Abstract
Non-alcoholic fatty liver disease (NAFLD) is a common cause of chronic liver disease. A single nucleotide polymorphism (SNPs), rs6834314, was associated with serum liver enzymes in the general population, presumably reflecting liver fat or injury. We studied rs6834314 and its nearest gene, HSD17B13 (17-beta hydroxysteroid dehydrogenase 13) to identify associations with histological features of NAFLD, and to characterize the functional role of HSD17B13 in NAFLD pathogenesis. The minor allele of rs6834314 was significantly associated with increased steatosis, but decreased inflammation, ballooning, Mallory-Denk bodies, and liver enzyme levels in 768 adult Caucasians with biopsy-proven NAFLD, and with cirrhosis in the general population. We found two plausible causative variants in the HSD17B13 gene. rs72613567, a splice-site SNP in high linkage with rs6834314 (r2=0.94) generates novel splice variants and shows a similar pattern of association with NAFLD histology. Its minor allele generates simultaneous expression of exon 6-skipping and G-nucleotide insertion variants. Another SNP, rs62305723 (encoding a P260S mutation) is significantly associated with decreased ballooning and inflammation. Hepatic expression of HSD17B13 is 5.9-fold higher (p=0.003) in patients with NAFLD. HSD17B13 is targeted to lipid droplets, requiring the conserved AA22–28 sequence and AA71–106 region. The protein has a retinol dehydrogenase (RDH) activity, with enzymatic activity dependent on lipid droplet targeting and co-factor binding site. The exon 6-deletion, G-insertion, and newly-described naturally-occurring P260S mutation, all confer loss of enzymatic activity. In conclusion, we demonstrate the association of variants in HSD17B13 with specific features of NAFLD histology, and identify the enzyme as a lipid droplet-associated RDH. Our data suggest that HSD17B13 plays a role in NAFLD through its enzymatic activity.
Keywords: Single nucleotide polymorphism, 17-beta hydroxysteroid dehydrogenase 13, Lipid droplet, Alternative splicing, Genetic variability
INTRODUCTION
The rise of non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) has sparked considerable interest in pathogenic mechanisms of the disease. Racial and ethnic variability and a known heritable component (1,2) hinted at a genetic contribution. Genome-wide association studies (GWAS) identified loci associated with NAFLD, most predominantly rs738409, a single nucleotide polymorphism (SNP) in the PNPLA3 gene (3–5). Subsequently, we and others (6–8) demonstrated that rs738409 is strongly associated with histological features and severity of NAFLD. Although the association is highly significant statistically, the effect size with regards to liver fat accumulation (9) is quite weak. Similarly, the major allele of the TM6SF2 SNP rs58542926 is associated with decreased liver fat (10), but conversely, with higher serum lipid levels and increased risk of atherosclerosis and cardiovascular disease (11). In vivo and in vitro studies provided evidence that TM6SF2 regulates VLDL secretion and acts as a determinant of liver damage or cardiovascular disease under metabolic stress (10–14). Of other genes associated with NAFLD by GWAS only a few (such as MBOAT7 (15–17)) were studied in large histologically-characterized NAFLD cohorts, and in even fewer has their mechanism been elucidated.
A recent large-scale GWAS identified new loci associated with plasma liver enzyme levels (18). In unselected population surveys, elevated transaminases occure concomitantly with metabolic syndrome risk factors (19,20) and are presumed to reflect hepatic fat accumulation and related injury. However, a direct association between these SNPs and NAFLD has not yet been shown. We hypothesized that plasma transaminase activity in the GWAS reflected the presence of fatty liver and associated injury, and that genetic associations with transaminases could identify novel genes involved in NAFLD pathogenesis. Our goals were to establish which of these newly-described SNPs is associated with histological features of NAFLD, identify the genes associated with these SNPs, and elucidate the mechanism by which these genes affect NAFLD.
In this work we demonstrate that SNPs in or near HSD17B13 are associated with histological features of NAFLD, identify HSD17B13 as a novel retinol dehydrogenase, and characterize the impact of genetic variants on its enzymatic activity. Our study provides a link between genetic variants, retinoid metabolism, and NAFLD pathogenesis, and identifies HSD17B13 as a potentially druggable target in NAFLD.
METHODS
STUDY DESIGN
We initially genotyped in a cohort of subjects with biopsy-proven NAFLD 38 SNPs described by Chambers (18), to identify loci associated with histological features. After identifying a locus of interest, we genotyped additional SNPs in the candidate gene locus in the same cohort and confirmed associations in additional cohorts. Finally, we performed functional analyses to characterize the gene product and identify the functional impact of gene variants (Supplementary Figure 1).
STUDY POPULATIONS
The NAFLD cohort was described in detail previously (6). Briefly, we included patients from the NASH Clinical Research Network (NASH-CRN) observational database (21) and PIVENS (22) studies, as well as patients with biopsy-proven NAFLD seen at the NIH Clinical Center. Analysis was limited to Caucasians aged ≥18 years at the time of liver biopsy. All subjects had histological evidence of NAFLD or NASH on liver biopsy. Liver histology was scored semi-quantitatively using the NASH-CRN scoring system (23).
Confirmatory analyses were performed in a cohort of patients with chronic hepatitis C and available liver histology, and in two population-based cohorts – the Michigan Genomics Initiative (MGI) and the UK Biobank.
HEPATIC GENE EXPRESSION
Deidentified normal and NASH human liver samples were obtained from the Liver Tissue Cell Distribution System and from clinical trials at the NIH Clinical Center. Gene expression was quantified by qPCR.
INTRA-CELLULAR LOCALIZATION
Cells were stained with primary antibodies against organelle markers, LipidTox Red (Life Technologies, Carlsbad, CA) for lipid droplets, and Hoechst 33342 for nuclei. Staining was visualized using confocal microscope.
RETINOL DEHYDROGENASE (RDH) ACTIVITY
RDH activity was tested in a cell-based assay as previously described (24). Briefly, HEK293 cells were transfected with protein expression vectors and treated with all-trans-retinol for 8 hours, followed by retinaldehyde and retinoic acid (RA) quantification by HPLC and protein quantification by western blot.
STATISTICAL ANALYSIS
Associations between genotypes and histology were determined by a two-step method. Univariate analysis was performed using the Jonkheere-Terpstra test for ordered differences. SNPs with univariate significance level <0.1 were selected for multivariate ordinal regression, with age, BMI, gender and type 2 diabetes as covariates. We utilized the Benjamini-Hochberg (25) false discovery rate (FDR) to correct for multiple comparisons, using the calculator by Pike (26), with q<0.05 considered significant. P-values are reported from the multivariate ordinal regression. Odds ratios (OR) of associations with histology were calculated using binary logistic regression. An additive inheritance model was used. Student’s t-test was used for 2-group differences and one-way ANOVA followed by Dunnett’s multiple comparison test were used for >2 groups in the in vitro enzymatic assays. All significance testing was two-sided. Statistical analyses were performed using SPSS Statistics V.19 (IBM) and Prism V.7 (GraphPad).
ETHICS
Human subject data was obtained from studies conducted according to the Decleration of Helsinki principles that were reviewed by Institutional Review Boards. All subjects gave written informed consent prior to inclusion.
Additional details are found in the Supplementary Methods.
RESULTS
We obtained DNA from 768 adult Caucasians with biopsy-proven NAFLD (mean age 49, mean BMI 34.6kg/m2, 28% diabetic). Histologically, 59.5% had NASH and 12.1% were cirrhotic (Supplementary Table 1). We genotyped in these subjects 38 SNPs described by Chambers (18) (Supplementary Table 2). The associations between SNPs and histological features of NAFLD are detailed in Supplementary Table 3. Multivariate ordinal logistic regression adjusting for age, gender, diabetes and BMI found 9 SNPs to be significantly associated with steatosis grade, 4 SNPs associated with degree of ballooning degeneration, and 3 SNPs associated with Mallory-Denk bodies. No SNP was associated with severity of inflammation or fibrosis. Associations were independent of the PNPLA3 rs738409 genotype (Supplementary Table 3). Of the SNPs associated with NAFLD histology, only rs6834314 was also associated with serum transaminases in patients with NAFLD (Supplementary Table 4) and thus was the focus of subsequent study.
rs6834314 IS ASSOCIATED WITH NAFLD HISTOLOGY
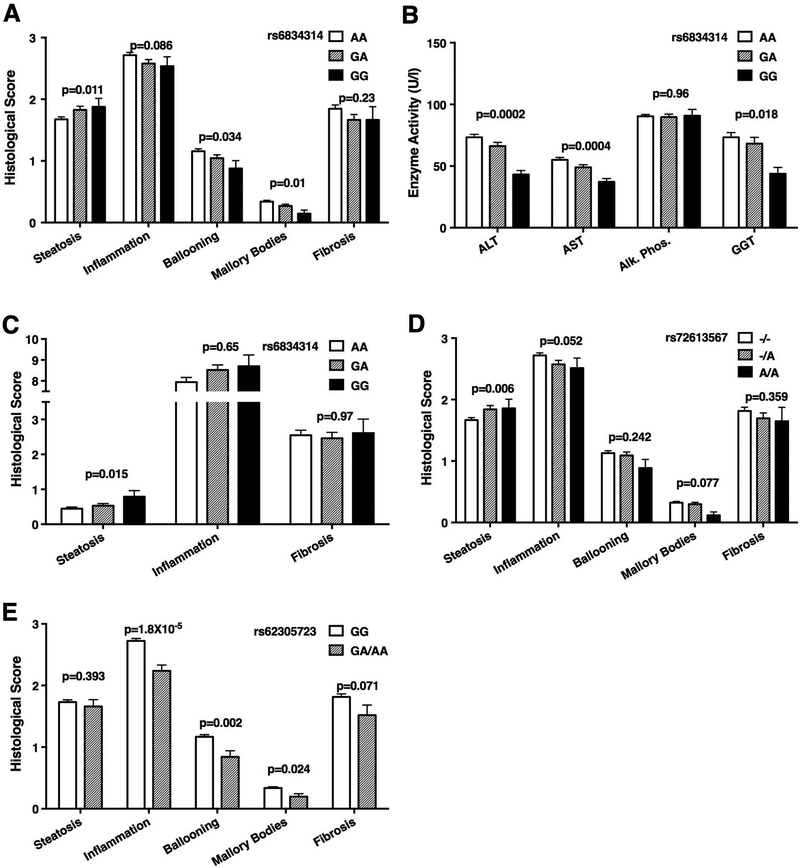
rs6834314 is located on chromosome 4, 11kb downstream of the 17-beta hydroxysteroid dehydrogenase 13 (HSD17B13) gene. Each copy of the minor G-allele increased the risk of having liver fat content ≥33% (OR=1.32, CI 1.02–1.70, Figure 1A, Supplementary Figure 2A). The mean steatosis grade in subjects with rs6834314G/G genotype was 0.2 units higher than in those with A/A (1.88±0.87 vs. 1.68±0.89), an effect similar to the impact of the PNPLA3 rs738409 in the same subjects (0.21 units; 1.59±0.84 for rs738409C/C vs. 1.80±0.89 for G/G) (6). In contrast to its association with increased liver fat, the minor G-allele was significantly associated with lower risks for inflammation (OR=0.77 for total inflammatory score ≥3, CI 0.60–0.99), significant ballooning (OR=0.67 for ballooning score >1, CI 0.51–0.87), and the presence of Mallory-Denk bodies (OR=0.68, CI 0.51–0.91) and showed a non-significant trend for reduced fibrosis (OR=0.79, CI 0.60–1.05) on binary logistic regression. The associations with inflammation, ballooning, and Mallory-Denk bodies were independent of the association with steatosis and in fact, were stronger when steatosis grade was included in the regression model (Supplementary Table 5). The minor allele of rs6834314 was also associated with lower serum transaminases and GGT (Figure 1B). NAFLD has been previously associated with genetic variants in PNPLA3 (rs738409), TM6SF2 (rs58542926), MBOAT7 (rs626283), and GCKR (rs1260326). After adjusting for these variants, rs6834314 remained significantly associated with steatosis (p=0.013) ballooning (p=0.031) and Mallory-Denk bodies (p=0.030), and trended for association with inflammation (p=0.067, Supplementary Table 6). rs6834314 genotype did not differ between patients with NASH and those with steatosis only.
Figure 1.
HSD17B13-related SNPs are associated with liver disease. Association of rs6834314 genotype with (A) liver histology scores and (B) blood levels of liver enzymes in subjects with NAFLD. (AA, n=485; GA, n=226; GG, n=41). (C) Association of rs6834314 genotype with liver histology scores in subjects with chronic hepatitis C. (AA, n=165; GA, n=125; GG, n=23). (D) Association of rs72613567 genotype with liver histology scores in subjects with NAFLD. (--, n=503; -A, n=220; AA, n=35). (E) Association of rs62305723 genotype with liver histology scores in subjects with NAFLD. (GG, n=615; GA/AA, n=76). Mean±SEM. P-values for histology from multivariate ordinal regression adjusted for age, gender, and BMI. P-values for enzymes from linear regression of log-transformed enzyme activity, adjusted for age, gender, and BMI.
To validate the association of rs6834314 with steatosis, we genotyped it in an independent sample of 317 Caucasian patients with chronic hepatitis C and available liver histology (Supplementary Table 7). Liver fat was present in 179 (57.2%) subjects. As with the NAFLD cohort, rs6834314 genotype was significantly associated with the degree of steatosis in univariate (p=0.032, Jonkheere-Terpstra test) and multivariate (p=0.028, ordinal regression adjusted for BMI, age, and gender) analyses. Each copy of the minor G-allele was significantly associated with the presence of liver fat (OR=1.67, CI 1.11–2.51, p=0.015; Figure 1C, Supplementary Figure 2B). As expected, rs6834314 genotype was not associated with HCV-driven inflammation or fibrosis.
As further confirmation, we determined the association of rs6834314 with liver enzymes and diagnoses in the MGI cohort (Supplementary Table 8). rs6834314-G was associated with lower ALT (p=3.3×10−4) and AST (p=5.1×10−5) and with a lower risk of cirrhosis diagnosis (OR=0.79, p=7.5×10−4, Supplementary Figure 2C). The MGI cohort is population-based and histological data is not available for detailed analyses.
OTHER VARIANTS IN HSD17B13 ARE ASSOCIATED WITH NAFLD
rs6834314 is downstream of HSD17B13 and in high LD with multiple adjacent SNPs, overlapping parts of the HSD17B13 gene. To determine whether the associations of rs6834314 reflect a biological effect of HSD17B13, we selectively genotyped additional variants in the gene region, including 2 non-synonymous coding SNPs, an insertion/deletion (indel) polymorphism in a splice site, and a frame-shift coding SNP, as well as 7 SNPs that capture most of the common variants in the gene region (Table 1). The tag SNP, rs6834314, is in strong LD (D’=0.995, r2=0.93) with the indel rs72613567, located immediately adjacent to the 3’-end of exon 6 (Supplementary Figure 3). As expected, this indel had a similar pattern of association with NAFLD histology and liver enzymes as rs6834314 (Figure 1D). The minor allele, rs72613567-A was associated with increased steatosis (OR=1.36, CI 1.04–1.77) but with decreased inflammation (OR=0.74, CI 0.57–0.96), and trended towards decreased ballooning (OR=0.78, CI 0.60–1.03), Mallory-Denk bodies (OR=0.77, CI 0.57–1.03), and fibrosis (OR=0.77, CI 0.58–1.03, Supplementary Figure 4A).
Table 1 –
P-Values for Association of SNPs in the HSD17B13 Gene with Histological Features and ALT in the NAFLD Cohort. Arrows Denote Direction of Effect for the Minor Allele
| SNP | SNP Type | Alleles | Minor Allele FrequencyA | Genotype FrequencyA | R2 vs. rs6834314 genotypeB | FatC | InflammationC | BallooningC | Mallory-Denk BodiesC | FibrosisC | ALTD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs72613567 | Splice site | -/A | 0.194 | 0.66/0.29/0.05 | 0.938 | 0.008* ↑ | 0.063 ↓ | 0.202 | 0.062 ↓ | 0.373 | 4.2×10−4*↓ |
| rs62305723 | Non-synonymous coding | G/A | 0.056 | 0.89/0.11/0.003 | 0.022 | 0.326 | 5×10−5* ↓ | 0.002* ↓ | 0.025 ↓ | 0.065 ↓ | 0.217 |
| rs10433937 | Non coding | T/G | 0.199 | 0.65/0.29/0.05 | 0.979 | 0.019* ↑ | 0.062 ↓ | 0.042 ↓ | 0.013 ↓ | 0.323 | 2.3×10−4*↓ |
| rs6531975 | Non coding | G/A | 0.202 | 0.64/0.32/0.05 | 0.019 | 0.016* ↓ | 0.098 | 0.728 | 0.641 | 0.392 | 0.744 |
| rs10022237 | Non coding | C/T | 0.428 | 0.33/0.49/0.18 | 0.403 | 0.004* ↑ | 0.021 ↓ | 0.05 ↓ | 0.100 | 0.090 | 0.033 ↓ |
| rs6531973 | Non coding | C/A | 0.238 | 0.57/0.39/0.05 | 0.091 | 0.535 | 0.621 | 0.853 | 0.321 | 0.402 | 0.207 |
| rs6531972 | Non coding | A/C | 0.404 | 0.35/0.49/0.16 | 0.171 | 0.067 | 0.648 | 0.532 | 0.993 | 0.603 | 0.333 |
| rs7692397 | Non coding | C/A | 0.241 | 0.58/0.36/0.06 | 0.104 | 0.074 | 0.068 | 0.257 | 0.231 | 0.227 | 0.215 |
| rs9647458E | Non coding | A/G | 0.096 | 0.82/0.18/0.008 | 0.035 | 0.646 | 0.696 | 0.137 | 0.472 | 0.661 | 0.579 |
Allele and genotype frequencies from the NAFLD cohort
R2 for linkage between SNPs and rs6834314 from LDLink, limited to populations of European ancestry
Multivariate ordinal logistic regression adjusted for age, BMI, gender and diabetes
Linear regression of log-transformed enzyme activity adjusted for age, BMI, gender and diabetes
rs76926692 (non-synonymous coding) and rs75049937 (frameshift) were genotyped but omitted for minor allele frequency ≤ 0.3%
FDR q-value < 0.05
rs72613567 was not genotyped in the MGI cohort but was genotyped in the UK Biobank cohort (Supplementary Table 9). The minor allele A-insertion was significantly less common in the NAFLD cohort (19.1%) compared to the population-based UK Biobank (27.9%, p<0.001) and was associated with a reduction in the risk of fibrosis and cirrhosis (OR=0.83, p=9×10−4, Supplementary Figure 4B). rs72613567-A was also associated with lower risk of diagnoses of liver disease (OR=0.93, p=8×10−4), other inflammatory liver diseases (a diagnostic code that includes NASH, OR=0.82, p=0.0012) and alcoholic liver disease (OR=0.87, p=0.015). Histological data or liver enzymes are not available for this cohort.
Another SNP, the non-synonymous coding rs62305723G>A in exon 6, encodes a P260S substitution and is not in LD with rs6834314 (r2=0.02). The rs62305723 minor A-allele is significantly associated with decreased inflammation (OR=0.46, CI 0.28–0.74), ballooning (OR=0.48, CI 0.30–0.76), and Mallory-Denk bodies (OR=0.51, CI 0.28–0.91), but, in contrast to rs72613567/rs6834314, is not associated with steatosis (Figure 1E, Supplementary Figure 5A). The minor A-allele was associated with a lower risk of NASH compared to steatosis (OR=0.39, CI=0.23–0.68, p=0.001). In the UK Biobank cohort, rs62305723 was not associated with liver-related diagnoses. Interestingly, the minor A-allele was associated with a diagnosis of blindness or low vision (OR=1.3, p=0.007, Supplementary Figure 5B).
Conditional analysis found rs62305723 and rs72613567 to associate with NAFLD histology independently of each other (Supplementary Table 10), suggesting both are causal variants. Including the PNPLA3 rs738409 in the model did not impact significance (Supplementary Table 10) and the association of HSD17B13 SNPs with histology and ALT was seen for all rs738409 genotypes (Supplementary Figure 6). We found no significant interaction between rs738409 and HSD17B13 SNPs, although our study was not powered to detect such an effect. Two additional non-coding SNPs (rs6531975, rs10022237), neither in LD with rs6834314, were associated with histological features and rs10022237 was also associated with ALT. The detailed mapping supports a functional role for HSD17B13 in the pathogenesis of NAFLD.
HEPATIC HSD17B13 EXPRESSION IS ELEVATED IN NASH PATIENTS
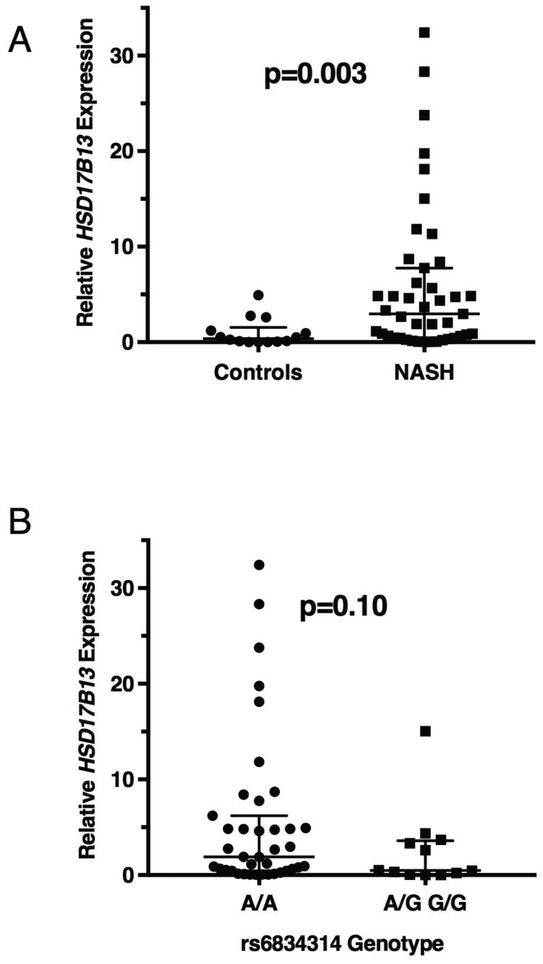
To determine whether HSD17B13 is expressed differentially in NAFLD, we quantified its expression in human NASH (n=43) or normal (n=14) liver samples. Hepatic expression of HSD17B13 is 5.9-fold higher in NASH patients compared with controls (p=0.003, Figure 2A).
Figure 2.
Hepatic expression of HSD17B13. (A) Expression levels of HSD17B13 by qPCR in liver samples from healthy controls (n=14) and NASH patients (n=43). (B) Expression levels of HSD17B13 by qPCR according to rs6834314 genotype in liver samples from control and NASH patients (A/A genotype: n=45; and A/G or G/G genotype: n=12). rs6834314A/G and G/G were pooled together due to lower frequency. Lines denote median and interquartile range and are normalized to controls. Mann-Whitney test was used for significance testing.
In the GTEx database, rs6834314 genotype did not affect hepatic HSD17B13 expression (Supplementary Figure 7A, p=0.6) and neither did rs72613567 nor rs62305723 (data not shown). We confirmed this in a pooled analysis of our liver tissue samples (Figure 2B). Since HSD17B13 expression is upregulated in NAFLD, we examined whether rs6834314 modulates gene expression only in these subjects but found no significant impact of genotype on hepatic HSD17B13 expression in either NASH (Supplementary Figure 7B) or normal liver (Supplementary Figure 7C), although nearly all high-expression samples had A/A genotype. To confirm that rs6834314 does not affect HSD17B13 expression in hepatocytes, as liver tissue samples contain other cell types, we quantified HSD17B13 mRNA in primary human hepatocytes from 22 donors and genotyped them for rs6834314. No donor was homozygous for the minor G-allele and there was no difference in HSD17B13 expression between A/A and A/G genotypes (Supplementary Figure 7D). Thus, hepatic HSD17B13 expression is upregulated in NAFLD but the association between rs6834314 genotype and NAFLD phenotype is not due to changes in expression levels and is more likely reflecting a change in protein function.
HSD17B13 LOCALIZES TO LIPID DROPLETS
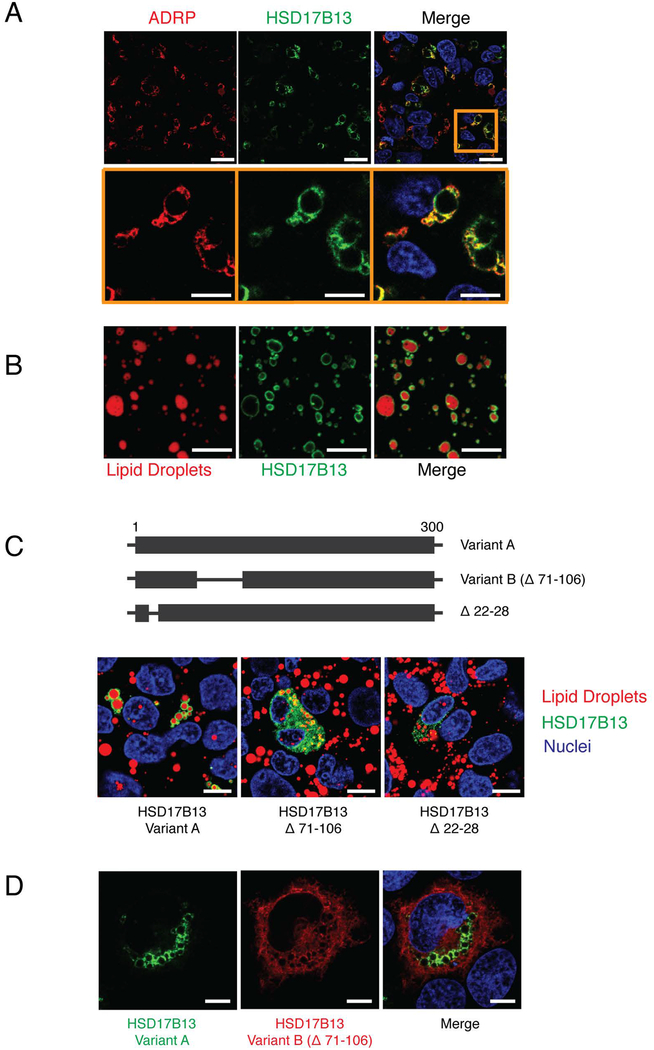
To better understand the function of HSD17B13, we determined its subcellular localization by transfecting HSD17B13-GFP into HepG2 cells and staining for cellular organelle markers. HSD17B13-GFP co-localizes with the lipid droplets (Figure 3A) and is detected on the surface of purified droplets from transfected cells (Figure 3B) but does not co-localize with other organelles (Supplementary Figure 8A).
Figure 3.
HSD17B13 localizes with lipid droplets. (A) HepG2 cells were transfected with HSD17B13(A)-GFP and stained for the lipid droplet marker protein ADRP (Perilipin-2). (B) Lipid droplets were isolated from HSD17B13(A)-GFP transfected cells and stained with the lipid droplet specific dye LipidTox. (C) Mutations were generated and tested for cellular localization with LipidTox used to stain lipid droplets. (D) HSD17B13(A)-GFP and HSD17B13(B)-Flag were co-transfected into HEK293 cells. Immunofluorescence staining was used to visualize HSD17B13(B)-Flag. Nuclei were counterstained with Hoechst 33342. One representative image from repeated experiments is shown. Bars indicate 10 μm.
There are two described transcript variants for the human HSD17B13 gene, formed by alternative splicing: variant A contains all 7 exons while variant B is missing exon 2 (Supplementary Figure 3) resulting in deletion of amino acids 71–106 (Figure 3C). Transfected HSD17B13(B)-GFP localized to the cytoplasm but not to lipid droplets (Figure 3C–D). By site-directed mutagenesis, we identified the N-terminal amino acids 22–28 of HSD17B13 as essential for lipid droplet targeting (Figure 3C). This 7-amino acid sequence is conserved within ADRP and TIP47 which are also lipid droplet-associated proteins (27) (Supplementary Figure 8B) and appears to be crucial for protein stability, as its deletion results in low protein expression.
HSD17B13 DOES NOT AFFECT HEPATOCYTE LIPID CONTENT
To assess the functional role of HSD17B13 in the liver, we investigated whether its expression level affects fat content of HepG2 cells incubated with a high concentration of fatty acids. The expression of HSD17B13 protein was dose-dependently induced by doxycycline using Tet-on inducible expression system (Supplementary Figure 9A) with no effect on lipid content (Supplementary Figure 9B–C). Similarly, stable HSD17B13 overexpression or knockout in HepG2 cells did not affect their lipid content (Supplementary Figure 9D–E), suggesting HSD17B13 does not regulate hepatocyte lipid content in a direct manner.
HSD17B13 HAS RETINOL DEHYDROGENASE ACTIVITY IN VITRO
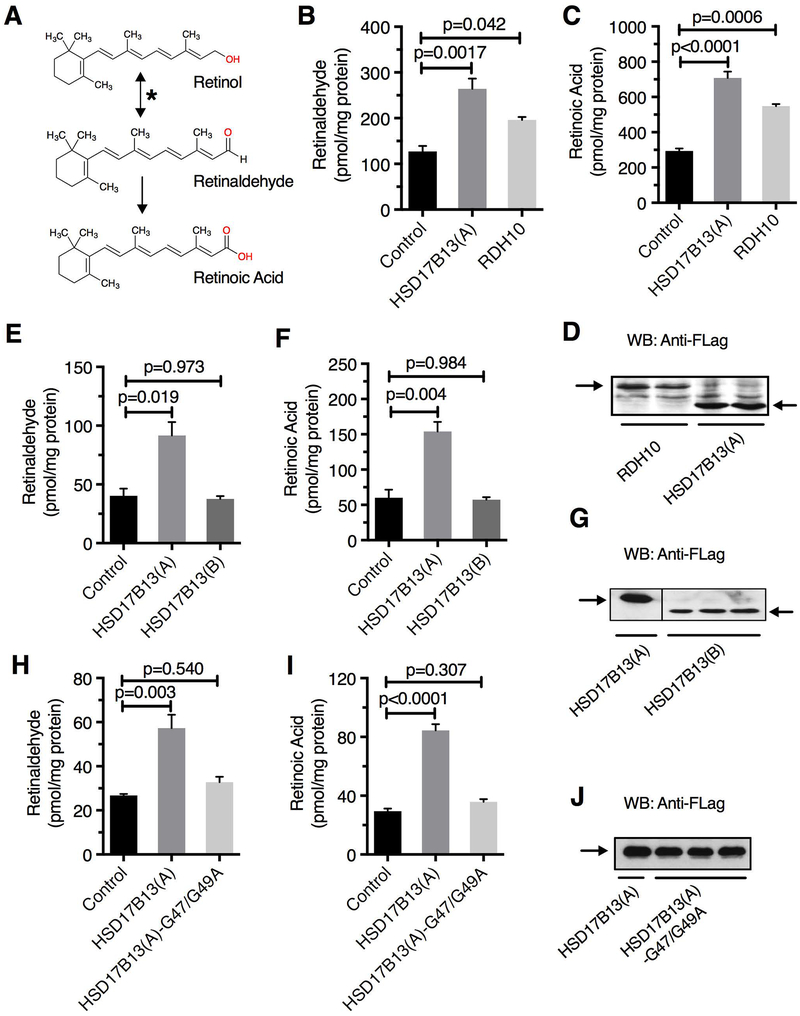
HSD17B13 is a member of the 17β hydroxysteroid dehydrogenase (HSD) family, 14 structurally-related enzymes implicated in steroid and fatty acid metabolism (28); the substrate of HSD17B13 is currently unknown. Given the association of HSD17B13 with NAFLD, we hypothesized that its functional role is due to its enzymatic activity and that SNPs modulate that activity. Beyond its similarity to the 17β-HSD family, HSD17B13 is structurally similar to other members of the short-chain dehydrogenase/reductases (SDR16C) family (29), known to play a critical role in retinoid metabolism through their retinol dehydrogenase enzymatic activity, catalyzing the oxidation of retinol to retinaldehyde, the rate-limiting step in all-trans-RA biosynthesis (Figure 4A). Cells transfected with HSD17B13(A) (variant A) demonstrated RDH activity comparable to that of the positive control RDH10 (Figure 4B–D). In contrast, HSD17B13(B) had no RDH activity (Figure 4E–G). HSD17B13-Δ22–28, which does not target lipid droplets, also lacked RDH activity although its expression was markedly lower than variants A and B, possibly because of low protein stability (Supplementary Figure 10). Thus, HSD17B13 demonstrates in vitro RDH activity that requires lipid droplet targeting for its stability and enzymatic function.
Figure 4.
HSD17B13 is a retinol dehydrogenase. (A) Oxidation of retinol to retinaldehyde by retinol dehydrogenases (asterisk) is the rate limiting step of retinoic acid synthesis. HEK293 cells transfected with HSD17B13(A), empty vector, or RDH10 as positive control were treated with retinol (5 μM) and levels of synthesized retinal (B) and retinoic acid (C) were measured by HPLC. Transfection of HSD17B13(B) variant does not lead to increased retinaldehyde (E) nor retinoic acid (F) synthesis. Co-factor binding site mutation protein HSD17B13(A)-G47/49A has no RDH activity (H and I) in HEK293 cells treated with 2 μM retinol. Anti-Flag Western blot used to determine protein transfection efficiency in each experiment (D, G, and J). Data are presented as Mean ± SEM.
HSD17B13 contains the conserved TGXXXGXG NAD(P)(H) co-factor binding site (31) common to SDRs (32). We generated a mutation within this motif (HSD17B13-G47/G49A) which resulted in loss of RDH activity (Figure 4H–J), confirming the need for cofactor binding for enzymatic activity.
THE IMPACT OF GENETIC VARIANTS IN HSD17B13 ON ENZYMATIC ACTIVITY
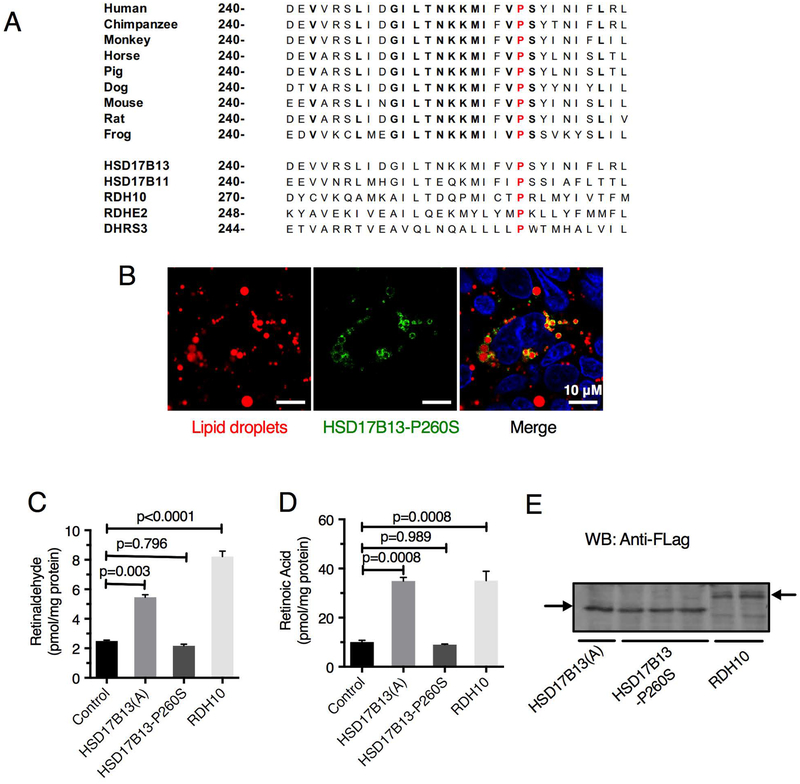
The non-synonymous coding SNP rs62305723G>A encodes a P260S substitution. P260 is highly conserved in vertebrate HSD17B13 orthologs and in other SDR16C family members (Figure 5A), even though it is outside the predicted catalytic or cofactor binding sites, suggesting its importance for protein function. We generated and tested this mutant form for enzymatic activity. Although the mutant protein HSD17B13(A)-P260S retains lipid droplet localization (Figure 5B) similarly to wild-type protein, it lacks RDH activity (Figure 5C–E). Thus, the minor allele of rs62305723 is generating a loss-of-function mutation that is associated with decreased hepatic inflammation and injury in patients with NAFLD.
Figure 5.
The P260S mutation in HSD17B13 (rs62305723G>A) is associated with loss of RDH activity despite lipid droplet targeting. (A) The P260S substitution is located within a highly conserved region of HSD17B13 orthologues and human homologues. (B) In HepG2 cells transfected with HSD17B13-P260S-GFP, the mutant protein, similar to the wild type protein, can be detected in lipid droplets. Transfection of HEK293 cells with HSD17B13-P260S-Flag does not lead to increased retinaldehyde (C) or retinoic acid (D) synthesis in the presence of 2 μM retinol despite adequate transfection efficiency (E). Data are presented as Mean ± SEM.
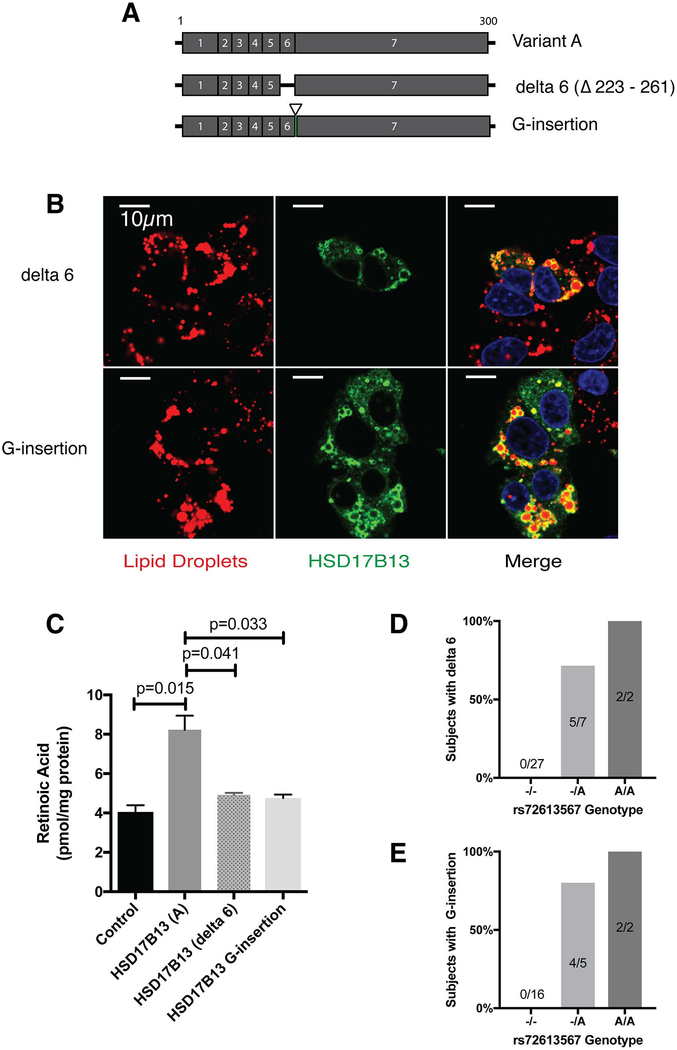
The indel rs72613567 was associated with histological and serological features of hepatic fat and injury (Table 1). The indel is located in the 5’ end of the intron between exon 6–7, with the minor allele (A-insertion) modifying a conserved splice donor sequence (GTAAGT to GTAAAGT). To determine whether this SNP affects splicing, we amplified and sequenced the surrounding regions from DNA and RNA of 36 human liver samples. We identified two novel HSD17B13 splice variants – one with a single G-nucleotide insertion (G-insertion) between exons 6–7 causing a frameshift and premature stop-codon, and another with exon 6 skipping (delta-6) (Figure 6A). Both the delta-6 and the G-insertion variants retain localization to lipid droplets (Figure 6B) but have no RDH activity (Figure 6C) and were only found in subjects with the minor rs72613567A allele (Figure 6D–E), who, as expected, also carried the rs6834314 minor G-allele (Supplementary Figure 11A–B). Delta-6 was expressed simultaneously with exon 6-containing variants at a relatively fixed ratio (Supplementary Figure 11C–D).
Figure 6.
HSD17B13 delta 6 and G-insertion variants localize with lipid droplets, lose RDH activity, and are associated with rs72613567. (A) Schema of HSD17B13 variants A, delta 6, and G-insertion. (B) In HepG2 cells transfected with HSD17B13-delta 6-Flag or HSD17B13-G insertion-Flag the mutant proteins can be detected in lipid droplets stained by LipidTox. (C) Transfection of HEK293 cells with HSD17B13-delta 6 or HSD17B13-G insertion does not lead to increased retinoic acid levels. The delta 6 (D)and G-insertion (E) variants are only detected in subjects who carry the rs72613567 minor A allele.
DISCUSSION
In otherwise-normal subjects, elevated plasma liver enzymes are often considered a surrogate for the presence of liver fat but can also reflect other liver processes, derive from non-hepatic sources, or reflect variability in the genes for the enzymes themselves. Chambers et al (18) found at the population level several genetic variants that associate with liver enzyme levels. We aimed to identify which of these associations actually reflects fatty liver or injury by assessing the association of the variants with histological features of NAFLD. We identified several SNPs that associated with NAFLD features and by focusing on one of them, rs6834314, identified HSD17B13 as a lipid-droplet enzyme associated with the disease.
Other 17β-HSDs act as molecular switches, modulating the activation of steroid hormone receptors in target tissues (28). In vivo, some 17β-HSDs may have targets other than steroid hormones, such as prostaglandins, bile acids, and cholesterol and some have been implicated in fatty acid metabolism (34). Compared to other family members that are restricted to gonads or ubiquitously expressed, HSD17B13 is predominantly expressed in the liver, and the protein is targeted from the ER to lipid droplets by its N-terminal region (27,31,35). We confirmed HSD17B13’s lipid-droplet localization and identified a conserved 7 amino acids sequence (AA22–28) that is crucial for lipid droplet targeting. Interestingly, a naturally occurring splice variant, HSD17B13(B) that lacks exon 2 is not targeted to lipid droplets, despite the presence of AA22–28. In overexpression experiments using equal amounts of plasmid, the Δ22–28 variant and HSD17B13(B) demonstrate lower protein levels than wild-type, suggesting that proper targeting is crucial for protein stability and protection from degradation; a similar phenotype was described for TM6SF2 variants (10).
We found HSD17B13 to be highly expressed in NAFLD patients, consistent with a recent report (35). However, its specific target and physiological function have not been defined so far and neither was its role in NAFLD pathogenesis. We establish that HSD17B13 has RDH activity, and that its enzymatic activity depends on lipid-droplet targeting and cofactor binding. HSD17B13 clearly has RDH activity in vitro and this assay can be used to assess the impact of genetic variations on enzymatic function. Although in vivo the enzyme may have other substrates in addition to retinol, retinol is still a plausible in vivo substrate. Retinoids are stored as retinyl esters in the lipid droplets of hepatic stellate cells that, when activated, play critical roles in hepatic fibrogenesis. There is significant evidence that vitamin A metabolites (retinaldehyde and RA) and retinol binding protein (RBP4) are associated with pathogenesis of hepatic steatosis, fibrosis, adipogenesis, and insulin resistance (36–41). Enzymes involved in retinol metabolism are upregulated in hepatic lipid droplets in NAFLD (35). Furthermore, PNPLA3, strongly associated with NAFLD (3), acts as a retinyl-palmitate lipase to control serum retinol and RBP4 level in NAFLD patients and to modulate stellate cell fibrogenesis (42–45). This supports the critical role retinoid metabolism plays in NAFLD progression. The association of the P260S substitution with a diagnosis of blindness or low vision further supports a role for HSD17B13 in retinoid metabolism. In addition, the putative substrate binding sites (L153/L156, K208, L199/E202), homodimer interaction sites, and the P260 residue found in this study are all crucial for enzymatic activity and are conserved within RDH10, DHRS3, and HSD17B11, but not with other 17β-HSDs (Supplementary Figure 12). In addition, a recent study in mice heterozygous for Rdh10 knockout found increased steatosis on high fat diet despite only a modest decrease in hepatic RA (46). This suggests unique substrate specificity and is consistent with retinoids being the substrate for HSD17B13. Ongoing in vivo experiments are aimed at confirming these findings. Hepatic HSD17B13 expression appears limited to hepatocytes, with no detectable expression in quiescent or activated stellate cells (data not shown). Whether HSD17B13 acts directly on the hepatocyte via modulation of hepatocyte RA synthesis, or indirectly through a paracrine or autocrine effect, remains to be shown. Our data suggest that HSD17B13 is a hitherto undescribed hepatic RDH and that its effect on NAFLD pathogenesis relates to its enzymatic activity and possibly to its impact on retinoid homeostasis. Further characterization of the enzymatic activity of HSD17B13 in vitro and in vivo can help clarify its role. The increased levels of HSD17B13 in NAFLD patients may reflect a separate pathological process in the multifactorial pathogenesis of NAFLD.
The genetic association of variants in HSD17B13 with features of NAFLD is complex, with different SNPs associated with different phenotypic patterns. We identified two different and independent SNPs that result in loss-of-function variants and reduction in NAFLD-associated injury and inflammation and are likely causal variants. The rs62305723 SNP encodes a P260S mutation that abolishes the RDH activity in vitro. Conservation of P260 among vertebrates in the HSD17B13 gene and in the SDR16C family indicates this site is essential for protein function. A second SNP, rs72613567, generates two simultaneously-expressed novel splice variants that are also devoid of enzymatic function, and are also associated with decreased injury. This SNP is in high LD with the tag SNP rs6834314, and likely explains the associations found with it, as well as the association with ALT reported by Chambers (18) and Xu (33), especially since we demonstrate that rs6834314 is not an expression quantitative trait locus (eQTL). Association with fibrosis stage, the major determinant of prognosis in NASH, was weaker, but rs6834314 and rs72613567 are associated with cirrhosis in the MGI and UK Biobank cohorts, respectively, consistent with their impact on liver injury. Interestingly, both rs62305723 and rs72613567 are located in or near exon 6 of the gene, away from the conserved catalytic and co-factor binding motifs of the 17β-HSD family (28). Overall, our data supports a role for HSD17B13 in mediating NAFLD-associated injury through its enzymatic activity.
Some variants in HSD17B13 were associated with histological degree of steatosis, including rs6834314, its associated splice SNP rs72613567, and other independent SNPs. The association of rs6834314 with steatosis was confirmed in an independent cohort of patients with chronic hepatitis C. The alleles that associate with increased hepatic fat are those that associate with decreased injury, and the association with injury was independent of the association with steatosis. However, the loss-of-function P260S variant is not associated with steatosis. This suggests that the association with hepatic fat may be unrelated to the enzymatic activity and could be driven by another mechanism. This is also supported by our in vitro data, where knocking-out or overexpressing HSD17B13 in hepatoma cells does not affect their fat-storage capacity. Consistent with our data, Abul-Husn (47) reported that HSD17B13 overexpression does not change cellular lipid content. In contrast, Adam (48) recently found that Hsd17b13 knock-out mice showed hepatic steatosis only at an older age, indicating HSD17B13 might affect hepatic fat metabolism in vivo; the underlying mechanism is yet to be explored.
In a proteomic analysis of hepatic lipid droplets, Su (35) recently identified upregulation of HSD17B13 in subjects with NAFLD, consistent with our results. They demonstrated an increase in lipid droplet size in cells over-expressing HSD17B13 tagged with GFP; we obtained similar results but were not able to replicate them when transfecting with HSD17B13 with a smaller tag (Flag epitope), suggesting the large GFP-tag on lipid-droplet associated HSD17B13 causes non-specific lipid aggregation (data not shown). HSD17B13 expression appears to be regulated by LXR and SREBP-1c (49), which can explain our finding of its upregulation in patients with NASH and is consistent with a functional role in NAFLD pathogenesis. Additional evidence supporting a role for HSD17B13 in lipid metabolism comes from a recent GWAS (50), in which rare variants in HSD17B13 were found to impact the effect of fenofibrate on plasma triglycerides and high-density lipoprotein.
Recently, Abul-Husn (47) also reported that the splice-site SNP rs72613567 is associated with levels of ALT and AST and with the risk of chronic liver disease. Their findings, including the presence of loss-of-function splice variants associated with rs72613567, are consistent with ours and together highlight the role of HSD17B13 in liver injury. Whether the association with injury is specific to fatty liver disease is unclear. A major strength of our study compared to (47) is our focus on genetic associations with the separate specific histological components of NAFLD, allowing us to highlight the different impact on hepatic fat and injury. Our findings suggest that HSD17B13 affects injury and inflammation in NAFLD differently than in viral hepatitis, and that the association with steatosis, not previously reported, may be due to a different mechanism. Furthermore, beyond the data on the impact of truncated or unstable forms generated by the splicing SNP, also demonstrated by Abul-Husn (47), we describe for the first time a novel single amino acid mutation of HSD17B13 that confers loss of enzymatic activity despite stable protein expression and proper cellular localization, and associate this novel mutation with histological severity of NAFLD. This confirms the link between enzymatic function and phenotype.
In conclusion, we confirmed that the association of rs6834314 with ALT reflects its association with NAFLD and that HSD17B13 represents the affected gene of interest. Our data establish HSD17B13 as a lipid-droplet associated protein and point to its RDH activity as a key aspect of function, with a potential pathogenic link to NAFLD. Genetic variations in HSD17B13 have complex consequences that vary by type and locus, and further investigation into the genotype-phenotype association is warranted.
Supplementary Material
Acknowledgements:
The authors thank Ms. Ronda K. Sapp for technical assistance; Christopher O’Donnell, MD and Caroline Fox, MD, MPH from the Framingham Heart Study for collaboration during study initiation; the University of Michigan Medical School Central Biorepository for providing biospecimen processing, storage, management, and distribution services in support of the research reported in this publication; and Haiqing Fu from NCI, NIH for sharing of plasmid.
The authors are grateful to the NASH CRN investigators, coordinators and patients for allowing data access.
Financial Support:
This study was supported by the Intramural Research Programs of the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and NIH, National Cancer Institute. N.Y.K. is supported by an NIH, National Institute of Alcohol Abuse and Alcoholism (NIAAA) grant AA012153. E.K.S., X.D., and Y.C. are supported by NIH grants R01 DK106621, R01 DK107904. S.K.H. is supported by NIH grant R01 DK106621. E.K.S., X.D., Y.C., S.K.H., and V.C. are supported by The University of Michigan Department of Internal Medicine.
The Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) is supported by the NIDDK (grants U01DK061718, U01DK061728, U01DK061731, U01DK061732, U01DK061734, U01DK061737, U01DK061738, U01DK061730, U01DK061713). Additional support is received from the National Center for Advancing Translational Sciences (NCATS) (grants UL1TR000439, UL1TR000436, UL1TR000006, UL1TR000448, UL1TR000100, UL1TR000004, UL1TR000423, UL1TR000058).
The Liver Tissue and Cell Distribution System, Department of Pediatric Gastroenterology, Hepatology, and Nutrition, University of Minnesota Medical School was funded by NIH Contract #HHSN276201200017C.
Data for UK Biobank analyses was provided under approved protocol 18120.
List of Abbreviations:
- HSD17B13
17-beta hydroxysteroid dehydrogenase 13
- ALT
Alanine aminotransferase
- CI
Confidence interval
- LD
Linkage disequilibrium
- GWAS
Genome wide association studies
- HPLC
High pressure liquid chromatography
- NAFLD
Non-alcoholic fatty liver disease
- NASH
Non-alcoholic steatohepatitis
- NASH CRN
NASH Clinical Research Network
- OR
Odds ratio
- RA
Retinoic acid
- RDH
Retinol dehydrogenase
- RBP4
Retinol binding protein 4
- SNP
Single nucleotide polymorphism
REFERENCES
- 1.Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology 2009;136:1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 2004;40:1387–1395. [DOI] [PubMed] [Google Scholar]
- 3.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS genetics 2011;7:e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 2008;83:520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010;52:894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res 2009;50:2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 2010;52:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology 2009;137:865–872. [DOI] [PubMed] [Google Scholar]
- 10.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjaerg-Hansen A, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmen OL, Zhang H, Fan Y, Hovelson DH, Schmidt EM, Zhou W, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet 2014;46:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A 2014;111:8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anstee QM, Day CP. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin Liver Dis 2015;35:270–290. [DOI] [PubMed] [Google Scholar]
- 14.Kahali B, Liu YL, Daly AK, Day CP, Anstee QM, Speliotes EK. TM6SF2: catch-22 in the fight against nonalcoholic fatty liver disease and cardiovascular disease? Gastroenterology 2015;148:679–684. [DOI] [PubMed] [Google Scholar]
- 15.Buch S, Stickel F, Trepo E, Way M, Herrmann A, Nischalke HD, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443–1448. [DOI] [PubMed] [Google Scholar]
- 16.Simons N, Isaacs A, Koek GH, Kuc S, Schaper NC, Brouwers M. PNPLA3, TM6SF2, and MBOAT7 Genotypes and Coronary Artery Disease. Gastroenterology 2017;152:912–913. [DOI] [PubMed] [Google Scholar]
- 17.Mancina RM, Dongiovanni P, Petta S, Pingitore P, Meroni M, Rametta R, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016;150:1219–1230 e1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol 2003;98:960–967. [DOI] [PubMed] [Google Scholar]
- 20.Ioannou GN, Boyko EJ, Lee SP. The prevalence and predictors of elevated serum aminotransferase activity in the United States in 1999–2002. Am J Gastroenterol 2006;101:76–82. [DOI] [PubMed] [Google Scholar]
- 21.Neuschwander-Tetri BA, Clark JM, Bass NM, Van Natta ML, Unalp-Arida A, Tonascia J, et al. Clinical, laboratory and histological associations in adults with nonalcoholic fatty liver disease. Hepatology 2010;52:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanyal AJ, Chalasani N, Kowdley KV, McCullough A, Diehl AM, Bass NM, et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med 2010;362:1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313–1321. [DOI] [PubMed] [Google Scholar]
- 24.Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, Kedishvili NY. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry 2005;44:7035–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 26.Pike N Using false discovery rates for multiple comparisons in ecology and evolution. Methods in Ecology and Evolution 2011;2:278–282. [Google Scholar]
- 27.Horiguchi Y, Araki M, Motojima K. Identification and characterization of the ER/lipid droplet-targeting sequence in 17beta-hydroxysteroid dehydrogenase type 11. Arch Biochem Biophys 2008;479:121–130. [DOI] [PubMed] [Google Scholar]
- 28.Marchais-Oberwinkler S, Henn C, Moller G, Klein T, Negri M, Oster A, et al. 17beta-Hydroxysteroid dehydrogenases (17beta-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. The Journal of steroid biochemistry and molecular biology 2011;125:66–82. [DOI] [PubMed] [Google Scholar]
- 29.Belyaeva OV, Chang C, Berlett MC, Kedishvili NY. Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem Biol Interact 2015;234:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belyaeva OV, Johnson MP, Kedishvili NY. Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem 2008;283:20299–20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Huang C, Li D, Ren W, Zhang H, Qi M, et al. Molecular cloning and expression analysis of a new gene for short-chain dehydrogenase/reductase 9. Acta Biochim Pol 2007;54:213–218. [PubMed] [Google Scholar]
- 32.Duax WL, Pletnev V, Addlagatta A, Bruenn J, Weeks CM. Rational proteomics I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family. Proteins 2003;53:931–943. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Jiang CQ, Lam TH, Zhang WS, Zhu F, Jin YL, et al. Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Hum Mol Genet 2017;26:430–437. [DOI] [PubMed] [Google Scholar]
- 34.Shafqat N, Marschall HU, Filling C, Nordling E, Wu XQ, Bjork L, et al. Expanded substrate screenings of human and Drosophila type 10 17beta-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3alpha/7alpha/7beta/17beta/20beta/21- HSD. The Biochemical journal 2003;376:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su W, Wang Y, Jia X, Wu W, Li L, Tian X, et al. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci U S A 2014;111:11437–11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G The link between Hepatic Vitamin A Metabolism and Nonalcoholic Fatty Liver Disease. Curr Drug Targets 2015;16:1281–1292. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya H, Ikeda Y, Ebata Y, Kojima C, Katsuma R, Tsuruyama T, et al. Retinoids ameliorate insulin resistance in a leptin-dependent manner in mice. Hepatology 2012;56:1319–1330. [DOI] [PubMed] [Google Scholar]
- 38.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med 2007;13:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SC, Kim CK, Axe D, Cook A, Lee M, Li T, et al. All-trans-retinoic acid ameliorates hepatic steatosis in mice by a novel transcriptional cascade. Hepatology 2014;59:1750–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356–362. [DOI] [PubMed] [Google Scholar]
- 41.Graham TE, Yang Q, Bluher M, Hammarstedt A, Ciaraldi TP, Henry RR, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 2006;354:2552–2563. [DOI] [PubMed] [Google Scholar]
- 42.Mondul A, Mancina RM, Merlo A, Dongiovanni P, Rametta R, Montalcini T, et al. PNPLA3 I148M Variant Influences Circulating Retinol in Adults with Nonalcoholic Fatty Liver Disease or Obesity. J Nutr 2015;145:1687–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruschi FV, Claudel T, Tardelli M, Caligiuri A, Stulnig TM, Marra F, Trauner M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017. [DOI] [PubMed] [Google Scholar]
- 44.Pirazzi C, Valenti L, Motta BM, Pingitore P, Hedfalk K, Mancina RM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet 2014;23:4077–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenti L, Romeo S. Destined to develop NAFLD? The predictors of fatty liver from birth to adulthood. J Hepatol 2016;65:668–670. [DOI] [PubMed] [Google Scholar]
- 46.Yang D, Vuckovic MG, Smullin CP, Kim M, Lo CP, Devericks E, et al. Modest Decreases in Endogenous All-trans-Retinoic Acid Produced by a Mouse Rdh10 Heterozygote Provoke Major Abnormalities in Adipogenesis and Lipid Metabolism. Diabetes 2018;67:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med 2018;378:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adam M, Heikela H, Sobolewski C, Portius D, Maki-Jouppila J, Mehmood A, et al. Hydroxysteroid (17beta) dehydrogenase 13 deficiency triggers hepatic steatosis and inflammation in mice. FASEB J 2018:fj201700914R. [DOI] [PubMed] [Google Scholar]
- 49.Su W, Peng J, Li S, Dai YB, Wang CJ, Xu H, et al. Liver X receptor alpha induces 17beta-hydroxysteroid dehydrogenase-13 expression through SREBP-1c. Am J Physiol Endocrinol Metab 2017;312:E357–E367. [DOI] [PubMed] [Google Scholar]
- 50.Rotroff DM, Pijut SS, Marvel SW, Jack JR, Havener TM, Pujol A, et al. Genetic variants in HSD17B3, SMAD3, and IPO11 impact circulating lipids in response to fenofibrate in individuals with type 2 diabetes. Clin Pharmacol Ther 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.