Greengenes project compatible with the PICRUSt package [78]. The additional sample of pink sponge (Sp2011pink/454) collected in 2011 and analyzed earlier by Denikina et al. [69] was included in the aggregate analysis. This sample was sequenced using the 454 GS technology, and the calculation of the numbers for this additional sample is included in S1 Table for reference. All types of analysis are presented in the “Results” section based on the samples shown in Table 1.
Downstream analysis
A suite of scripts based on Python and JavaScript were developed for downstream analysis and presentation of results. Tools provided by scikit-bio Python package were used extensively for calculations of biodiversity, PERMANOVA analysis and correspondence analysis. In addition, tools from 'NumPy' and 'SciPy' Python packages were used to expand the approaches used in the study; functions stats.f_oneway and stats.mannwhitneyu from SciPy package were used for ANOVA and ranked tests, stats.kendalltau was used for Kendall correlation. The approaches used for data processing are partly based on the methods described by Feranchuk et al. [79]. The functional annotation of microbiomes was implemented using PICRUSt package [78] using a conventional pipeline, as it was described in PICRUSt documentation.
Tools used for data presentation were incorporated into an interactive system developed with the use of d3 JavaScript library running in the front-end and Python scripts running in the back-end. In particular, the stats.f_oneway and stats_mannwhitney Python functions are called in the back-end of the interactive system to present the significant changes of relative abundance in specific phylotypes in two or more groups of samples, following the ANOVA and ranked tests. The heatmap charts represent the estimated significance is represented as–ln (p-value). The source scripts of the interactive system are available at https://github.com/sferanchuk/d3b_charts. A manuscript describing the interactive system for data visualization is under preparation. Finally, the graphics charts generated by the interactive system were manually edited in Inkscape package.
For the additional support of the compatibility between two sequencing technologies, a third-party dataset was combined from the surveys where symbiotic communities of marine sponges were used. Two surveys in the dataset were sequenced with the Illumina (BioProject PRJNA454201) and 454 GS (BioProject Id PRJNA216132) technologies. The dataset was processed with the same pipeline of OTU picking, for a comparative verification of the methodology. The validation of statements about the changes in sponge microbiomes and issues of compatibility between the two technologies are extensively discussed in S1 Text.
Results
This study focuses on determining the shift in the microbial community of sponges collected in different areas of Lake Baikal in 2015 during the period of their illness and mass death. Differences in microbiomes of sponges were shown using the approach based on the pyrosequencing of the 16S rRNA gene fragment. Data on the composition of the microbiome of one sponge collected before the onset of the disease in 2010, and two samples of sponges collected in 2011, during the appearance of the first symptoms of the disease, were used as a control. These control data were obtained in 2018 using the Illumina sequencing platform. The analysis of the control sample was necessary due to the search for the causes of abrupt changes in the ecosystem of Lake Baikal and the possible relation between the appearance of pink sponges in 2011 and the current state of sponges in 2015. We are very interested in the possible connection between the appearance of pink sponges in 2011 and the current state of sponges in 2015, as well as the composition of microbiomes of a healthy sponge collected before the onset of the disease.
The number of reads included into the OTU table ranged from 1059 (diseased-OV-r3) to 5385 (healthy-T-r3) for the 454 GS samples, and from 11412 (diseased-2011-515) to 61354 (healthy-2010-357) for the Illumina samples. The estimates of sampling depth using Michaelis-Menten fit to rarefaction curves show that in the samples sequenced on Illumina platform the composition of microbiomes at the family level is presented almost completely (underestimate 2.4%). For the 2015 samples of sequenced on 454 GS platform, the average underestimate at the family level is 13.2%.
The challenge of comparative analysis of samples for 3 years was to reduce the biases introduced when the two sequencing technologies were integrated into a single table of abundances. The problems of incompatibility between the two sequencing technologies are well known, as it was extensively demonstrated in Barb et al. [80]. However, despite that, the values of abundances for the same group could be drastically different based on the two approaches; the relative changes in abundance within the same method are known to be consistent for several methods [81].
To focus the data processing on a best compatibility between the two sequencing technologies, the closed-reference OTU picking strategy was used, since it results in the most stable taxonomic units [82]. The comparison of samples from several years was mostly presented at high levels of bacterial taxonomy, as it was expected to be more robust to outline the changes in sponge microbiomes. The chloroplast species were intentionally included into the analysis, since unicellular symbiotic algae of L. baicalensis sponge were considered as an important part of sponge hologenome in a healthy state.
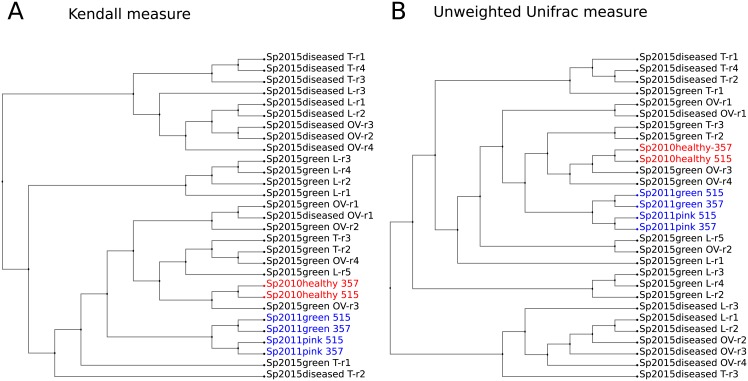
A possible way to compare these data, when the abundance values are biased is to use the presence / absence of a phylotype or to use the phylotype rank, according to Fig 1, which shows dendrograms of proximity between samples. There, the Kendall correlation measure, based on the ranks of the rows, is converted into distances between samples by a simple transformation: distance = 1—correlation (Fig 1A). Unweighted UniFrac measure less clearly separates groups of healthy and sick sponges (Fig 1B).
Fig 1. Dendrograms representing a degree of proximity between microbiomes compositions.
(A) Rank-based Kendall measure. (B) Presence-based unweighted UniFrac measure. Abundance values in the samples were used at the family level; dendrograms were constructed using UPGMA clustering.
The proximity between samples estimated by both rank-based and presence-based approaches demonstrates that 2010 healthy sponge is separated from the 2011 samples, and is close to some of the healthy sponges, collected in 2015 in the same area of Baikal near Olkhon Island. A detailed description of the possibility to compare the two technologies is provided in the additional material S1 Text. The consistency of rank comparisons allows us to compare the number of individual families after applying quantile normalization [83].
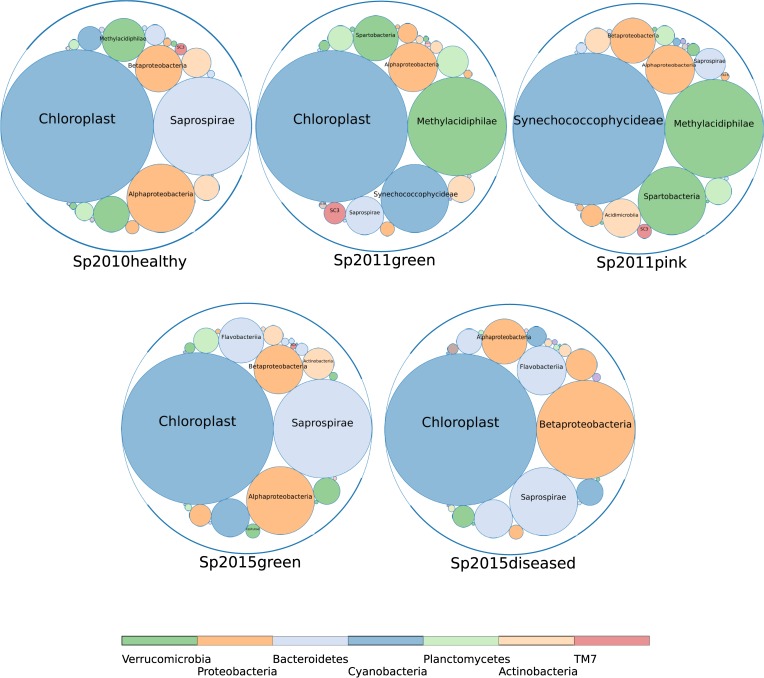
Comparison of the composition of microbiomes for samples collected in 2010, 2011 and 2015 is shown in the form of a bubble diagram in Fig 2. The abundance values for this diagram were transformed by quantile normalization in order to reduce the deviations caused by various sequencing technologies. The sizes of the circles have the same proportions in all five charts due to normalization.
Fig 2. Relative abundances of bacterial groups in the microbiomes of healthy and diseased sponges.
Bubble chart is shown at the class level for the 2010, 2011 and 2015 samples of sponges 2010, 2011 and 2015.
Obviously, the healthy in appearance Sp2011green sponge has a high content of Verrucomicrobia (class Methylacidiphilae), which is characteristic of the diseased Sp2011pink sponge. Thus, sponges with no visible signs of the disease may have changes in the microbiomes that are characteristic of sick sponges. Moreover, the content of abundant phylotypes in the microbiome of a healthy sponge Sp2011green has changed dramatically. The increased abundance of Methylacidiphilae is typical only for 2011 sponges, while these Verrucomicrobia are absent in the 2015 sponges. A characteristic feature of Sp2011pink is also the replacement of chloroplasts with cyanobacteria Synechococcophycideae. The most abundant symbionts of healthy sponges of 2010 and 2015 are the phylum Bacteroidetes (Saprospirae / Saprospiria), but they are replaced by Betaproteobacteria in diseased sponges.
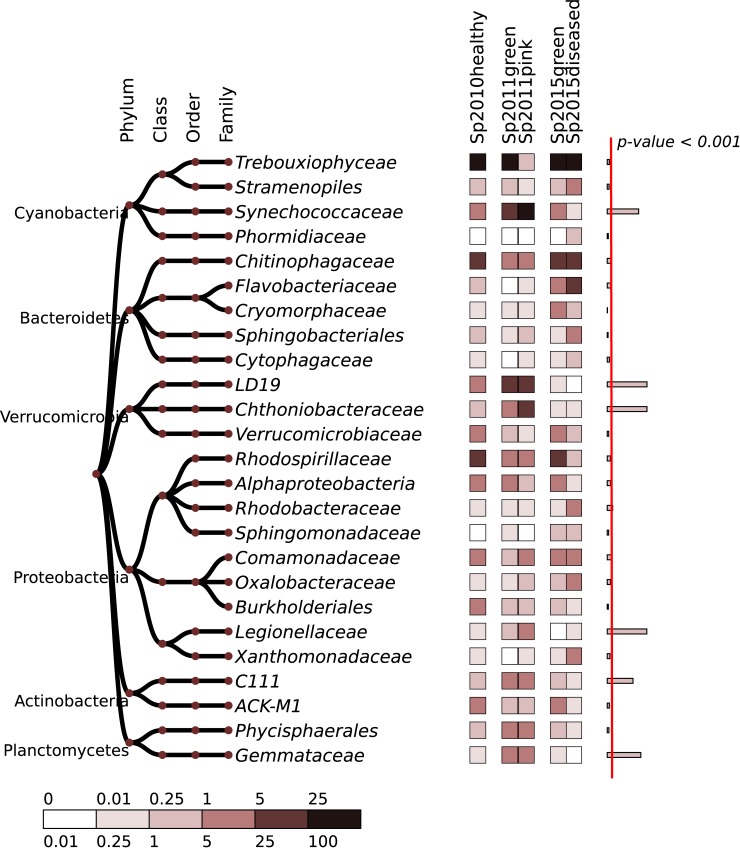
Similar changes in the number of bacterial groups are shown in heatmap (Fig 3) and in S1 Table. Bacteria abundance values were transformed by normalizing quantiles to reduce the biases introduced by sequencing technology. Evidently, the heterogeneity of microbiomes increases in diseased in sponges, and the average content of chloroplasts in the 2015 sponges does not change significantly.
Fig 3. Heatmap of 25 most abundant bacterial families.
The relative abundances for individual 2015 samples are averaged. The abundance values were transformed by the quantile normalization, to reduce the biases introduced by sequencing technology. The right column shows a significance of variation between groups, estimated using ANOVA test; width of the bars corresponds to–ln (p-value). Red line separates the significance level of 0.999 (p-value < 0.001).
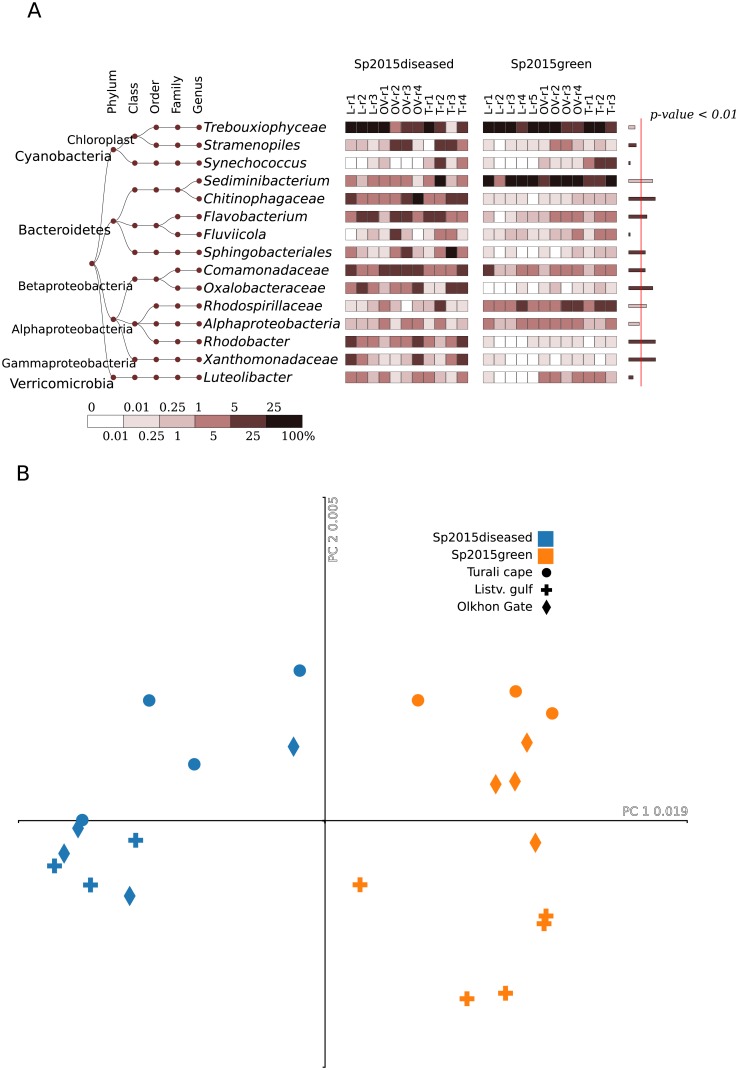
Combining data on the composition of microbiomes in the groups of healthy and sick sponges of 2015 is not correct, since significant changes are observed in individual samples, and the number of bacteria varies randomly between sponge samples (Fig 4, S2 Fig).
Fig 4. A comparative presentation of the 2015 samples.
Result given for (A) heatmap of the 15 most abundant bacterial groups, at the genus level. The right column on the right shows a significant difference between the healthy and diseased samples, estimated using Mann-Whitney test; width of the bars corresponds to–ln (p-value). Red line separates the significance level of 0.99 (p-value < 0.01). (B) Correspondence analysis at the OTU level based on the unweighted UniFrac measure as a distance.
The differences in the composition of microbiomes are observed in more detailed taxonomic annotations and in the distribution of the minor components of the microbiome. At the genus level, many unclassified Xanthomonadaceae, Comamonadaceae, Oxalobacteraceae, Chitinophagaceae, Flavobacterium and Rhodobacter should be distinguished in diseased sponges (Fig 4A). The numbers of these groups and OTU in these groups vary from sample to sample, and no single bacterium can be closely associated with signs of disease.
The distribution of chloroplasts in the samples indicates that unicellular symbiotic algae are an important component of sponge microbiomes, but the reasons for such drastic changes remain unknown and require further study. The abundance of chloroplasts contributes to the uniformity of the microbiomes, but the average proportions of the microbial species that separate the healthy and diseased sponge remain almost the same even without taking into account chloroplasts, as explained in S1 Text.
The dependence of microbiomes on the condition of sponge disease is also clearly seen in the graph of compliance analysis (Fig 4B). There is a separation of samples by geographic location, but the separation of sponges for health reasons of sponges is more significant. Estimates of species richness based on the direct amount of OTU and extrapolation of Chao and Ace indices show that the number of species in Northern Baikal (Turali Cape) is greater than in the regions of Southern Baikal, where the anthropogenic load is higher (p-value <0.003 for Ace index) (S2 Table).
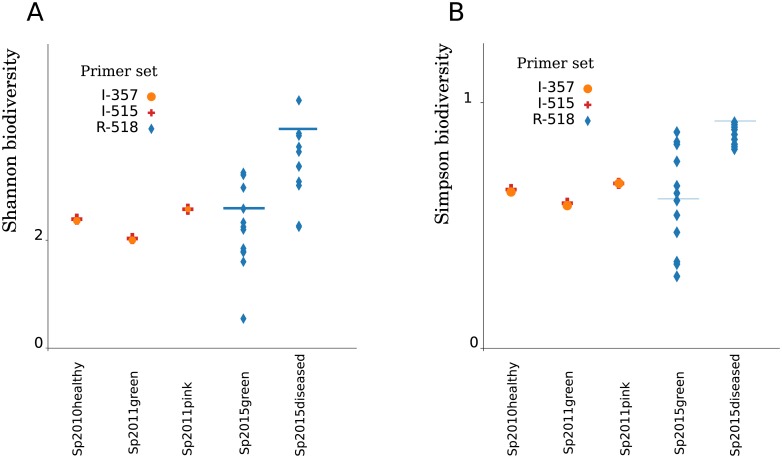
Shannon's biological diversity increases in diseased sponges of 2015 even more than in 2011 (Fig 5). Both Shannon (Fig 5A) and Simpson (Fig 5B) estimates of biodiversity are indicators of community unevenness; hence, microbiomes of diseased sponges are more heterogeneous and that dysbiosis increases with the development of the disease.
Fig 5. An increase in heterogeneity of diseased samples.
Biodiversity values for the samples used in the study (A) Shannon distribution and (B) Simpson distribution. Bars indicate the values for a composition of aggregated communities.
We evaluated at a first approximation the properties of a dysbiotic state in diseased and relatively healthy sponges by a functional annotation of functional annotation of relative changes in metabolic pathways. S2 Fig shows the heat map with the results of functional annotation. We have not identified an increase in carbon fixation, methane and nitrogen metabolism and a decrease in the biosynthesis of antibiotics have been noted. A detailed analysis of food chains of microbial communities developing in dying sponges is beyond the scope of this study, but requires a detailed consideration.
Discussion
Lake Baikal is the deepest freshwater lake in the world. It has a volume of 23,000 km3, a depth of 1,637 m and an age exceeding 25 million years [84,85]. Endemic freshwater Baikal sponges of the Lubomirskiidae family dominate the littoral zone of the Lake and their biomass is more than 700 g/m2 [86–88]. This sponge biomass is unusually high for freshwater body [89], but it is comparable to coastal Antarctic benthic communities [90] and some reefs [91].
According to the current systematics, the family Lubomirskiidae of endemic sponges in Lake Baikal is represented by 4 genera (Lubomirskia, Baikalospongia, Rezinkovia and Swartschewskia) and by 14 species [92,93]. Baikal sponges inhabit depths from one meter down to the maximum depth, but they are most concentrated at depths between 5 and 40 meters, where their biomass exceeds all other benthic organisms taken together [86,87].
Healthy sponges L. baicalensis sponges have a saturated green color due to the presence of a large number of symbiotic green alga Choricystis sp. (Trebouxiophyceae). Disease of freshwater Baikal sponges was recorded for the first time in 2011 by Bormotov [67], and it was accompanied by the change of green color into pink. No other signs of the disease, such as a change in the consistency of the spongin or tissue necrosis were registered. L. baicalensis is the most susceptible to the disease. It is distributed throughout the lake at depths from 1 to 50 m and the most numerous between depths of 5 and 20 meters [71,94]. Sponges with abnormal coloring were found only in the central basin of Baikal at depths of 25–55 meters (S1 Fig). This distribution of pink sponges calls into question some of the hypotheses about the possible causes of the sponge disease, such as global warming and the anthropogenic impact. In these places, the anthropogenic impact is minimal, and large depths determine the constant water temperature [68,71].
The comparisons of the microbial community of sponges revealed that the symbiotic algae Choricystis sp. (class Trebouxiophyceae) is completely absent in the Sp2011pink and is replaced by the cyanobacterium Synechococcaceae (Fig 2). The Synechococcaceae are the family of Cyanobacteria and are typical picoplanktonic Cyanoprokaryota in the littoral and deep-water areas of Lake Baikal [95].
Together with the appearance of Cyanobacteria in the pink sponge, there was a relative abundance of the bacteria of the family Chthoniobacteraceae and LD19 (phylum Verrucomicrobia) as well as of the minor families C111 (Actinobacteria), Legionellaceae (Gammaproteobacteria), Gemmataceae and Phycisphaerales order (Planctomycetes) had increased (Fig 3, S1 Table). A similar increase in the relative abundance of the same family of bacteria is found in Sp2011green without visible signs of the disease. Thus, sponges with the absence of visible signs of the disease may have changes in microbiomes typical for diseased sponges. The principal (core) microbiome of a healthy sponge has changed significantly, and the relative content of Chitinophagaceae, Rhodospirillacea and Comamonadaceae which was most abundant in the healthy sponge Sp2010healthy, has decreased dramatically.
In order to identify a possible relation between the sponge disease and differences in habitats, we collected samples of healthy and diseased sponges in the southern, central and northern basins of Baikal, in which the anthropogenic load decreases from south to north. We found that the number of the most abundant phyla in diseased and healthy sponges did not differ substantially (Fig 2). In addition, the composition of bacteria in sponge samples near Listvyanka, where a high concentration of biogenic elements was detected [71], is very similar to the composition of sponge microbiomes from cleaner areas of Baikal. The difference between these locations could be detected only by the absence of several minor components of microbiomes in the samples from Listvyanka. At the same time, the mortality of sponges in this region is the highest, which apparently indicates a lack of a direct correlation between the composition of microbiomes and the mortality of sponges.
Changes in the bacterial composition of sponges in 2015 differ significantly from changes in the sponge microbiomes in 2011. The pink sponge microbial samples collected in 2011 differ from the healthy sponge microbiome collected in 2010 by replacing the chloroplast with the cyanobacterium Synechococcus sp. and the emergence of uncultivated Verrucomicrobia (Chthoniobacteraceae and LD19). Previously, we assumed that an increase in Verrucomicrobia LD19 might be associated with an increased concentration of methane in the Baikal water [69].
We found that Verrucomicrobia LD19 and Chthoniobacteraceae are absent in the sponge microbiomes of 2015, in contrast to 2011, as was shown previously [69]. A decrease in methane concentration was not noted in the 2015 samples, which indicates that our assumption was erroneous. In addition, Verrucomicrobia LD19 does not contain genes for methane monooxygenase and, therefore it probably cannot oxidize methane, unlike its phylogenetically closely related acidophilic methanotroph Methylacidiphilum infernorum [96]. In addition, in the diseased sponges of year 2015, the relative abundance of Chthoniobacteraceae was significantly lower.
However, “healthy or green” sponges of 2015, with no visible signs of disease, have significant shifts in the microbiomes compared to Sp2010healthy (Fig 5), although the content of chloroplast is high in most samples. The relative abundance of Trebouxiophyceae, Procabacteraceae and unclassified Alphaproteobacteria is lower in sponges of 2015, but the content of Rhodobacteraceae, Comamonadaceae, Flavobacteriaceae, Oxalobacteraceae, Xanthomonacacaces and others has increased (Fig 4A, S1 Table). These changes in the composition of microbiomes are distributed randomly in individual samples of sponges collected in 2015 (Fig 4A, S2 Fig). The representatives of Flavobacterium, Comamonadaceae and Rhodobacteraceae have been observed to have adaptive survival strategies and transformations to opportunistic pathogens [97–99]. This suggests that the sponges in the diseased state are suffering the most from opportunistic pathogens of different origins.
Most of the bacteria, whose abundance has grown in diseased sponges, are Bacteroidetes and Proteobacteria, many of which may have quorum sensing activity, that affecting pathogenicity and virulence factors, as well as the ability to form biofilms [100–102]. The presence of QS activity can probably lead to the coordinated joint action of several opportunistic pathogens and may be the cause of the rapid death of the Baikal sponges. Although this relationship between the QS activity in bacteria and disease of sponges is not known, it has been described in corals infected with the white band disease [103,104].
Thus, our research shows that sponge diseases in 2011 and 2015 are fundamentally different, as shown by the PERMANOVA statistical tests described in S1 Text.
A common sign uniting the events of 2011 and 2015 is the inhibition of the growth of symbiotic green algae. Based on this data, we can assume two different hypotheses for the development of the disease. In the first scenario, an unknown aggressive factor arose in the central basin of Lake Baikal, which led to the rapid and complete destruction of green symbionts and the subsequent dying off sponges. Then the concentration of this factor decreased due to the mixing with water, which reduced its aggressive effect and led to a partial, rather than complete, inhibition of the development of symbiotic algae in sponges. On the second hypotheses, from 2013 to the present time, another factor (quorum sensing inducer?) came into the game.
This may cause significant inhibition of the immune system of the sponges and can triggers the development of coordinated action of opportunistic pathogens. Baikal sponge death in 2014–2018 is explained mainly by the development of opportunistic pathogens, which abundance differ from sample to sample. Determining the cause of the disease and the study activities of quorum sensing and quorum quenching in the diseased and convalescent sponges will be the goal of our further research.
Conclusions
We considered the possibility of using various technologies for sequencing microbiomes of diseased and healthy Baikal sponges. This study allowed us to expand our understanding of changes in the consortium of microorganisms and their complex symbiotic relationships in freshwater Baikal sponges. We found large-scale changes in the microflora of Baikal sponges and an increase in the number of Bacteroidetes and Proteobacteria, and found several opportunistic microorganisms in diseased sponges, which probably act in concert. Understanding how diseases arise and spread in freshwater sponges, and finding their etiological agents, is key to supporting healthy aquatic biodiversity in the Lake Baikal ecosystem.
Supporting information
(MP4)
Relative abundancies of microbial groups and degree of separation between healthy and diseased samples are evaluated using two independent sequencing experiments for a sample of pink sponge collected in 2011.
(PDF)
Four pointed stars—places of detection of pink sponges in 2011; 5 pointed stars—places for collecting samples of healthy and sick sponges in 2015. 1—cape Hoboy, July 2011, 30–45 meters; 2 –cape Orso, July 2011, 30–45 meters; 3 –cape Ukhan, October 2011, 25–35 meters; 4– cape Izhimey, November 2011, 45–55 meters; 5 –Ushkany Islands, November 2011, 25–35 meters.
(TIF)
The relative presence of functional groups in microbiomes of sponges from 2015 are presented as heatmap chart, in units of Z-score, for 'Energy metabolism' and 'Biosynthesis of Other Secondary Metabolites' KEGG ontology terms.
(TIF)
(XLS)
(PDF)
Acknowledgments
We thank Adelshin R.V. for his help in the 16S rRNA gene amplicon pyrosequencing and Dr. Colin H Brown for a versatile support of our research activity.
Editor: Brenda A Wilson, PhD Academic Editor.
Data Availability
The raw sequencing reads are available under BioProject ID: PRJNA369024, PRJNA503292.
Funding Statement
This research was supported by Funding by budget project of Federal Agency of Scientific Organizations (no 0345-2016-0002 to SB); Russian Foundation for Basic Research (grants no 16-54-150007 to SB); Russian Foundation for Basic Research (grants no 18-04-00224 to SB); and Russian Foundation for Basic Research (grants no 16-04-00065 to LC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vargas S, Schuster A, Sacher K, Büttner G, Schätzle S, Läuchli B. Barcoding sponges: an overview based on comprehensive sampling. PLoS ONE. 2012;7(7):e39345 10.1371/journal.pone.0039345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van RWM, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, et al. Global Diversity of Sponges (Porifera). PLoS ONE. 2012;7:e35105 10.1371/journal.pone.0035105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manconi R., Pronzato R. Suborder Spongillina subord. nov.: Freshwater Sponges In: Hooper JNA, Van Soest RWM, Willenz P, editors. Systema Porifera. Springer, Boston, MA: 2002. pp. 921–1019. 10.1007/978-1-4615-0747-5_97 [DOI] [Google Scholar]
- 4.Manconi R, Pronzato R. Global diversity of sponges (Porifera: Spongillina) in freshwater. Hydrobiologia. 2008;595: 27–33. 10.1007/978-1-4020-8259-7_3 [DOI] [Google Scholar]
- 5.Bell JJ, McGrath E, Biggerstaff A, Bates T, Crdenas CA, Bennett H. Global conservation status of sponges. Conserv Biol. 2015;29(1): 42–53. 10.1111/cobi.12447 [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson CR. Nutrient translocation from green algal symbionts to the freshwater sponge Ephydatia fluviatilis. Hydrobiologia. 1980;75(3): 241–250. 10.1007/BF00006488 [DOI] [Google Scholar]
- 7.Alex A, Vasconcelos V, Tamagnini P, Santos A, Antunes A. Unusual symbiotic cyanobacteria association in the genetically diverse intertidal marine sponge Hymeniacidon perlevis (Demospongiae, Halichondrida). PLoS ONE. 2012;7(12):e51834 10.1371/journal.pone.0051834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernogor L, Denikina N, Kondratov I, Solovarov I, Khanaev I, Belikov S, et al. Isolation and identification of the microalgal symbiont from primmorphs of the endemic freshwater sponge Lubomirskia baicalensis (Lubomirskiidae, Porifera). Eur J Phycol. 2013;48(4): 497–508. 10.1080/09670262.2013.862306 [DOI] [Google Scholar]
- 9.Raharinirina NA, Brandt G, Merico A. A trait-based model for describing the adaptive dynamics of coral-algae symbiosis. Front Ecol Evol. 2017;5: 31 10.3389/fevo.2017.00031 [DOI] [Google Scholar]
- 10.Ziegler M, Arif C, Burt JA, Dobretsov S, Roder C, LaJeunesse TC, et al. Biogeography and molecular diversity of coral symbionts in the genus Symbiodinium around the Arabian Peninsula. J Biogeogr. 2017;44(3): 674–686. 10.1111/jbi.12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, et al. Microbial diversity of marine sponges. Prog Mol Subcell Biol. 2003;37: 59–88. [DOI] [PubMed] [Google Scholar]
- 12.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71(2): 295–347. 10.1128/MMBR.00040-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisz JB, Lindquist N, Martens CS. Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia. 2008;155(2): 367–376. 10.1007/s00442-007-0910-0 [DOI] [PubMed] [Google Scholar]
- 14.Richardson C, Hill M, Marks C, Runyen-Janecky L, Hill A. Experimental manipulation of sponge/bacterial symbiont community composition with antibiotics: sponge cell aggregates as a unique tool to study animal/microorganism symbiosis. FEMS Microbiol Ecol. 2012;81(2): 407–418. 10.1111/j.1574-6941.2012.01365.x [DOI] [PubMed] [Google Scholar]
- 15.Pita L, Fraune S, Hentschel U. Emerging sponge models of animal-microbe symbioses. Front Microbiol. 2016;7: 2102 10.3389/fmicb.2016.02102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster NS, Thomas T. The sponge hologenome. MBio. 2016;7(2): 00135–16. 10.1128/mBio.00135-16 PMCID: PMC4850255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol. 2016;70: 317–340. 10.1146/annurev-micro-102215-095440 [DOI] [PubMed] [Google Scholar]
- 18.Butina TV, Potapov SA, Belykh OI, Belikov SI. Genetic Diversity of Cyanophages of the Myoviridae Family as a Constituent of the Associated Community of the Baikal Sponge Lubomirskia baicalensis. Russian Journal of Genetics. 2015;51(3): 384–388. [PubMed] [Google Scholar]
- 19.Proksch P. Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon. 1994;32(6): 639–655. [DOI] [PubMed] [Google Scholar]
- 20.Sand-Jensen K, Pedersen MF. Photosynthesis by symbiotic algae in the freshwater sponge Spongilla lacustris. Limnol Oceanogr. 1994;39: 551–561. [Google Scholar]
- 21.Hentschel U, Usher KM, Taylor MW. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 2006;55(2): 167–177. 10.1111/j.1574-6941.2005.00046.x [DOI] [PubMed] [Google Scholar]
- 22.Della G, Hochmuth T, Costantino V, Teta R, Gerwick W, Gerwick L, et al. Polyketide genes in the marine sponge Plakortis simplex: a new group of mono-modular type I polyketide synthases from sponge symbionts. Environ Microbiol Rep. 2013;5(6): 809–818. 10.1111/1758-2229.12081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, et al. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci U S A. 2012;109(27): 1878–87. 10.1073/pnas.1203287109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupont S, Corre E, Li Y, Vacelet J, Bourguet-Kondracki ML. First insights into the microbiome of a carnivorous sponge. FEMS Microbiol Ecol. 2013;86(3): 520–531. 10.1111/1574-6941.12178 [DOI] [PubMed] [Google Scholar]
- 25.Cuvelier ML, Blake E, Mulheron R, McCarthy PJ, Blackwelder P, Thurber RL, et al. Two distinct microbial communities revealed in the sponge Cinachyrella. Front Microbiol. 2014;5: 581 10.3389/fmicb.2014.00581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gernert C, Glckner FO, Krohne G, Hentschel U. Microbial diversity of the freshwater sponge Spongilla lacustris. Microb Ecol. 2005;50(2): 206–212. 10.1007/s00248-004-0172-x [DOI] [PubMed] [Google Scholar]
- 27.Costa R, Keller-Costa T, Gomes NC, Rocha UN, Overbeek L, Elsas JD. Evidence for selective bacterial community structuring in the freshwater sponge Ephydatia fluviatilis. Microb Ecol. 2013;65(1): 232–244. 10.1007/s00248-012-0102-2 [DOI] [PubMed] [Google Scholar]
- 28.Keller-Costa T, Jousset A, Overbeek L, Elsas JD, Costa R. The freshwater sponge Ephydatia fluviatilis harbours diverse Pseudomonas species (Gammaproteobacteria, Pseudomonadales) with broad-spectrum antimicrobial activity. PLoS ONE. 2014;9(2):e88429 10.1371/journal.pone.0088429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gladkikh AS, Kaluyzhnaya OV, Belykh OI, Ahn TS, Parfenova VV. Analysis of bacterial communities of two Lake Baikal endemic sponge species. Mikrobiologiia. 2014;83(6): 682–693. [PubMed] [Google Scholar]
- 30.Seo EY, Jung D, Belykh OI, Bukshuk NA, Parfenova VV, Joung Y, et al. Comparison of bacterial diversity and species composition in three endemic Baikalian sponges. Ann Limnol. 2016;52: 27–32. 10.1051/limn/2015035 [DOI] [Google Scholar]
- 31.Crdenas CA, Bell JJ, Davy SK, Hoggard M, Taylor MW. Influence of environmental variation on symbiotic bacterial communities of two temperate sponges. FEMS Microbiol Ecol. 2014;88(3): 516–527. 10.1111/1574-6941.12317 [DOI] [PubMed] [Google Scholar]
- 32.Antunes A. Pyrosequencing characterization of the microbiota from Atlantic intertidal marine sponges reveals high microbial diversity and the lack of co-occurrence patterns. PLoS ONE. 2015;10(5):e0127455 10.1371/journal.pone.0127455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigel BL, Erwin PM. Intraspecific variation in microbial symbiont communities of the sun sponge, Hymeniacidon heliophila, from inter- tidal and subtidal habitats. Appl Environ Microbiol. 2016;82(2): 650–658. 10.1128/AEM.02980-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pisera A, Manconi R, Siver PA, Wolfe AP. The sponge genus Ephydatia from the high-latitude middle Eocene: environmental and evolutionary significance. Palaontol Z. 2016;90(4): 673–680. 10.1007/s12542-016-0328-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinert G, Taylor MW, Deines P, Simister RL, Voogd NJ, Hoggard M, et al. In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity. PeerJ. 2016;4:e1936 10.7717/peerj.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webster NS. Sponge disease: a global threat? Environ Microbiol 2007;9(6):1363–1375. 10.1111/j.1462-2920.2007.01303.x [DOI] [PubMed] [Google Scholar]
- 37.Cebrian E, Uriz MJ, Garrabou J, Ballesteros E. Sponge mass mortalities in a warming Mediterranean Sea: are cyanobacteria-harboring species worse off? PLoS ONE 2011;6(6):e20211 10.1371/journal.pone.0020211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luter HM, Webster NS. Sponge disease and climate change In. Carballo JL, Bell JJ, editors. Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization. Springer, 2017. pp. 411–428. 10.1007/978-3-319-59008-0_1 [DOI] [Google Scholar]
- 39.Gaino E, Pronzato R, Corriero G, Buffa P. Mortality of commercial sponges: incidence in two Mediterranean areas. Boll Zool. 1992;59(1): 79–85. 10.1080/11250009209386652 [DOI] [Google Scholar]
- 40.Olson JB, Gochfeld DJ, Slattery M. Aplysina red band syndrome: a new threat to Caribbean sponges. Dis Aquat Organ. 2006;71(2): 163–168. 10.3354/dao071163 [DOI] [PubMed] [Google Scholar]
- 41.Webster NS, Xavier JR, Freckelton M, Motti CA, Cobb R. Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environ Microbiol. 2008;10(12): 3366–3376. 10.1111/j.1462-2920.2008.01734.x [DOI] [PubMed] [Google Scholar]
- 42.Garrabou J, Comaz R, Bensoussan N, Bally M, Chevaldonne P, et al. Mass mortality in Northwestern Mediterranean rocky benthic communities: effects of the 2003 heat wave. Global Change Biol. 2009;15: 1090–1103. 10.1111/j.1365-2486.2008.01787.x [DOI] [Google Scholar]
- 43.Luter HM, Whalan S, Webster NS. Exploring the role of microorganisms in the disease-like syndrome affecting the sponge Ianthella basta. Appl Environ Microbiol. 2010;76(17): 5736–5744. 10.1128/AEM.00653-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado M, Sanchez-Tocino L, Navarro C. Recurrent disease outbreaks in corneous demosponges of the genus Ircinia: epidemic incidence and defense mechanisms. Mar Biol. 2010;157(7): 1577–1590. 10.1007/s00227-010-1431-7 [DOI] [Google Scholar]
- 45.Angermeier H, Kamke J, Abdelmohsen UR, Krohne G, Pawlik JR, Lindquist NL, et al. The pathology of sponge orange band disease affecting the Caribbean barrel sponge Xestospongia muta. FEMS Microbiol Ecol. 2011;75(2): 218–230. 10.1111/j.1574-6941.2010.01001.x [DOI] [PubMed] [Google Scholar]
- 46.Angermeier H, Glckner V, Pawlik JR, Lindquist NL, Hentschel U. Sponge white patch disease affecting the Caribbean sponge Amphimedon compressa. Dis Aquat Organ. 2012;99(2): 95–102. 10.3354/dao02460 [DOI] [PubMed] [Google Scholar]
- 47.Gao ZM, Wang Y, Tian RM, Lee OO, Wong YH, Batang ZB, et al. Pyrosequencing revealed shifts of prokaryotic communities between healthy and disease-like tissues of the Red Sea sponge Crella cyathophora. PeerJ. 2015;3: e890 10.7717/peerj.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sweet M, Bulling M, Cerrano C. A novel sponge disease caused by a consortium of microorganisms. Coral Reefs. 2015;34(3): 871–883. 10.1007/s00338-015-1284-0 [DOI] [Google Scholar]
- 49.Blanquer A, Uriz MJ, Cebrian E, Galand PE. Snapshot of a bacterial microbiome shift during the early symptoms of a massive sponge die-off in the Western Mediterranean. Front Microbiol. 2016;7: 752 10.3389/fmicb.2016.00752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wulff JL. Rapid diversity and abundance decline in a Caribbean coral reef sponge community. Biol Conserv. 2006;127(2): 167–176. 10.1016/j.biocon.2005.08.007 [DOI] [Google Scholar]
- 51.Lejeusne C, Chevaldonne P, Pergent-Martini, Boudouresque CF, Perez T. Climate change effects on a miniature ocean: the highly diverse, highly impacted Mediterranean Sea. Trends Ecol Evol. 2010;25(4): 250–260. 10.1016/j.tree.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 52.Stabili L, Cardone F, Alifano P, Tredici SM, Piraino S, Corriero G, et al. Epidemic mortality of the sponge Ircinia variabilis (Schmidt, 1862) associated to proliferation of a Vibrio bacterium. Microb Ecol. 2012;64(3): 802–813. 10.1007/s00248-012-0068-0 [DOI] [PubMed] [Google Scholar]
- 53.Carballo JL, Bautista E, Nava H, Cruz-Barraza JA, Chvez JA. Boring sponges, an increasing threat for coral reefs affected by bleaching events. Ecol Evol. 2013;3(4): 872–886. 10.1002/ece3.452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez T, Vacelet J. Effect of climatic and anthropogenic disturbances on sponge fisheries In: Goffedro S., Baader H., Dubinsky Z, editors. The Mediterranean Sea: Its History and Present Challenges. Chapter: Effect of climatic and anthropogenic disturbances on sponge fisheries. Springer, 2014. pp. 577–587. 10.1007/978-94-007-6704-1_35 [DOI] [Google Scholar]
- 55.Webster NS, Negri AP, Webb RI, Hill RT. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar Ecol Prog Ser. 2002;232: 305–309. 10.3354/meps232305 [DOI] [Google Scholar]
- 56.Cervino JM, Winiarski-Cervino K, Poison SW, Goreau T, Smith GW. Identification of bacteria associated with a disease affecting the marine sponge Ianthella basta in New Britain, Papua New Guinea. Mar Ecol Prog Ser. 2006;324: 139–150. 10.1007/s10126-015-9627-y [DOI] [PubMed] [Google Scholar]
- 57.Mukherjee J, Webster N, Llewellyn LE. Purification and characterization of a collagenolytic enzyme from a pathogen of the great barrier reef sponge, Rhopaloeides odorabile. PLoS ONE. 2009;4(9):e7177 10.1371/journal.pone.0007177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choudhury JD, Pramanik A, Webster NS, Llewellyn LE, Gachhui R, Mukherjee J. The pathogen of the Great Barrier Reef sponge Rhopaloeides odorabile is a new strain of Pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar Biotechnol. 2015;17: 463–478. 10.1007/s10126-015-9627-y [DOI] [PubMed] [Google Scholar]
- 59.Smith FGW. Sponge disease in British Honduras, and its transmission by water currents. Ecology. 1941;22: 415–421. 10.2307/1930719 [DOI] [Google Scholar]
- 60.Vacelet J, Gallissian MF. Virus-like particles in cells of the sponge Verongia cavernicola (Demospongiae, Dictyoceratida) and accompanying tissue changes. J Invertebr Pathol. 1978;31: 246–254 10.1016/0022-2011(78)90014-9 [DOI] [Google Scholar]
- 61.Rutzler K. Mangrove sponge disease induced by cyanobacterial symbionts: failure of a primitive immune system? Dis Aquat Org. 1988;5: 143–149. [Google Scholar]
- 62.Sweet M, Bum D, Croquer A, Leary P. Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS ONE. 2013;8(4):e62580 10.1371/journal.pone.0062580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Camillo CG, Cerrano C. Mass mortality events in the NW Adriatic Sea: phase shift from slow- to fast-growing organisms. PLoS ONE. 2015;10(5):e0126689 10.1371/journal.pone.0126689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Correa AM, Ainsworth TD, Rosales SM, Thurber AR, Butler CR, Vega RL. Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front Microbiol. 2016;7: 127 10.3389/fmicb.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luter HM, Bannister RJ, Whalan S, Kutti T, Pineda MC, Webster NS. Microbiome analysis of a disease affecting the deep-sea sponge Geodia barretti. FEMS Microbiol Ecol. 2017;93(6). 10.1093/femsec/fix074 [DOI] [PubMed] [Google Scholar]
- 66.Deignan LK, Pawlik JR, Erwin P. Agelas Wasting Syndrome Alters Prokaryotic Symbiont Communities of the Caribbean Brown Tube Sponge, Agelas tubulata. Microb Ecol. 2018;3: 00248–017. 10.1007/s00248-017-1135-3 [DOI] [PubMed] [Google Scholar]
- 67.Bormotov AE. What has happened to Baikal sponges? Science First Hand 2012;32(2): 20–23. [Google Scholar]
- 68.Kravtsova LS, Izhboldina LA, Khanaev IV, Pomazkina GV, Rodionova EV, Domysheva VM, et al. Nearshore benthic blooms of filamentous green algae in Lake Baikal. J Great Lakes Res. 2014;40: 441–448. 10.1016/j.jglr.2014.02.019 [DOI] [Google Scholar]
- 69.Denikina NN, Dzyuba EV, Belkova NL, Khanaev IV, Feranchuk SI, Makarov MM, et al. The First Case of Disease of the Sponge Lubomirskia baicalensis: Investigation of Its Microbiome. Translated in Biology Bulletin. 2016;43(3): 263–270. 10.1134/S106235901603002X [DOI] [Google Scholar]
- 70.Timoshkin OA, Samsonov DP, Yamamuro M, Moore MV, Belykh OI, Malnik VV, et al. Rapid ecological change in the coastal zone of Lake Baikal (East Siberia): Is the site of the world’s greatest freshwater bio- diversity in danger? J Great Lakes Res. 2016;42: 487–497. 10.1016/j.jglr.2016.02.011 [DOI] [Google Scholar]
- 71.Khanaev IV, Kravtsova LS, Maikova OO, Bukshuk NA, Sakirko MV, Kulakova NV, et al. Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge disease and the problem of conservation of diversity. J Great Lakes Res. 2018;44(1): 77–85. 10.1016/j.jglr.2017.10.004 [DOI] [Google Scholar]
- 72.Kulakova NV, Sakirko MV, Adelshin RV, Khanaev IV, Nebesnykh IA, Prez T. Brown Rot Syndrome and Changes in the Bacterial community of the Baikal Sponge Lubomirskia baicalensis. Microb Ecol. 2018;75(4): 1024–1034. 10.1007/s00248-017-1097-5 [DOI] [PubMed] [Google Scholar]
- 73.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18(5): 1403–1414. 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- 74.Huber JA, Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. Microbial population structures in the deep marine biosphere. Science. 2007;318: 97–100. 10.1126/science.1146689 [DOI] [PubMed] [Google Scholar]
- 75.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23): 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5): 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kopylova E, Navas-Molina JA, Mercier C, Xu ZZ, Mahé F, He Y, et al. Open-Source Sequence Clustering Methods Improve the State Of the Art. mSystems. 2016;1(1). 10.1128/mSystems.00003-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mukherjee A, Chettri B, Langpoklakpam JS, Basak P, Prasad A, Mukherjee AK, et al. Bioinformatic Approaches Including Predictive Metagenomic Profiling Reveal Characteristics of Bacterial Response to Petroleum Hydrocarbon Contamination in Diverse Environments. Sci Rep. 2017;7(1): 1108 10.1038/s41598-017-01126-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feranchuk S, Belkova N, Potapova U, Kuzmin D, Belikov S. Evaluating the use of diversity indices to distinguish between microbial communities with different traits. Res Microbiol. 2018;169(4–5): 254–261. 10.1016/j.resmic.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 80.Barb JJ, Oler AJ, Kim HS3 Chalmers N, Wallen GR, Cashion A, Munson PJ, Ames NJ. Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples. PLoS ONE. 2016;11(2):e0148047 10.1371/journal.pone.0148047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lebret K, Schroeder J, Balestreri C, Highfield A, Cummings D, Smyth T3, Schroeder D. Choice of molecular barcode will affect species prevalence but not bacterial community composition. Mar Genomics. 2016;29: 39–43. 10.1016/j.margen.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He Y, Caporaso JG, Jiang XT, Sheng HF, Huse SM, Rideout JR, Edgar RC, Kopylova E, Walters WA, Knight R, Zhou HW. Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome. 2015;3: 20 10.1186/s40168-015-0081-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics, 2003;19 (2): 185–193. [DOI] [PubMed] [Google Scholar]
- 84.Petit C, Deverchere J. Structure and evolution of the Baikal rift: A synthesis. Geochem. Geophys. Geosyst. 2006;7: Q11016 10.1029/2006GC001265 [DOI] [Google Scholar]
- 85.Mats VD, Perepelova TI. A new perspective on evolution of the Baikal Rift. Geoscience Frontiers. 2011;2(3): 349–365. 10.1016/j.gsf.2011.06.002 [DOI] [Google Scholar]
- 86.Kozhov MM Essays on Baikal studies. Irkutsk: East-Sib Press; 1972. (in Russian) [Google Scholar]
- 87.Pile AJ, Patterson MR, Savarese M, Chernykh V, Fialkov V. Trophic effects of sponge feeding within Lake Baikal's littoral zone. 2. Sponge abundance, diet, feeding efficiency, and carbon flux. Limnol Oceanogr. 1997;42(1): 178–184. 10.4319/lo.1997.42.1.0171 [DOI] [Google Scholar]
- 88.Semiturkina NA, Efremova SM, Timoshkin OA State-of-the art of biodiversity and ecology of spongiofauna of lake Baikal with special attention to the diversity, peculiarities of ecology and vertical distribution of Porifera on Berezovy ecology test site In: Index of animal species inhabiting lake Baikal and its catchment area, Novosibirsk: Nauka; 2009. pp. 891–901. [Google Scholar]
- 89.Bailey RC, Day KE, Norris RH, Reynoldson TB. Macroinvertebrate community structure and sediment bioassay results from nearshore areas of North American Great Lakes. J Great Lakes Res. 1995;21: 42–52. 10.1016/S0380-1330(95)71019-X [DOI] [Google Scholar]
- 90.Dayton IK, Robilliard GA, Paine RT, Dayton LB Biological accommodation in the benthic community at McMurdo Sound, Antarctica. Ecol. Monogr. 1974;44: 105–128. 10.2307/1942321 [DOI] [Google Scholar]
- 91.Wilkinson CR. Productivity and abundance of large sponge populations on Flinders Reef flats, Coral Sea. Coral Reefs. 1987;5: 183 10.1007/BF00300961 [DOI] [Google Scholar]
- 92.Efremova S M. New genus and new species of sponges from family Lubomirskiidae Rezvoj, 1936 In Timoshkin O.A. editor. Index of animal species inhabiting lake Baikal and its catchment. Novosibirsk: Nauka; 2004. pp. 1261–1278. (in Russian) [Google Scholar]
- 93.Itskovich V, Glyzina O, Kaluzhnaya O. Intraspecific and interspecific sequence variability in the ITS region of the rDNA of freshwater sponges of Lake Baikal and East Siberia. Inland Waters. 2017;7(3): 259–266 10.1080/20442041.2017.1320507 [DOI] [Google Scholar]
- 94.Masuda Y. Studies on the taxonomy and distribution of freshwater sponges in Lake Baikal. Prog Mol Subcell Biol. 2009;47: 81–110 10.1007/978-3-540-88552-8_4 [DOI] [PubMed] [Google Scholar]
- 95.Belykh OI, Tikhonova IV, Sorokovikova EG, Sherbakova TA, Kureishevich-Lishchuk A. Picoplankton Cyanoprokaryota of Genera Synechococcus Nägeli and Cyanobium Rippka et Cohen-Baz. from Lake Baikal (Russia). International Journal on Algae. 2011;13(2): 149–163. 10.1615/InterJAlgae.v13.i2.50 [DOI] [Google Scholar]
- 96.Hugerth LW, Larsson J, Alneberg J, Lindh MV, Legrand C, Pinhassi J et al. Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 2015;16: 279 10.1186/s13059-015-0834-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pulkkinen K, Suomalainen LR, Read AF, Ebert D, Rintamäki P, Valtonen ET. Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proceedings of the Royal Society B: Biological Sciences. 2009;277: 593–600. 10.1098/rspb.2009.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jezberová J, Jezbera J, Znachor P. The Limnohabitans genus harbors generalistic and opportunistic subtypes: evidence from spatiotemporal succession in a canyon-shaped reservoir. Applied and Environmental Microbiology 2017;83:e01530–17. 10.1128/AEM.01530-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baldani JI, Videira SS, dos Santos Teixeira KR. et al. The family Rhodospirillaceae In: Rosenberg E, DeLong EF, Lory S et al. editors. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Berlin: Springer-Verlag; 2014. pp. 533–618. 10.1007/978-3-642-30197-1_300. [DOI] [Google Scholar]
- 100.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. Journal of Bacteriology. 1970;104: 313–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Whitehead NA, Anne ML, Slater BH, Natalie J.L. Simpson NJL , Salmond GPC. Quorum-sensing in Gram-negative bacteria. FEMS Microbiology Reviews. 2001;25: 365–404. 10.1111/j.1574-6976.2001.tb00583.x [DOI] [PubMed] [Google Scholar]
- 102.Tan CH, Koh KS, Xie C, Tay M, Zhou Y, Williams R, et al. The role of quorum sensing signalling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 2014;8: 1186–1197. 10.1038/ismej.2013.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gignoux-Wolfsohn SA, Vollmer SV. Identification of Candidate Coral Pathogens on White Band Disease-Infected Staghorn Coral. PLoS ONE. 2015; 10(8):e0134416 10.1371/journal.pone.0134416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Certner RH, Vollmer SV. Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ Microbiol. 2018;20: 645–657. 10.1111/1462-2920.13991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(MP4)
Relative abundancies of microbial groups and degree of separation between healthy and diseased samples are evaluated using two independent sequencing experiments for a sample of pink sponge collected in 2011.
(PDF)
Four pointed stars—places of detection of pink sponges in 2011; 5 pointed stars—places for collecting samples of healthy and sick sponges in 2015. 1—cape Hoboy, July 2011, 30–45 meters; 2 –cape Orso, July 2011, 30–45 meters; 3 –cape Ukhan, October 2011, 25–35 meters; 4– cape Izhimey, November 2011, 45–55 meters; 5 –Ushkany Islands, November 2011, 25–35 meters.
(TIF)
The relative presence of functional groups in microbiomes of sponges from 2015 are presented as heatmap chart, in units of Z-score, for 'Energy metabolism' and 'Biosynthesis of Other Secondary Metabolites' KEGG ontology terms.
(TIF)
(XLS)
(PDF)
Data Availability Statement
The raw sequencing reads are available under BioProject ID: PRJNA369024, PRJNA503292.