Abstract
Bottom-up proteomics relies on the proteolytic or chemical cleavage of proteins into peptides, the identification of those peptides via mass spectrometry, and the mapping of the identified peptides back to the reference proteome to infer which possible proteins are identified. Reliable mapping of peptides to proteins still poses substantial challenges when considering similar proteins, protein families, splice isoforms, sequence variation, and possible residue mass modifications, combined with an imperfect and incomplete understanding of the proteome. The ProteoMapper tool enables a comprehensive and rapid mapping of peptides to a reference proteome. The indexer component creates a segmented index for an input proteome from a FASTA or PEFF file. The ProMaST component provides ultra-fast mapping of one or more input peptides against the index. ProteoMapper allows searches that take into account known sequence variation encoded in PEFF files. It also enables fuzzy searches to find highly similar peptides with residue order changes or other isobaric or near-isobaric substitutions within a specified mass tolerance. We demonstrate an example of a one-hit-wonder identification in PeptideAtlas that may be better explained by a combination of catalogued and uncatalogued sequence variation in another highly observed protein. ProteoMapper is free and open source, available for local use after downloading, embedding in other applications, as an on-line web tool at http://www.peptideatlas.org/map, and as a web service.
Keywords: Human Proteome Project, mass spectrometry, peptides, single amino acid variations, PEFF, ProteoMapper, CLIPS, ProMaST, proteomics
Introduction
Mass spectrometry-based proteomics is currently the most prevalent technique for identifying and quantifying the abundance of proteins in biological samples at almost complete proteome scales1–3. In typical workflows, proteins extracted from a sample are digested with trypsin, fractionated by liquid chromatography, ionized, and introduced into a mass spectrometer for analysis4. After measuring the intact mass-to-charge ratio of the charged peptide precursors, the instrument fragments these by methods such as collisional dissociation and records the resultant product ion mass spectra generated. Current instrumentation collects millions of fragment ion spectra generated per experiment which are then subjected to computational analysis to identify the peptidoforms present in the sample and quantify their abundances5,6. The peptidoform identification process involves substantial uncertainty, but the statistics are generally well understood and thresholds can be set such that nearly all identifications are correct, with the tolerance of uncertainty selectable by the researcher.
However, the following step in which the putatively identified peptide sequences, even if nearly all are correct, are subsequently mapped to their original proteins also introduces substantial uncertainty, which must be properly handled, such as with tools like ProteinProphet7 and IDPicker8. Approximately 90% of peptides in the Human PeptideAtlas map uniquely to one gene and are generally termed uniquely mapping peptides. However, there remain a substantial number of complications. For example, one gene often yields multiple splice isoforms, several of which may be present in a sample; distinguishing which of several isoforms are present may or may not be important for a particular experiment. In addition, many different genes and their proteins have sequences similar enough that some peptides map to multiple proteins, both among genes related to each other in a family as well as due to short repeated motifs among unrelated proteins. An often-overlooked complication is that for many species there is a tremendous amount of genetic variation, a fraction of which will yield changes in protein sequence. Indeed our overall understanding of the proteome of a species or an individual continues to change. The shorter the peptide, the greater the chance that it will map to multiple proteins, and is often termed a multiply mapping peptide. The result of this ambiguity results in additional uncertainty in the identification of proteins, which must be modeled in combination with the uncertainty of peptide identifications.
The international Human Proteome Project9,10 (HPP) of the Human Proteome Organization11 (HUPO) has defined one of its primary goals as advancing our understanding of the many aspects that comprise the human proteome, including a more complete understanding of the protein “parts list”, the full complement of splice isoforms, functions, sequence variations, and post-translational modifications (PTMs) for each protein. The knowledge being generated as part of the concerted effort of the international community of biomedical research, both within the HPP and external to it, is being assembled into the neXtProt12 knowledge base, which itself derives from UniProtKB13. A part of this effort is to classify protein entries in terms of the evidence for their bona fide translation into a protein in vivo. The protein evidence (PE) score denotes PE=1 for proteins with high confidence evidence of translation, PE=2 for evidence of transcription (but not yet translation), PE=3 for orthologs of predicted proteins in other species, and PE=4 for solid predictions but no other evidence. PE=5 is reserved for entries that are likely to be pseudogenes and are not translated, but there remains some past questionable translation evidence that hinders their deletion from the proteome; a few may be real proteins and await better evidence before an upgrade to PE=1. The HPP has defined PE=2–4 proteins as “missing proteins” (MPs), a set for which solid translation evidence is being actively sought in order to upgrade them to PE=1 or delete them from the proteome, as appropriate. As of neXtProt version 2018–01, there remain only 2186 missing proteins, a mere 10% of the expected entire human proteome based on the agreed identification of open reading frames resulting in gene calls. However, gathering the necessary evidence for these MPs is becoming progressively harder as most of these proteins are low-abundance, highly tissue specific, and/or membrane-bound proteins, and are therefore difficult to isolate, detect and identify via current mass spectrometry workflows. The HPP has written a set of mass spectrometry data interpretation guidelines14 to aid in the analysis and presentation of mass spectrometry data that purport to provide evidence for these MPs or other translation products not even in the current neXtProt proteome. One of these guidelines requests that a high confidence peptide that appears to map uniquely to a MP be carefully checked to determine if a mapping to a commonly detected PE=1 protein with a type of variation that is not trivially accounted for is possible. This is a laudable and important guideline, but one that is quite difficult for many to address.
The neXtProt knowledge base has already take some steps toward addressing the challenge posed by this guideline. A first step taken by neXtProt is to collect all reports of human protein variation and disseminate those results in a form that can be easily leveraged in software tools. These variations are included on neXtProt’s web site, in its custom API, in its custom XML export format, and in an emerging format in the final stages of ratification by the HUPO Proteomics Standards Initiative (PSI). This PSI Extended FASTA Format (PEFF) is similar to the common FASTA format, but enables a consistent mechanism for parsing annotations about each protein entry, including annotations that denote single amino acid variations (SAAVs), PTMs, disulfide bonds, and more. The neXtProt team has also provided a software tool called the neXtProt uniqueness checker15 that enables the searching of peptides against neXtProt in order to aid in compliance with the HPP guidelines. However, the uniqueness checker tool only partially implements a solution for fully conforming to HPP guidelines about peptide mapping, and additional work in this area is needed.
Here we describe a new tool called ProteoMapper that provides a more extensive solution to the needs for comprehensively fulfilling the HPP guidelines. In the following sections, we first provide a general overview of the tool, and then describe details about the proteome indexing and peptide searching functionalities. We describe the many ways to use this tool and provide some examples of its application.
Overview
There are two primary components to the ProteoMapper application, the indexer and the search tool. The indexer component (CLIPS) reads a FASTA or PEFF file and organizes the sequences (including variations) therein into a highly efficient index that enables ultra-fast searching of peptides against that index. The ProteoMapper search tool (ProMaST) component takes as input one or more peptides and maps those peptides to the designated proteome index modulated by various user-selectable options.
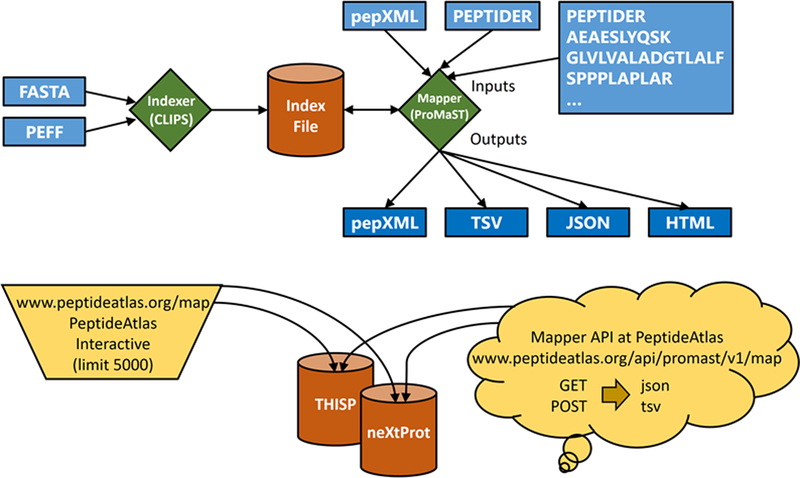
Figure 1 provides a graphical overview of the general workflow of the various ProteoMapper components. While the indexer component is a command-line tool that need only be run when new proteomes are incorporated, the ProMaST component has several usage modes from a command-line tool to embeddable source code that can be included in other applications to web services that can be easily called from web-enabled applications.
Figure 1:
Graphical overview of all ProteoMapper components. The indexer takes as input a FASTA or PEFF file and creates a segmented index. The mapper takes as input one or more peptides as a list or a pepXML file, a previously created index, and other optional parameters. It then provides its output in pepXML, TSV, JSON, or HTML. In addition to a downloadable form for local use, an interactive web page and a web service are available for a predefined set of reference proteomes at PeptideAtlas.
Proteome Indexing
The ProteoMapper indexer component (CLIPS) transforms an input FASTA file or PEFF file into an index format needed by ProMaST. The basic workflow is to iterate over each protein entry in the input file, splitting each entry into all possible segments of “n” amino acids in length, irrespective of any protease. All discovered segments are written in alphabetical order to the index file along with a listing of all proteins and locations to which the segments map. The user may select the desired segment size. The longer the segment size, the more entries there are in the index, albeit with few protein mappings per segment. For example with a segment size of two with 20 amino acids, there would only be 400 entries in the index (AA, AC, AD, etc.), but each segment would map to nearly every protein. At a segment size of 10, there are 20^10 entries, albeit with most entries containing no matches. Figure 2 plots the overall size of the index, the time to build the index, and the time required to search a uniform test set of peptides against each index of segment sizes ranging from 3 to 6, both for the human proteome without variants and for the neXtProt human proteome with all SAAVs considered. Based on the trade-offs depicted in Figure 2, we define a default segment size of 5, although the user can override this as desired.
Figure 2:
Bar chart of the size of the index, time to build the index, and mapping time for a reference set of 3700 peptides, all as a function of segment sizes 3–6, and running 1, 2, 4, or 8 threads (parallel executions within the same process).
When an input file is a PEFF file that contains sequence variations (as specified by the VariantSimple keyword), all possible permutations of SAAVs are encoded into the index by default. If the -V flag is set, then SAAVs are ignored.
Another default setting is to treat all isoleucines (I) as leucines (L) in the index, since these two amino acids have identical m/z and cannot be distinguished with most current mass spectrometry workflows. This enables a smaller index and ensures that I and L are interchangeable without fuzzy searching, as is usually appropriate. This option can be disabled if desired for use with workflows which are able to distinguish between I and L16. In order to reduce processing time and storage costs for input files with duplicate entries, all input proteins are checked for cases of identical sequence and, where appropriate, identical SAAVs, and the mapping of duplicate entries is stored in a separate section of the index file, while only being segmented once. Instances of duplicate identifiers are flagged as an error.
The index building does incur an overhead, both in terms of size on disk and CPU time. However, these costs are quite modest by modern standards. A 7 MiB FASTA file of the baker’s yeast proteome of 13,368 proteins, including contaminants and decoys but with no variations, expands to a 57 MiB index in 10 seconds on average hardware. A 124 MiB neXtProt PEFF file with 43,000 isoform sequences and 4.3 million SAAVs expands to a 1.4 GiB index in 9 minutes on average hardware. The indexing is only single threaded (serial execution) since the indexer is run rather infrequently at times that do not delay a user experience.
Peptide Mapping
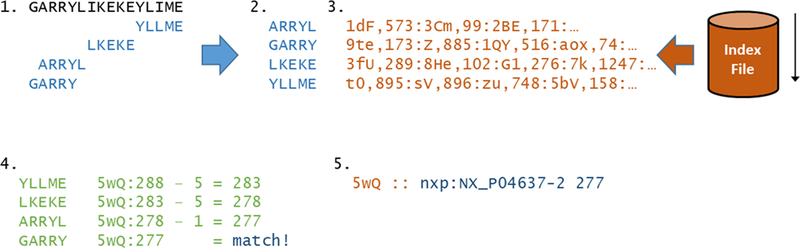
The peptide mapping component, ProMaST, takes as input one or more peptide sequences to map, an index that has already been created by the indexer, and a set of user-selectable options that control several aspects of the mapping. The basic workflow of ProMaST is to execute the following steps for the set of input peptides, as depicted in Figure 3. First, all input peptides are decomposed into an approximately minimal set of segments of the same segment size used for the reference input index. Two segments for a peptide may overlap if the peptide is not a multiple of the segment size. For example, an input peptide of PEPTIDER would decompose to PEPTI and TIDER for an index size of five. Next, the sorted list of input segments is searched in order as a single pass through the index.
Figure 3:
Graphical overview of the detailed workflow of the searching process. 1. Every input peptide is split into segments, taking into account I->L substitutions; 2. All input segments are sorted alphabetically; 3. In a single pass, segment entries (encoded as a colon-separated list of protein key,offsets) are extracted from the index, which is also alphabetically sorted for efficiency; 4. For each peptide, its segment entries are matched based on protein entry and position within it; 5. The protein alias is resolved via lookup and reported along with position
Next, with a complete list of the mapping of the segments in hand, the contiguity of the mappings is checked. In the above example it is not sufficient that both PEPTI and TIDER map to a given protein, but also that the mapping position of TIDER is 3 amino acids after the mapping position of PEPTI for the mapping to be complete. Complications where some or all segments map multiply to the same protein are also handled by selecting only the segments that can form contiguous sets. The final step is to report the final list of mapping locations for each input peptide, along with a few additional attributes of the mapping such as the preceding and following amino acids, and the number of simultaneous sequence variations required to enable the mapping. There is no upper bound to the number of peptide sequences that may be passed into the command-line program, although large lists require greater computer resources. A set of ~1.4 million peptide sequences was passed and required 4 GB of RAM. Doubling the number of input peptides will less than double the required RAM because many segment mappings will be reused.
A key advanced feature of ProMaST over previous similar tools is to enable fuzzy searches in addition to exact searches. In this mode the search tool can find all mappings of one or more peptides where each amino acid may be substituted for any other. For instance, reusing our previous example, a 1-wildcard search would search for XEPTIDER, PXPTIDER, PEXTIDER, etc. throughout the index, where X represents any amino acid. A 2-wildcard search scans the index for all instances of the 1-wildcard case plus XXPTIDER, XEXTIDER, XEPXIDER, etc. ProMaST supports up to three wildcards per peptide, although with three wildcards, they are only considered as a consecutive group. Naturally, this dramatically increases the search space and search time required, and potentially the output list. When fuzzy mode is enabled, only one peptide may be passed at a time because the single peptide is expanded into a list of all possible permutations and this list of permutations becomes the effective input. Future versions will allow a list input in fuzzy mode.
Importantly, the user may also specify a mass tolerance with which to filter the candidate list, such that the reported wildcard matches must not alter the mass of the new peptide by more than the specified tolerance to be reported. A mass tolerance of 0 easily finds cases of amino-acid position swapping with two or more wildcards. Although I/L is the only single isobaric amino acid pair, there are many double and triple isobaric groups (e.g., SL=TV, AM=CV). A specified mass tolerance of 0.1 Da reveals many more near equivalences such as K ≈ Q.
Along with the above fuzzy search and mass tolerance settings, ProMaST can also consider a subset of common mass modifications present in UniMod, a public database of known mass modifications available at http://unimod.org/. Currently implemented potential mass modifications are acetyl, carbamidomethyl, carboxymethyl, deamidation, methyl, hydroxylation, and phospho (Unimod names).
ProteoMapper implements several performance and capability improvements over the current neXtProt uniqueness checker and the pepx program on which it is based. ProteoMapper creates a smaller index in a single file, provides context information such as position offsets and flanking amino acids, is about twice as fast while providing more context information (about 4 times as fast when gathering context information is skipped), and encodes all permutations of annotated variants. ProteoMapper ensures complete mapping when used at the command line and supports fuzzy matching capability to search for highly similar sequences not encoded as known variations.
Usability
The ProteoMapper tools are implemented in the Perl language and run well on any platform for which the Perl interpreter is installed, which is available by default or as an additional package on nearly all versions of GNU/Linux, Microsoft Windows, and Apple OS X. The source code can be downloaded from the web site http://tppms.org/pm as a zip file. Further documentation is also available the same site. In addition to standalone use, the application may be bundled with or embedded in other applications that need to be able to map identified peptides to all proteins. ProteoMapper is an embedded component of the Trans-Proteomic Pipeline17–19 as of version 5.2.0, enabling it to map discovered peptides in pepXML17 files against complex databases with variants in PEFF files such as the neXtProt proteome. ProteoMapper is licensed under the LGPL license, which permits its use in a wide variety of open and closed source scenarios.
The input set of peptides to search may be a single peptide, a list of peptides, or a pepXML file containing a set of matched peptides from the output of a search engine such as Comet20, X!Tandem21, MSFragger22, or any other for which output to or conversion to pepXML is possible. If the input was pepXML, the output may also be a refreshed pepXML file with the alternative mappings encoded. For all inputs, the output may be a tab-separated value format containing the alternative mapping information.
In addition to downloading the application and using it locally, ProMaST is also available for remote use at the PeptideAtlas23–25 server. ProMaST at PeptideAtlas can be used via the interactive web page at http://www.peptideatlas.org/map, where single peptides and peptide lists may be run through the tool and the output explored interactively via a web browser. The output is documented with extensive column descriptions and any variations and substitutions are highlighted with colors. The Indexer component is not available for use remotely. However, the PeptideAtlas server automatically regenerates indexes for a variety of sequence databases, including a subset of THISP databases26, neXtProt PEFF with variants, mouse and yeast proteomes, and more. The indexes are refreshed on the first day of every month automatically.
ProMaST is also available as a web service at PeptideAtlas at the endpoint http://www.peptideatlas.org/api/promast/v1/map. Documentation for the endpoint is available by pointing a browser to the same URL. The web service allows mapping of a single peptide and several of the most common options via an HTTP GET call. The output may be selected as JSON or TSV as described in the documentation.
In order to facilitate its use, several tutorials are available to demonstrate the use of ProteoMapper. One tutorial demonstrates interactive use of the tool at the ProMaST web page. A second tutorial demonstrates downloading the toolkit, indexing a FASTA file, and searching a list of peptides locally.
Applications
Several needs drove the development of ProteoMapper, including the need to map the millions of peptide sequences catalogued in PeptideAtlas to continually advancing reference proteomes and the need to be able to understand cases where very high scoring peptide-spectrum-matches that appear to implicate only very rare proteins may instead be mappable to common proteins with typically unconsidered variations.
The Human PeptideAtlas 2018–01b build contains ~1.4 million distinct peptide sequences derived from over 1000 datasets. These peptides are mapped to the THISP PeptideAtlas Mapping proteome26, which contains all variations from neXtProt as well as proteins from UniProtKB/TrEMBL13, ENSEMBL27, RefSeq28, and many more sources with 372,934 sequences in all, with substantial redundancy. Building the index for this very large database takes 25 minutes on standard hardware. Then the mapping of the ~1.4 million peptides takes 12 minutes total, a huge improvement over previous techniques, which took over 5 hours.
The HPP has written a set of mass spectrometry data interpretation guidelines14 to aid in the analysis and presentation of data that purport to provide evidence for the MPs or other translation products not even in the current neXtProt proteome. In version 2.1, guideline #14 states: “Even when very high confidence peptide identifications are demonstrated, consider alternate mappings of the peptides to proteins other than the claimed extraordinary result. Consider isobaric sequence/mass modification variants, all known SAAVs, and unreported SAAVs.” This guideline has been challenging for authors to meet because tools were lacking to assist with consideration of alternate mappings. The uniqueness checker tool at neXtProt (https://www.nextprot.org/tools/peptide-uniqueness-checker) was an important first step in assisting authors with meeting this guideline. However, it does not provide position offset information, works only against the most recently released neXtProt version, and does not provide options for exploring unannotated SAAVs or any mass modifications. These features are now available in ProteoMapper, which can thus assist in more thorough compliance with HPP guideline #14.
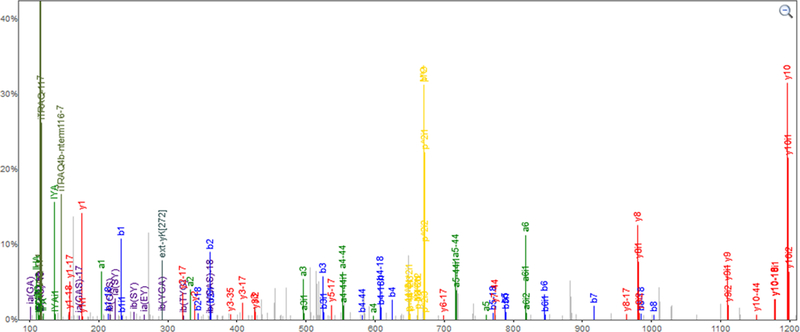
As an example of how to use ProMaST to address cases where complex ambiguity in mapping casts doubt on what otherwise would seem like a high confidence detection, we consider here the case of the Homeobox protein DLX-3 (O60479). In the latest build of the Human PeptideAtlas (2018–01b), this protein has a single peptide identification from one of the CPTAC29 datasets (https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/GetProtein?action=QUERY&atlas_build_id=472&protein_name=O60479). The PeptideAtlas protein classification is “weak” because it does not have the required two uniquely-mapping peptides of length ≥9 residues (from HPP guideline #15). The single peptide identification has an exquisite PSM, with all y ions y1 – y10 observed as well as b1 – b8 observed, with many neutral loss ions, internal fragmentation ions, and other corroborating peaks, as displayed in Figure 4. The iProphet probability is 1.000. It is difficult to imagine this PSM being anything but correct or very nearly correct (e.g. an isobaric residue substitution). But, is it conclusive evidence for O60479? A search for this peptide SEYTYGASYR with ProMaST with no fuzzy matching reveals no alternatives. However, when a single substitution is permitted with a null mass tolerance, a potential mapping to P23083 is revealed. P23083 is an immunoglobulin heavy chain V-I region V35 sequence, which has been observed a very large number of times via other peptides in these CPTAC data and dozens of other experiments. The precise sequence of this peptide is not included among the listed variations of P23083 in the neXtProt PEFF file, but after considering the listed variations plus one additional fuzzy substitution of 49T to 49G (position 49 has four annotated variations, but does not have T to G), this peptide can match to an immunoglobulin with over 60,000 PSMs in PeptideAtlas rather than a homeobox protein never otherwise detected with sufficient confidence in over 1000 other experiments in PeptideAtlas.
Figure 4.
PSM for SEYTYGASYR in PeptideAtlas. With an iProphet probability of 1.000, all y ions y1 – y10 are observed, as well as b1 – b8, with many neutral loss ions, internal fragmentation ions, and other corroborating peaks. With a simple mapping search, this peptide appears to be uniquely mapping to a protein not seen anywhere else in PeptideAtlas, a one-hit wonder. With the complex fuzzy mapping capabilities of ProMaST, a potential alternative mapping to a highly variable region of an immunoglobulin is revealed.
Another important application of ProteoMapper is the comprehensive mapping of peptide identifications made within the TPP when using a PEFF database that includes SAAV variants. Previous versions of the TPP could comprehensively map peptides to a FASTA file without modifications, but were incapable of handling SAAVs in any format.
We also use ProteoMapper to explore the mapping of all ~1.4 million Human PeptideAtlas peptides to all immunoglobulins (Igs) in neXtProt (using the collapseIsoforms option, which treats any mapping to any splice isoform as a mapping to the parent entry). Without any variations, we find that 7882 peptide sequence map to Igs. When we include all known PEFF-encoded variations already annotated in neXtProt, 24,038 peptides map to Igs. When we further allow one additional fuzzy-match substitution anywhere in the sequence on top of current annotations (no mass delta constraint), the number of mappings jumps to a remarkable 136,743 peptides.
We then compare these lists with the list of “weak” proteins in PeptideAtlas; these are proteins with a single peptide of 9+ amino acids that appears to be uniquely mapping. In addition to the above example SEYTYGASYR, we find several additional examples of peptides with excellent PSMs that appear to map uniquely to a neXtProt protein with no other evidence in PeptideAtlas, but also map to an immunoglobulin variable region with PEFF-encoded variations and a single substitution. These include TTETLLLLSR, AAYLSTLSK, ETGLETSSGGK, RNSLESVEFVK, and YSLNSTTWK. Searches of these peptides with ProteoMapper with one fuzzy match and a null tolerance setting reveal the original one-hit-wonder mapping and at least one additional fuzzy mapping to an immunoglobulin with many other hits. Hyperlinks into PeptideAtlas reveal additional information including the annotated spectra. With only half a dozen high quality cases apparent, the problem does not appear to be highly pervasive for peptides of 9+ amino acids. However, this underscores the need to mitigate this problem by requiring multiple peptides of substantial length (currently 2 peptides at a length of 9+ are required by the HPP guidelines) for detection claims of newly detected proteins, particularly in samples where Igs are present.
Conclusion
The ProteoMapper software enables large-scale (several million sequences in one pass) mapping of peptides to proteomes. We have described the inner workings of both the indexer component (CLIPS) as well as the searching component (ProMaST). The tools are very fast, enabling exact searches of a thousand peptides against the neXtProt proteome in under a second, and fuzzy wildcard searching of a single peptide in just seconds. The ProteoMapper tools can be downloaded and run locally, embedded in other applications, used interactively at the PeptideAtlas web site, or run programmatically as a web service.
As the HPP nears completion of the protein parts list by demonstrating the confident detection of nearly all human proteins, the final shrinking list of missing proteins will become increasingly difficult to reduce. Yet, it has been shown that only 22 human proteins cannot generate any fully protease-specific peptides suitable for current mass spectrometry workflows using a handful of different common proteases for their positive detection30. Other estimates that take into account other factors such as previous detections of transcripts place the number of inaccessible proteins close to 100031,32. To ensure correct identification, the HPP guidelines will become increasingly important for achieving confidence in the detection claims. Broad availability of this tool enables easier compliance with the HPP MS dataset interpretation guideline #14, so that authors, reviewers, and readers who are exploring the implications of very high quality peptide identifications can readily see alternatives to the default peptide-to-protein mapping interpretations.
Acknowledgements
This work was funded in part by the National Institutes of Health, National Institute of General Medical Sciences grants: R01GM087221, R24GM127667, P50GM076547, the National Institute of Allergy And Infectious Diseases grant: R21AI133335, the National Institute of Biomedical Imaging and Bioengineering grant U54EB020406, and the National Heart Lung and Blood Institute grant: R01HL133135.
Footnotes
The authors declare no competing financial interest.
Supporting Information
None
References
- (1).Nilsson T; Mann M; Aebersold R; Yates JR; Bairoch A; Bergeron JJM Mass Spectrometry in High-Throughput Proteomics: Ready for the Big Time. Nat. Methods 2010, 7 (9), 681–685. [DOI] [PubMed] [Google Scholar]
- (2).Mann M; Kulak NA; Nagaraj N; Cox J The Coming Age of Complete, Accurate, and Ubiquitous Proteomes. Mol. Cell 2013, 49 (4), 583–590. [DOI] [PubMed] [Google Scholar]
- (3).Meier F; Geyer PE; Virreira Winter S; Cox J; Mann M BoxCar Acquisition Method Enables Single-Shot Proteomics at a Depth of 10,000 Proteins in 100 Minutes. Nat. Methods 2018, 15, 440–448. [DOI] [PubMed] [Google Scholar]
- (4).Aebersold R; Mann M Mass Spectrometry-Based Proteomics. Nature 2003, 422 (6928), 198–207. [DOI] [PubMed] [Google Scholar]
- (5).Nesvizhskii AI A Survey of Computational Methods and Error Rate Estimation Procedures for Peptide and Protein Identification in Shotgun Proteomics. J. Proteomics 2010, 73 (11), 2092–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Deutsch EW; Lam H; Aebersold R Data Analysis and Bioinformatics Tools for Tandem Mass Spectrometry in Proteomics. Physiol. Genomics 2008, 33 (1), 18–25. [DOI] [PubMed] [Google Scholar]
- (7).Nesvizhskii AI; Keller A; Kolker E; Aebersold R A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem 2003, 75 (17), 4646–4658. [DOI] [PubMed] [Google Scholar]
- (8).Ma Z-Q; Dasari S; Chambers MC; Litton MD; Sobecki SM; Zimmerman LJ; Halvey PJ; Schilling B; Drake PM; Gibson BW; et al. IDPicker 2.0: Improved Protein Assembly with High Discrimination Peptide Identification Filtering. J. Proteome Res 2009, 8 (8), 3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Legrain P; Aebersold R; Archakov A; Bairoch A; Bala K; Beretta L; Bergeron J; Borchers CH; Corthals GL; Costello CE; et al. The Human Proteome Project: Current State and Future Direction. Mol. Cell. Proteomics MCP 2011, 10 (7), M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Omenn GS; Lane L; Lundberg EK; Overall CM; Deutsch EW Progress on the HUPO Draft Human Proteome: 2017 Metrics of the Human Proteome Project. J. Proteome Res 2017, 16 (12), 4281–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hanash S; Celis JE The Human Proteome Organization: A Mission to Advance Proteome Knowledge. Mol. Cell. Proteomics MCP 2002, 1 (6), 413–414. [DOI] [PubMed] [Google Scholar]
- (12).Gaudet P; Michel P-A; Zahn-Zabal M; Cusin I; Duek PD; Evalet O; Gateau A; Gleizes A; Pereira M; Teixeira D; et al. The NeXtProt Knowledgebase on Human Proteins: Current Status. Nucleic Acids Res. 2015, 43 (Database issue), D764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Breuza L; Poux S; Estreicher A; Famiglietti ML; Magrane M; Tognolli M; Bridge A; Baratin D; Redaschi N; UniProt Consortium. The UniProtKB Guide to the Human Proteome. Database 2016, 2016, bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Deutsch EW; Overall CM; Van Eyk JE; Baker MS; Paik Y-K; Weintraub ST; Lane L; Martens L; Vandenbrouck Y; Kusebauch U; et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. J. Proteome Res 2016, 15 (11), 3961–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schaeffer M; Gateau A; Teixeira D; Michel P-A; Zahn-Zabal M; Lane L The NeXtProt Peptide Uniqueness Checker: A Tool for the Proteomics Community. Bioinforma. Oxf. Engl 2017, 33 (21), 3471–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xiao Y; Vecchi MM; Wen D Distinguishing between Leucine and Isoleucine by Integrated LC-MS Analysis Using an Orbitrap Fusion Mass Spectrometer. Anal. Chem 2016, 88 (21), 10757–10766. [DOI] [PubMed] [Google Scholar]
- (17).Keller A; Eng J; Zhang N; Li X; Aebersold R A Uniform Proteomics MS/MS Analysis Platform Utilizing Open XML File Formats. Mol. Syst. Biol 2005, 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Deutsch EW; Mendoza L; Shteynberg D; Farrah T; Lam H; Tasman N; Sun Z; Nilsson E; Pratt B; Prazen B; et al. A Guided Tour of the Trans-Proteomic Pipeline. Proteomics 2010, 10 (6), 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Deutsch EW; Mendoza L; Shteynberg D; Slagel J; Sun Z; Moritz RL Trans-Proteomic Pipeline, a Standardized Data Processing Pipeline for Large-Scale Reproducible Proteomics Informatics. Proteomics Clin. Appl 2015, 9 (7–8), 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Eng JK; Jahan TA; Hoopmann MR Comet: An Open-Source MS/MS Sequence Database Search Tool. Proteomics 2013, 13 (1), 22–24. [DOI] [PubMed] [Google Scholar]
- (21).Craig R; Beavis RC TANDEM: Matching Proteins with Tandem Mass Spectra. Bioinforma. Oxf. Engl 2004, 20 (9), 1466–1467. [DOI] [PubMed] [Google Scholar]
- (22).Kong AT; Leprevost FV; Avtonomov DM; Mellacheruvu D; Nesvizhskii AI MSFragger: Ultrafast and Comprehensive Peptide Identification in Mass Spectrometry-Based Proteomics. Nat. Methods 2017, 14 (5), 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Desiere F; Deutsch EW; Nesvizhskii AI; Mallick P; King NL; Eng JK; Aderem A; Boyle R; Brunner E; Donohoe S; et al. Integration with the Human Genome of Peptide Sequences Obtained by High-Throughput Mass Spectrometry. Genome Biol 2005, 6 (1), R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Desiere F; Deutsch EW; King NL; Nesvizhskii AI; Mallick P; Eng J; Chen S; Eddes J; Loevenich SN; Aebersold R The PeptideAtlas Project. Nucleic Acids Res. 2006, 34 (Database issue), D655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Deutsch EW; Sun Z; Campbell D; Kusebauch U; Chu CS; Mendoza L; Shteynberg D; Omenn GS; Moritz RL State of the Human Proteome in 2014/2015 As Viewed through PeptideAtlas: Enhancing Accuracy and Coverage through the AtlasProphet. J. Proteome Res 2015, 14 (9), 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Deutsch EW; Sun Z; Campbell DS; Binz P-A; Farrah T; Shteynberg D; Mendoza L; Omenn GS; Moritz RL Tiered Human Integrated Sequence Search Databases for Shotgun Proteomics. J. Proteome Res 2016, 15 (11), 4091–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Aken BL; Achuthan P; Akanni W; Amode MR; Bernsdorff F; Bhai J; Billis K; Carvalho-Silva D; Cummins C; Clapham P; et al. Ensembl 2017. Nucleic Acids Res. 2017, 45 (D1), D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).O’Leary NA; Wright MW; Brister JR; Ciufo S; Haddad D; McVeigh R; Rajput B; Robbertse B; Smith-White B; Ako-Adjei D; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44 (D1), D733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rivers RC; Kinsinger C; Boja ES; Hiltke T; Mesri M; Rodriguez H Linking Cancer Genome to Proteome: NCI’s Investment into Proteogenomics. Proteomics 2014, 14 (23–24), 2633–2636. [DOI] [PubMed] [Google Scholar]
- (30).Kusebauch U; Campbell DS; Deutsch EW; Chu CS; Spicer DA; Brusniak M-Y; Slagel J; Sun Z; Stevens J; Grimes B; et al. Human SRMAtlas: A Resource of Targeted Assays to Quantify the Complete Human Proteome. Cell 2016, 166 (3), 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Omenn GS; Lane L; Overall CM; Corrales FJ; Schwenk JM; Paik Y-K; Van Eyk JE; Liu S; Snyder M; Baker MS; et al. Progress on Identifying and Characterizing the Human Proteome: 2018 Metrics from the HUPO Human Proteome Project. J. Proteome Res 2018, in press, this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Duek P; Gateau A; Bairoch A; Lane L Exploring the Uncharacterized Human Proteome Using NeXtProt. J. Proteome Res 2018, in press, this issue. [DOI] [PubMed] [Google Scholar]