Abstract
Rye (Secale cereale L.) is known for its wide adaptation due to its ability to tolerate harsh environments in semiarid areas. To assess the diversity in rye we genotyped a panel of 178 geographically diverse accessions of four Secale sp. from U.S. National Small Grains Collection using 4,037 high-quality SNPs (single nucleotide polymorphisms) developed by genotyping-by-sequencing (GBS). PCA and STRUCTURE analysis revealed three major clusters that separate S. cereale L. from S. strictum and S. sylvestre, however, genetic clusters did not correlate with geographic origins and growth habit (spring/winter). The panel was evaluated for response to Pyrenophora tritici-repentis race 5 (PTR race 5) and nearly 59% accessions showed resistance or moderate resistance. Genome-wide association study (GWAS) was performed on S. cereale subsp. cereale using the 4,037 high-quality SNPs. Two QTLs (QTs.sdsu-5R and QTs.sdsu-2R) on chromosomes 5R and 2R were identified conferring resistance to PTR race 5 (p < 0.001) that explained 13.1% and 11.6% of the phenotypic variation, respectively. Comparative analysis showed a high degree of synteny between rye and wheat with known rearrangements as expected. QTs.sdsu-2R was mapped in the genomic region corresponding to wheat chromosome group 2 and QTs.sdsu-5R was mapped to a small terminal region on chromosome 4BL. Based on the genetic diversity, a set of 32 accessions was identified to represents more than 99% of the allelic diversity with polymorphic information content (PIC) of 0.25. This set can be utilized for genetic characterization of useful traits and genetic improvement of rye, triticale, and wheat.
Introduction
Rye (Secale cereale L.) belongs to the Triticeae tribe of the family Poaceae [1] and is believed to share a common ancestor with wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) [2]. Most of the species of genus Secale originated in the Middle East, particular in Turkey [3]. Later along with the dissemination of wheat and barley to Europe and the Western Mediterranean regions, rye first came as a weed to these places. From the weedy species of rye, farmers consciously or unconsciously selected variants with a non-brittle rachis and larger seeds [3], which became current Secale cereale, the only cultivated species of rye. Due to its resilience, rye first adapted as a secondary crop in the areas with harsh environments (cold and heat stress), where other staple crops like wheat and barley were not able to survive [3]. Eventually, seeing its versatility, farmers started cultivating rye. Now, it is grown in Canada and northern parts of the United States, Russia, Japan, Australia, and South Africa [4].
In general, the genus Secale is classified into four species: S. cereale—an annual allogamous species, S. sylvestre and vavilovii—an annual autogamous species and S. strictum—a perennial wild-type allogamous species [5]. Around the globe, rye is cultivated mainly for food, feed, and pasture; and also, as a cover crop or green manure crop. Rye-based products are a rich source of nutritionally essential compounds such as minerals (Zn, Fe, and P), β-glucan (1.3–2.7%), starch, and dietary fibers [6,7]. In Europe, rye grain provides a substantial portion of the human (as bread) and animal diet. In North America, rye is preferably grown as a cover crop or pasture, and its grain is used as livestock feed and for alcohol distillation. In dry lands of southern Australia, it is grounded to prevent wind erosion. Furthermore, due to its sturdiness, it is also considered as a good pioneer crop to restore the fertility of waste lands [4].
Triticale (X Triticosecale Wittmack), a cross between durum wheat (AABB) and rye (RR) further signifies the stress tolerance ability of rye by producing relatively higher biomass and grain yield over the other cereals in dry and cold environments [8]. Through chromosome substitutions or translocations, important genes from rye have been exploited for the improvement of other cereals especially wheat [9]. One of the important examples signifying the pest resistance of rye is 1BL.1RS translocation in wheat. Rye chromosome arm 1RS carries savior genes conferring resistance to stem rust (Sr31), leaf rust (Lr26), powdery mildew (Pm8), and yellow rust (Yr9) [10–12]. Likewise, there are many other wheat-rye translocations harboring stress-resistance genes that aided in increasing the grain yield and the adaptation potential of bread wheat [13–16]. Rye offers great potential for wheat improvement and should be further explored [17].
Assessing the genetic diversity in rye can help broaden the genetic base of rye, enhance accessibility to the important genes and improve management of gene bank resources.[18]. Genetic diversity analysis involves the comparison of the accessions for their dissimilarities at the molecular level to determine the variation present in a set of accessions. Genetically manipulating a large collection of accessions could be costly and laborious, therefore extracting a core set that represents the genetic diversity of the entire set is a promising methodology [19–21]. A core set of germplasm can make the available germplasm resources to be more systematically and easily utilized in breeding programs by eliminating redundancy [19].
Among the diploid species of Poaceae family, rye has the largest genome (~7.9 Gbps) [22] with about 90% of repetitive sequences [23]. Due to the genome complexity and its regional cultivation, the rye genome has not been extensively studied. Several studies on rye genetic diversity have been conducted using different marker systems including SSR (Simple sequence repeats) [5,24–28], AFLP (Amplified Fragment Length Polymorphism) [29], DArT (Diversity Arrays Technology) [21,30], and recently SNPs (Single nucleotide polymorphism) [31]. Majority of these studies either used a limited number of markers covering a small portion of a genome or may have ascertainment bias. GBS (genotyping-by-sequencing) provides an opportunity for simultaneous SNP discovery across a genome and analysis of the genetic diversity, population structure and evolution processes of any species. Recently a draft sequence of the rye genome has been released, which will facilitate SNP marker development and the molecular characterization of economically important traits in rye.
Identification of linked molecular markers to genes of interest may help understand the molecular mechanisms of gene actions and facilitate marker-assisted selection of important traits in breeding and wide hybridization. Several genetic linkage maps have been developed in rye [32–35]. A number of genes/QTLs have been mapped in rye including plant height [36,37], spike length [37], number of spikelets per spike [37], benzoxazinoid content, rust resistance, α-amylase activity, and preharvest sprouting resistance [38]. However, linkage mapping has limitations in capturing the available genetic diversity and has a lower capacity to detect polygenic traits. With the availability of large-scale SNP data, genome-wide association studies (GWAS) provide an opportunity to characterize and map genes for important traits. GWAS is based on linkage disequilibrium (LD), and tightly linked genes theoretically have high LD which is maintained over generations [39]. GWAS has been used to characterize several economically important traits such as yield, disease, pest resistance, and abiotic stress tolerance in many crop species including rice [40–44], maize [45–51], barley [52–58], and wheat [59,60,69–71,61–68].
Rye has been reported to carry several important resistance genes and QTLs [9,10,12,13]. However, GWAS has not been widely used to identify genes/QTLs and linked markers in rye. In this study, we used GWAS approach to characterize the rye tan spot, an economically important disease caused by a necrotrophic fungus Pyrenophora tritici-repentis (PTR) and has caused up to 49% yield loss under favorable conditions [72]. Rye is known to be infected with tan spot and but QTLs for resistance to PTR race 1 and PTR race 5 has not been reported. Identification of additional genes/QTLs for tan spot resistance in rye could facilitate the development of tan spot resistant wheat, rye, and triticale varieties.
The goals of the current study were to assess the genetic diversity in cultivated rye and develop a smaller diverse representative set to facilitate genetic improvement in rye, triticale and wheat through the development of new germplasm and to demostrate the potential of GWAS in rye by identifying QTL(s) conferring resistance to tan spot PTR race 5.
Materials and methods
Plant materials
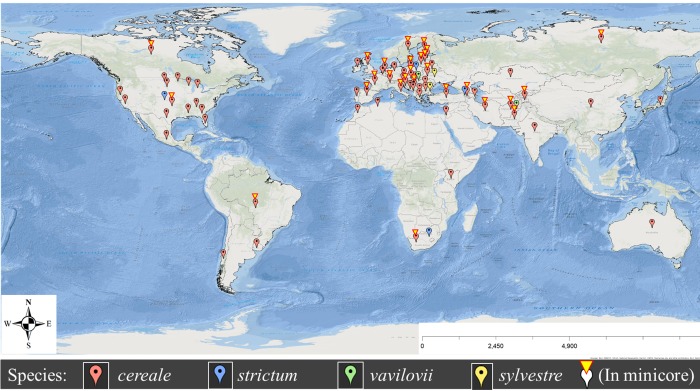
A panel of 178 geographically diverse (70 countries) accessions of Secale sp. was selected from the USDA National Small Grains Collection (NSGC). The majority of the accessions are from the Middle East (primary center of origin) and Europe (secondary center of origin) (Fig 1). The panel includes 160 cultivated rye (S. cereale subsp. cereale), nine wild S. cereale subsp., five S. strictum, and two each of S. sylvestre and S. vavilovii (S1 and S2 Tables). Only S. cereale subsp. cereale accessions were used as an association panel and to extract a smaller diverse representative set.
Fig 1. Geographic diversity of the panel of 178 accessions and the smaller diverse representative set of 32 accessions of Secale sp.
Color code: red, blue, green, and yellow map pins, correspond to Secale cereale subsp., S. strictum subsp., S. vavilovii, and S. sylvestre, respectively. Overlaid yellow triangles refer to accessions in smaller diverse representative set, which was selected from 160 cultivated rye (S. cereale subsp. cereale) accessions.
Genotyping and SNP discovery
Young leaf tissues were collected from three-week-old plants of each accession for DNA isolation using the hexadecyltrimethylammonium bromide (CTAB) method [73]. The DNA was quantified and normalized to 20ng/μl. GBS was performed following the double-restriction enzyme digestion protocol for library construction and Ion Proton system sequencing [74]. Briefly, 10μl of the normalized DNA from each accession was double-digested with PstI and MspI restriction enzymes (New England BioLabs, Inc., Ipswich, MA, USA) and then ligated to two adapters with barcodes [74–76]. After the adapters were ligated, the samples were pooled together for PCR amplification and subsequently size-selected for 250-300bp amplicons using the E-gel system (Thermo Fisher Scientific, Waltham, MA, USA). Libraries (80 pM) were prepared and sequenced in an Ion Proton system using Ion PI Hi-Q Chef and Sequencing 200 kits with two Ion P1v3 chips (Thermo Fisher Scientific, Waltham, MA, USA). SNPs were called using a reference-based SNP calling pipeline of TASSEL 5 [77]. A rye custom reference genome was obtained from the Plant Genome and Systems Biology (PGSB) website (http://pgsb.helmholtz-muenchen.de/plant/rye/gz/download/) [78] and used as a reference to locate the SNPs within the rye genome.
Population structure and genetic diversity
Basic genetic diversity indices: polymorphic information content (PIC) and Shanon’s diversity index (I-index) were calculated for each SNP using the formula:
Where, p and q correspond to the major and minor allele frequency, respectively [79], and pi is the allele frequency of the ith allele at a particular locus [80]. Percentage dissimilarity based principal coordinate analysis (PCA) among and between the species was performed using a R-package prcomp [81]. For comparison among accessions, a pairwise genetic dissimilarity (GD) matrix was computed using a R-package ape [82]. GD was employed for hierarchical clustering and a neighbor-joining (NJ) tree was constructed using R-package fastcluster [83]. Finally, the tree was pictographically developed using an online tool, Tree of life (iTOL) [84].
Population structure among all Secale sp. accessions was analyzed using STRUCTURE software [85]. DeltaK, the estimated likelihood [LnP (D)], method described by Evano et al. [86] was used to determine the optimum number of clusters. This method is based on a change in the log probability of the data in question, moving from successive K values. The number of clusters (K) with the highest value of DeltaK was selected. Accessions with > 60% ancestry from one particular cluster were grouped into corresponding population e.g. accessions in P1 population carries > 60% ancestry from cluster 1 (C1). Accessions with < 60% ancestry from any single cluster were declared admixture of two clusters with which they share > 20% ancestry. For example, accessions in the cluster P12 shared ancestries of cluster 1 (C1) and cluster 2 (C2).
Selection of the smaller diverse representative set of rye
To identify a diverse representative set to represent the diversity of 160 accessions of S. cereale subsp. cereale, the association panel was first classified into distance-based clusters. From the clusters containing less than 10 accessions, a single accession that represents the corresponding cluster was selected as a member of the smaller set. If a cluster had larger than 10 accessions, it was further sub-divided such that each sub-cluster had less than 10 accessions. Then, the most representative accession was selected from each of the sub-clusters to ensure the PIC value of the smaller diverse represenative set was maximized.
Evaluation of reaction to Pyrenophora tritici-repentis (PTR) race 5
Nine seeds of each genotype were planted in three cones (3.8 cm in diameter and 20 cm in length) with three seeds per cone as one replication. Wheat genotypes 6B662 and Salamouni were used as susceptible and resistant checks, respectively. Plants were grown in a greenhouse at temperatures ranging from 20 to 23°C with 16 h photoperiod till inoculation. At the second leaf stage, plants were inoculated with PTR race 5 using a spore suspension of 2,500 spores/ml. Inoculated plants were moved to a mist chamber (18°C) for 24 h and then grown for 7 d in a greenhouse at 21°C with 16 h photoperiod. Disease lesions were rated in a 1–5 scale [87] with 1 as resistant, 2 as moderately resistant, 3 moderately susceptible, and 4 and 5 as susceptible at the 7th-day post-inoculation (S1 Fig). The experiment was repeated once under the same growing conditions. The average reaction score of each accession from the two experiments was used for GWAS analysis (S2 Table).
GWAS analysis
Genome-wide association for resistance to PTR race 5 was conducted using R package GAPIT (Genome Association and Prediction Integrated Tool) [88]. Three linear models, namely GLM (generalized linear model), MLM (mixed linear model), and CMLM (compressed mixed linear model), were evaluated. GLM is based on the least square fixed effects [88], whereas, MLM includes both fixed and random effects. The fixed effects in our case were the SNP effect and population structure (Q), and the random effect was for relatedness among the individuals (kinship). MLM model is mathematically denoted as:
Where, “y” is the vector of phenotypic values (categorical values in our case), “β” is the vector containing fixed effects namely SNP effects and Q, “u” is the vector of the random effects, which in our case is random genetic effects from multiple background QTL that were not controlled by markers (kinship). “X” and “Z” are known incidence matrixes for corresponding vectors. Kinship matrix was calculated using GAPIT’s kinship algorithm–VanRaden method [89] and Q matrix was obtained using principal component analysis [90]. CMLM is an extension of MLM, in which the individuals were clustered into groups using the group-based kinship matrix rather than individual based [91]. We primarily focused on MLM. The markers with a p-value < 1.0 ×10−3 or log10 (p-value) > 3 were considered to be significantly associated with the trait. We performed 5-fold jackknife validation to confirm the accuracy of significant marker-trait association (MTA) [92]. Briefly, the entire panel of 160 accessions was divided into five random sub-groups and four of them were used for association analysis, each time leaving one random group out. Results were also compared with the results from TASSEL 5.0 [77].
Comparative analysis of rye and wheat
To study the synteny between wheat and rye genomes, specifically for genomic regions conferring resistance against PTR race 5 in rye, a comparative analysis was conducted between the two genomes. Flanking sequence (150 base pair) of each 4,037 SNPs including the candidate SNPs identified from MTAs were retrieved from the rye reference genome. A 300bp long sequence for each SNP was compared with IWGSC wheat genome assembly RefSeq v1.0 [93] using BLASTn (E-value cut off of 1E-50 with an identity higher than 75%.) [94]. The comparative analysis results were visualized using a Perl based software Circos [95].
Results
Genotype-by-sequencing for genome-wide SNP discovery
We obtained a total of 178,598,329 sequence reads from a GBS library prepared from 178 rye accessions. Using the reference-based pipeline, 27,882 SNPs with 80% or fewer missing genotypes were identified (NCBI SRA accession: PRJNA512245). On an average, each chromosome has 4,000 SNPs (Table 1), with maximum (5,505) on chromosome 5R and minimum (2,536) on the chromosome 6R (S4 Table). To keep only the most informative SNPs, we removed 7,113 indel markers. A total of 4,037 high-quality SNPs that have less than 20% missing genotypes, less than 10% heterozygotes and above 5% MAF (minimum allele frequency) were used for further analysis. The final selected set of SNPs showed a similar trend of distribution on the seven chromosomes with an average of 577 SNPs per chromosome, maximum of 734 SNPs on chromosome 5R and a minimum of 358 SNPs on chromosome 6R (Table 1).
Table 1. Number and chromosome location of SNPs discovered by genotyping-by-sequencing of 178 rye accessions.
| Chromosome | Total SNPs | Filtered SNPs* |
|---|---|---|
| 1R | 3,468 | 504 |
| 2R | 3,914 | 600 |
| 3R | 3,916 | 605 |
| 4R | 5,505 | 685 |
| 5R | 4,774 | 734 |
| 6R | 2,536 | 358 |
| 7R | 3,892 | 551 |
| Total | 28,005 | 4,037 |
*Number of SNPs with 20% or less missing genotypes, heterozygotes less than 10% and MAF >5%
Genetic variability in the rye germplasm collection
The average PIC value for the 4,037 SNPs present in 160 S. cereale subsp. cereale accessions was 0.26, ranging from 0.09 to 0.5. About 38% SNPs had PIC values ranging from 0.1 to 0.2, 26% SNPs had PIC values of 0.2 to 0.3, 19% had PIC values of 0.3 to 0.4, and only 14% had PIC values 0.4 to 0.5. The PIC values for SNPs on each chromosome followed the similar pattern of distribution. Average PIC values were 0.25 for 6R, 7R, and 4R; 0.26 for 2R; and 0.27 for 1R, 3R, and 5R (S2 Fig). The average I-index for 4,037 SNPs in 160 S. cereale subsp. cereale accessions was 0.48. Among 18 accessions of wild species, average PIC value and I-index were 0.25 and 0.57, respectively.
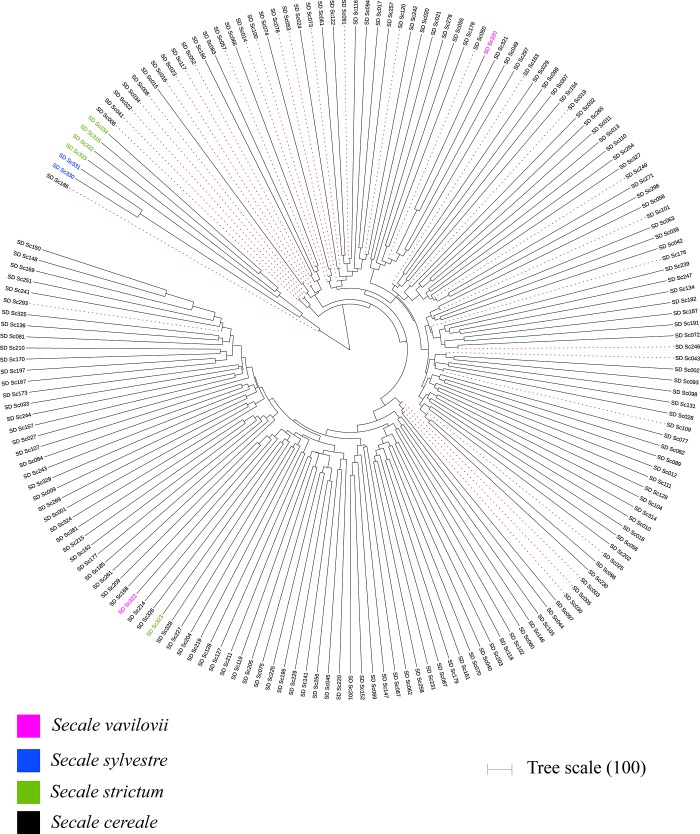
The average percentage dissimilarity (GD) among the entire panel of S. cereale subsp. cereale was 0.48, ranging from 0.26 to 0.63. The lowest GD (0.26) was found between the accessions SD_Sc150 and SD_Sc148, and the highest GD (0.63) was found between SD_Sc195 and SD_Sc186. Average GD for individual chromosomes ranged from 0.46 to 0.49 (S3 Fig). The average GD among wild species (18 accessions) was 0.51, ranging from 0.15 to 0.66. Among the wild species, SD_Sc330 (S. sylvestre) and SD_Sc322 (S. vavilovii) were the most distant accessions, and SD_Sc330 (S. sylvestre) and SD_Sc331 (S. sylvestre) were the most similar accessions with 0.66 and 0.15 GD, respectively. GD matrix-based farthest Neighbor-joining phylogenetic tree (Fig 2 & S5 Fig) accurately clustered the three species, S. cereale, S. strictum, and S. sylvestre, into three different clusters, except for SD_Sc323, the only spring type accession of S. strictum, which falls in a cluster of S. cereale. On the contrary, S. vavilovii clades were found scattered within a cluster of S. cereale. The spring type accessions of S. vavilovii (SD_Sc322) was found in the same cluster as spring type accession of S. strictum. S. sylvestre and S. strictum were found to be closer to each other as compared to S. cereale.
Fig 2. Pairwise dissimilarity-based neighbor-joining tree.
Smaller diverse representative set (red-dash clades) represents all the major clusters of Secale cereale subsp. cereale. S. strictum (green) and S. sylvestre (blue) and S. vavilovii (pink) are also shown.
Population structure and principal component analysis (PCA)
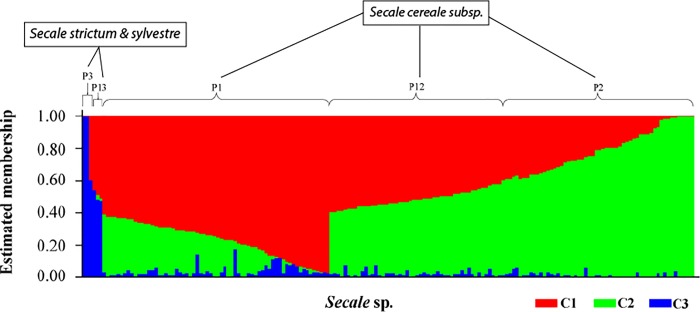
Bayesian clustering (STRUCTURE) analysis was performed on the 178 Secale sp. accessions and the estimated likelihood [LnP (D)] was found to be greatest at K = 3, suggesting three major populations that explain most of the genetic variation (Fig 3). Among all the accessions, 67% (120) belongs to one of the three unique populations (P1, P2, and P3) with > 60% ancestry contributed by anyone corresponding cluster. The populations P1, P2, and P3 contain 66, 51, and three accessions, respectively. Other 32% (58) of the accessions have admixtures in their ancestries (> 20% contribution by two clusters), in which 55 accessions in P12 have ancestries of both C1 and C2, whereas, P13 has only three accessions having ancestries of both C1 and C3. No accession shared significant ancestry (> 20%) between C2 and C3. Accessions of S. cereale subsp. were mainly found in P1, P2, and P12, whereas, P3 and P13 consisted of S. strictum and S. sylvestre.
Fig 3. Structure analysis on (K = 3) 178 Secale sp. accessions.
Y-axis represents the estimated membership of individuals from populations and X-axis represents 178 Secale sp. accessions. Accessions are ordered according to the population ancestry.
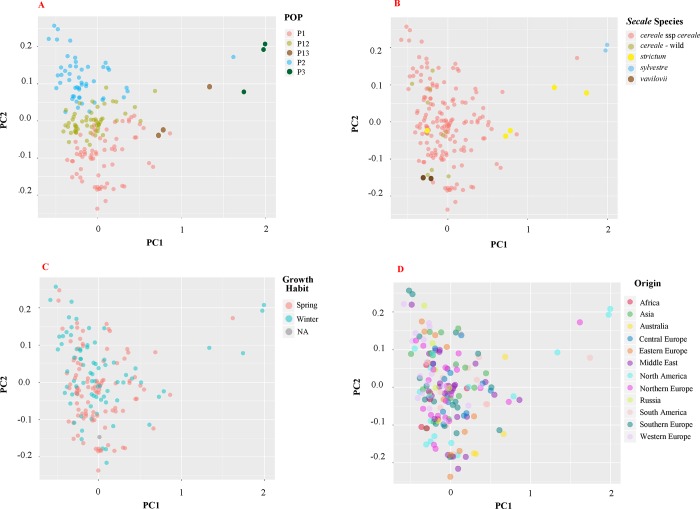
GD-based PCA results were basically consistent with the model-based population structure (Fig 4A). First and second PCA explained 40% and 3% of the genetic diversity, respectively. Three main populations (P1, P2, and P3) are clearly separated. The two admixtures (P12 and P23) lie between the corresponding populations with which they share ancestry. P3 mostly consist of wild species of S. strictum and S. sylvestre and is separated from rest of the evaluated accessions (Fig 4B). One accession of S. strictum was found in the population of S. cereale subsp. Interestingly, this accession is the only spring type accession of S. strictum. We also found S. vavilovii accessions mixed with the S. cereale. Correlation between genetic clusters and growth habit (spring vs winter, Fig 4C) or geographic origin (Fig 4D) was not observed. Geographic regions were divided according to Bolibok-Bragoszewska et al. (2014), in which Europe is divided into five regions: east, west, south, north, and central; and other countries are combined into corresponding broad geographic regions such as Middle East, Asia, South America, North America, Australia, and Russia [21].
Fig 4. Principal component analysis (PCA) on 178 Secale sp. accessions to show the relationship of genetic clusters with population structure.
(A), Secale species investigated (B), spring or winter type habit (C), and the geographic origin (D).
Selection of the smaller diverse representative set of rye
A total of 32 accessions were extracted as the smaller diverse representative set from 160 accessions of S. cereale subsp. cereale (PIC = 0.2518). The smaller diverse representative set contains only 20% of the accessions evaluated, but it covered 99% of the allelic diversity in the entire set. The smaller diverse representative set includes all the main clusters in NJ tree, with a minimum of one accession from each cluster (Fig 2). Accessions within a cluster are closer to each other as compared to accessions between clusters. The smaller diverse representative also captured a large portion of the geographic diversity (27 countries) of the entire collection (70 countries) of S. cereale subsp. cereale to represent the major geographic regions (Fig 1). The average PIC value and I-index of the smaller diverse representative set are not significantly (p < 0.01) different from the original whole set (Table 2). Average GD is significantly (p < 0.01) higher among accessions within the smaller diverse representative as compared to the original whole set (Table 2).
Table 2. Comparison of the diversity indices between smaller diverse representative set and geographically diverse set of 160 accessions of Secale cereale subsp. cereale.
| Size | Average PIC | Average I-index† | Average GD‡ | |
|---|---|---|---|---|
| Geographically diverse Set | 160 | 0.26 | 0.60 | 0.48 |
| Smaller representative set | 32 | 0.25 | 0.59 | 0.51 |
| T-test (p-value) | 0.02 | 0.11 | 1.90e-90* |
†Shannon’s diversity index
‡Pairwise genetic dissimilarity
*Significant at α <0.01.
Reactions of rye accessions to Pyrenophora tritici-repentis race 5
A set of 178 accessions of S. cereale were evaluated for resistance to tan spot (PTR race 5), however, only 160 accessions of S. cereale subsp. cereale were used for GWAS analysis. We observed a range of responses to PTR race 5 inoculations with 31.8% (51) resistant accessions (R—category 1), 26.9% (43) moderately resistant accessions (MR—category 2), 24.4% (39) moderately susceptible (MS—category 3), and 16.8% (27) of susceptible accessions (S—categories 4 and 5) (S4 Fig). As expected, the resistant check (Salamouni) showed resistant (Score—1) response and the susceptible check (6B662) produced chlorosis reaction with a score of 4 to 5. All these results were consistent in both experiments.
Marker-trait association (MTA) for tan spot (PTR race 5) resistance in rye
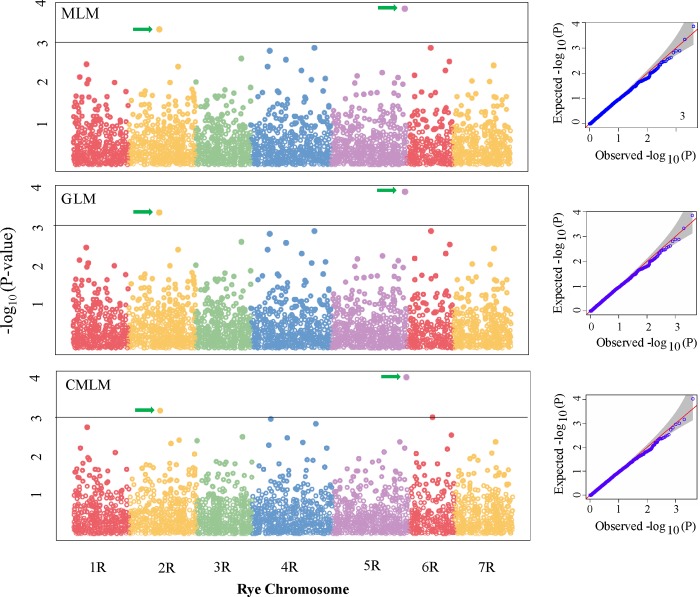
Among the tested linear models, we focused on MLM, because of obvious kinship and population structure detected for the population. Two genomic regions were associated with tan spot (PTR race 5) resistance in rye, with one region on chromosome 2R (QTs.sdsu-2R) and the other on 5R (QTs.sdsu-5R). The SNPs “S5R_16433036” (p = 1.4 × 10−4) on chromosome 5R and “S2R_6856816” (p = 4.5 × 10−4) on chromosome 2R explained 13.1% and 11.6% of the phenotypic variation, respectively (Fig 5). The two SNPs associated with tan spot resistance identified with the MLM algorithm were consistent with the results derived from GLM, and CMLM algorithms (Fig 5). The candidate SNPs were also validated using 5K jackknife approach, and both were consistent in the five repetitions with a p-value ≤ 1.0 ×10−3.
Fig 5. Genome-wide association scan for tan spot (PTR race 5) resistance in rye.
Three different model-based Manhattan plots represent–log10 (p-value) for SNPs distributed across all seven chromosomes of rye. Y-axis:–log10 (p-value) and x-axis: rye chromosomes. The dashed line stands as a threshold for significant markers with–log10 (p-value) of > 3 which correspond to a p-value <1 × 10−3. The arrows pointed two significant SNPs. On the right side of each model, Quantile-Quantile (QQ) plots represent expected null distribution of p-values vs observed p-values.
Comparative analysis of rye and wheat
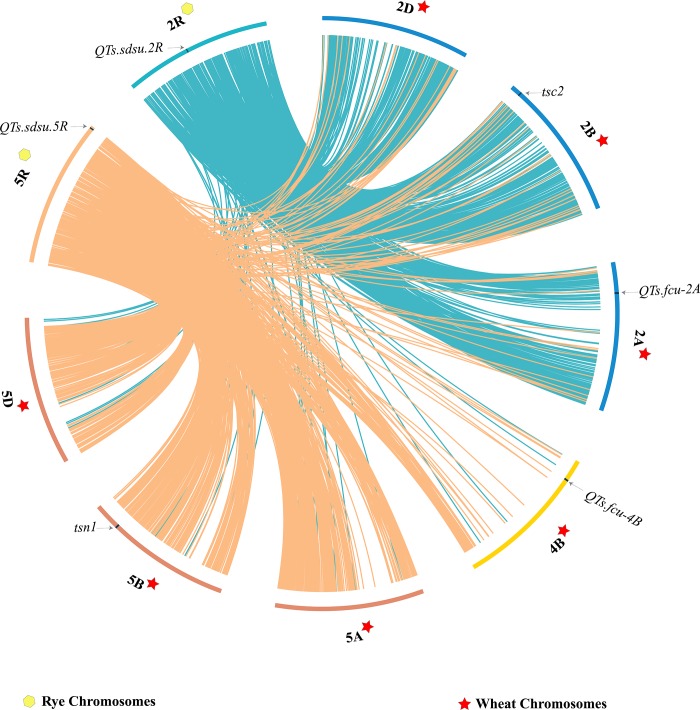
Syntenic analysis was conducted to compare the marker sequences of the identified QTL regions between wheat and rye. The SNPs flanking QTS.sdsu-2R had a hit on wheat group 2 which has a tan spot (PTR race 5) insensitivity gene (tsc2) on chromosome 2B and a QTL (QTs.fcu-2A) on chromosome 2A (Fig 6). S5R_16433036 in the QTs.sdsu-5R region showed a hit on a small segment on the terminal (tip) region of the long arm of wheat chromosome 4B although 5R is mainly syntenic with group 5 of wheat (Fig 6). No tan spot related QTL has been reported on the long arm of 4B, however, QTs.fcu-4B has been reported on the chromosome arm 4BS. Comparative analysis between rye genome (7 chromosomes) and their corresponding wheat homeologous groups (group 1–7) showed a general trend of synteny between the two genomes (S6 Fig). Majority of chromosomes 1R, 2R, 3R, and 5R are syntenic to wheat corresponding homeologous groups (1, 2, 3, and 5), respectively. However, blocks of rye chromosome 4R showed synteny with wheat groups 4, 6, and 7. Chromosome 6R is syntenic to wheat groups 6 and 3, because of fewer markers were found on 6R, the synteny with 6R was not very clear. Chromosome 7R shared syntenic blocks with wheat groups 5, 4, and 7.
Fig 6. Circular genome data visualization of synteny between wheat homoeologous groups (Group 2 – 2A, 2B, & 2D; Group 5 – 5A, 5B, & 5D; chromosome 4B) and rye chromosomes (2R and 5R) harboring tan spot (PTR race 5) resistance QTLs discovered in our study.
Each chromosome clockwise–short arm to long arm. QTs.sdsu-5R and QTs.sdsu-2R are shown on their corresponding rye chromosomes. QTs.sdsu-2R has a hit on wheat chromosome group 2 which harbors tan spot insensitivity gene—tsc 2 (2B) and tan spot resistance QTL (QTs.fcu-2A) (2A). QTs.sdsu-5R hits a small segment on wheat chromosome arm 4BL which harbors no tan spot related QTL, however, QTs.fcu-4B has been reported on the chromosome arm 4BS in wheat.
Discussion
Rye genome coverage by GBS-SNPs
Assessing the genetic diversity in germplasm resources using DNA markers can help in better exploitation of germplasm for crop improvement. In rye, several diversity studies have been conducted using a limited number of DNA markers ranging from few dozens of markers [5,24,27] to hundreds [28,29,31], probably due to laborious genotyping methods and technological limitations. Furthermore, chromosomal positions of these markers were not reported. To address this issue of anonymous marker positions and low genome coverage, one study has reported using 1,054 DArT markers to cover seven chromosomes of rye [21]. In the present study, we employed more than 4000 GBS-SNPs that provided much better coverage of the genome than in all the previous studies.
To our knowledge, this is the first report of double enzyme digestion-based GBS used in rye. GBS is a next-generation-sequencing based method that generates a large number of SNPs and shows advantages for high diversity species like rye. We found more than 4,000 polymorphic SNPs distributed evenly across all the seven chromosomes of rye except chromosome 6R, which has lower number (358) than the average number of SNPs per chromosome (576) (Table 1). With GBS, the number of SNPs discovered in a genomic region is directly correlated to the level of its genetic diversity [96], This suggests that chromosome 6R is likely less diverse than the other rye chromosomes, which is in line with several previous studies [1,2,21,30].
Genetic diversity in rye
Among 178 accessions investigated in this study, 160 are cultivated accessions of S. cereale subsp. cereale that cover most of the geographic diversity of the cultivated rye. In this rye collection, the average PIC value for all the SNPs is 0.26, which is lower than several previously reported values [21, 96–98). High PIC values from previous studies might be due to the use of carefully and deliberately selected markers [97], or due to the use of multi-allelic markers as for multi-allelic markers PIC values range from 0 to 1 but 0 to 0.5 for bi-allelic markers such as SNPs. However, lower PIC of SNPs can be overweighed by their enormous number and genome-wide distribution, thus giving a similar or even better estimation of genetic diversity. The similar PIC values across chromosomes in this study indicate that polymorphic SNPs were evenly distributed on all the seven rye chromosomes.
Average GD value among S. cereale subsp. cereale was 0.48 with a range from 0.26 to 0.63, which is comparable with other studies in rye [5,21,98]. Accessions SD_Sc195 and SD_Sc186 have highest dissimilarity index of 0.63. These accessions being most diverse may be of future interest for exploiting heterosis. Average GD among wild species (0.51) was higher as compared to cultivated species, which is in accordance with the expectation that wild species conserve larger diversity [21]. Therefore, wild species can further be exploited to infuse diversity into cultivated germplasm. SD_Sc330 (S. sylvestre) and SD_Sc322 (S. vavilovii) were the most diverse accessions.
Results among all three clustering approaches (Bayesian clustering, PCA, and Neighbor-Joining clustering) were consistent. Bayesian clustering predicted three populations: P1, P2, and P3. P1 and P2 both consisted of S. cereale subsp. and S. vavilovii accessions; P3 consisted of S. sylvestre and S. strictum accessions, reported in previous studies as well [30,98]. These clusters were also seen with PCA. Genome composition of S. sylvestre was 100% from the P3 population, whereas, S. strictum had about 10 to 20% from P1. Shared ancestry among some of the accessions of S. strictum and S. cereale subsp. group (P1) supports the hypothesis that S. strictum is the potential ancestor of S. cereale [3,99–101]. Unlike other wild Secale sp., S. vavilovii accessions were found in the clusters of S. cereale subsp, which is in accordance with previous reports [30,98], suggesting its classification may need to be revisited. Wild species of S. cereale cannot be separated out of the clusters of the S. cereale subsp. cereale in our study similar to previous studies [21], suggesting an active gene transfer among these species.
After reviewing the geographic origin of accessions in different genetic clusters, we did not find any correlation between genetic cluster and geographic origin. This may be due to the sharing of the common genetic background among the accessions being analyzed in each study as it is also reported by other independent studies [5,21,27,29]. It has been reported that vernalization requirement can lead to population divergence in rye [98], triticale [102], and wheat [103]. We examined growth habit (winter vs spring) of the genetic clusters and did not see a substantial association between clusters and growth habit or vernalization requirement.
Use of the smaller diverse set in representing the geograhically diverse set of cultivated rye
Mini core sets have been established for number of crops including wheat [104,105], rice [106], maize [107], soybean [108], and rye [109]. Adding one more collection to that list, we identified a smaller diverse representative set of 32 accessions representing genetic (99% alleles) and geographic diversity (all major regions) of 160 accessions of S. cereale subsp. cereale. The PIC value and I-index of the smaller diverse representative is comparable to those of the whole set while average GD is significantly higher than the whole set. Thus, the smaller diverse representative carries similar genetic information as the whole set, which can be easily and efficiently used for rye gene mining and transferring important genes into X. Triticosecale and wheat. Also, our smaller diverse representative set contains only one-fifth of the accessions from the original set but retain 99% of the allelic diversity, thus demonstrating that germplasm banks can significantly reduce the germplasm conservation cost without the loss of genetic information if they genotype entire available germplasm and develop core diversity sets.
Identification of putative genomic regions conferring tan spot (PTR race 5) resistance
Rye is known for its resilience to the abiotic and biotic stress tolerance [109] and it has contributed a number of important genes into wheat germplasm [9,11,12,110]. Previously, we reported that rye might carry resistance genes to tan spot [111], but QTLs have not been defined. Using a GWAS panel of 160 accessions of S. cereale subsp. cereale we identified two putative loci conferring resistance to PTR race 5. The two SNPs (S2R_6856816” on chromosome 2 and “S5R_16433036” on chromosome 5) collectively explained 24.7% of the phenotypic variation.
Comparative analysis between rye and wheat revealed that the significant marker linked to tan spot resistance on chromosome 2R is homologous to chromosome group 2 in wheat. Interestingly, chromosome group 2 of wheat carries many tan spot resistance or insensitivity related genes including tsc2 (PTR race 5) on chromosome 2B, and QTs.fcu-2A (PTR race 5) and another resistance QTL (PTR race 1) on chromosome 2A [112–114]. However, the incomplete genome assembly of rye and no information on the sensitivity of rye accessions to Ptr ToxB, we don't know if the QTL on 2R is syntenic to the wheat gene tsc2. The QTL QTs.sdsu-5R had a most significant hit on wheat chromosome 4B. Though most of the chromosome 5R of rye is syntenic to chromosome group 5 of wheat, a small 5R segment hits a region on the long arm of chromosome 4B which includes our candidate SNP. Though no tan spot resistance QTL is reported so far on the terminal segment of chromosome 4BL (region hit by QTs.sdsu-5R), however, a QTL, QTs.fcu-4B, has been reported on chromosome arm 4BS [115]. Thus, QTs.sdsu-5R may harbor novel genes for PTR race 5 resistance. The QTLs identified in our study can be easily transferred using linked SNPs into wheat and triticale for improving tan spot resistance in these crops. Using similar approach, genes/QTLs controlling agronomic traits and tolerance to biotic and abiotic stress can be mapped in rye and transferred to triticale and wheat.
Conclusions
Our study reports the rye genetic diversity analysis using GBS and identifies a smaller diverse representative set of 32 accessions that retains ~99% of the allelic diversity. This set of 32 accessions can be used as parents for rye, triticale, and wheat improvement. Genetic clustering was neither linked with geographic origins and nor with growth habit, suggesting individuals shared a common genetic background due to germplasm exchange and no major genomic changes happened due to vernalization requirements. Further, using GWAS we identified two genomic regions conferring resistance to tan spot (PTR race 5) in rye and the tightly linked SNPs S5R_16433036 (QTs.sdsu-5R) and S2R_6856816 (QTs.sdsu-2R) can be used for marker-assisted selection of the tan spot resistance genes.
Supporting information
1 –Resistant wheat Salamouni (check), 2 –Resistant rye, 3 –Moderately susceptible rye, 4 –Susceptible rye.
(TIF)
X-axis: PIC value and Y-axis rye chromosomes. Violin plots show the density distribution of SNPs for the chromosome corresponding PIC values. Box plots represent first and third quartiles. Horizontal white bars are corresponding median PIC value and yellow dot stands for average PIC value.
(TIF)
X-axis: GD–pairwise genetic dissimilarity–percentage and Y-axis rye chromosomes. Violin plots show the density distribution of pairwise dissimilarities values. Box plots represent first and third quartiles. Horizontal white bars are corresponding median pairwise dissimilarity and yellow dot stands for average pairwise dissimilarity corresponding to each chromosome.
(TIF)
1 = resistant, 2 = moderately resistant, 3 = moderately susceptible and 4 & 5 = susceptible.
(TIF)
Smaller diverse representative set (red-dash clades) represents all the major clusters of Secale cereale subsp. cereale. S. strictum (green) and S. sylvestre (blue) and S. vavilovii (pink) are also shown.
(TIF)
Black bars on rye chromosomes denotes SNP density (SNPs/10Mb) on the rye chromosomes. QTs.sdsu-5R and QTs.sdsu-2R are presented adjacent to their corresponding rye chromosomes. Red italics denotes the mapped tan spot insensitivity genes (tsn1, tsc1, and tsc2) and resistance genes (tsr2, tsr3, tsr4, and tsr5) adjacent to their corresponding wheat chromosomes. Wheat is an allohexaploid species (2n = 6x = 42) with three (A, B, and D) homoeologous chromosome sets and rye is a diploid species (2n = 2x = 14).
(TIF)
These accessions represent 70 different countries around the globe.
(DOCX)
For each accession, its country of origin, corresponding PI number, species, population, and response to tan spot (PTR race 5) is given. Populations are based on structure results.
(DOCX)
(DOCX)
Information on 4,037 GBS SNPs and genotype of 178 accessions at each of these SNP locations in HapMap format.
(TXT)
Acknowledgments
The authors would like to thank the South Dakota Agriculture Experimental Station (Brookings, SD, USA) for providing the resources to conduct the experiments. This project was collectively funded by the USDA hatch project SD00H538-15, USDA-CAP 2016-68004-24768 and South Dakota Wheat Commission 3X7262 and 3X8258. The funders had no role in the study design, data collection, and analysis, decision to publish or preparation of the manuscript.
Data Availability
All the raw sequence data has been submitted to NCBI SRA under SRA accession: PRJNA512245. In addition, the SNP data genotyping with 4,037 SNPs from all 178 accessions utilized in our study has been made available as S4 Table.
Funding Statement
SS - This project was collectively funded by the USDA hatch project SD00H538-15, USDA-CAP 2016-68004-24768 and South Dakota Wheat Commission 3X7262 and 3X8258. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bauer E, Schmutzer T, Barilar I, Mascher M, Gundlach H, Martis MM, et al. Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 2016; 10.1111/tpj.13436 [DOI] [PubMed] [Google Scholar]
- 2.Martis MM, Zhou R, Haseneyer G, Schmutzer T, Vrána J, Kubaláková M, et al. Reticulate Evolution of the Rye Genome. Plant Cell. 2013;25: 3685–3698. 10.1105/tpc.113.114553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sencer HA, Hawkes JG. On the origin of cultivated rye. Biol J Linn Soc. 1980;13: 299–313. 10.1111/j.1095-8312.1980.tb00089.x [DOI] [Google Scholar]
- 4.Bushuk W. Rye production and uses worldwide. Cereal Foods World. 2001;46: 70–73. [Google Scholar]
- 5.Shang HY, Wei YM, Wang XR, Zheng YL. Genetic diversity and phylogenetic relationships in the rye genus Secale L. (rye) based on Secale cereale microsatellite markers. Genet Mol Biol. 2006; 10.1590/S1415-47572006000400018 [DOI] [Google Scholar]
- 6.Andersson R, Fransson G, Tietjen M, Åman P. Content and molecular-weight distribution of dietary fiber components in whole-grain rye flour and bread. J Agric Food Chem. 2009;57: 2004–2008. 10.1021/jf801280f [DOI] [PubMed] [Google Scholar]
- 7.El Khoury D, Cuda C, Luhovyy BL, Anderson GH. Beta glucan: health benefits in obesity and metabolic syndrome. J Nutr Metab. Hindawi Publishing Corporation; 2011;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ammar K., Mergoum M., Rajaram S. The history and evolution of triticale. Triticale Improv Prod. 2004; 1–10. doi: 92-5-105182-8 [Google Scholar]
- 9.Crespo-Herrera LA, Garkava-Gustavsson L, Åhman Inger. A systematic review of rye (Secale cereale L.) as a source of resistance to pathogens and pests in wheat (Triticum aestivum L.). Hereditas. Hereditas; 2017;154: 1–9. 10.1186/s41065-016-0023-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mago R, Spielmeyer W, Lawrence GJ, Lagudah ES, Ellis JG, Pryor A. Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet. 2002;104: 1317–1324. 10.1007/s00122-002-0879-3 [DOI] [PubMed] [Google Scholar]
- 11.Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. Springer; 1996;91: 59–87. [Google Scholar]
- 12.Mohler V, Hsam SLK, Zeller FJ, Wenzel G. An STS marker distinguishing the rye-derived powdery mildew resistance alleles at the Pm8/Pm17 locus of common wheat. Plant Breed. Wiley Online Library; 2001;120: 448–450. [Google Scholar]
- 13.Friebe B, Hatchett JH, Sears RG, Gill BS. Transfer of Hessian fly resistance from ‘Chaupon’rye to hexaploid wheat via a 2BS/2RL wheat-rye chromosome translocation. Theor Appl Genet. Springer; 1990;79: 385–389. 10.1007/BF01186083 [DOI] [PubMed] [Google Scholar]
- 14.Carver BF, Rayburn AL. Comparison of related wheat stocks possessing 1B or 1RS. 1BL chromosomes: agronomic performance. Crop Sci. Crop Science Society of America; 1994;34: 1505–1510. [Google Scholar]
- 15.McKendry AL, Tague DN, Miskin KE. Effect of 1BL. 1RS on agronomic performance of soft red winter wheat. Crop Sci. Crop Science Society of America; 1996;36: 844–847. [Google Scholar]
- 16.Kim W, Johnson JW, Baenziger PS, Lukaszewski AJ, Gaines CS. Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci. 2004;44: 1254–1258. 10.2135/cropsci2004.1254 [DOI] [Google Scholar]
- 17.Saulescu NN, Ittu G, Ciuca M, Ittu M, Serban G, Mustatea P, et al. Transferring useful rye genes to wheat, using triticale as a bridge. Czech J Genet Plant Breed. 2011;47: 56–62. [Google Scholar]
- 18.Von Mark VC, Kilian A, Dierig DA. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS One. Public Library of Science; 2013;8: e64062 10.1371/journal.pone.0064062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upadhyaya HD, Ortiz R. A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor Appl Genet. 2001;102: 1292–1298. 10.1007/s00122-001-0556-y [DOI] [Google Scholar]
- 20.YAO Q, FAN P, ZOU S. Constructing a Core Collection for Maize (Zea mays L.) Landrace from Wuling Mountain Region in China. Agric Sci China. Chinese Academy of Agricultural Sciences; 2008;7: 1423–1432. 10.1016/S1671-2927(08)60398-3 [DOI] [Google Scholar]
- 21.Bolibok-Brągoszewska H, Targońska M, Bolibok L, Kilian A, Rakoczy-Trojanowska M. Genome-wide characterization of genetic diversity and population structure in Secale. BMC Plant Biol. 2014;14: 184 10.1186/1471-2229-14-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartoš J, Paux E, Kofler R, Havránková M, Kopecký D, Suchánková P, et al. A first survey of the rye (Secale cereale) genome composition through BAC end sequencing of the short arm of chromosome 1R. BMC Plant Biol. 2008;8: 95 10.1186/1471-2229-8-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flavell RB, Bennett MD, Smith JB, Smith DB. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem Genet. Springer; 1974;12: 257–269. [DOI] [PubMed] [Google Scholar]
- 24.Bolibok H, Rakoczy-Trojanowska M, Hromada A, Pietrzykowski R. Efficiency of different PCR-based marker systems in assessing genetic diversity among winter rye (Secale cereale L.) inbred lines. Euphytica. 2005;146: 109–116. 10.1007/s10681-005-0548-0 [DOI] [Google Scholar]
- 25.Burger JC, Lee S, Ellstrand NC. Origin and genetic structure of feral rye in the western United States. Mol Ecol. 2006;15: 2527–2539. 10.1111/j.1365-294X.2006.02938.x [DOI] [PubMed] [Google Scholar]
- 26.Fu S, Tang Z, Ren Z, Zhang H, Yan B. Isolation of rye-specific DNA fragment and genetic diversity analysis of rye genus Secale L. using wheat SSR markers. J Genet. 2010;89: 489–492. 10.1007/s12041-010-0070-6 [DOI] [PubMed] [Google Scholar]
- 27.Isik Z, Parmaksiz I, Coruh C, Geylan-Su YS, Cebeci O, Beecher B, et al. Organellar genome analysis of rye (Secale cereale) representing diverse geographic regions. Genome. 2007;50: 724–34. 10.1139/g07-052 [DOI] [PubMed] [Google Scholar]
- 28.Matos M, Pinto-Carnide O, Benito C. Phylogenetic Relationships among Portuguese Rye Based on Isozyme, RAPD and ISSR Markers. Hereditas. Munksgaard International Publishers; 2004;134: 229–236. 10.1111/j.1601-5223.2001.00229.x [DOI] [PubMed] [Google Scholar]
- 29.Chikmawati T, Skovmand B, Gustafsson JP. Phylogenetic relationships among Secale species revealed by amplified fragment length polymorphisms. Genoma. 2005;48: 792–801. 10.1139/g05-043 [DOI] [PubMed] [Google Scholar]
- 30.Bolibok-Bragoszewska H, Heller-Uszyńska K, Wenzl P, Uszyński G, Kilian A, Rakoczy-Trojanowska M. DArT markers for the rye genome—genetic diversity and mapping. BMC Genomics. 2009;10: 578 10.1186/1471-2164-10-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagenblad J, Oliveira HR, Forsberg NEG, Leino MW. Geographical distribution of genetic diversity in Secale landrace and wild accessions. BMC Plant Biol. BioMed Central; 2016;16: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X-F, Wanous MK, Houchins K, Milla MAR, Goicoechea PG, Wang Z, et al. Molecular linkage mapping in rye (Secale cereale L.). Theor Appl Genet. Springer; 2001;102: 517–523. [Google Scholar]
- 33.ilczarski Pawełand Bolibok-Br\kagoszewska H, Myśków B, Stojałowski S, Heller-Uszyńska K, Góralska M, Br\kagoszewski P, et al. A high density consensus map of rye (Secale cereale L.) based on DArT markers. PLoS One. Public Library of Science; 2011;6: e28495 10.1371/journal.pone.0028495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milczarski P, Hanek M, Tyrka M, Stoja??owski S. The application of GBS markers for extending the dense genetic map of rye (Secale cereale L.) and the localization of the Rfc1 gene restoring male fertility in plants with the C source of sterility-inducing cytoplasm. J Appl Genet. 2016;57: 439–451. 10.1007/s13353-016-0347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korzun V, Malyshev S, Kartel N, Westermann T, Weber WE, Börner A. A genetic linkage map of rye (Secale cereale L.). Theor Appl Genet. Springer; 1998;96: 203–208. [Google Scholar]
- 36.Börner A, Korzun V, Voylokov A V, Worland AJ, Weber WE. Genetic mapping of quantitative trait loci in rye (Secale cereale L.). Euphytica. Springer; 2000;116: 203–209. [Google Scholar]
- 37.Świ\kecka S, Berdzik M, Myśków B. Genetic mapping of the ScHd1 gene in rye and an assessment of its relationship with earliness per se and plant morphology. J Appl Genet. Springer; 2014;55: 469–473. 10.1007/s13353-014-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milczarski Pawełand Masojć P, Krajewski Pawełand Stochmal A, Kowalczyk M, Angelov M, Ivanova V, Schollenberger M, et al. QTL mapping for benzoxazinoid content, preharvest sprouting, $α$-amylase activity, and leaf rust resistance in rye (Secale cereale L.). PLoS One. Public Library of Science; 2017;12: e0189912 10.1371/journal.pone.0189912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jannink J-L, Walsh B. Association mapping in plant populations Quant Genet genomics plant Breed. CAB International: New York, NY, USA; 2002; 59–68. [Google Scholar]
- 40.Huang X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. Nature Publishing Group; 2010;42: 961 10.1038/ng.695 [DOI] [PubMed] [Google Scholar]
- 41.Famoso AN, Zhao K, Clark RT, Tung C-W, Wright MH, Bustamante C, et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. Public Library of Science; 2011;7: e1002221 10.1371/journal.pgen.1002221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCouch SR, Wright MH, Tung C-W, Maron LG, McNally KL, Fitzgerald M, et al. Open access resources for genome-wide association mapping in rice. Nat Commun. 2016;7: 10532 10.1038/ncomms10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raboin L-M, Ballini E, Tharreau D, Ramanantsoanirina A, Frouin J, Courtois B, et al. Association mapping of resistance to rice blast in upland field conditions. Rice. Springer; 2016;9: 59 10.1186/s12284-016-0131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2: 467 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Liu H, Wu L, Warburton M, Yan J. Genome-wide association studies in maize: praise and stargaze. Mol Plant. Elsevier; 2017;10: 359–374. 10.1016/j.molp.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 46.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. The American Society of Human Genetics; 2012;90: 7–24. 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beló A, Zheng P, Luck S, Shen B, Meyer DJ, Li B, et al. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol Genet Genomics. Springer; 2008;279: 1–10. 10.1007/s00438-007-0289-y [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Luo X, Niu L, Xiao Y, Chen L, Liu J, et al. Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol Plant. Elsevier; 2017;10: 414–426. 10.1016/j.molp.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Li K, Hu X, Liu Z, Wu Y, Huang C. Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biol. BioMed Central; 2016;16: 227 10.1186/s12870-016-0919-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farfan IDB, De La Fuente GN, Murray SC, Isakeit T, Huang P-C, Warburton M, et al. Genome Wide Association Study for Drought, Aflatoxin Resistance, and Important Agronomic Traits of Maize Hybrids in the Sub-Tropics. Lukens L, editor. PLoS One. 2015;10: e0117737 10.1371/journal.pone.0117737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Shrestha R, Ding J, Zheng H, Mu C, Wu J, et al. Genome-Wide Association Study and QTL Mapping Reveal Genomic Loci Associated with Fusarium Ear Rot Resistance in Tropical Maize Germplasm. G3: Genes|Genomes|Genetics. 2016; 10.1534/g3.116.034561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy JK, Smith KP, Muehlbauer GJ, Chao S, Close TJ, Steffenson BJ. Association mapping of spot blotch resistance in wild barley. Mol Breed. 2010;26: 243–256. 10.1007/s11032-010-9402-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, et al. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012;12: 16 10.1186/1471-2229-12-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gawenda I, Thorwarth P, Günther T, Ordon F, Schmid KJ. Genome-wide association studies in elite varieties of German winter barley using single-marker and haplotype-based methods. Bürstmayr H, editor. Plant Breed. 2015;134: 28–39. 10.1111/pbr.12237 [DOI] [Google Scholar]
- 55.Bellucci A, Tondelli A, Fangel JU, Torp AM, Xu X, Willats WGT, et al. Genome-wide association mapping in winter barley for grain yield and culm cell wall polymer content using the high-throughput CoMPP technique. Perovic D, editor. PLoS One. Public Library of Science; 2017;12: e0173313 10.1371/journal.pone.0173313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinert S, Kortz A, Léon J, Naz AA. Genome-Wide Association Mapping in the Global Diversity Set Reveals New QTL Controlling Root System and Related Shoot Variation in Barley. Front Plant Sci. 2016;7 10.3389/fpls.2016.01061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan Y, Zhou G, Shabala S, Chen Z-H, Cai S, Li C, et al. Genome-Wide Association Study Reveals a New QTL for Salinity Tolerance in Barley (Hordeum vulgare L.). Front Plant Sci. Frontiers; 2016;7: 946 10.3389/fpls.2016.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wójcik-Jagła M, Fiust A, Kościelniak J, Rapacz M. Association mapping of drought tolerance-related traits in barley to complement a traditional biparental QTL mapping study. Theor Appl Genet. 2018;131: 167–181. 10.1007/s00122-017-2994-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmad I, Ali N, Ahmad H, Inamullah. Association Mapping of Root Traits for Drought Tolerance in Bread Wheat. Wheat Improvement, Management and Utilization. InTech; 2017. 10.5772/67242 [DOI] [Google Scholar]
- 60.Mwadzingeni L, Shimelis H, Rees DJG, Tsilo TJ. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS One. Public Library of Science; 2017;12: e0171692 10.1371/journal.pone.0171692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ain Q, Rasheed A, Anwar A, Mahmood T, Imtiaz M, Mahmood T, et al. Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Front Plant Sci. 2015;6 10.3389/fpls.2015.00743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tadesse W, Ogbonnaya FC, Jighly A, Sanchez-Garcia M, Sohail Q, Rajaram S, et al. Genome-Wide Association Mapping of Yield and Grain Quality Traits in Winter Wheat Genotypes. Wu R, editor. PLoS One. 2015;10: e0141339 10.1371/journal.pone.0141339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lozada DN, Mason RE, Babar MA, Carver BF, Guedira G-B, Merrill K, et al. Association mapping reveals loci associated with multiple traits that affect grain yield and adaptation in soft winter wheat. Euphytica. 2017;213: 222 10.1007/s10681-017-2005-2 [DOI] [Google Scholar]
- 64.Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet. 2014;127: 791–807. 10.1007/s00122-013-2257-8 [DOI] [PubMed] [Google Scholar]
- 65.Liu J, He Z, Rasheed A, Wen W, Yan J, Zhang P, et al. Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum L.). BMC Plant Biol. 2017;17: 220 10.1186/s12870-017-1167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi W, Hao C, Zhang Y, Cheng J, Zhang Z, Liu J, et al. A Combined Association Mapping and Linkage Analysis of Kernel Number Per Spike in Common Wheat (Triticum aestivum L.). Front Plant Sci. 2017;8 10.3389/fpls.2017.01412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, et al. Whole Genome Association Mapping of Plant Height in Winter Wheat (Triticum aestivum L.). Hernandez P, editor. PLoS One. 2014;9: e113287 10.1371/journal.pone.0113287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arruda MP, Brown P, Brown-Guedira G, Krill AM, Thurber C, Merrill KR, et al. Genome-Wide Association Mapping of Fusarium Head Blight Resistance in Wheat using Genotyping-by-Sequencing. Plant Genome. 2016;9: 0 10.3835/plantgenome2015.04.0028 [DOI] [PubMed] [Google Scholar]
- 69.Gurung S, Mamidi S, Bonman JM, Xiong M, Brown-Guedira G, Adhikari TB. Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS One. Public Library of Science; 2014;9: e108179 10.1371/journal.pone.0108179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kidane YG, Hailemariam BN, Mengistu DK, Fadda C, Pè ME, Dell’Acqua M. Genome-Wide Association Study of Septoria tritici Blotch Resistance in Ethiopian Durum Wheat Landraces. Front Plant Sci. 2017;8 10.3389/fpls.2017.01586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ayana GT, Ali S, Sidhu JS, Gonzalez-Hernandez JL, Turnipseed B, Sehgal SK. Genome-Wide Association Study for Spot Blotch Resistance in Hard Winter Wheat. Front Plant Sci. Frontiers; 2018;9: 926 10.3389/fpls.2018.00926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dinglasan E, Godwin ID, Mortlock MY, Hickey LT. Resistance to yellow spot in wheat grown under accelerated growth conditions. Euphytica. Springer Netherlands; 2016;209: 693–707. 10.1007/s10681-016-1660-z [DOI] [Google Scholar]
- 73.Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull Bot Soc Am. 1987;19: 11–15. [Google Scholar]
- 74.Mascher M, Wu S, Amand PS, Stein N, Poland J. Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley. PLoS One. Public Library of Science; 2013;8: e76925 10.1371/journal.pone.0076925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alipour H, Bihamta MR, Mohammadi V, Peyghambari SA, Bai G, Zhang G. Genotyping-by-Sequencing (GBS) Revealed Molecular Genetic Diversity of Iranian Wheat Landraces and Cultivars. Front Plant Sci. Frontiers; 2017;8: 1293 10.3389/fpls.2017.01293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. 2012;7 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 78.helmholtz Zentrum Munchen. PGSB Plant Genome and Systems Biology [Internet]. [cited 1 Jan 2016]. Available: http://pgsb.helmholtz-muenchen.de/plant/rye/gz/download/
- 79.Alheit K V, Maurer H, Reif JC, Tucker MR, Hahn V, Weissmann EA, et al. Genome-wide evaluation of genetic diversity and linkage disequilibrium in winter and spring triticale (x Triticosecale Wittmack). BMC Genomics. 2012;13: 235 10.1186/1471-2164-13-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherwin WB, Jabot F, Rush R, Rossetto M. Measurement of biological information with applications from genes to landscapes. Mol Ecol. 2006;15: 2857–2869. 10.1111/j.1365-294X.2006.02992.x [DOI] [PubMed] [Google Scholar]
- 81.Team RC, others. R: A language and environment for statistical computing. Citeseer; 2013; [Google Scholar]
- 82.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. Oxford University Press; 2004;20: 289–290. [DOI] [PubMed] [Google Scholar]
- 83.Müllner D. fastcluster: Fast hierarchical, agglomerative clustering routines for R and Python. J Stat Softw. Foundation for Open Access Statistics; 2013;53: 1–18. [Google Scholar]
- 84.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. Oxford University Press; 2016;44: W242—W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155: 945–959. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14: 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 87.Lamari L, Bernier CC. Evaluation of wheat lines and cultivars to tan spot [Pyrenophora tritici-repentis] based on lesion type. Can J Plant Pathol. Taylor & Francis; 1989;11: 49–56. [Google Scholar]
- 88.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. Oxford University Press; 2012;28: 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- 89.VanRaden PM, Olson KM, Wiggans GR, Cole JB, Tooker ME. Genomic inbreeding and relationships among Holsteins, Jerseys, and Brown Swiss. J Dairy Sci. Elsevier; 2011;94: 5673–5682. 10.3168/jds.2011-4500 [DOI] [PubMed] [Google Scholar]
- 90.Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. Public Library of Science; 2007;3: e4 10.1371/journal.pgen.0030004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore MA, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. Nature Publishing Group; 2010;42: 355 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kohavi R, others. A study of cross-validation and bootstrap for accuracy estimation and model selection. Ijcai. 1995. pp. 1137–1145. [Google Scholar]
- 93.Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (80-). American Association for the Advancement of Science; 2018;361: eaar7191. [DOI] [PubMed] [Google Scholar]
- 94.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. Elsevier; 1990;215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 95.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. Cold Spring Harbor Lab; 2009;19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6: 1–10. 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Varshney RK, Beier U, Khlestkina EK, Kota R, Korzun V, Graner A, et al. Single nucleotide polymorphisms in rye (Secale cereale L.): Discovery, frequency, and applications for genome mapping and diversity studies. Theor Appl Genet. 2007;114: 1105–1116. 10.1007/s00122-007-0504-6 [DOI] [PubMed] [Google Scholar]
- 98.Ma R, YLI-MATTILA T, Pulli S. Phylogenetic relationships among genotypes of worldwide collection of spring and winter ryes (Secale cereale L.) determined by RAPD-PCR markers. Hereditas. Wiley Online Library; 2004;140: 210–221. 10.1111/j.1601-5223.2004.01844.x [DOI] [PubMed] [Google Scholar]
- 99.Zohary D. Orgin of south-west Asiatic cereals: wheats, barley, oats and rye. Plant life south-west Asia. 1971; 235–263. [Google Scholar]
- 100.Khush GS, Stebbins GL. Cytogenetic and evolutionary studies in Secale. I. Some new data on the ancestry of S. cereale. Am J Bot. JSTOR; 1961; 723–730. [Google Scholar]
- 101.Bustos a De, Jouve N. Phylogenetic relationships of the genus Secale based on the characterisation of rDNA ITS sequences. Plant Syst Evol. 2002;235: 147–154. 10.1007/s00606-002-0215-z [DOI] [Google Scholar]
- 102.Niedziela A, Orłowska R, Machczyńska J, Bednarek PT. The genetic diversity of triticale genotypes involved in Polish breeding programs. Springerplus. Springer International Publishing; 2016;5: 355 10.1186/s40064-016-1997-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chao S, Dubcovsky J, Dvorak J, Luo M-C, Baenziger SP, Matnyazov R, et al. Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genomics. BioMed Central Ltd; 2010;11: 727 10.1186/1471-2164-11-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong YS, Cao YS, Zhang XY, Liu SC, Wang LF, You GX, et al. Establishment of candidate core collections in Chinese common wheat germplasm. J Plant Genet Resour. 2003;1: 1. [Google Scholar]
- 105.Hao C, Dong Y, Wang L, You G, Zhang H, Ge H, et al. Genetic diversity and construction of core collection in Chinese wheat genetic resources. Chinese Sci Bull. 2008;53: 1518–1526. 10.1007/s11434-008-0212-x [DOI] [Google Scholar]
- 106.Zhang H, Zhang D, Wang M, Sun J, Qi Y, Li J, et al. A core collection and mini core collection of Oryza sativa L. in China. Theor Appl Genet. 2011;122: 49–61. 10.1007/s00122-010-1421-7 [DOI] [PubMed] [Google Scholar]
- 107.Li Y, Shi Y, Cao Y, Wang T. Establishment of a core collection for maize germplasm preserved in Chinese National Genebank using geographic distribution and characterization data. Genet Resour Crop Evol. Springer; 2005;51: 845–852. [Google Scholar]
- 108.Guo Y, Li Y, Hong H, Qiu LJ. Establishment of the integrated applied core collection and its comparison with mini core collection in soybean (Glycine max). Crop J. Crop Science Society of China and Institute of Crop Sciences, CAAS; 2014;2: 38–45. 10.1016/j.cj.2013.11.001 [DOI] [Google Scholar]
- 109.Targońska M, Bolibok-Brągoszewska H, Rakoczy-Trojanowska M. Assessment of Genetic Diversity in Secale cereale Based on SSR Markers. Plant Mol Biol Report. 2016;34: 37–51. 10.1007/s11105-015-0896-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mago R, Spielmeyer W, Lawrence GJ, Lagudah ES, Ellis JG, Pryor A. Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet. 2002;104: 1317–1324. 10.1007/s00122-002-0879-3 [DOI] [PubMed] [Google Scholar]
- 111.Abdullah S, Sehgal SK, Glover KD, Ali S. Reaction of Global Collection of Rye (Secale cerealeL.) to Tan Spot andPyrenophora tritici-repentisRaces in South Dakota. plant Pathol J. The Korean Society of Plant Pathology; 2017;33: 229–237. 10.5423/PPJ.OA.12.2016.0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Friesen TL, Faris JD. Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor Appl Genet. Springer; 2004;109: 464–471. 10.1007/s00122-004-1678-9 [DOI] [PubMed] [Google Scholar]
- 113.Faris JD, Liu Z, Xu SS. Genetics of tan spot resistance in wheat. Theor Appl Genet. 2013;126: 2197–2217. 10.1007/s00122-013-2157-y [DOI] [PubMed] [Google Scholar]
- 114.Juliana P, Singh RP, Singh PK, Poland JA, Bergstrom GC, Huerta-Espino J, et al. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor Appl Genet. Springer Berlin Heidelberg; 2018; 1–18. 10.1007/s00122-018-3086-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Virdi SK, Liu Z, Overlander ME, Zhang Z, Xu SS, Friesen TL, et al. New insights into the roles of host gene-necrotrophic effector interactions in governing susceptibility of durum wheat to tan spot and Septoria nodorum blotch. G3 Genes, Genomes, Genet. G3: Genes, Genomes, Genetics; 2016;6: 4139–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1 –Resistant wheat Salamouni (check), 2 –Resistant rye, 3 –Moderately susceptible rye, 4 –Susceptible rye.
(TIF)
X-axis: PIC value and Y-axis rye chromosomes. Violin plots show the density distribution of SNPs for the chromosome corresponding PIC values. Box plots represent first and third quartiles. Horizontal white bars are corresponding median PIC value and yellow dot stands for average PIC value.
(TIF)
X-axis: GD–pairwise genetic dissimilarity–percentage and Y-axis rye chromosomes. Violin plots show the density distribution of pairwise dissimilarities values. Box plots represent first and third quartiles. Horizontal white bars are corresponding median pairwise dissimilarity and yellow dot stands for average pairwise dissimilarity corresponding to each chromosome.
(TIF)
1 = resistant, 2 = moderately resistant, 3 = moderately susceptible and 4 & 5 = susceptible.
(TIF)
Smaller diverse representative set (red-dash clades) represents all the major clusters of Secale cereale subsp. cereale. S. strictum (green) and S. sylvestre (blue) and S. vavilovii (pink) are also shown.
(TIF)
Black bars on rye chromosomes denotes SNP density (SNPs/10Mb) on the rye chromosomes. QTs.sdsu-5R and QTs.sdsu-2R are presented adjacent to their corresponding rye chromosomes. Red italics denotes the mapped tan spot insensitivity genes (tsn1, tsc1, and tsc2) and resistance genes (tsr2, tsr3, tsr4, and tsr5) adjacent to their corresponding wheat chromosomes. Wheat is an allohexaploid species (2n = 6x = 42) with three (A, B, and D) homoeologous chromosome sets and rye is a diploid species (2n = 2x = 14).
(TIF)
These accessions represent 70 different countries around the globe.
(DOCX)
For each accession, its country of origin, corresponding PI number, species, population, and response to tan spot (PTR race 5) is given. Populations are based on structure results.
(DOCX)
(DOCX)
Information on 4,037 GBS SNPs and genotype of 178 accessions at each of these SNP locations in HapMap format.
(TXT)
Data Availability Statement
All the raw sequence data has been submitted to NCBI SRA under SRA accession: PRJNA512245. In addition, the SNP data genotyping with 4,037 SNPs from all 178 accessions utilized in our study has been made available as S4 Table.