Abstract
One of the unresolved questions in modern medicine is why certain individuals develop a disorder such as rheumatoid arthritis or lupus, while others do not. Contemporary science holds genetics partly responsible and blames the remainder on environmental and stochastic factors. Among the many genes that increase the risk of autoimmune conditions, the risk allele encoding the W620 variant of PTPN22 is shared between multiple rheumatologic diseases, suggesting a fundamental role in the development of immune dysfunction. Here, we discuss how the presence of the PTPN22 risk allele may shape the signs and symptoms of these diseases. Besides the emerging clarity in how PTPN22 tunes T and B cell antigen receptor signaling, we discuss recent discoveries of important functions of PTPN22 in myeloid cell lineages. Taken together, these new insights are revealing important clues to the molecular mechanisms of prevalent diseases like rheumatoid arthritis and lupus and may open new avenues for the development of personalized therapies that spare the normal function of our immune system.
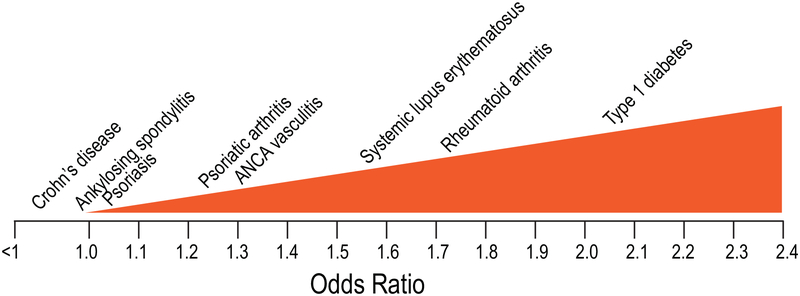
In 2004, we and others published the discovery that a single-nucleotide polymorphism C1858T (rs2476601) in the PTPN22 gene is associated with type 1 diabetes [1], rheumatoid arthritis (RA) [2] and systemic lupus erythematosus (SLE) [3]. These initial findings have been extensively replicated, and today there are close to 1,000 published papers confirming and refining the genetic association of PTPN22 with numerous autoimmune diseases. Indeed, PTPN22 is now recognized as the most influential non-major histocompatibility complex (MHC) gene to promote the development of autoimmunity, including most rheumatologic conditions. Here we summarize the progress in our understanding of the role of PTPN22 in rheumatologic diseases, highlighting how PTPN22 likely operates through multiple mechanisms of action. The strength and direction of the association between PTPN22 and various diseases (Fig. 1) reflects the differential contribution of these mechanisms and introduces a vision for novel pathogenetic disease classification.
Fig. 1. The spectrum of PTPN22 1858T-associated diseases by magnitude of the risk.
The x-axis indicates odds ratio, 1.0 being no association and <1 meaning protection. The diseases are placed above the approximate average odds ratio for heterozygotic carriers from all published papers and meta-analyses.
PTPN22 in rheumatoid and juvenile idiopathic arthritis
A statistically significant association of the PTPN22 1858T allele (encoding the W620 variant) with rheumatoid arthritis (RA) was first reported by Begovich and co-workers in 2004 [2]. The odds ratio was 1.65 in their discovery cohort (n=475 RA, 475 controls) and as high as 2.63 for the homozygous TT genotype versus CC in their replication cohort (n=840 from 463 white families). Rheumatoid factor (RF)-positive RA patients had a higher odds ratio than RF-negative patients. Numerous papers have validated these findings [4] and documented an association with anti-citrullinated protein antibodies (ACPA) [5–9] as well as smoking [10, 11], erosive disease [12], and earlier onset [8, 13]. However, the presence of the PTPN22 risk allele 1858T did not correlate with response to anti-TNF agents [9] or methotrexate [14].
Despite the now well-established geographical gradient in T allele frequency with the highest prevalence in white Caucasians of northern European descent and considerably lower frequencies in southern Europeans [15] and Hispanics, particularly low frequencies (<1%) in Asians, and near absence in people of African origin, a significant association of the T allele with RA has been documented in populations world-wide, including RA patients from South Asia [16], Tunisia [17], and Turkey [18]. However, in some populations the T allele frequency is too low (<1%) for a meaningful analysis [19]. Interestingly, another polymorphism (rs2488457) located in the promoter region of PTPN22 is instead associated with RA in a Chinese population [20].
The PTPN22 1858T allele is also associated with juvenile idiopathic arthritis (JIA) [4, 21] with a somewhat lower odds ratio of ~1.3 in a meta-analysis that included over 4,000 patients and 6,000 controls [22], as well as in a more recent meta-analysis [23], in which RF-positive polyarticular JIA had a stronger odds ratio of 2.12. Interestingly, this subtype of JIA is most similar to RA. Other association studies have treated all seven recognized forms of JIA as a single entity.
PTPN22 in spondylarthropathies
While most susceptibility loci identified in psoriasis (PsO) tend to be equally associated with skin psoriasis and with psoriatic arthritis (PsA), PTPN22 is an exception: the association of the 1858T allele with general skin psoriasis is weak or absent whereas its association with PsA is statistically highly significant with odds ratios of up to 1.32 [24, 25]. This suggests that PsA has additional components in its pathogenesis compared to skin-restricted disease and more involvement of cells and pathways influenced by PTPN22. Also somewhat surprising considering the partially shared pathogenic mechanisms between PsA and ankylosing spondylitis (AS), the latter does not associate with PTPN22. The known role of PTPN22 in CD8 memory T cell function [26] and interleukin-17-producing T helper (Th17) cell differentiation [27] suggest the possibility that the differential association of PTPN22-W620 with PsA vs. PsO or AS depends on alterations in the function of CD8 T cells -which are thought to play a more prominent role in PsA vs. PsO or AS [28]- or that PTPN22-W620 contributes to differential phenotypes of Th17 in PsA vs. PsO or AS [29].
PTPN22 in SLE
The first reported association of the PTPN22 1858T allele with SLE by Kyogoku and co-workers [3] found that a single copy of the T allele increases risk of SLE with an odds ratio of 1.37 (95% confidence interval 1.07–1.75), while TT homozygotes have a much higher odds ratio of 4.37 (95% confidence interval 1.98–9.65). The association with SLE has now been replicated in nearly 20 studies and two recent meta-analyses give an overall odds ratio of ~1.5 [30, 31]. SLE patients with an 1858T allele were noted in different studies to have higher IFNα levels and lower TNFα [32], somewhat more prevalent nephritis [33], and a higher incidence of anti-phospholipid syndrome with anti-cardiolipin autoantibodies [34].
PTPN22 in vasculitides
The rs2476601 PTPN22 polymorphism is also positively associated with ANCA-associated vasculitis (AAV) [35–38]. Specifically, PTPN22 1858T is associated with two of the three distinct autoimmune vasculitides associated with ANCAs [39]: microscopic polyangiitis (MPA) and granulomatosis with polyangiitis (GPA, formerly known as Wegener’s granulomatosis), but has not been reported in eosinophilic granulomatosis with polyangiitis (eGPA, formerly known as Churg-Strauss syndrome). PTPN22 association was equal between patients of both the anti-proteinase 3 and anti-myeloperoxidase serotypes. Although an initial paper [40] reported a lack of association, several later studies have documented and replicated a significant association of 1858T with biopsy-proven giant cell arteritis (GCA) [41, 42] with an odds ratio of 1.62 (95% confidence interval 1.29–2.04). These findings have been replicated in Spanish, Scandinavian, British, American, and Australian patient samples. There was no difference in risk between patients with or without polymyalgia rheumatica or visual ischemic manifestations. In contrast, PTPN22 did not associate with Takayasu’s arteritis, Behcet’s disease, or IgA vasculitis.
PTPN22 in other rheumatologic and autoimmune conditions
PTPN22 1858T is also strongly associated with type 1 diabetes, autoimmune thrombocytopenia, vitiligo, idiopathic inflammatory myopathies, Graves’ disease, myasthenia gravis, and Addison’s disease. On the other hand, the association with systemic sclerosis (SSc) is weaker and has only been found in larger cohorts and meta-analyses [43]. Other diseases that lack an association with PTPN22 1858T include multiple sclerosis, pemphigus vulgaris, ulcerative colitis (UC), primary sclerosing cholangitis, primary biliary cholangitis (formerly known as primary biliary cirrhosis), and acute anterior uveitis, all of which represent diseases in epithelial, mucosal, or immune privileged organs [44]. Intriguingly, in Crohn’s disease the direction of association is reversed, and PTPN22 1858T plays a protective role. Fig. 1 summarizes the spectrum of association between PTPN22 and various rheumatologic/autoimmune diseases. The observed variability likely reflects fundamental differences in disease pathogenesis, although it is possible that in some diseases, PTPN22 1858T promotes both pathogenic and disease-protective pathways, ultimately attenuating the strength of the association.
Other polymorphisms in PTPN22
Besides the C1858T polymorphism, two additional PTPN22 SNPs have been found to have disease associations, particularly in populations with a low frequency of the 1858T allele. The G-1123C polymorphism (rs2488457) is located in the 5’ promoter region of PTPN22 and associates with RA [20], JIA [45], and UC [46] in Chinese populations–in which the rs2476601 SNP does not associate with autoimmunity. The impact of this non-coding SNP on the transcription, stability, or translation of the mRNA remains to be fully clarified. Interestingly, both SNPs rs2476601 and rs2488457 were recently reported as potential cis-expression quantitative trait loci (eQTLs) in whole blood from Spanish RA patients [47], and another study demonstrated that PTPN22 expression is significantly decreased in whole blood from RA patients carrying the risk alleles of SNPs rs2476601 and rs2488457 compared to healthy controls [48].
The second (rs33996649) is a missense G788A mutation that encodes an R263Q substitution in the catalytic domain of the protein. This variant changes the conformation of the PTPN22 active-site, and therefore, unlike the 1858T allele, results in reduced catalytic activity of PTPN22 [49]. It is therefore interesting that in European populations, the 788A allele displays a pattern of autoimmune disease association that is distinct from the 1858T allele. In contrast to 1858T, the 788A allele protects against both SLE and RA [50]. The 788A allele also protects against UC, which 1858T does not associate with, and the 788A does not associate with Crohn’s disease, which the 1858T is protective against [50, 51]. Single studies have so far shown no associations with SSc, GCA, IgA vasculitis, uveitis, or Graves’ disease [50].
Structure and molecular functions of PTPN22
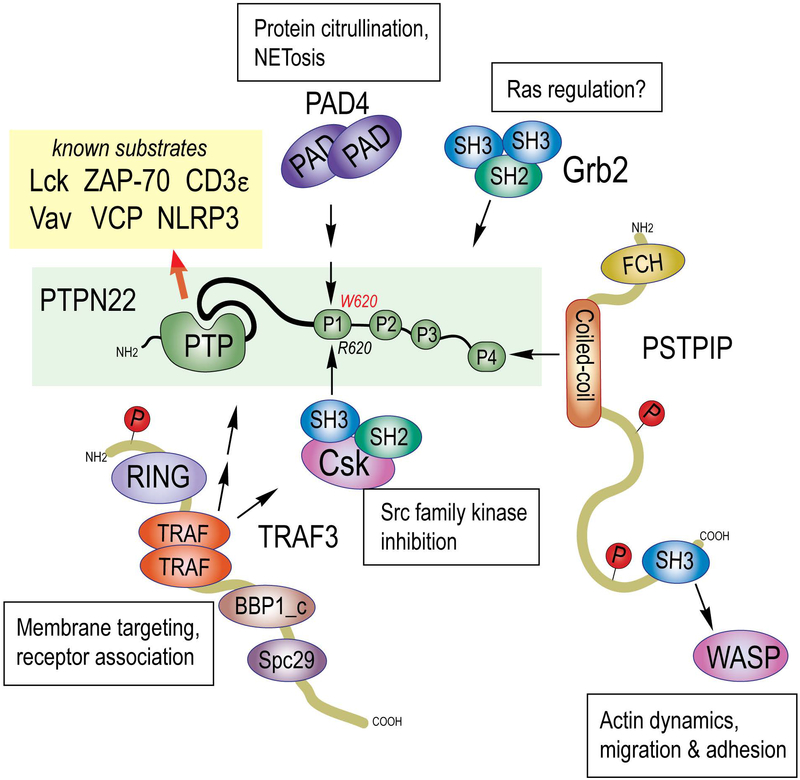
The PTPN22 gene encodes a 110-kDa protein (Fig. 2) known as the lymphoid tyrosine phosphatase (LYP) [52], although now more commonly referred to simply as PTPN22. It is found in all leukocyte lineages, its mRNA being particularly abundant in neutrophils, natural killer (NK) cells, and B cells. The PTPN22 protein has a classical and strictly phosphotyrosine-specific protein tyrosine phosphatase (PTP) domain in its N-terminus followed by a linker region and a long C-terminus of unknown structure. Within the last 200 residues of PTPN22, there are four proline-rich sequence motifs, termed P1 - P4, the first of which (PPPLPERTPESFIVV) binds with high affinity to the Src homology 3 (SH3) domain of the C-terminal c-Src kinase, Csk [53]. While Csk phosphorylates the negative regulatory C-terminus of Lck [54] and other Src family kinases that mediate signaling from a variety of immune receptors, including the T cell receptor (TCR) and B cell antigen receptors and Fc receptors [55], PTPN22, in a complementary manner, dephosphorylates the same kinases at their activation loop tyrosines [56] (Fig. 2). This gives PTPN22 a strong inhibitory function in the T cell receptor and Fc receptor signaling pathways, while the role of PTPN22 in B cell receptor (BCR) signaling remains less defined. In the context of TCR signaling, PTPN22 can also dephosphorylate subunits of the receptor (e.g. CD3ε) and other proximal signaling molecules, such as Vav, zeta chain-associated protein of 70 kDa (ZAP-70), and valosin-containing protein (VCP). The elimination of PTPN22 in the mouse results in accumulation of effector/memory T cells later in life [57]. These cells show enhanced responsiveness to TCR engagement. PTPN22 knockout (KO) mice also display enhanced numbers of follicular helper T cells and germinal centers, and KO CD4+ T cells provide increased T cell help to B cells. [58] Additionally, PTPN22 deletion in mice results in expansion of CD4+CD25+Foxp3+ regulatory T cells, which display enhanced suppressive and adhesive functions [59]. No effects on B cell receptor signaling or B cell development were reported [57].
Fig. 2. Structure, interactors, and substrates of PTPN22.
PTPN22 (green box) contains a catalytic PTP domain, a linker (thicker black line) and 4 proline rich regions (P1 – P4). The location of the R620W (C1858T) missense mutation in the P1 motif is indicated. Protein domains of interacting proteins are: protein arginine deiminase (PAD), protein tyrosine phosphatase (PTP), Src homology 2 domain (SH2), Src homology 3 domain (SH3), really interesting new gene zinc finger domain (RING), TNF receptor-associated factor domain (TRAF), Spindle pole body component 1, C-terminal homology (BBP1_c), Spindle pole component 29 homology (Spc29), Fes-CIP homology domain (FCH), and Wiscott-Aldrich protein (WASP).
The C1858T polymorphism switches amino acid residue 620 in PTPN22 from Arg (R) to Trp (W) [1], changing the P1 motif to PPPLPEWTPESFIVV. This change is significant because R620 is a key residue for binding the Csk SH3 domain. Indeed, the PTPN22*W620 protein no longer binds Csk [1]. We reported that PTPN22*W620 isolated from primary T cells from healthy or diabetes patients had higher catalytic activity than PTPN22*R620 [60]. The consequences of increased catalytic activity, lost Csk binding, and other mechanisms (described below) are likely different and potentially all important for increasing the risk of autoimmunity. Mutagenic and chemical biology approaches–for example, generation of chemical activators of PTPN22 catalytic activity or probes that would disrupt the interaction between PTPN22 and CSK- would aid in the dissection of the impact of these mechanisms on PTPN22 function.
Indicative of even more complexity (Fig. 2), PTPN22 has also been reported to associate with the TNF receptor associated protein TRAF3 [61], the adapter protein Grb2 [62], the cytoskeletal protein PSTPIP [63], and the citrullinating enzyme protein deiminase 4 (PAD4) [64]. The physiological relevance of these interactions remain to be fully elucidated, but binding to TRAF3 has been shown to restrict the physical location of PTPN22 and Csk to enhance signaling from the T cell antigen receptor [61] and the interleukin-6 receptor [65]. The binding of PTPN22 to proline, serine, threonine phosphatase-interacting protein 1 (PSTPIP1), the gene for which is mutated in familial recurrent arthritis (FRA) [66, 67] and pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA) syndrome [68], may direct PTPN22 to its recently recognized role in dephosphorylating the inflammasome subunit NACHT, LRR and PYD domains-containing protein 3 (NLRP3) [69]. This dephosphorylation was found to stimulate inflammasome activation and subsequent interleukin-1β production [69]. Importantly, PTPN22*W620 also bound NLRP3, but was more effective in dephosphorylating it, leading to increased IL-1β production. Finally, the binding of PTPN22 to PAD4 reportedly only involved the major allele PTPN22*R620, while the disease-predisposing variant did not bind [64]. The authors propose that this difference may result in increased protein citrullination in RA patients carrying the 1858T allele.
Another likely important aspect of the biology of PTPN22 is its regulation by mechanisms that modulate T cell function, such as its direct transcriptional suppression by the regulatory T cell transcription factor Foxp3 [70] and by inhibition of translation of its mRNA by the T cell modulating micro-RNA mir-181a [71]. These findings emphasize the importance of PTPN22 fine-tuning in the complex, but important, regulation of T cell immunity.
T and B cell mechanisms by which PTPN22*W620 may drive autoimmunity
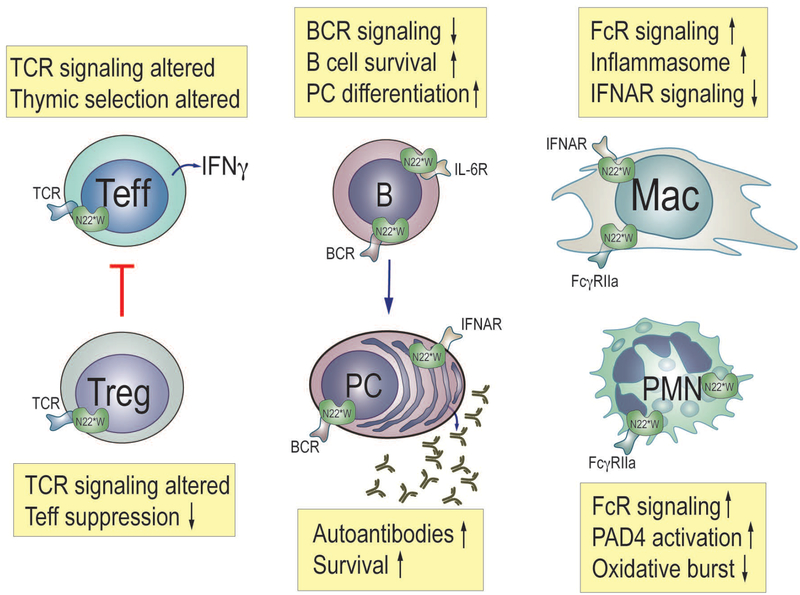
The scientific community has not yet reached a consensus on how exactly PTPN22*W620 increases the risk of autoimmunity. Most investigators have focused on T cells. Some have proposed that PTPN22*W620 alters T cell antigen receptor signaling during thymic selection to promote the survival of autoreactive T cells that later participate in self-reactivity [72, 73]. One study suggested that Treg are less effective in 1858T carriers at suppressing activation of effector T cells [27] (Fig. 3). Several in vitro studies with human T cells have showed that PTPN22*W620 decreases TCR signaling, as a gain-of-function effect would predict [60, 74]. This was seen as diminished tyrosine phosphorylation of early signaling molecules and reduced calcium mobilization [60]. In contrast, experiments in mouse T cells largely came to the opposite conclusion [75]. Despite these differences in TCR signaling effects between the two species, the disease-predisposing allele causes many of the same perturbations in T and B cell immunity in both species [75]. In addition, other pathways activated by TCR signaling in human T cells (including the extracellular signal-regulated kinase [ERK] and protein kinase B [PKB/AKT] pathways) were found to be enhanced in 1858T carriers. This suggests that the PTPN22 W620 mechanism of action in T cells and the consequent immunopathogenic effects are complex. Indeed, inhibition of autoimmune-protective IL-10 release, enhancement of IL-2 and IFNγ release, and expansion of the memory T cell compartment have been variably reported in studies of W620 in human T cells [60, 74, 76, 77].
Fig. 3. Effects of the PTPN22 risk allele on the function of important immune cells.
Effector T cells (Teff), regulatory T cells (Treg), B cells (B), plasma cells (PC), macrophages (Mac), and neutrophils (PMN, polymorphonuclear cells). N22*W denotes the disease-associated PTPN22*W620 variant.
Intriguingly, while loss of PTPN22 in B cells does not seem to impact BCR signaling [78], multiple reports suggest that the W620 variation results in inhibition of human B cell activation. Although the underlying molecular mechanism remains to be further clarified, these observations suggest that PTPN22 W620 can impinge on the pathogenesis of autoimmunity by impairing the elimination of autoreactive B cells [78–80]. This conclusion is also supported by experiments in humanized mice [79]. Furthermore, overexpression of Ptpn22 W619 only in B cells was sufficient to cause spontaneous features of autoimmunity on a mixed C57BL/6J-129/Sv mouse background [81].
Cell-type specific contributions of PTPN22 to disease
Since PTPN22 is expressed in all leukocyte lineages, it is plausible that PTPN22*W620 contributes to the pathogenesis of different rheumatological conditions through a mix of effects on different immune cell lineages, but perhaps with a different weight of each lineage in each disease. We extrapolate this assumption from the varying roles that different immune cell lineages are thought to play in the pathogenesis of each disease. For example, in diseases where autoantibodies are recognized to be particularly important (e.g., SLE), dysfunction of PTPN22*W620-expressing B cells is likely to play a larger role. Similarly, in diseases where a primary dysfunction of neutrophils is considered instrumental for pathogenesis (e.g. AAV or RA), PTPN22*W620 is more likely to contribute to pathogenesis through neutrophils. However, in AAV, anti-neutrophil autoantibodies are also important, suggesting that PTPN22*W620 may also contribute to this disease through B cell dysfunction. As we learn more about the effects of PTPN22*W620 in other immune cells, such as myeloid or plasmacytoid dendritic cells, macrophages, monocytes, or eosinophils, we will be able to hypothesize how dysfunction of these cell lineages may contribute to diseases where they are critically involved in the pathogenesis. These concepts will require lineage-specific knockout or knock-in mice for a deeper mechanistic analysis.
How PTPN22*W620 may corrupt myeloid cells in rheumatological diseases
Macrophages, dendritic cells, and neutrophils are emerging as key players in many autoimmune conditions, for example in disposition of apoptotic or necrotic cells and immune complexes in SLE [82], protein citrullination in RA, autoantigen exposure through neutrophil extracellular traps (NETs) [83], nucleic acid sensing and type I interferon production [84]. All of these cells express PTPN22, but there are relatively few papers addressing its role in these cells.
PTPN22 selectively promotes type I interferon responses after activation of myeloid-cell pattern-recognition receptors. In contrast to TCR signaling, this function of PTPN22 is not mediated by PTPN22 catalytic activity; rather PTPN22 binds to the E3 ubiquitin ligase TRAF3 and selectively promotes its Lys63-linked autoubiquitination in myeloid cells after engagement of toll-like receptors (TLRs). This leads to production of type I interferon without an effect on expression of proinflammatory cytokines, such as IL-1β and TNF. PTPN22*W620 displays reduced binding to TRAF3, and myeloid cells carrying this variant display deficient type 1 interferon production following TLR stimulation. Other studies carried out in mice suggest that macrophages carrying PTPN22*W620 have hyperreactive phagocytic and pro-inflammatory abilities and/or skewed polarization [85, 86].
In neutrophils, PTPN22*W620 appears to play a strikingly different role than in T cells. Vermeren and co-workers [87] found that loss of PTPN22 impaired (rather than augmented!) FcγRII signaling. They measured receptor-triggered Ca2+ mobilization, the oxidative burst, and NETosis, all of which were reduced in PTPN22−/− neutrophils. Bayley and co-workers [88] went a bit further and isolated neutrophils from genotyped RA patients or healthy volunteers and measured neutrophil activation in heterozygous (*W/*R) and homozygous (*R/*R) individuals; there was only one homozygous carrier of the disease-predisposing allele (*W/*W) in the control group and 2 in the RA group, precluding much experimentation with neutrophils of this genotype. They found that PTPN22*W620 enhanced neutrophil activation, oxidative burst, and NETosis in RA donors [88]. In contrast, Cao et al [89] reported that leukocytes (PBMC) from AAV patients with *W620 had reduced Erk activity and IL10 transcription. However, they observed elevated activity of p38 kinase [89]. It should be noted that many experiments in this paper used mixed leukocytes – hence a difference between neutrophils and lymphocytes may have gone unnoticed.
Perhaps the rheumatological condition with the most likely manifestation of neutrophil dysfunction potentially caused by PTPN22*W620 is AAV. It is clear that ANCA play a key role in driving disease by activating neutrophils to degranulate [90], produce reactive oxygen species (ROS) [90], and extrude NETs [91]. In patients [92], vasculitis begins with local accumulation and activation of neutrophils, which rapidly undergo NETosis, apoptosis, or necrosis while driving a necrotizing inflammation that results in endothelial cell death, vascular leakage, fibrin deposition, and a subsequent monocyte and macrophage recruitment [39]. This phase eventually evolves into a fibrin and collagen-rich lesion, which may resolve if the initial inflammation was limited, or become permanent scar tissue with lingering chronic mononuclear cell infiltrates with B and T cells in ectopic germinal center-like structures. In these instances, the inflamed artery may be permanently occluded. The events leading to the formation of a non-vascular granuloma are less understood but appear to be similar. The margins of a granuloma consist of monocytes and epithelioid macrophages that wall off a center of necrotic neutrophil-derived and fibrinous debris.
PTPN22 is involved in several steps of this pathogenesis and may influence: 1) ANCA-mediated neutrophil activation via regulation of FcγRIIa signaling; 2) neutrophil priming by IL-6 via TRAF3 association; 3) NET extrusion via regulation of PAD4; and 4) inflammasome-mediated production of IL-1β, IL-18, and gasdermin D activation, leading to neutrophil cell death by pyroptosis. All of these possible points of influence may synergize with each other and with the increased numbers of autoreactive T and B cells that produce ANCA in a vicious circle of disease propagation.
Remaining key questions
While it is intellectually gratifying that a risk allele such as that of PTPN22 is associated with so many different autoimmune diseases, it also is thought-provoking that the risk it confers varies from strong (second only to MHC) to weak or non-existent between different diseases. What can we learn from this? Are there patterns of disease manifestations that segregate the PTPN22-associated diseases from those that are not associated? Why do subjects who carry one or two risk alleles come down with a very specific autoimmune disease instead of another one? Clearly, other genetic and external factors must play a role, some of them in concert with PTPN22, some independently.
By examining the spectrum of diseases on a scale of their magnitude of association (e.g., by odds ratio) (Fig. 1), one can draw a few tentative conclusions: 1) strongly associated diseases tend to have autoantibodies, the presence or titer of which correlate with the presence of the 1858T allele (e.g., RA, T1D, AAV); 2) strongly associated diseases tend to have a prominent role of autoreactive T and B cells (RA, SLE, AAV); 3) diseases with a central role of neutrophils tend to associate strongly (RA, AAV); 4) diseases with a key role of Th17 cells, but little involvement of autoantibodies, tend to associate poorly (AS, PsO); 5) perhaps related to the previous point, diseases of mucosal sites tend to associate poorly, or even be protected by the 1858T allele.
Another curious observation is that arthritis is strongly associated with the 1858T allele in two diseases, RA and PsA, but not in ankylosing spondylitis. The association is weaker in sero-negative RA, suggesting that it is not the manifestation of joint disease per se that correlates with PTPN22, but the underlying immune dysfunction that can be seen as autoantibodies against immunoglobulins.
Therapeutic implications?
If indeed the degree of PTPN22 association with a disease, or a subset of patients with the disease, tells us more about the critical mechanisms of pathogenesis, then these insights should be helpful for the selection of new drug targets, perhaps even PTPN22 itself. One can envision using PTPN22 expression or genetic variation as a biomarker for disease predisposition or responsiveness to specific therapeutic agents. Therapeutic targeting of PTPN22 might also be useful in prevention or control of rheumatic diseases. For example, “molecular glue” compounds that re-establish the interaction between CSK and PTPN22 W620 might be able to rescue most of the immune abnormalities induced by the PTPN22 risk allele. A simpler approach using small-molecule inhibitors of PTPN22 might be sufficient to correct key pathogenic mechanisms and still exert sufficient preventative or therapeutic action. For example, inhibition of PTPN22 activity with the small-molecule compound LTV-1 was effective at rescuing B cell selection and preventing the development of autoreactive W620 B cells in a humanized mouse model [79]. Considering that in a mouse model of RA, PTPN22 promoted Th17 cell differentiation, a PTPN22 inhibitor might also be useful in treatment of the large spectrum of diseases characterized by enhanced Th17 presence/activation.
Acknowledgments
This work was supported by NIH grant R01AI070544 and R01AR066053 to N.B.
REFERENCES
- 1.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M et al. : A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004, 36(4):337–338. [DOI] [PubMed] [Google Scholar]
- 2.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM et al. : A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 2004, 75(2):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM et al. : Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 2004, 75(3):504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinks A, Barton A, John S, Bruce I, Hawkins C, Griffiths CE, Donn R, Thomson W, Silman A, Worthington J: Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum 2005, 52(6):1694–1699. [DOI] [PubMed] [Google Scholar]
- 5.Johansson M, Arlestig L, Hallmans G, Rantapaa-Dahlqvist S: PTPN22 polymorphism and anti-cyclic citrullinated peptide antibodies in combination strongly predicts future onset of rheumatoid arthritis and has a specificity of 100% for the disease. Arthritis Res Ther 2006, 8(1):R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feitsma AL, Toes RE, Begovich AB, Chokkalingam AP, de Vries RR, Huizinga TW, van der Helm-van Mil AH: Risk of progression from undifferentiated arthritis to rheumatoid arthritis: the effect of the PTPN22 1858T-allele in anti-citrullinated peptide antibody positive patients. Rheumatology (Oxford) 2007, 46(7):1092–1095. [DOI] [PubMed] [Google Scholar]
- 7.Kokkonen H, Johansson M, Innala L, Jidell E, Rantapaa-Dahlqvist S: The PTPN22 1858C/T polymorphism is associated with anti-cyclic citrullinated peptide antibody-positive early rheumatoid arthritis in northern Sweden. Arthritis Res Ther 2007, 9(3):R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goeb V, Dieude P, Daveau R, Thomas-L’otellier M, Jouen F, Hau F, Boumier P, Tron F, Gilbert D, Fardellone P et al. : Contribution of PTPN22 1858T, TNFRII 196R and HLA-shared epitope alleles with rheumatoid factor and anti-citrullinated protein antibodies to very early rheumatoid arthritis diagnosis. Rheumatology (Oxford) 2008, 47(8):1208–1212. [DOI] [PubMed] [Google Scholar]
- 9.Potter C, Hyrich KL, Tracey A, Lunt M, Plant D, Symmons DP, Thomson W, Worthington J, Emery P, Morgan AW et al. : Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann Rheum Dis 2009, 68(1):69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallberg H, Padyukov L, Plenge RM, Ronnelid J, Gregersen PK, van der Helm-van Mil AH, Toes RE, Huizinga TW, Klareskog L, Alfredsson L et al. : Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet 2007, 80(5):867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costenbader KH, Chang SC, De Vivo I, Plenge R, Karlson EW: Genetic polymorphisms in PTPN22, PADI-4, and CTLA-4 and risk for rheumatoid arthritis in two longitudinal cohort studies: evidence of gene-environment interactions with heavy cigarette smoking. Arthritis Res Ther 2008, 10(3):R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lie BA, Viken MK, Odegard S, van der Heijde D, Landewe R, Uhlig T, Kvien TK: Associations between the PTPN22 1858C->T polymorphism and radiographic joint destruction in patients with rheumatoid arthritis: results from a 10-year longitudinal study. Ann Rheum Dis 2007, 66(12):1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlson EW, Chibnik LB, Cui J, Plenge RM, Glass RJ, Maher NE, Parker A, Roubenoff R, Izmailova E, Coblyn JS et al. : Associations between human leukocyte antigen, PTPN22, CTLA4 genotypes and rheumatoid arthritis phenotypes of autoantibody status, age at diagnosis and erosions in a large cohort study. Ann Rheum Dis 2008, 67(3):358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majorczyk E, Pawlik A, Kusnierczyk P: PTPN22 1858C>T polymorphism is strongly associated with rheumatoid arthritis but not with a response to methotrexate therapy. Int Immunopharmacol 2010, 10(12):1626–1629. [DOI] [PubMed] [Google Scholar]
- 15.Totaro MC, Tolusso B, Napolioni V, Faustini F, Canestri S, Mannocci A, Gremese E, Bosello SL, Alivernini S, Ferraccioli G: PTPN22 1858C>T polymorphism distribution in Europe and association with rheumatoid arthritis: case-control study and meta-analysis. PLoS One 2011, 6(9):e24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastana S, Gilmour A, Ghelani A, Smith H, Samanta A: Association of PTPN22 with rheumatoid arthritis among South Asians in the UK. J Rheumatol 2007, 34(10):1984–1986. [PubMed] [Google Scholar]
- 17.Sfar I, Dhaouadi T, Habibi I, Abdelmoula L, Makhlouf M, Ben Romdhane T, Jendoubi-Ayed S, Aouadi H, Ben Abdallah T, Ayed K et al. : Functional polymorphisms of PTPN22 and FcgR genes in Tunisian patients with rheumatoid arthritis. Arch Inst Pasteur Tunis 2009, 86(1–4):51–62. [PubMed] [Google Scholar]
- 18.Ates A, Karaaslan Y, Karatayli E, Ertugrul E, Aksaray S, Turkyilmaz A, Ozet G: Association of the PTPN22 gene polymorphism with autoantibody positivity in Turkish rheumatoid arthritis patients. Tissue Antigens 2011, 78(1):56–59. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Korman BD, Le JM, Kastner DL, Remmers EF, Gregersen PK, Bae SC: Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum 2009, 60(2):364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng X, Li YZ, Zhang Y, Bao SM, Tong DW, Zhang SL, Hu CJ: Association of the PTPN22 gene (−1123G > C) polymorphism with rheumatoid arthritis in Chinese patients. Tissue Antigens 2010, 76(4):297–300. [DOI] [PubMed] [Google Scholar]
- 21.Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, Lilleby V, Sollid LM, Forre OT, Thorsby E, Smerdel A et al. : Association analysis of the 1858C>T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun 2005, 6(3):271–273. [DOI] [PubMed] [Google Scholar]
- 22.Lee YH, Bae SC, Song GG: The association between the functional PTPN22 1858 C/T and MIF −173 C/G polymorphisms and juvenile idiopathic arthritis: a meta-analysis. Inflamm Res 2012, 61(5):411–415. [DOI] [PubMed] [Google Scholar]
- 23.Kaalla MJ, Broadaway KA, Rohani-Pichavant M, Conneely KN, Whiting A, Ponder L, Okou DT, Angeles-Han S, Rouster-Stevens K, Brown MR et al. : Meta-analysis confirms association between TNFA-G238A variant and JIA, and between PTPN22-C1858T variant and oligoarticular, RF-polyarticular and RF-positive polyarticular JIA. Pediatr Rheumatol Online J 2013, 11(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowes J, Loehr S, Budu-Aggrey A, Uebe S, Bruce IN, Feletar M, Marzo-Ortega H, Helliwell P, Ryan AW, Kane D et al. : PTPN22 is associated with susceptibility to psoriatic arthritis but not psoriasis: evidence for a further PsA-specific risk locus. Ann Rheum Dis 2015, 74(10):1882–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YF, Chang JS: PTPN22 C1858T and the risk of psoriasis: a meta-analysis. Mol Biol Rep 2012, 39(8):7861–7870. [DOI] [PubMed] [Google Scholar]
- 26.Mehlhop-Williams ER, Bevan MJ: Memory CD8+ T cells exhibit increased antigen threshold requirements for recall proliferation. J Exp Med 2014, 211(2):345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vang T, Landskron J, Viken MK, Oberprieler N, Torgersen KM, Mustelin T, Tasken K, Tautz L, Rickert RC, Lie BA: The autoimmune-predisposing variant of lymphoid tyrosine phosphatase favors T helper 1 responses. Hum Immunol 2013, 74(5):574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowes J, Budu-Aggrey A, Huffmeier U, Uebe S, Steel K, Hebert HL, Wallace C, Massey J, Bruce IN, Bluett J et al. : Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun 2015, 6:6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benham H, Norris P, Goodall J, Wechalekar MD, FitzGerald O, Szentpetery A, Smith M, Thomas R, Gaston H: Th17 and Th22 cells in psoriatic arthritis and psoriasis. Arthritis Res Ther 2013, 15(5):R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lima SC, Adelino JE, Crovella S, de Azevedo Silva J, Sandrin-Garcia P: PTPN22 1858C > T polymorphism and susceptibility to systemic lupus erythematosus: a meta-analysis update. Autoimmunity 2017, 50(7):428–434. [DOI] [PubMed] [Google Scholar]
- 31.Hu LY, Cheng Z, Zhang B, Yin Q, Zhu XW, Zhao PP, Han MY, Wang XB, Zheng HF: Associations between PTPN22 and TLR9 polymorphisms and systemic lupus erythematosus: a comprehensive meta-analysis. Arch Dermatol Res 2017, 309(6):461–477. [DOI] [PubMed] [Google Scholar]
- 32.Kariuki SN, Crow MK, Niewold TB: The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum 2008, 58(9):2818–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy MV, Johansson M, Sturfelt G, Jonsen A, Gunnarsson I, Svenungsson E, Rantapaa-Dahlqvist S, Alarcon-Riquelme ME: The R620W C/T polymorphism of the gene PTPN22 is associated with SLE independently of the association of PDCD1. Genes Immun 2005, 6(8):658–662. [DOI] [PubMed] [Google Scholar]
- 34.Ostanek L, Ostanek-Panka M, Bobrowska-Snarska D, Binczak-Kuleta A, Fischer K, Kaczmarczyk M, Ciechanowicz A, Brzosko M: PTPN22 1858C>T gene polymorphism in patients with SLE: association with serological and clinical results. Mol Biol Rep 2014, 41(9):6195–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jagiello P, Aries P, Arning L, Wagenleiter SE, Csernok E, Hellmich B, Gross WL, Epplen JT: The PTPN22 620W allele is a risk factor for Wegener’s granulomatosis. Arthritis Rheum 2005, 52(12):4039–4043. [DOI] [PubMed] [Google Scholar]
- 36.Carr EJ, Niederer HA, Williams J, Harper L, Watts RA, Lyons PA, Smith KG: Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis. BMC Med Genet 2009, 10:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martorana D, Maritati F, Malerba G, Bonatti F, Alberici F, Oliva E, Sebastio P, Manenti L, Brugnano R, Catanoso MG et al. : PTPN22 R620W polymorphism in the ANCA-associated vasculitides. Rheumatology (Oxford) 2012, 51(5):805–812. [DOI] [PubMed] [Google Scholar]
- 38.Cao Y, Liu K, Tian Z, Hogan SL, Yang J, Poulton CJ, Falk RJ, Li W: PTPN22 R620W polymorphism and ANCA disease risk in white populations: a metaanalysis. J Rheumatol 2015, 42(2):292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jennette JC, Falk RJ : Pathogenesis of antineutrophil cytoplasmic autoantibody-mediated disease. Nat Rev Rheumatol 2014, 10(8):463–473. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Gay MA, Oliver J, Orozco G, Garcia-Porrua C, Lopez-Nevot MA, Martin J: Lack of association of a functional single nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with susceptibility to biopsy-proven giant cell arteritis. J Rheumatol 2005, 32(8):1510–1512. [PubMed] [Google Scholar]
- 41.Serrano A, Marquez A, Mackie SL, Carmona FD, Solans R, Miranda-Filloy JA, Hernandez-Rodriguez J, Cid MC, Castaneda S, Morado IC et al. : Identification of the PTPN22 functional variant R620W as susceptibility genetic factor for giant cell arteritis. Ann Rheum Dis 2013, 72(11):1882–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lester S, Hewitt AW, Ruediger CD, Bradbury L, De Smit E, Wiese MD, Black R, Harrison A, Jones G, Littlejohn GO et al. : PTPN22 R620W minor allele is a genetic risk factor for giant cell arteritis. RMD Open 2016, 2(1):e000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dieude P, Guedj M, Wipff J, Avouac J, Hachulla E, Diot E, Granel B, Sibilia J, Cabane J, Meyer O et al. : The PTPN22 620W allele confers susceptibility to systemic sclerosis: findings of a large case-control study of European Caucasians and a meta-analysis. Arthritis Rheum 2008, 58(7):2183–2188. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J, Ibrahim S, Petersen F, Yu X: Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun 2012, 13(8):641–652. [DOI] [PubMed] [Google Scholar]
- 45.Fan ZD, Wang FF, Huang H, Huang N, Ma HH, Guo YH, Zhang YY, Qian XQ, Yu HG: STAT4 rs7574865 G/T and PTPN22 rs2488457 G/C polymorphisms influence the risk of developing juvenile idiopathic arthritis in Han Chinese patients. PLoS One 2015, 10(3):e0117389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Zhang H, Xia B, Wang P, Jiang T, Song M, Wu J: Association of PTPN22 gene (rs2488457) polymorphism with ulcerative colitis and high levels of PTPN22 mRNA in ulcerative colitis. Int J Colorectal Dis 2013, 28(10):1351–1358. [DOI] [PubMed] [Google Scholar]
- 47.Walsh AM, Whitaker JW, Huang CC, Cherkas Y, Lamberth SL, Brodmerkel C, Curran ME, Dobrin R: Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol 2016, 17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remuzgo-Martinez S, Genre F, Castaneda S, Corrales A, Moreno-Fresneda P, Ubilla B, Mijares V, Portilla V, Gonzalez-Vela J, Pina T et al. : Protein tyrosine phosphatase non-receptor 22 and C-Src tyrosine kinase genes are down-regulated in patients with rheumatoid arthritis. Sci Rep 2017, 7(1):10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, Dasgupta J, Hartiala J, Zhao L, Ortego-Centeno N, D’Alfonso S et al. : A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Hum Mol Genet 2009, 18(3):569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae SC, Lee YH: Association between the functional PTPN22 G788A (R263Q) polymorphism and susceptibility to autoimmune diseases: A meta-analysis. Cell Mol Biol (Noisy-le-grand) 2018, 64(5):46–51. [PubMed] [Google Scholar]
- 51.Diaz-Gallo LM, Espino-Paisan L, Fransen K, Gomez-Garcia M, van Sommeren S, Cardena C, Rodrigo L, Mendoza JL, Taxonera C, Nieto A et al. : Differential association of two PTPN22 coding variants with Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis 2011, 17(11):2287–2294. [DOI] [PubMed] [Google Scholar]
- 52.Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM: Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood 1999, 93(6):2013–2024. [PubMed] [Google Scholar]
- 53.Cloutier JF, Veillette A: Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J 1996, 15(18):4909–4918. [PMC free article] [PubMed] [Google Scholar]
- 54.Bergman M, Mustelin T, Oetken C, Partanen J, Flint NA, Amrein KE, Autero M, Burn P, Alitalo K: The human p50csk tyrosine kinase phosphorylates p56lck at Tyr-505 and down regulates its catalytic activity. EMBO J 1992, 11(8):2919–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mustelin T, Tasken K: Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem J 2003, 371(Pt 1):15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T: Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol 1999, 29(12):3845–3854. [DOI] [PubMed] [Google Scholar]
- 57.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC: PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 2004, 303(5658):685–689. [DOI] [PubMed] [Google Scholar]
- 58.Maine CJ, Marquardt K, Cheung J, Sherman LA: PTPN22 controls the germinal center by influencing the numbers and activity of T follicular helper cells. J Immunol 2014, 192(4):1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brownlie RJ, Miosge LA, Vassilakos D, Svensson LM, Cope A, Zamoyska R: Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Science signaling 2012, 5(252):ra87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F et al. : Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 2005, 37(12):1317–1319. [DOI] [PubMed] [Google Scholar]
- 61.Wallis AM, Wallace EC, Hostager BS, Yi Z, Houtman JCD, Bishop GA: TRAF3 enhances TCR signaling by regulating the inhibitors Csk and PTPN22. Sci Rep 2017, 7(1):2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B: The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol 2002, 30(3):237–244. [DOI] [PubMed] [Google Scholar]
- 63.Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA: PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol 1997, 138(4):845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang HH, Dwivedi N, Nicholas AP, Ho IC: The W620 Polymorphism in PTPN22 Disrupts Its Interaction With Peptidylarginine Deiminase Type 4 and Enhances Citrullination and NETosis. Arthritis Rheumatol 2015, 67(9):2323–2334. [DOI] [PubMed] [Google Scholar]
- 65.Lin WW, Yi Z, Stunz LL, Maine CJ, Sherman LA, Bishop GA: The adaptor protein TRAF3 inhibits interleukin-6 receptor signaling in B cells to limit plasma cell development. Sci Signal 2015, 8(392):ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wise CA, Bennett LB, Pascual V, Gillum JD, Bowcock AM: Localization of a gene for familial recurrent arthritis. Arthritis Rheum 2000, 43(9):2041–2045. [DOI] [PubMed] [Google Scholar]
- 67.Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL: Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A 2003, 100(23):13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S, Lovett M: Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum Mol Genet 2002, 11(8):961–969. [DOI] [PubMed] [Google Scholar]
- 69.Spalinger MR, Kasper S, Gottier C, Lang S, Atrott K, Vavricka SR, Scharl S, Gutte PM, Grutter MG, Beer HD et al. : NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest 2016, 126(5):1783–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA: Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 2007, 445(7130):931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P et al. : miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007, 129(1):147–161. [DOI] [PubMed] [Google Scholar]
- 72.Vang T, Miletic AV, Bottini N, Mustelin T: Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity 2007, 40(6):453–461. [DOI] [PubMed] [Google Scholar]
- 73.Bottini N, Vang T, Cucca F, Mustelin T: Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol 2006, 18(4):207–213. [DOI] [PubMed] [Google Scholar]
- 74.Aarnisalo J, Treszl A, Svec P, Marttila J, Oling V, Simell O, Knip M, Korner A, Madacsy L, Vasarhelyi B et al. : Reduced CD4+T cell activation in children with type 1 diabetes carrying the PTPN22/Lyp 620Trp variant. J Autoimmun 2008, 31(1):13–21. [DOI] [PubMed] [Google Scholar]
- 75.Zheng J, Petersen F, Yu X: The role of PTPN22 in autoimmunity: learning from mice. Autoimmun Rev 2014, 13(3):266–271. [DOI] [PubMed] [Google Scholar]
- 76.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH: Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol 2007, 179(7):4704–4710. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Zahir N, Jiang Q, Miliotis H, Heyraud S, Meng X, Dong B, Xie G, Qiu F, Hao Z et al. : The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet 2011, 43(9):902–907. [DOI] [PubMed] [Google Scholar]
- 78.Menard L, Saadoun D, Isnardi I, Ng YS, Meyers G, Massad C, Price C, Abraham C, Motaghedi R, Buckner JH et al. : The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest 2011, 121(9):3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schickel JN, Kuhny M, Baldo A, Bannock JM, Massad C, Wang H, Katz N, Oe T, Menard L, Soulas-Sprauel P et al. : PTPN22 inhibition resets defective human central B cell tolerance. Sci Immunol 2016, 1(1):aaf7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Metzler G, Dai X, Thouvenel CD, Khim S, Habib T, Buckner JH, Rawlings DJ: The Autoimmune Risk Variant PTPN22 C1858T Alters B Cell Tolerance at Discrete Checkpoints and Differentially Shapes the Naive Repertoire. J Immunol 2017, 199(7):2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, Moon RT, Liggitt D, Wolf-Yadlin A, Buckner JH et al. : A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest 2013, 123(5):2024–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elkon KB: Cell Death, Nucleic Acids and Immunity: Inflammation beyond the Grave. Arthritis and rheumatism 2018, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gupta S, Kaplan MJ: The role of neutrophils and NETosis in autoimmune and renal diseases. Nature reviews Nephrology 2016, 12(7):402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lood C, Blanco LP, Purmalek MM, Carmona-Rivera C, De Ravin SS, Smith CK, Malech HL, Ledbetter JA, Elkon KB, Kaplan MJ: Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nature medicine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li M, Beauchemin H, Popovic N, Peterson A, d’Hennezel E, Piccirillo CA, Sun C, Polychronakos C: The common, autoimmunity-predisposing 620Arg > Trp variant of PTPN22 modulates macrophage function and morphology. J Autoimmun 2017, 79:74–83. [DOI] [PubMed] [Google Scholar]
- 86.Chang HH, Miaw SC, Tseng W, Sun YW, Liu CC, Tsao HW, Ho IC: PTPN22 modulates macrophage polarization and susceptibility to dextran sulfate sodium-induced colitis. J Immunol 2013, 191(5):2134–2143. [DOI] [PubMed] [Google Scholar]
- 87.Vermeren S, Miles K, Chu JY, Salter D, Zamoyska R, Gray M: PTPN22 Is a Critical Regulator of Fcgamma Receptor-Mediated Neutrophil Activation. J Immunol 2016, 197(12):4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bayley R, Kite KA, McGettrick HM, Smith JP, Kitas GD, Buckley CD, Young SP: The autoimmune-associated genetic variant PTPN22 R620W enhances neutrophil activation and function in patients with rheumatoid arthritis and healthy individuals. Ann Rheum Dis 2015, 74(8):1588–1595. [DOI] [PubMed] [Google Scholar]
- 89.Cao Y, Yang J, Colby K, Hogan SL, Hu Y, Jennette CE, Berg EA, Zhang Y, Jennette JC, Falk RJ et al. : High basal activity of the PTPN22 gain-of-function variant blunts leukocyte responsiveness negatively affecting IL-10 production in ANCA vasculitis. PLoS One 2012, 7(8):e42783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falk RJ, Terrell RS, Charles LA, Jennette JC: Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci U S A 1990, 87(11):4115–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kessenbrock K, Krumbholz M, Schonermarck U, Back W, Gross WL, Werb Z, Grone HJ, Brinkmann V, Jenne DE: Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med 2009, 15(6):623–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mark EJ, Matsubara O, Tan-Liu NS, Fienberg R: The pulmonary biopsy in the early diagnosis of Wegener’s (pathergic) granulomatosis: a study based on 35 open lung biopsies. Hum Pathol 1988, 19(9):1065–1071. [DOI] [PubMed] [Google Scholar]