Abstract
Chronic hepatitis B (CHB) is one of the major etiological causes of liver failure, cirrhosis, and hepatocellular carcinoma worldwide, and it cannot be completely cured by currently available drugs due to the persistent existence of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA), the bona fide transcription template for HBV RNAs, in the infected hepatocytes. Since quantifying cccDNA per se requires an invasive procedure, serum biomarkers reflecting the intrahepatic cccDNA activity are warranted. Recently, a growing body of research suggests that the circulating HBV RNA may serve as a new serum biomarker for HBV infection, treatment and prognosis. In order to delineate the molecular and clinical characteristics of serum HBV RNA, we systematically reviewed the available literature on serum HBV RNA dating back to early 1990s. In this review, we will summarize the reported serum HBV RNA quantification methods and discuss the potential HBV RNA species in patient serum, and compare the reported correlations of serum HBV RNA with other serological markers, including HBV DNA, hepatitis B surface antigen (HBsAg), e antigen (HBeAg), and core-related antigen (HBcrAg), as well as their correlations with the intrahepatic cccDNA, to assess its potential in clinical applications. The future directions for serum HBV RNA research will also be discussed.
Keywords: HBV, chronic hepatitis B, serum marker, cccDNA, antiviral therapy
1. INTRODUCTION
Currently, nucleos(t)ide analogues (NAs) and peginterferon are being used to treat patients with chronic hepatitis B (CHB), leading to suppression of HBV replication, improved histology, reversed histologic cirrhosis and reduced risk of hepatocellular carcinoma (HCC) (1). However, HBV infection cannot be completely eliminated due to the persistence of cccDNA in the nuclei of infected hepatocytes (2). Due to the invasive nature of liver biopsy, it is impractical to quantify intrahepatic cccDNA as a routine diagnosis, making it necessary to develop noninvasive surrogate markers to monitor the quantity or activity of cccDNA. In the past, several serological markers, including HBV DNA, hepatitis B surface antigen (HBsAg) and hepatitis B core-related antigens (HBcrAgs), have been shown to correlate with intrahepatic cccDNA (3–5). Recently, serum HBV RNA has been considered as a new biomarker for cccDNA (6), especially in virally suppressed patients with low detectable HBV DNA under NA therapy. In this review, we will discuss the importance and scope of using serum HBV RNA as a potential biomarker for hepatitis B infection.
2. SERUM HBV RNA IN VIRAL LIFE CYCLE
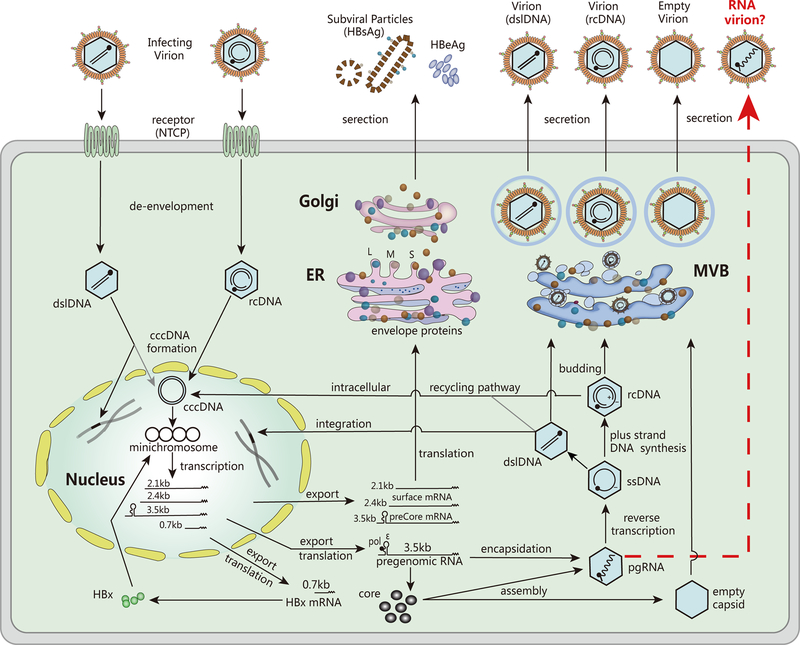
As shown in Figure 1, HBV life cycle starts with the engagement of virion particles to the hepatocyte receptor, termed sodium taurocholate cotransporting polypeptide (NTCP), and then the release of nucleocapsid containing viral relaxed circular DNA (rcDNA) into hepatocytes. rcDNA is then transported to the nucleus, where it is converted into an episomal, nucleosome-decorated cccDNA minichromosome, through a series of mechanisms not yet fully understood (7). The cccDNA serves as a transcription template for all viral transcripts, including the 3.5kb precore mRNA (pcRNA) and pregenomic RNA (pgRNA), the 2.4kb and 2.1kb surface mRNAs, and a 0.7kb X mRNA(8). The pgRNA is the template for both reverse transcription and translation of viral polymerase (pol) and core proteins (9). The pol binds to the 5’-epsilon (ε) region of pgRNA, and together they are incorporated into the viral capsid (10). Inside the capsid, the pol converts pgRNA into rcDNA through reverse transcription, followed by envelopment by HBV envelope proteins on the endoplasmic reticulum (ER) membrane for virion secretion through multivesicular bodies (MVBs) (11), or re-entry into the nucleus to replenish the cccDNA pool. In addition to rcDNA, HBV reverse transcription produces a minor viral DNA species, the double-stranded linear DNA (dslDNA), which can be secreted as virion DNA and redirected into the nucleus to form cccDNA (12). However, dslDNA is often randomly integrated into host genome through non-homologous end joining (NHEJ) (13), which may promote HCC development by inducing chromosomal instability, insertional mutagenesis of HBV genes and HCC-associated host genes (14). Unlike cccDNA, the integrated HBV genome does not produce functional pgRNA for viral DNA replication, but may serve as an additional source for surface mRNA transcription and HBsAg expression (15).
Figure 1. HBV life cycle.
The major steps in HBV life cycle including entry, de-envelopment, cccDNA formation, mRNA transcription, protein translation, pgRNA encapsidation, DNA replication, viral particle assembly and secretion are shown. See text for details. (Abbreviations: NTCP: sodium taurocholate cotransporting polypeptide; rcDNA: relaxed circular DNA; cccDNA: covalently closed circular DNA; pgRNA: pregenomic RNA; ssDNA: single-stranded DNA; dslDNA: double-stranded linear DNA; pol: polymerase; L: large surface protein; M: middle surface protein; S: small surface protein; HBsAg: hepatitis B surface antigen; HBeAg: hepatitis B e antigen; HBx: hepatitis B X protein; ER: endoplasmic reticulum; MVB: multivesicular body.)
According to the dogma of HBV life cycle, only the mature, DNA-containing nucleocapsid interacts with viral surface proteins to undergo envelopment and virion secretion from hepatocytes (16). Nonetheless, a large amount of HBV genome-free (“empty”) virions have been found in patient blood and HBV cell culture fluid (17) (Figure 1), and Ning et al. have coined a “Single Strand Blocking” model to explain such selective HBV morphogenesis whereby the single-stranded nucleic acids (ssRNA or ssDNA) within the viral capsids prevent HBV envelopment (18). However, this model does not apply to several exceptions, such as the ssDNA-containing virion of snow goose hepatitis B virus (SGHBV) (19), and the circulating HBV pgRNA-containing virus particles found in patient blood (6, 20). Theoretically, serum HBV RNA comes from the infected hepatocytes. Wang et al. deep-sequenced HBV RNA in serum and the corresponding regions of paired viral RNA in the liver, and found that the complexity and mean genetic distance between quasispecies of the two were comparable (21). However, the mechanism underlying the release of HBV RNA into circulation from infected hepatocytes remains unclear. Previous studies have demonstrated that HBV cell cultures noncytolytically secrete a large amount of nonenveloped (“naked”) capsids which contain all types of HBV DNA replicative intermediates including pgRNA (22), it is thus unclear whether the detected serum HBV RNA, or at least part of them, are from the “naked” capsid released into the blood stream from HBV infected hepatocytes in vivo or in vitro (6).
If the pgRNA-containing virions do exist, it is interesting to know whether they are infectious and could establish a new round of infection. Wang et al. reported that HBV RNA virion-like particles produced by NA treatment are infection-deficient (23). However, because NA-treated pgRNA-virion is a dead-end due to the irreversible chain termination effect of NA, the above study does not rule out the potential infectivity of untreated pgRNA-virion. Due to the similar protein composition and density between pgRNA- and DNA-virions, it is technically challenging to purify pgRNA virions for infectivity test in cell cultures. Therefore, it remains debatable to incorporate HBV RNA virion-like particles into the canonical HBV replication cycle (24). It is well known that the encapsidated pgRNA is converted into rcDNA through reverse transcription, enveloped by viral surface proteins and secreted into the peripheral blood as Dane particles (25). However, it is possible that a certain amount of pgRNA-containing capsids may prematurely acquire envelope and get released into the peripheral blood (24). On the other hand, the secretion of immature pgRNA-virions and “naked” capsids may represent a host defense mechanism to combat with HBV infection. Hence, further investigation is needed to accurately portray the biogenesis and function of HBV RNA virion-like particles in chronic HBV infection.
3. SERUM HBV RNA SPECIES AND MEASUREMENT
HBV is known as a blood-borne, enveloped, double-stranded DNA virus. In 1996, Kock et al., for the first time, detected the polyadenylated HBV RNA in the serum of CHB patients by using rapid amplification of complementary DNA (cDNA)-ends (RACE) (26). After extracting HBV RNA from patient serum, a special primer consisting of an oligo(dT) stretch and a unique artificial anchored sequence was used to generate cDNA. The cDNA was PCR amplified with the upstream HBV-specific primer and downstream primers identical to the anchored sequence, thereby ensuring a high specificity for HBV polyA RNA amplification (27). Thereafter, similar method was used to detect serum HBV RNA in patients with HBV-related glomerulonephritis or CHB with or without successful treatment (28–30). RACE-based real-time quantitative PCR (RACE-qPCR) using specific primers designed for reverse transcription according to Kairat et al. (31), was later developed to selectively quantify serum 3’ full-length polyadenylated HBV RNA (flRNA) and 3’ internally truncated polyadenylated HBV RNA (trRNA). The RACE-qPCR-based flRNA quantification was widely employed in recent studies (32–35). In addition to RACE-qPCR, regular RT-qPCR methods with HBV-specific primers targeting X, C, or S region of HBV genome were also developed to quantify serum HBV RNA (6, 20, 36–38). However, in order to avoid DNA contamination during RT-qPCR, DNase I treatment of the nucleic acids extracted from serum is required. Alternatively, quantification of serum HBV RNA has been conducted through total HBV nucleic acid measurement by one step real-time RT-qPCR without removing HBV DNA by DNase digestion, followed by subtracting HBV DNA copy numbers determined by qPCR (39–41). QuantiGene assay developed by Lam et al. uses HBV-specific probes, designed to hybridize with the X ORF, to enable a direct quantification of RNA without cDNA synthesis or PCR amplification (42). Recently, Butler et al. developed an automated high-throughput assay to quantify serum HBV RNA, which HBV RNA was isolated using RNA selective extraction chemistry (Abbott mSample Preparation System), followed by a multiplex RT-qPCR procedure for detecting amplicons of HBV X and core region on the m2000 platform(Abbott Molecular) (43). To further evaluate the automated prototype assay performance, they conducted a comparative testing of flRNA and found a good correlation between these two methods. All the published methods for quantifying serum HBV RNA are summarized in Table 1 and Figure 2 for comparison.
Table 1.
Methods for quantifying serum HBV RNA
| Methods | RT Primera | Primer sites | References |
|---|---|---|---|
| RT-qPCR | HBV specific primer | X or C or S region | [6], [20], [36], [37], [38], [43] |
| ddPCRb | HBV specific primer | X or C region | [21], [38], [52] |
| 3’RACE-basedc qPCR | Oligo(dT) primer | poly A tail | [32], [33], [34], [35] |
| QuantiGene assaysd | n/af | n/af | [42] |
| Indirect quantificatione | HBV specific primer | PreC and C region | [39], [40], [41] |
The primer for reverse transcription of serum HBV RNA into cDNA;
Droplet digital PCR;
A rapid amplification of complimentary DNA (cDNA)-ends (RACE)-based real-time polymerase chain reaction (PCR) technique;
QuantiGene Plex assays are hybridization-based and use branched DNA (bDNA) signal amplification technology;
Serum HBV RNA values equal to HBV nucleic acid determined by real-time RT-PCR minus HBV DNA determined by real-time PCR;
Information not available.
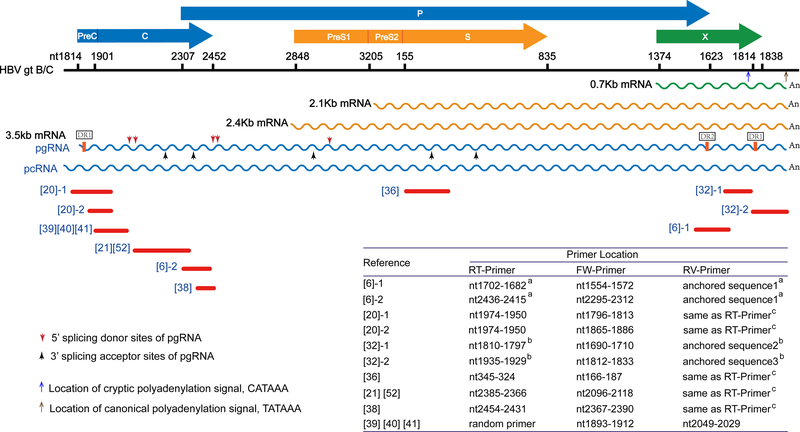
Figure 2. Representatives of published PCR methods for detecting serum HBV RNA.
HBV ORFs are shown as horizontal arrows and their nucleotide positions are marked on the scale bar which represents the length of HBV 3.5kb mRNA (genotype B/C). The blue and brown upward arrows pointing to scale bar indicate the location of canonical polyadenylation signal TATAAA and a cryptic polyadenylation signal CATAAA, respectively. The overlapping full-length HBV mRNAs are shown in wave lines. The direct repeat (DR) sequence DR1 and DR2 are denoted on pgRNA, the vertical red and black arrows pointing to pgRNA indicate the major 5’ splicing donor sites and 3’ splicing acceptor sites of pgRNA. The red lines underneath HBV mRNAs represent the PCR amplicon in the referenced studies (denoted with reference numbers in square brackets). Multiple PCR amplicons from the same reference are numbered. The primer information is provided in the embedded table, and additional information can be found in Table 1. “RT-Primer”, FW-Primer”, and “RV-Primer” refer to reverse transcription primer, forward primer and reverse primer of PCR, respectively. (Note: a The HBV-specific RT-primer in reference [6] is composed of a HBV-specific sequence and a unique anchored sequence at the 5’-end. The anchored sequence is used as RV-primer in PCR cycles. b RT-primer in reference [32] contains a 3’-terminal sequence complementary to the HBV flRNA or trRNA sequence adjacent to polyA tail, a middle stretch of oligo(dT), and a 5’ artificial anchored sequence. The anchored sequence is same as RV-primer. c The sequences of RV-primers in reference [20], [21], [36], [38] and [52] are identical to RT-primers.)
In earlier studies, the species and existence form of serum HBV RNA have not been rigorously characterized. Rokuhara et al. speculated that the serum HBV RNA is incorporated into viral particles, but did not specify what kind of particles harbor HBV RNA (36). In 2015, Jansen et al. reported that HBV RNA is present in virions in CHB patients’ plasma by immunoprecipitating HBcAg with anti-core antibodies after treatment with Nonidet P-40 and dithiothreitol, however, they failed to detect HBV RNA after HBsAg immunoprecipitation (20). In 2016, Wang et al. conducted a series of experiments in HBV stable cell line HepAD38 and claimed that the supernatant HBV RNA is encapsidated pgRNA (6). However, as mentioned above, their study did not distinguish virions from “naked” capsids, both of which may contain pgRNA. Interestingly, “naked” capsids are commonly detected in supernatant of HBV cell cultures but are barely detected in blood of CHB patients (44, 45), indicating a possible presence of RNA-virion in patient sera. Therefore, more reliable assays and convincing evidence are needed to verify the existence of enveloped RNA-containing HBV particles.
As mentioned above, HBV produces the 3.5kb pcRNA and pgRNA, 2.4/2.1kb surface mRNAs, and 0.7kb X mRNA intracellularly, which share the same 3’-termimal sequences overlapping with X mRNA (Figures 1–2). In order to determine the serum HBV RNA species, Wang et al. conducted RT-qPCR assays to quantify serum total HBV RNA and 3.5kb RNA separately and obtained similar copy numbers. They further conducted PCR with 3.5kb HBV RNA-specific primers and pcRNA-specific primers and ruled out the presence of pcRNA in serum (6). However, Prakash et al. performed super-sensitive droplet digital PCR (ddPCR) with similar primers as used by Wang et al. and detected a small amount of pcRNA in serum (38). Nonetheless, although the discrepancy among different studies might be due to the variable specificity and/or sensitivity of different assays, it seems that the detected serum HBV RNA sequences are predominantly derived from pgRNA, Then, a critical question that needs to be answered is whether the serum pgRNA is 3.5kb genome-length or not. Prakash et al. reported that serum HBV RNA is polyadenylated and of genomic length (38), however, the multiple-round PCR method used in their study might have selectively amplified the genome-length polyadenylated pgRNA but neglected the presence of polyA-free serum HBV RNA, which has been identified by Hacker et al. (30). Furthermore, Lam et al. detected both full-length HBV pgRNA (minor species) and the spliced variants (major species) in the supernatant of HBV-infected HepaRG cells treated with or without lamivudine, as well as in CHB patient sera (42). Another study by Wang et al. reported that numerous short pgRNA species were detected by PCR and identified as pgRNA splicing variants and/or 3’-terminal truncations induced by NA treatment (23). The NA-induced accumulation of 3’-truncated polyA-free pgRNA, catalyzed by the RNase H activity of HBV polymerase, have also been demonstrated intracellularly before (46). Therefore, serum HBV RNA is likely a mixture of intact, spliced, and polyA-free pgRNA. Among polyadenylated serum HBV RNA species, the previously reported flRNA and trRNA species can be discriminated by aforementioned 3’-RACE-qPCR with specific RT-primers (32). The flRNA refers to HBV transcripts terminating at the canonical polyadenylation signal TATAAA, which is downstream of the HBx ORF (29, 30, 32, 47–49). A trRNA species, first identified by Hilger et al. in paired liver tumor tissue and peritumor tissues in 1991, it terminates at a cryptic polyadenylation signal within the HBx ORF (49). This signal, a CATAAA motif, is present in most HBV DNA integrants without the canonical polyadenylation signal (47), which may be used to polyadenylate the surface mRNA or HBx mRNA transcribed from the integrated HBV DNA. Thus, whether the encapsulated, polyadenylated serum HBV RNA is equal to flRNA or contains such trRNA species awaits further investigations. Collectively, the available data suggest that the species of serum HBV RNA are heterogeneous and the compositions of these RNA species may vary depending on different stages of chronic HBV infection and different antiviral treatments, and more importantly, the detection methods with different primers (Figure 2, Table 1).
Therefore, considering that the molecular characteristics of serum HBV RNA has not been well defined and a standardized method for serum HBV RNA detection is unavailable, further basic research and assays, such as Northern blot and single-molecule direct RNA sequencing, are needed to achieve a better understanding of the molecular biology of serum HBV RNA, which will aid the development of an accurate and reliable serum HBV RNA measurement method.
4. CORRELATION BETWEEN SERUM HBV RNA AND OTHER HBV MARKERS
4.1. Correlation with serum HBV DNA and HBeAg
Theoretically, both serum HBV DNA and RNA can be useful markers for HBV cccDNA activity in treatment-naïve patients. However, in CHB patients under NAs therapy, HBV DNA is suppressed due to halted pgRNA reverse transcription, making RNA a more direct marker for cccDNA. Consistently, van Bommel et al. demonstrated that the levels of both serum HBV flRNA and trRNA strongly correlate with serum HBV DNA before treatment, but the correlation becomes weaker upon receiving NAs (32) (Table 2). For untreated patients, the correlation coefficient of serum HBV pgRNA and DNA is higher in HBeAg-negative patients than HBeAg-positive patients (r=0.741 and r=0.532, P<0.001, respectively) (37). However, a different study by van Campenhout et al. reported a comparable correlation of serum HBV flRNA to DNA between HBeAg-positive and -negative patients (r=0.72 and r=0.78, P<0.001, respectively) (35). Such discrepancy may be due to the differences in the number of subjects and/or patient characteristics. Moreover, the serum HBV RNA quantitation methods used in these two studies are different (Table 1), which may have resulted in different quantification results as well as different correlation with serum HBV DNA. Thus, the correlation between serum HBV RNA and DNA in HBeAg-positive and -negative patients with or without antiviral treatment is contingent on further investigations.
Table 2.
Correlation between serum HBV RNA and serum HBV DNA
| No. of subjects | Correlation | References | |||
|---|---|---|---|---|---|
| Before treatment P values | P values | on treatment P values | P values | ||
| 273 | flRNAa : r= 0.79 | <0.05 | n/ac | n/ac | [29] |
| trRNAb : r= 0.14 | <0.05 | n/ac | n/ac | ||
| 62 | flRNAa : r= 0.59 | <0.001 | 3rd month: flRNAa: r=0.53 | n/ac | [32] |
| trRNAb : r= 0.75 | <0.001 | trRNAb: r=0.61 | n/ac | ||
| n/ac | n/ac | 6th month: flRNAa: r= 0.53 | n/ac | ||
| n/ac | n/ac | trRNAb: r=0.62 | n/ac | ||
| 52 | n/ac | n/ac | C RNAd: 3rd month: R2= 0.321 | n/ac | [41] |
| 86 | PreC-C RNAd: HBeAg(+):strongly | n/ac | n/ac | n/ac | [20] |
| HBeAg(–):strongly | n/ac | n/ac | n/ac | ||
| 24 | S RNAd: r=0.801 | <0.001 | S RNAd: 2nd month: r=0.837 | <0.001 | [36] |
| 95 | C RNAd: HBeAg(+): R2=0.69 | <0.0001 | n/ac | n/ac | [38] |
| HBeAg(−): R2=0.62 | <0.0001 | n/ac | n/ac | ||
| 102 | C RNAd: r=0.928 | <0.001 | n/ac | n/ac | [52] |
| 84 | C RNAd: HBeAg(+): r= 0.532 | <0.001 | n/ac | n/ac | [37] |
| HBeAg(−): r= 0.741 | <0.001 | n/ac | n/ac | ||
| 488 | flRNAa: HBeAg(+): r=0.72 | <0.001 | n/ac | n/ac | [35] |
| HBeAg(−): r=0.78 | <0.001 | n/ac | n/ac | ||
| 102/16e | X RNAd: R2=0.7756 | n/ac | X RNAd: R2=0.4963 | n/ac | [43] |
| C RNAd: R2=0.8198 | n/ac | C RNAd: R2=0.5434 | n/ac | ||
| 131 | flRNAa: r=0.72 | <0.05 | n/ac | n/ac | [34] |
flRNA refers to the HBV transcripts terminating at the canonical polyadenylation signal TATAAA motif downstream of HBx ORF;
trRNA refers to the HBV transcripts terminating at a cryptic polyadenylation signal, CATAAA, within the HBx ORF;
Information not available;
Primers for quantifying serum HBV RNA target at the X/PreC/C/S region of HBV genome;
102 untreated patients and 16 on-treatment patients.
4.2. Correlation with serum HBsAg and HBcrAg
HBsAg is a widely used serological marker for chronic HBV infection, which not only reflects the intrahepatic HBV activity, but predicts the efficacy of antiviral agents (50). Thus far, only two studies have shown that serum HBV RNA levels of on-treatment patients strongly correlated with HBsAg titers with a correlation coefficient around 0.7 (21, 41), but other studies reported much moderate correlation with a correlation coefficient around 0.4 for pretreatment patients (32, 35, 37) (Table 3). However, for HBeAg-negative patients, there is a weak or no correlation between serum HBV RNA and HBsAg (20, 35, 37). One plausible explanation is that the serum HBsAg in HBeAg-negative patients is largely derived from the integrated HBV genome, rather than cccDNA (15).
Table 3.
Correlation between serum HBV RNA and serum HBsAg
| No. of subjects | Correlation | References | |||
|---|---|---|---|---|---|
| Before treatment | P values | on treatment | P values | ||
| 47 | n/ac | n/ac | C RNAd: >1 year: r=0.665 | <0.001 | [21] |
| 62 | flRNAa: r=0.4 | n/ac | 3rd month: flRNAa: r=0.39 | n/ac | [32] |
| trRNAb: r=0.37 | n/ac | trRNAb: r=0.34 | n/ac | ||
| n/ac | n/ac | 6th month: flRNAa: r=0.36 | n/ac | ||
| n/ac | n/ac | trRNAb: r=0.31 | n/ac | ||
| 52 | n/ac | n/ac | C RNAd: 3rd month: R 2=0.407 | n/ac | [41] |
| 86 | PreC-C RNAd: HBeAg(+):moderate | n/ac | n/ac | n/ac | [20] |
| HBeAg(−):no correlation | n/ac | n/ac | n/ac | ||
| 102 | C RNAd: r=0.703 | <0.001 | n/ac | n/ac | [52] |
| 84 | C RNAd: HBeAg(+): r= 0.537 | <0.001 | n/ac | n/ac | [37] |
| HBeAg(−): r=0.151 | 0.503 | n/ac | n/ac | ||
| 488 | flRNAa: HBeAg(+): r= 0.54 | <0.001 | n/ac | n/ac | [35] |
| HBeAg(−): r=0.19 | 0.04 | n/ac | n/ac | ||
| 131 | flRNAa: r=0.71 | <0.05 | n/ac | n/ac | [34] |
flRNA refers to the HBV transcripts terminating at the canonical polyadenylation signal TATAAA motif downstream of HBx ORF;
trRNA refers to the HBV transcripts terminating at a cryptic polyadenylation signal, CATAAA, within the HBX ORF;
Information not available.
Primers for quantifying serum HBV RNA target at the X/PreC/C/S region of HBV genome.
In addition, HBcrAg, a mixture of HBcAg, HBeAg and the less characterized p22cr, all of which are predominantly cccDNA-derived, has been advocated as a potential surrogate marker for cccDNA activity (51). One study by Akinori et al. reported that serum HBV RNA levels correlate well with serum HBcrAg in patients at the start of treatment (r = 0.841, P<0.001) and during treatment (r = 0.777, P<0.001) (36). However, more studies are needed to validate and establish the correlation between serum HBV RNA and HBcrAg.
4.3. Correlation with cccDNA and other viral markers
NAs therapy can reduce the serum HBV DNA down to undetectable level but does not directly influence the production of HBsAg and HBcrAg, or serum HBV RNA from cccDNA. In this regard, HBsAg, HBcrAg, and particularly the serum HBV RNA, appear to be better surrogate markers for cccDNA than serum HBV DNA. Huang et al. reported that serum HBV pgRNA weakly correlated with cccDNA in the untreated CHB patients with a correlation coefficient of 0.363 (P<0.001) (37), which is similar with the result from Gao et al. (r=0.25, P<0.01) who quantified the serum polyadenylated HBV RNA by RACE-qPCR (33). In a CHB natural history study, Wang et al. found a moderate overall correlation between serum 3.5Kb RNA and intrahepatic cccDNA (r=0.596, P<0.001) among all the recruited patients; however, when classifying the patients into immune tolerant, HBeAg-positive immune active, inactive carrier, and HBeAg-negative immune active phases, the correlation disappeared in all phases (52). For the patients under NAs treatment, Wang et al. found no significant correlation between serum HBV RNA and cccDNA copy numbers (21). In addition, Gao et al. reported that serum HBV polyA RNA has no correlation with intrahepatic cccDNA at 96 weeks after NAs treatment (33). Despite there being no correlation between cccDNA and serum HBV pgRNA, the latter strongly correlates with intrahepatic HBV pgRNA, as well as with the ratio of intrahepatic HBV pgRNA to cccDNA, which reflects intrahepatic viral transcription activity (21). In addition to the intrahepatic viral replicative markers, serum HBV pgRNA also correlated with liver injury or histopathology (21, 52), which raise a question on whether the serum HBV RNA, or at least part of them, are released through hepatocyte destruction.
Thus, it can be concluded that the correlation between serum HBV RNA and intrahepatic cccDNA is not well justified, and may differ between untreated and treated patients. Additionally, the correlation may vary based on different CHB stages, and even different detection methods for serum HBV RNA and cccDNA. Therefore, developing an accurate and standardized protocol for serum HBV RNA and intrahepatic cccDNA quantitation are urgently needed to determine the correlation between serum HBV RNA and intrahepatic cccDNA.
5. CLINICAL SIGNIFICANCE OF SERUM HBV RNA
5.1. Serum HBV RNA and antiviral treatment efficacy
A number of studies have inferred that serum HBV RNA can be a useful marker for monitoring the efficacy of antiviral therapy, such as NAs and interferon (IFN-α) (20, 32, 34, 41). A study on 52 CHB patients receiving NAs therapy revealed that low serum HBV RNA levels at week 12 of treatment can predict the initial virological response, which is defined as the interval from detectable to undetectable HBV DNA level <16 weeks (41). However, the ability of serum HBV RNA to predict the long-term outcome of NAs treatment remains unclear. Due to this fact, additional longitudinal studies are needed to assess the potential predictive role of serum HBV RNA in virological response. If it holds up, “virological response” can be redefined by complementing the current criteria with serum HBV RNA. Another study of 50 HBeAg-positive CHB patients treated with NAs reported that serum HBV flRNA levels at baseline can predict HBeAg seroconversion with superior accuracy to that of HBV DNA levels (AUROC=0.73 versus 0.67), and the decline of serum HBV RNA also showed a better prediction of HBeAg seroconversion than HBV DNA and HBsAg during antiviral treatment (32). This study indicates a quite good predictive value of serum HBV RNA for HBeAg seroconversion, which should be further validated in future studies with large sample size of HBeAg seroconverted patients.
It has been reported that the effect of NAs and IFN-α treatment on serum HBV RNA is different (53). Mechanistically, while NAs specifically block the reverse transcription of encapsidated HBV pgRNA to DNA, the pleiotropic IFN-α can induce cccDNA degradation and inhibit cccDNA transcription (54), promote the decay of HBV pgRNA by interferon-stimulated ribonucleases (55), and prevent pgRNA encapsidation (56). Therefore, when compared to NAs treatment, IFN-α treatment is expected to achieve a stronger decline of serum HBV RNA. Van Bömmel et al. reported that there was a greater reduction of serum HBV RNA in patients receiving PEGylated IFN-2a (peg-IFN) + LAM combination therapy compared to those receiving peg-IFN monotherapy (P<0.05) (34). However, to the best of our knowledge, no research has been done so far to compare the kinetics of serum HBV RNA decline between NAs monotherapy and IFN-α monotherapy. In addition, Jansen et al. found a more pronounced decline of HBV RNA load at week 30 in responders to peg-IFN and adefovir combination therapy than non-responders (P=0.01) (20). Regarding patients under peg-IFN therapy, van Bömmel et al. reported that the cutoff of serum HBV RNA at 5.5-log10 copies/mL at week 12 can identify a higher proportion of non-responders (30%) than the HBV DNA cutoff of 8.9-log10 IU/mL (22%) or HBeAg cutoff of 2.7-log10 IU/mL (29%). However, the HBsAg cutoff of 2.8-log10 IU/mL could identify the largest proportion (41%) of non-responders (34).
All these above-mentioned studies indicate that serum HBV RNA may serve as an additional biomarker for monitoring the efficacy of antiviral therapy. However, it remains unclear whether serum HBV RNA is better than current biomarkers, or whether it can replace other markers for such clinical applications.
5.2. Serum HBV RNA and viral rebound after treatment cessation
Currently, for HBeAg-positive patients being treated with NAs, treatment discontinuation can be considered after at least 1 year of consolidation therapy if patients achieve undetectable HBV DNA, ALT normalization and HBeAg seroconversion (57). Despite these criteria, virological relapse is common after treatment cessation. Therefore, better biomarker(s) for predicting safe withdrawal of NAs treatment in CHB patients is warranted. As serum HBV RNA is considered as a potential biomarker for cccDNA activity, the loss of serum HBV RNA may reflect the transcription silencing of cccDNA and may be an indicator for safe withdrawal of antiviral treatment (58, 59). In line with this notion, a study on 36 patients treated with NAs showed that, after 3 months of treatment, serum HBV DNA+RNA titers were tightly associated with HBV DNA rebound (P=0.043, OR 9.474, 95% CI (1.069–83.957)) and ALT relapse (P=0.050, OR 8.032, 95% CI 0.997–64.683) (40). However, the criteria for discontinuing NAs therapy in this study were not uniform. Another cohort study on 33 CHB patients withdrawing NAs revealed that viral rebound occurred in all patients with HBV RNA positive at the end of treatment, whereas, only in 25% of patients with HBV RNA negative (6). Theoretically, the most promising area for clinical application of serum HBV RNA is the prediction of relapse and sustained response, especially HBsAg loss after treatment discontinuation. However, additional studies with larger sample size are required to further evaluate the role of HBV RNA in predicting HBV relapse and sustained response after discontinuation of NAs treatment.
6. FUTURE RESEARCH ON SERUM HBV RNA
Before a widespread clinical applications of serum HBV RNA for CHB can be accepted and applied, certain fundamental questions need to be answered. 1) Basic and clinical studies are needed to further delineate the composition and molecular details of serum HBV RNA under different situations, such as different stages of CHB natural history, from baseline to different treatment time points, receiving NAs therapy or interferon therapy, as well as other new therapies for CHB. It is also essential to understand how the serum HBV RNA, specifically pgRNA, is released from hepatocytes. Is it in the virion or a naked capsid? Is the egress mechanism different between HBV DNA-containing particles and RNA-containing particles? 2) The methodology for detecting and quantifying serum HBV RNA, which may be related to the RNA species, as well as the intrahepatic cccDNA quantification, should be standardized to make the results of different studies comparable. Due to the potential high complexity of serum HBV RNA, an ideal PCR method would be expected to detect all possible serum HBV RNA species discussed in this review. 3) After characterizing serum HBV RNA species and perfecting the method for detection, their correlations with the clinical outcomes of CHB patients, including response to antiviral therapy especially HBsAg loss, hepatitis flare after withdrawal of NAs, need further investigation as different species of serum HBV RNA may have different clinical implications. 4) The general applicability of serum HBV RNA among CHB patients of different ethnic groups and genotypes should be studied, and a pan-genotypic serum HBV RNA detection is warranted. 5) It has been reported that the circulating HBV RNA may be a marker for hepatocarcinogenesis (47, 49). Furthermore, a resent study demonstrated that pgRNA in liver tumor tissue correlated with the absence of tumorous microvascular invasion and better patient survival (60). Hence, the association and correlation of serum HBV RNA with HCC development also deserve exploration.
7. CONCLUSION
In summary, serum HBV RNA possess potentials to be a new marker for chronic hepatitis B virus infection. Growing evidence supports its correlations with serum HBsAg and HBcrAg and also with HBV DNA before treatment, and it may serve as a better surrogate marker for cccDNA activity in virally suppressed patients receiving NAs therapy. Although the methodologies of serum HBV RNA detection varied from study to study, it has shown preliminary clinical significance. Therefore, it is envisaged that extensive research will be conducted to further characterize the molecular biology of serum HBV RNA and assess its clinical potentials.
ACKNOWLEDGMENT
We apologize to those investigators whose work we did not cite due to oversight or space limitations.
Financial Support: The study is supported by National Science and Technology Major Project of China (2017ZX10202202), Guangzhou Science and Technology Plan Project (201604020002, 201803040013, 201804020001), Guangdong Natural Science and Technology Grant (2016A030313550), National Natural Science Foundation of China (81600475 and U1401226), and the US National Institutes of Health (NIH) grants (R01AI094474, R01AI110762, R01AI123271, R01AI134818, and T32AI060519).
Abbreviations
- ALT
Alanine aminotransferase
- cccDNA
covalently closed circular DNA
- CHB
Chronic hepatitis B
- dslDNA
double-stranded linear DNA
- HBV
hepatitis B virus
- HBcrAg
hepatitis B core-related antigen
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HCC
hepatocellular carcinoma
- IFN-α
alpha-interferon
- LAM
Lamivudine
- NAs
nucleos(t)ide analogues
- NTCP
sodium taurocholate cotransporting polypeptide
- pcRNA
precore RNA
- pgRNA
pregenomic RNA
- qPCR
quantitative polymerase chain reaction
- RACE
rapid amplification of complementary DNA (cDNA)-ends
- rcDNA
relaxed circular DNA
- RT-qPCR
reverse transcription quantitative PCR
REFERENCES
- 1.Hou J, Wang G, Wang F, Cheng J, Ren H, Zhuang H, et al. Guideline of Prevention and Treatment for Chronic Hepatitis B (2015 Update). J Clin Transl Hepatol 2017;5:297–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nassal M HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015;64:1972–1984. [DOI] [PubMed] [Google Scholar]
- 3.Wong DKH, Seto WK, Cheung KS, Chong CK, Huang FY, Fung J, et al. Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver International 2017;37:995–1001. [DOI] [PubMed] [Google Scholar]
- 4.Cornberg M, Wong VW, Locarnini S, Brunetto M, Janssen HLA, Chan HL. The role of quantitative hepatitis B surface antigen revisited. J Hepatol 2017;66:398–411. [DOI] [PubMed] [Google Scholar]
- 5.Guner R, Karahocagil M, Buyukberber M, Kandemir O, Ural O, Usluer G, et al. Correlation between intrahepatic hepatitis B virus cccDNA levels and other activity markers in patients with HBeAg-negative chronic hepatitis B infection. Eur J Gastroenterol Hepatol 2011;23:1185–1191. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016;65:700–710. [DOI] [PubMed] [Google Scholar]
- 7.Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology 2017;66:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block TM, Guo H, Guo JT. Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis 2007;11:685–706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones SA, Hu J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention. Emerg Microbes Infect 2013;2:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahl M, Beck J, Nassal M. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J Virol 2007;81:13354–13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prange R Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol 2012;201:449–461. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Liu K. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A 2004;101:11135–11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol 2016;64:S84–S101. [DOI] [PubMed] [Google Scholar]
- 15.Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol 1996;70:4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luckenbaugh L, Kitrinos KM, Delaney WEt, Hu J. Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J Viral Hepat 2015;22:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, et al. Secretion of genome-free hepatitis B virus--single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog 2011;7:e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greco N, Hayes MH, Loeb DD. Snow goose hepatitis B virus (SGHBV) envelope and capsid proteins independently contribute to the ability of SGHBV to package capsids containing single-stranded DNA in virions. J Virol 2014;88:10705–10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B Virus Pregenomic RNA Is Present in Virions in Plasma and Is Associated With a Response to Pegylated Interferon Alfa-2a and Nucleos(t)ide Analogues. J Infect Dis 2016;213:224–232. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Yu Y, Li G, Shen C, Meng Z, Zheng J, et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J Hepatol 2018;68:16–24. [DOI] [PubMed] [Google Scholar]
- 22.Chou SF, Tsai ML, Huang JY, Chang YS, Shih C. The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion. PLoS Pathog 2015;11:e1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Sheng Q, Ding Y, Chen R, Sun X, Chen X, et al. HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication-deficient. J Hepatol 2017;68:847–849. [DOI] [PubMed] [Google Scholar]
- 24.Lu F, Wang J, Chen X, Xu D, Xia N. Potential use of serum HBV RNA in antiviral therapy for chronic hepatitis B in the era of nucleos(t)ide analogs. Front Med 2017;11:502–508. [DOI] [PubMed] [Google Scholar]
- 25.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015;479–480: 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kock J, Theilmann L, Galle P, Schlicht HJ. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology 1996;23:405–413. [DOI] [PubMed] [Google Scholar]
- 27.Sallie R Selective detection of hepatitis B virus RNA by PCR. PCR Methods Appl 1994;3:376–377. [DOI] [PubMed] [Google Scholar]
- 28.Lai KN, Ho RT, Tam JS, Lai FM. Detection of hepatitis B virus DNA and RNA in kidneys of HBV related glomerulonephritis. Kidney Int 1996;50:1965–1977. [DOI] [PubMed] [Google Scholar]
- 29.Su Q, Wang SF, Chang TE, Breitkreutz R, Hennig H, Takegoshi K, et al. Circulating hepatitis B virus nucleic acids in chronic infection : representation of differently polyadenylated viral transcripts during progression to nonreplicative stages. Clin Cancer Res 2001;7:2005–2015. [PubMed] [Google Scholar]
- 30.Hacker HJ, Zhang W, Tokus M, Bock T, Schroder CH. Patterns of circulating hepatitis B virus serum nucleic acids during lamivudine therapy. Ann N Y Acad Sci 2004;1022:271–281. [DOI] [PubMed] [Google Scholar]
- 31.Kairat A, Beerheide W, Zhou G, Tang ZY, Edler L, Schroder CH. Truncated hepatitis B virus RNA in human hepatocellular carcinoma: its representation in patients with advancing age. Intervirology 1999;42:228–237. [DOI] [PubMed] [Google Scholar]
- 32.van Bommel F, Bartens A, Mysickova A, Hofmann J, Kruger DH, Berg T, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015;61:66–76. [DOI] [PubMed] [Google Scholar]
- 33.Gao Y, Li Y, Meng Q, Zhang Z, Zhao P, Shang Q, et al. Serum Hepatitis B Virus DNA, RNA, and HBsAg: Which Correlated Better with Intrahepatic Covalently Closed Circular DNA before and after Nucleos(t)ide Analogue Treatment? J Clin Microbiol 2017;55:2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Bommel F, van Bommel A, Krauel A, Wat C, Pavlovic V, Yang L, et al. Serum HBV RNA as a Predictor of Peginterferon Alfa-2a (40KD) Response in Patients With HBeAg-Positive Chronic Hepatitis B. J Infect Dis 2018;218:1066–1074. [DOI] [PubMed] [Google Scholar]
- 35.van Campenhout MJH, van Bommel F, Pfefferkorn M, Fischer J, Deichsel D, Boonstra A, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018. March 7. doi: 10.1002/hep.29872. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rokuhara A, Matsumoto A, Tanaka E, Umemura T, Yoshizawa K, Kimura T, et al. Hepatitis B virus RNA is measurable in serum and can be a new marker for monitoring lamivudine therapy. J Gastroenterol 2006;41:785–790. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Wang J, Li W, Chen R, Chen X, Zhang F, et al. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naive HBV-infected individuals. J Clin Virol 2018;99–100: 71–78. [DOI] [PubMed] [Google Scholar]
- 38.Prakash K, Rydell GE, Larsson SB, Andersson M, Norkrans G, Norder H, et al. High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol J 2018;15:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatakeyama T, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, et al. Serum HBV RNA is a predictor of early emergence of the YMDD mutant in patients treated with lamivudine. Hepatology 2007;45:1179–1186. [DOI] [PubMed] [Google Scholar]
- 40.Tsuge M, Murakami E, Imamura M, Abe H, Miki D, Hiraga N, et al. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. J Gastroenterol 2013;48:1188–1204. [DOI] [PubMed] [Google Scholar]
- 41.Huang YW, Takahashi S, Tsuge M, Chen CL, Wang TC, Abe H, et al. On-treatment low serum HBV RNA level predicts initial virological response in chronic hepatitis B patients receiving nucleoside analogue therapy. Antivir Ther 2015;20:369–375. [DOI] [PubMed] [Google Scholar]
- 42.Lam AM, Ren S, Espiritu C, Kelly M, Lau V, Zheng L, et al. Hepatitis B Virus Capsid Assembly Modulators, but Not Nucleoside Analogs, Inhibit the Production of Extracellular Pregenomic RNA and Spliced RNA Variants. Antimicrob Agents Chemother 2017;61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butler EK, Gersch J, McNamara A, Luk KC, Holzmayer V, de Medina M, et al. HBV serum DNA and RNA levels in nucleos(t)ide analogue-treated or untreated patients during chronic and acute infection. Hepatology 2018. May 7. doi: 10.1002/hep.30082. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 44.Schormann W, Kraft A, Ponsel D, Bruss V. Hepatitis B virus particle formation in the absence of pregenomic RNA and reverse transcriptase. J Virol 2006;80:4187–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Possehl C, Repp R, Heermann KH, Korec E, Uy A, Gerlich WH. Absence of free core antigen in anti-HBc negative viremic hepatitis B carriers. Arch Virol Suppl 1992;4:39–41. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, Liu F, Guo F, Zhao Q, Chang J, Guo JT. Characterization of novel hepadnaviral RNA species accumulated in hepatoma cells treated with viral DNA polymerase inhibitors. Antiviral Res 2016;131:40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breitkreutz R, Zhang W, Lee M, Hoffmann A, Tokus M, Su Q, et al. Hepatitis B virus nucleic acids circulating in the blood: distinct patterns in HBs carriers with hepatocellular carcinoma. Ann N Y Acad Sci 2001;945:195–206. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Hacker HJ, Tokus M, Bock T, Schroder CH. Patterns of circulating hepatitis B virus serum nucleic acids during lamivudine therapy. J Med Virol 2003;71:24–30. [DOI] [PubMed] [Google Scholar]
- 49.Hilger C, Velhagen I, Zentgraf H, Schroder CH. Diversity of hepatitis B virus X gene-related transcripts in hepatocellular carcinoma: a novel polyadenylation site on viral DNA. J Virol 1991;65:4284–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinot-Peignoux M, Lapalus M, Asselah T, Marcellin P. HBsAg quantification: useful for monitoring natural history and treatment outcome. Liver Int 2014;34 Suppl 1:97–107. [DOI] [PubMed] [Google Scholar]
- 51.Mak LY, Wong DK, Cheung KS, Seto WK. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. 2018;47:43–54. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Yu Y, Li G, Shen C, Li J, Chen S, et al. Natural history of serum HBV-RNA in chronic HBV infection. J Viral Hepat 2018;25:1038–1047. [DOI] [PubMed] [Google Scholar]
- 53.Huang YW, Chayama K, Tsuge M, Takahashi S, Hatakeyama T, Abe H, et al. Differential effects of interferon and lamivudine on serum HBV RNA inhibition in patients with chronic hepatitis B. Antivir Ther 2010;15:177–184. [DOI] [PubMed] [Google Scholar]
- 54.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, et al. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest 2012;122:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, et al. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog 2017;13:e1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A 2005;102:9913–9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016;63:261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Du M, Huang H, Chen R, Niu J, Jiang J, et al. Reply to: “Serum HBV pgRNA as a clinical marker for cccDNA activity”: Consistent loss of serum HBV RNA might predict the “para-functional cure” of chronic hepatitis B. J Hepatol 2017;66:462–463. [DOI] [PubMed] [Google Scholar]
- 59.Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol 2017;66:460–462. [DOI] [PubMed] [Google Scholar]
- 60.Halgand B, Desterke C, Riviere L, Fallot G, Sebagh M, Calderaro J, et al. Hepatitis B Virus Pregenomic RNA in Hepatocellular Carcinoma: A Nosological and Prognostic Determinant. Hepatology 2018;67:86–96. [DOI] [PubMed] [Google Scholar]