Abstract
Cigarette smoking during pregnancy is a major public health concern. While there are well-described consequences in early child development, there is very little known about the effects of maternal smoking on human cortical biology during prenatal life. We therefore performed a genome-wide differential gene expression analysis using RNA sequencing (RNA-seq) on prenatal (N=33; 16 smoking-exposed) as well as adult (N=207; 57 active smokers) human post-mortem prefrontal cortices. Smoking exposure during the prenatal period was directly associated with differential expression of 14 genes; in contrast, during adulthood, despite a much larger sample size, only 2 genes showed significant differential expression (FDR<10%). Moreover, 1,315 genes showed significantly different exposure effects between maternal smoking during pregnancy and direct exposure in adulthood (FDR<10%) – these differences were largely driven by prenatal differences that were enriched for pathways previously implicated in addiction and synaptic function. Furthermore, prenatal and age-dependent differentially expressed genes were enriched for genes implicated in non-syndromic autism spectrum disorder (ASD) and were differentially expressed as a set between patients with ASD and controls in post-mortem cortical regions. These results underscore the enhanced sensitivity to the biological effect of smoking exposure in the developing brain and offer insight into how maternal smoking during pregnancy affects gene expression in the prenatal human cortex. They also begin to address the relationship between in utero exposure to smoking and the heightened risks for the subsequent development of neuropsychiatric disorders.
Introduction
Cigarette smoking and nicotine addiction continue to be major public health problems due to their established association with increased risk of cancer, respiratory disease, and many other disease outcomes in adults1. Cigarette smoking exposure also is highly deleterious to fetal development, having been associated with heightened risk of intrauterine growth restriction, low birth weight, stillbirth, and mortality2,3,4–8. Despite these adverse health outcomes for the smoker and the fetus carried by mothers who smoke, 8.4% of pregnant women continue to smoke while pregnant in the United States9. Accumulating evidence suggests in utero exposure to nicotine and other elements of cigarette smoking is associated with a greater risk of the subsequent development of mental illnesses10,11. Components of cigarette smoke, including nicotine, have neuroteratogenic effects12, and there is mixed epidemiological evidence for maternal smoking during pregnancy as a risk factor for neuropsychiatric disorders including schizophrenia10,11, attention-deficit / hyperactivity disorder13–17, Tourette syndrome17,18, impaired cognitive development19,20, obsessive-compulsive disorder17, and autism spectrum disorder21. Despite the potential ramifications of maternal smoking on the mental health of exposed offspring, little is known about the biological effects of smoke exposure on the developing human brain.
Possible molecular mechanisms underlying risk for neuropsychiatric disease from maternal smoking during pregnancy remain elusive. Previous studies have found epigenetic changes in cord blood22,23 and differential gene expression in placental tissue24. However, only one prior study has directly examined associations of maternal smoking during pregnancy with epigenetic changes in the prenatal human brain25, and no study, to our knowledge, has characterized transcriptional changes in the prenatal brain. Characterizing the gene regulatory changes associated with smoking exposure in prenatal life may identify potential mechanisms related to disease risks later in life especially with regard to neuropsychiatric disorders. We can better understand the link between maternal smoking during pregnancy and future behavioral and cognitive sequelae by directly studying molecular changes in prenatal human cortical tissue exposed to cigarette smoke.
To characterize molecular changes in the developing prenatal brain associated with in utero smoking exposure, we first analyzed RNA-seq data from post-mortem fetal human prefrontal cortex tissue, contrasting exposed to unexposed prenatal tissue. Then to assess the developmental specificity of these molecular changes in the prenatal cortex, we did an analogous comparison of cortical gene expression between adult smokers to non-smokers. Lastly to ascertain the potentially age-dependent effects of smoking exposure on the cortex, we formally tested whether the effects of smoking exposure on gene expression were different on the developing brain compared to the adult brain. The results of these investigations highlight dysregulation of the developing prenatal cortex’s transcriptome associated with in utero smoking exposure and their implications for possible mechanisms of risk for neurodevelopmental disorders later in life.
Results
Gene expression changes associated with smoking exposure in the developing brain
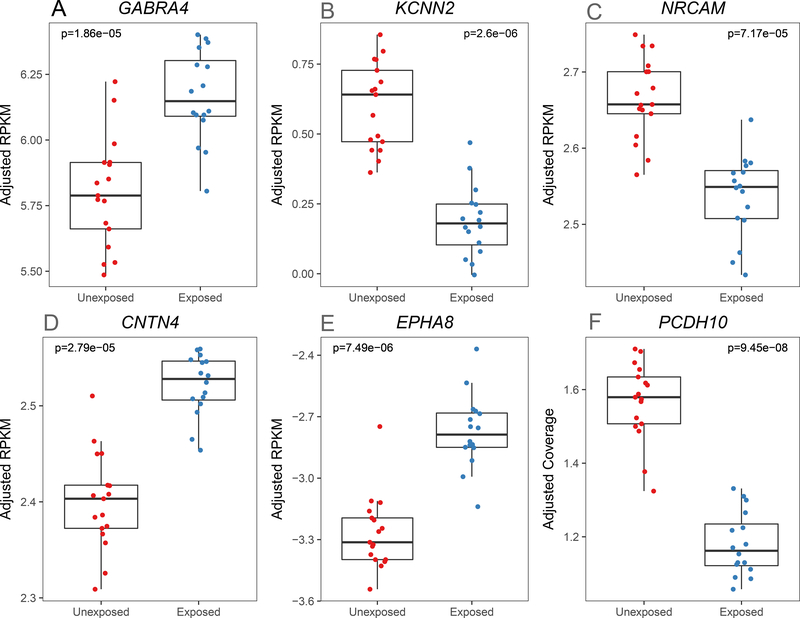
We performed differential expression analysis of genes and their transcript features comparing dorsolateral prefrontal cortex (DLPFC) tissue from 16 smoking-exposed compared to 17 smoking-unexposed prenatal samples (Table 1, for further details see Results S1 and Supplemental Table S1). After correcting for multiple testing (at false discovery rate, FDR<10%), 14 prenatal genes had evidence of differential expression (DE), mostly at the gene-level summarization (Table 2, median absolute log2 fold change [LFC] = 0.618). Some of these differentially expressed genes are potentially related to nicotine dependence or addiction more broadly: KCNN2, GABRA4, and NRCAM (Figure 1A–C). The most significant differentially expressed transcript feature at the gene level was KCNN2 which encodes a small-conductance Ca2+-activated K+ channel (Figure 1A, p=2.60×10−6, LFC = −0.694). GABRA4, a gene coding for the alpha-4 subunit of the GABA receptor, was also differentially expressed (Figure 1B, p=1.86×10−5, LFC = 1.06). NRCAM, a gene encoding neuronal cell adhesion molecule, was differentially expressed at the gene level (Figure 1C, p=7.17×10−5, LFC = −0.568) and six previously annotated NRCAM exon-exon splice junctions were also differentially expressed (pmin=2.29×10−7, LFCp-min= −0.689, Supplemental Table S2). Thus, prenatal smoking exposure is associated with changes in the expression of some genes previously associated with addiction in the literature.
Table 1.
Description of sample characteristics.
| Prenatal | Adult (non-psychiatric) | |||
|---|---|---|---|---|
| Smoking Unexposed | Smoking Exposed | Non-Smoker | Smoker | |
| N | 17 | 16 | 150 | 57 |
| Age | 17.36 (2.20) pcw† | 19.81 (5.19) pcw | 41.46 (15.79) yrs‡ | 42.48 (14.92) yrs |
| Race | ||||
| Caucasian | 0 (0.0%) | 3 (18.8%) | 76 (50.7%) | 22 (38.6%) |
| African American | 17 (100.0%) | 13 (81.2%) | 74 (49.3%) | 35 (61.4%) |
| Male | 10 (58.8%) | 5 (31.2%) | 108 (72.0%) | 40 (70.2%) |
| Source | ||||
| NIMH | 0 (0.0%) | 0 (0.0%) | 124 (82.7) | 55 (96.5) |
| Stanley | 0 (0.0%) | 0 (0.0%) | 12 (8.0) | 2 (3.5) |
| UMB | 17 (100.0%) | 16 (100.0%) | 14 (9.3) | 0 (0.0) |
| RIN * | 8.82 (1.35) | 8.94 (0.99) | 8.33 (0.67) | 8.52 (0.52) |
preconception weeks
years
RNA Integrity Number
Table 2.
Significant differentially expressed genes between smoke exposed and unexposed human brain cortices in prenatal and adult cohorts (FDR<10%).
| Prenatal | Adult | ||||||
|---|---|---|---|---|---|---|---|
| Cohort | Symbol | Feature Level | Log2 Fold Change | FDRa | Log2 Fold Change | FDRa | Interaction P |
| Prenatal | PCDH10 | Expressed Region | −1.367 | 0.026 | −0.114 | 1 | 5.87×10−4 |
| Prenatal | KCNN2 | Gene | −0.694 | 0.047 | −0.015 | 0.991 | 1.65×10−5 |
| Prenatal | EPHA8 | Gene | 1.546 | 0.048 | −0.019 | 0.993 | 2.54×10−5 |
| Prenatal | TENM3 | Gene | 0.804 | 0.048 | −0.006 | 0.996 | 2.79×10−11 |
| Prenatal | IL1RAPL2 | Gene | −1.036 | 0.048 | −0.06 | 0.991 | 2.43×10−4 |
| Prenatal | MPPED1 | Gene | 0.385 | 0.051 | −0.005 | 0.995 | 1.96×10−5 |
| Prenatal | GABRA4 | Gene | 1.059 | 0.056 | 0.031 | 0.987 | 8.82×10−16 |
| Prenatal | ECHDC2 | Gene | 0.864 | 0.056 | 0.015 | 0.993 | 3.21×10−7 |
| Prenatal | SDC1 | Gene | 0.367 | 0.056 | 0.092 | 0.978 | 0.455 |
| Prenatal | CNTN4 | Gene | 0.682 | 0.056 | 0.003 | 0.997 | 1.63×10−8 |
| Prenatal | CHSY3 | Gene | 0.55 | 0.063 | 0.02 | 0.991 | 2.82×10−8 |
| Prenatal | RNF13 | Gene | −0.28 | 0.063 | 0.015 | 0.991 | 0.191 |
| Prenatal | ZNF608 | Gene | 0.343 | 0.067 | 0.035 | 0.991 | 0.013 |
| Prenatal | NRCAM | Gene, Junctionb | −0.568 | 0.1 | 0.014 | 0.991 | 1.27×10−5 |
| Adult | MARCO | Gene | 0.896 | 0.432 | −1.603 | 8.44×10−5 | 1.53×10−4 |
| Adult | CEP85 | Junction | −0.054 | 0.939 | −0.253 | 0.061 | 0.042 |
False discovery rate.
NRCAM gene level results presented here. For exon-exon splice junction results see Supplemental Table S2.
Fig. 1.
Representative differentially expressed prenatal genes. [RPKM: reads per kilobase per million]. Normalized expression levels for six genes with significant (FDR<10%, moderated t-test) differential expression are shown for smoking unexposed (N=17) and exposed (N=16) prenatal prefrontal cortex samples. These representative genes have been previously implicated in neurodevelopment and neuropsychiatric disease.
Other differentially expressed genes between the exposed and unexposed prenatal groups play a role in neurodevelopment and may be linked to neuropsychiatric disorders: CNTN4, EPHA8, and PCDH10 (Figure 1D–F). Contactin 4 (CNTN4, Figure 1D, p=2.79×10−5, LFC=0.682) is a cell adhesion molecule involved in synaptic formation26. The association with PCDH10 was in a differentially expressed region (Figure 1E, p=9.45×10−8, LFC= −1.37, chr4:134,116,962–134,116,976), a 15 base pair strictly intronic region. Protocadherin-10 (PCDH10) encodes a cell adhesion molecule involved in synaptic elimination27. EPH Receptor A8 (EPHA8, Figure 1F, p=7.49×10−6, LFC=1.55) encodes a member of the Eph family of tyrosine protein kinase receptors, which has a functional role in axonal pathfinding during neurodevelopment28. As a sensitivity analysis to further assess confounding by ethnicity (all 3 prenatal Caucasian samples were smoke-exposed), we undertook a subgroup analysis of the significant differentially expressed genes using only African-American prenatal samples (N=30). The resulting effect sizes were consistent with those from the original model supporting the robustness of these findings (r=0.99, p=1.18×10−10, Supplemental Fig. S2). These findings suggest, perhaps not surprisingly, that genes involved in neurodevelopment may be disrupted by prenatal exposure to cigarette smoke, a putative neurodevelopmental insult.
More subtle gene expression changes associated with smoking exposure in the adult brain
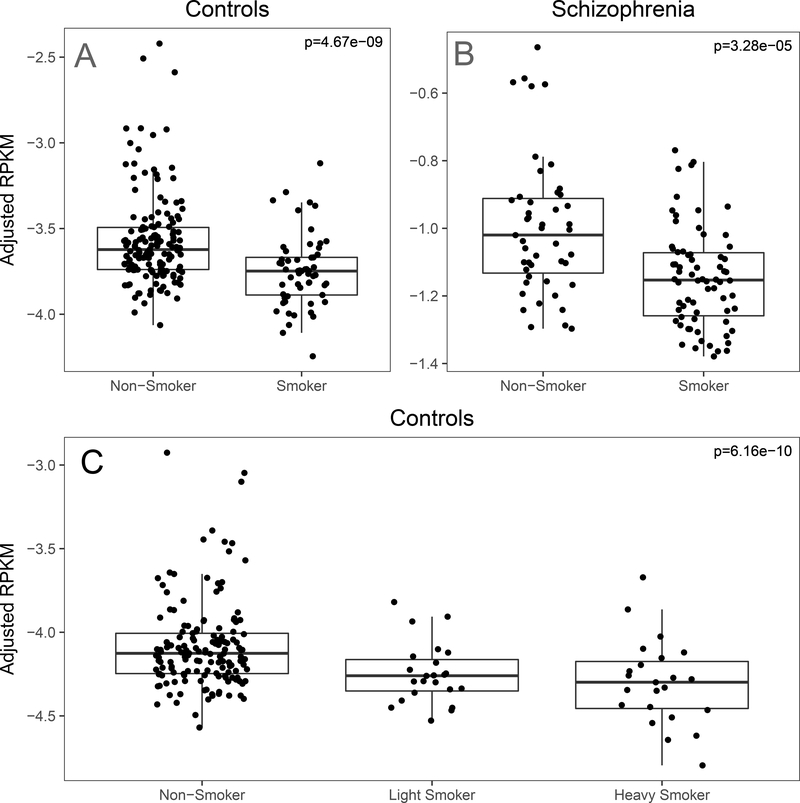
In the adult sample, only two genes were differentially expressed between active smoker (N=57) and non-smoker groups (N=150; Table 1). MARCO, an immune gene, was the most significantly differentially expressed gene among adults (Figure 2A; p=4.67×10−9, LFC = −1.603). We used a different cohort of DLPFC tissue from 107 patients with schizophrenia (65 active smokers, 42 non-smokers) to assess the replicability of this differential gene expression in adults. MARCO was similarly differentially expressed in this cohort (p=3.28×10−5, LFC = −1.34, Figure 2B). To assess the robustness of our model, we then stratified adult smokers into three categories: non-smokers (N=150), light-smokers (N=23), and heavy smokers (N=23), based on cotinine levels (200 ng/mL heavy-light threshold). Under this ordinal sensitivity model, MARCO remained significantly differentially expressed (p=6.16×10−10, LFC=−1.211, Figure 2C), and our global results were highly correlated across all transcript feature-levels confirming the robustness of our analysis (Supplemental Fig. S3). An exon-exon splice junction mapping to four transcripts of CEP85 (chr1:26,595,127–26,595,950) was also differentially expressed between adult smokers and non-smokers (p=4.46×10−7, LFC = −0.25), but this association did not replicate in our independent schizophrenia cohort (p=0.47, Supplemental Fig. S4). Together, these findings indicate cigarette smoke exposure has subtler effects on the adult prefrontal cortex’s transcriptome than on the developing brain, despite having the larger sample size compared with the fetal cohort.
Fig. 2.
A. MARCO expression in original model across development. [RPKM: reads per kilobase per million]. MARCO (Macrophage Receptor With Collagenous Structure) expression was reduced in adult smokers (N=57) compared to non-smokers (N=150). B. MARCO expression in the schizophrenia replication sample. MARCO expression was significantly reduced in smokers (N=65) compared to non-smokers (N=42) in a separate sample of schizophrenics collected, processed, analyzed with the same pipeline as the adult controls. C. MARCO expression under a sensitivity model in adult non-psychiatric controls (N=196). MARCO expression was inversely proportional to smoking intensity under our sensitivity ordinal model in which we stratified smokers into light (N=23) and heavy (N=23) smokers.
Significant changes in susceptibility to smoking exposure across development
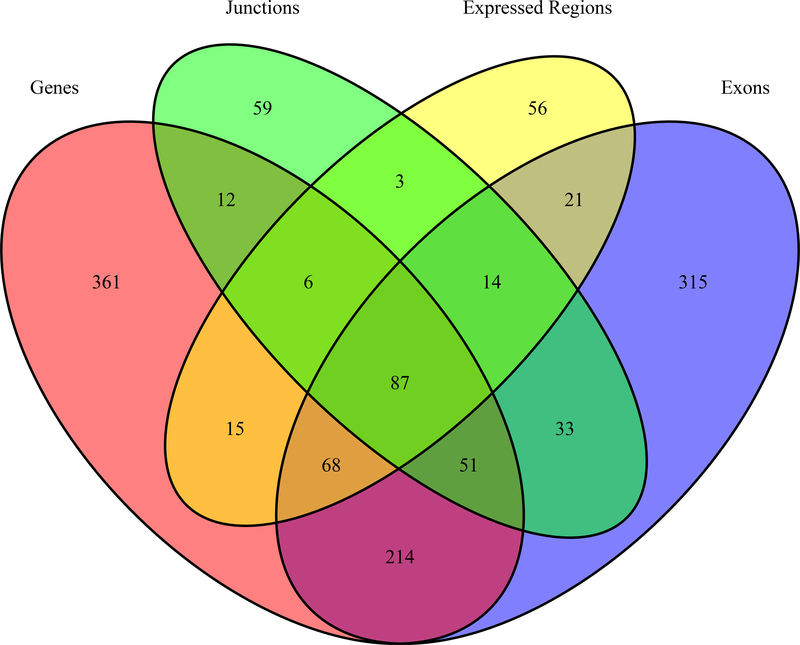
After observing non-overlapping genes within the differentially expressed transcript features across the adult and prenatal cohorts associated with smoking exposure, we performed an interaction analysis to more formally determine the potentially age-dependent effects of smoking exposure in prenatal versus adult samples. While the significant main effects of smoking exposure in adulthood were limited to MARCO and the CEP85 junction described above, there were 5,293 differentially expressed features corresponding to 1,315 Ensembl genes (Supplemental Table S3) associated with the interaction between developmental stage (pre- versus post-natal) and smoking exposure after adjusting for possible technical and biological confounds (FDR<10%). The different feature type analyses offer multiple lines of evidence for genes implicated as interaction genes (i.e., genes that respond differently to cigarette smoking in prenatal vs. adult cortical tissue), suggesting transcript-feature level analysis beyond the gene-level summary offer additional insight into changes in the transcriptomic landscape. For instance, 87 genes were shared as significant interaction genes across all feature types, but 315 genes were solely significant for an interaction effect at the exon-level summarization (Figure 3).
Fig. 3.
Venn diagram of differentially expressed gene overlap by feature level summarization. 87 genes are implicated across all feature level summarizations (genes, exon-exon junctions, expressed regions, and exons). Other genes are tagged solely by our feature level analysis such as the 56 genes that were only identified as differentially expressed regions and not by other feature-level summarizations.
We next asked whether smoking exposure’s differential effects on gene expression across the lifespan are mostly driven by significant changes in the prenatal cortical transcriptome, hypothesizing that the prenatal cortical transcriptome is more pliable than the adult one. Post-hoc analysis confirmed that more of these transcript features with a significant interaction were nominally differentially expressed (p<0.05) by smoking exposure in prenatal cortex (N=1,850, 35.0%) than in adult cortex (N=585, 11.1%; p=1.40×10−8). These findings underscore that smoking exposure exerts different effects on gene expression across the human lifespan and these effects occur more frequently in the prenatal cortex.
Enrichment of interaction genes in diverse processes and neurodevelopmental disorders
To explore the potential functional implications of the 1,315 genes with differential effects of cigarette smoke exposure in prenatal and adult life, we performed gene ontology and pathway analyses with the sets of genes tagged by differentially expressed age-dependent transcript features: genes (N=752), exons (N=746), junctions (N=257), and ERs (N=285). We uncovered significant enrichment of biological categories across all ontologies tested (Supplemental Table S4).
Here, “nicotine addiction” (hsa05033, p=2.04×10−7, q=3.15×10−5) was one of the most significantly enriched KEGG categories across multiple feature summarizations. Other top KEGG categories included “retrograde endocannabinoid signaling” (hsa04723, p=2.04×10−7, q=3.15×10−5) and “neuroactive ligand-receptor interaction” (hsa04080, p=2.04×10−7, q=3.15×10−5). The most significantly enriched biological processes involved “behavior” (GO:0007610, p=2.33×10−10, q=9.87×10−7), “positive regulation of nervous system development” (GO:0051962, p=1.31×10−7, q=2.32×10−4), and “synapse organization” (GO:0050808, p=2.84×10−7, q=2.57×10−4). Interestingly, using disease ontology (DO) enrichment analyses, where pre-defined gene sets are based on prior disease associations in the literature, rather than biological function/pathways, the top enriched disease ontologies (DO) were related to autism spectrum disorder (ASD), including gene sets “autism spectrum disorder” (DOID:0060041, p=8.45×10−8, q=2.57 ×10−5), “autistic disorder” (DOID:12849, p=8.45×10−8, q=2.57 ×10−5), and “pervasive developmental disorder” (DOID:0060040, p=1.25×10−7, q=2.57 ×10−5).
After observing the DO enrichment related to autism and relevant molecular processes, we performed targeted enrichment analyses using publicly available autism spectrum disorder (ASD) gene databases29 and previously constructed neurodevelopmental gene sets30 to better determine sensitivity and specificity of the ASD gene set enrichments. In prenatal samples, this ASD enrichment was driven by IL1RAPL2, GABRA4, CNTN4, NRCAM, and PCDH10 being differentially expressed (Table 1, N=5/14 SFARI genes, p=3.79×10−4, Supplemental Table S5A). We further observed enrichment of significant age-dependent interaction genes across multiple autism gene sets including SFARI Gene (p=7.20×10−8), AutDb (p=1.17×10−7), and the curated non-syndromic ASD gene database set (p=3.77×10−4, Supplemental Table S5B). We undertook a parallel analysis of smoking-unexposed cortical samples across the lifespan (prenatal vs. adult) to identify neurodevelopmentally regulated genes and contextualize the results (Results S3). We found that gene expression patterning over cortical development is similarly enriched for neuropsychiatric disease gene sets; mirroring the enrichment results with age-dependent interaction genes: the greatest enrichment was for ASD gene sets (Supplemental Table S5C). Interaction genes were less significantly enriched for schizophrenia genes implicated from single nucleotide variant (SNV) studies (p=0.00136), and only nominally significantly enriched for intellectual disability genes (p=0.0115) and syndromic neurodevelopmental disorder (NDD) genes (p=0.0175; Supplemental Table S5B). The enriched NDD (N=6) were a subset of the enriched ASD genes, but the enriched schizophrenia SNV and intellectual disability genes were mostly distinct (Supplemental Fig. S5). Therefore, genes affected differently by cigarette smoke exposure across the lifespan appear enriched in autism spectrum disorder and other neuropsychiatric disorders gene sets.
Smoking exposure’s transcriptomic pattern is similar to post-mortem brain ASD case-control differences
Next, to more fully interrogate these genes in ASD, we performed gene set enrichment analysis in post-mortem human brain tissue from a public database of patients with ASD compared to unaffected controls31 by comparing the gene expression differences from our significant differentially expressed prenatal and interaction genes against ASD-related gene expression differences (Supplemental Table S6, Supplemental Fig. S6A). Differentially expressed prenatal genes (N=14) were more highly ranked relative to other genes in terms of the ASD case-control differences in frontal cortex (BA9; p=1.35×10−6) and temporal cortex (BA41–42-22; p=6.49×10−5), but not vermis (p=0.646, Supplemental Fig. S6B). Likewise, the interaction genes tested in this ASD data (N=1,293) also showed significant enrichment for ASD case-control differences in frontal cortex (p=3.45×10−5) and temporal cortex (p=2.77×10−9), but not vermis (p=0.963, Supplemental Fig. S6C). We further found enrichment of these sets of genes among known protein-protein interactions (Results S2, Supplemental Figure S7). We did not observe significant enrichment for schizophrenia case-control differences32,33 with either the differentially expressed prenatal genes (p=0.14) or the interaction genes (p=0.23). These findings suggest molecular changes in the developing human cortex associated with maternal smoking during pregnancy are related at the transcriptomic scale to the cortical pathology of ASD (i.e. these genes are differentially expressed as a group in a comparison of ASD samples to controls from a separate study), and these differences are at least partially selective.
Discussion
This study is the first, to our knowledge, to perform an extensive interrogation of the effect of smoking exposure on the prenatal cortical transcriptome. The prenatal brain is more sensitive to many environmental agents in both human and animal studies than the adult brain34–36. Therefore, one might expect signatures of smoking exposure would be more pronounced in the developing human brain. Consistent with this hypothesis, we found 14 genes differentially expressed in the prenatal cortex compared to only 2 genes in the adult cortex. Likewise, developmentally regulated genes were more affected by smoke-exposure during prenatal life than adulthood. The transcriptomic effects of smoking exposure were more noticeable in the prenatal cortex despite the smaller sample size available, underscoring the effects of maternal smoking on the prenatal human brain’s sensitive developmental trajectory.
Prenatal exposure was associated with changes in gene expression relevant to neurodevelopment and addiction which some studies have linked to maternal smoking during pregnancy37–39. NRCAM encodes a neuronal cell adhesion molecule, and expression was reduced within smoking-exposed prenatal brain. This differential expression may have downstream consequences, as neuronal cell adhesion molecules are involved in neurodevelopment40–42 and have been linked to autism43–45 and schizophrenia46–48. KCNN2 levels were also reduced in the smoking-exposed prenatal group. KCNN2 encodes small conductance Ca2+-activated K+ channels, and is part of a family of proteins that has been linked to substance abuse49,50.
Smoking exposure was also associated with changes in prenatal expression of multiple genes important in neurodevelopment such as CNTN4 and EPHA8 as well as components of the GABAergic signaling pathway (Discussion S1). Genes that were differentially expressed by smoking exposure in prenatal but not adult donors and vice versa were preferentially involved in processes relevant to neurodevelopment, such as synapse organization. Consistent with the neurodevelopmental origins of ASD, smoking-associated age-dependent and prenatal genes were enriched in publicly available autism spectrum disorder (ASD) gene databases and were differentially expressed as a group in an ASD case-control31 quality-corrected33 analysis. This intriguing finding was statistically robust and relatively specific to ASD, which may reflect a relatively greater role for disruptions to early brain development in ASD etiology compared to other severe mental illnesses such as bipolar disorder. Moreover, these enrichment result are consistent with the greater enrichment for ASD genes in preferentially prenatal expressed genes compared to other neuropsychiatric disorders30, and a parallel analysis likewise indicates developmentally regulated genes are highly enriched for ASD. Given the limited findings of smoking-associated differential expression in adults and the fact that adults are the sample population in many ASD studies, we do not believe this enrichment reflects possible confounding in ASD studies of gene expression in brain by smoking at the time of death. Furthermore, schizophrenia patients smoke at a much higher rate than the non-psychiatric population—studies of schizophrenia are often more confounded by smoking than ASD studies—but we observed only modest enrichment for schizophrenia genes. Our findings add orthogonal molecular support to the hypothesis that maternal smoking during pregnancy may be a risk factor for neuropsychiatric disease later in life, especially for ASD.
Regardless of age, differentially expressed genes between the smoking exposed and unexposed donors could also relate to other hazardous chemicals besides nicotine, indirect effects of cigarette smoke exposure, and residual confounding by unmeasured factors associated with smoking. Post-mortem brain studies, such as this one, are liable to confounding by various factors (e.g. RNA quality, age, sex, and race), but our exploratory analysis suggests these confounds are unlikely to be responsible for the gene expression differences. Furthermore, we constructed surrogate variables that capture transcriptomic variance driven by hidden confounds and adjusted for these in our analyses which improved our power to detect differential expression. Cigarette smoke contains over 500 distinct chemicals of which 98 have been deemed dangerous to human health51 and the molecular correlates of these compounds in human brain range in characterization. The genes identified in this study as differentially expressed by smoking exposure likely include ones that predispose to cigarette smoking susceptibility as well as ones that are altered by smoking or other related exposures. Nonetheless, these differentially expressed genes highlight genes and pathways that represent smoking-related changes in the developing brain.
Our results in DLPFC may not translate to other brain regions, as the effects of cigarette smoke exposure on the developing human cortex may vary across brain regions which have distinct cell type compositions and unique molecular profiles52. Moreover, our prenatal cohort includes mostly first trimester fetal brains, and the effects of smoking-exposure may differ in later prenatal cortical development. Future studies should seek to determine the molecular signature of cigarette smoke exposure across multiple developing brain regions and later time periods to refine the transcriptomic footprint of exposure in the developing human cortex. Finally, the temporality and relative contributions of genes and the environment, particularly the intrauterine environment53, on the observed molecular differences deserves further attention. Untangling the extent by which maternal smoking during pregnancy relates to a greater genetic burden for nicotine dependence subsequently inherited by the neonate54–56 compared to direct environmental or epigenetic insults57 warrants further portioning. Here, comprehensive expression quantitative trait loci mapping may help disentangle the functional consequences of predisposing genetic risk for nicotine dependence from the environmental effect of cigarette smoke exposure.
In summary, we performed transcriptome-wide scans to search for disturbances in gene expression associated with in utero smoking exposure and to understand the possible implications of gene dysregulation. The effect of smoking exposure on gene expression was more prominent in the developing prenatal brain than the mature adult brain. Disrupted neurodevelopment due to early environmental insults confers risk for neuropsychiatric disorders later in life, and indeed maternal smoking during pregnancy has been suggested as a risk factor for neuropsychiatric disorders in affected offspring10,11. Our study provides molecular evidence that maternal smoking during pregnancy may disrupt pathways relevant in neurodevelopment, and potentially influence risk of neuropsychiatric disease in offspring—particularly autism spectrum disorder. Overall, these findings offer new molecular clues into how in utero smoking exposure may disrupt human cortex development and increase risk for the development of neuropsychiatric disorders in the offspring.
Materials and Methods
Sample information:
We used a subset of postmortem human brain samples with RNA-sequencing data (N=240) described in Jaffe el al.32 to investigate the molecular correlates of smoke exposure across the lifespan. Smoking exposure was determined by nicotine and cotinine levels quantified via LC/MS-MS for the majority of samples (SI Appendix).
RNA-sequencing data processing:
Four convergent measurements of expression (“feature summarizations”) were used to comprehensively assess the transcriptome: gene, exon, exon-exon splice junction, and expressed regions58 (SI Appendix). We retained genes, exons, and junctions with at least 10% or more of samples having at least 1 count per million (cpm), with at least partially annotated. Expressed regions (ERs) were defined by contiguous sequences with mean library-size normalized coverage > 5 reads per 80 million58.
Differential expression:
Features were tested under an empirical Bayes framework for differential expression using the moderated t-test implemented in the limma59. Surrogate variables (SVs) were constructed using sva60 and were included as covariates to mitigate latent confounding (Results S4). Tested features with a false discovery rate (FDR) below 10% were considered differentially expressed (DE).
for feature i and donor j and binary smoking exposure Smokej, adjusting for age, RNA Integrity Number (RIN), the proportion of reads aligning to chrM (a proxy for RNA quality), gene assignment rate (GAR, a proxy for RNA quality), the first MDS component (ancestry), surrogate variables (SVs), and the sex for each donor.
A sensitivity analysis was done with adult samples by stratifying smokers into heavy and light smokers (200 ng/mL cotinine cutpoint) and using testing the ordinal variable for differential expression (non-smoker < light smoker <heavy smoker). Replication, defined as p<0.05 and same directionality of effect, of DE in adults was assessed using an independent cohort of 107 patients diagnosed with schizophrenia.
Gene set enrichment:
For gene set enrichment, transcript features were mapped to Entrez IDs and subsequently to six different gene ontologies (GO): KEGG61, GO-BP62, GO-MF62, GO-CC62, DO63, and Reactome64,65. We tested for enrichment in each ontology with the hypergeometric test for overrepresentation implemented in clusterProfiler66. Enrichment of DE genes in curated gene sets was tested using Fisher’s Exact Test.
Publicly available autism data:
We reprocessed reads from Parikshak et al.31 and tested for autism spectrum disorder (ASD) case-control gene expression differences, analogous to our procedure here (SI Appendix). The geneSetTest function in limma was used to competitively test whether sets of genes differentially expressed by smoking exposure were differentially expressed by ASD status, separately by brain region.
Protein-protein interaction:
To create protein-protein interaction (PPI) networks, gene symbols were mapped to unique identifiers in the STRING (v. 10)67 database (STRING score>400, homo sapiens). Functional association networks were then constructed on the basis of corresponding PPI scores.
Supplementary Material
Acknowledgements:
S.A.S., L.C.T., C.A.M., A.D.S., L.J.B., B.M., E.O.J., D.B.H. and A.E.J were supported by R01DA042090.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Code availability: Code is deposited in the Github repository: https://github.com/LieberInstitute/Smoking_DLPFC_Devel
Data availability: raw data is available is available from Synapse.org at accession: doi:10.7303/syn12299750. Processed data (gene, exon, and junction counts) are available at http://eqtl.brainseq.org/phase1
For additional details please see Methods and Materials in the Supplemental Appendix.
References:
- 1.Organization, W. H. WHO report on the global tobacco epidemic, 2011: Warning about the dangers of tobacco., (2011).
- 2.in The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General Reports of the Surgeon General (2014).
- 3.in The Health Consequences of Smoking: A Report of the Surgeon General Reports of the Surgeon General (2004).
- 4.Niemela S et al. Prenatal Nicotine Exposure and Risk of Schizophrenia Among Offspring in a National Birth Cohort. Am J Psychiatry 173, 799–806, doi: 10.1176/appi.ajp.2016.15060800 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Biederman J, Martelon M, Woodworth KY, Spencer TJ & Faraone SV Is Maternal Smoking During Pregnancy a Risk Factor for Cigarette Smoking in Offspring? A Longitudinal Controlled Study of ADHD Children Grown Up. Journal of attention disorders, doi: 10.1177/1087054714557357 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Melchior M et al. Maternal tobacco smoking in pregnancy and children’s socio-emotional development at age 5: The EDEN mother-child birth cohort study. European psychiatry : the journal of the Association of European Psychiatrists 30, 562–568, doi: 10.1016/j.eurpsy.2015.03.005 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Talati A, Wickramaratne PJ, Wesselhoeft R & Weissman MM Prenatal tobacco exposure, birthweight, and offspring psychopathology. Psychiatry research 252, 346–352, doi: 10.1016/j.psychres.2017.03.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holbrook BD The effects of nicotine on human fetal development. Birth defects research. Part C, Embryo today : reviews 108, 181–192, doi: 10.1002/bdrc.21128 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Curtin SC & Matthews TJ Smoking Prevalence and Cessation Before and During Pregnancy: Data From the Birth Certificate, 2014. National vital statistics reports : from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System 65, 1–14 (2016). [PubMed] [Google Scholar]
- 10.Stathopoulou A, Beratis IN & Beratis S Prenatal tobacco smoke exposure, risk of schizophrenia, and severity of positive/negative symptoms. Schizophrenia research 148, 105–110, doi: 10.1016/j.schres.2013.04.031 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Niemelä S et al. Prenatal Nicotine Exposure and Risk of Schizophrenia Among Offspring in a National Birth Cohort. American Journal of Psychiatry 173, 799–806, doi: 10.1176/appi.ajp.2016.15060800 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Liao C-Y, Chen Y-J, Lee J-F, Lu C-L & Chen C-H Cigarettes and the developing brain: Picturing nicotine as a neuroteratogen using clinical and preclinical studies. Tzu Chi Medical Journal 24, 157–161, doi: 10.1016/j.tcmj.2012.08.003 (2012). [DOI] [Google Scholar]
- 13.Thapar A et al. Maternal smoking during pregnancy and attention deficit hyperactivity disorder symptoms in offspring. Am J Psychiatry 160, 1985–1989, doi: 10.1176/appi.ajp.160.11.1985 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Nomura Y, Marks DJ & Halperin JM Prenatal exposure to maternal and paternal smoking on attention deficit hyperactivity disorders symptoms and diagnosis in offspring. The Journal of nervous and mental disease 198, 672–678, doi: 10.1097/NMD.0b013e3181ef3489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joelsson P et al. Prenatal smoking exposure and neuropsychiatric comorbidity of ADHD: a finnish nationwide population-based cohort study. BMC psychiatry 16, 306, doi: 10.1186/s12888-016-1007-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motlagh MG et al. Severe psychosocial stress and heavy cigarette smoking during pregnancy: an examination of the pre- and perinatal risk factors associated with ADHD and Tourette syndrome. European child & adolescent psychiatry 19, 755–764, doi: 10.1007/s00787-010-0115-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Browne HA et al. Prenatal Maternal Smoking and Increased Risk for Tourette Syndrome and Chronic Tic Disorders. Journal of the American Academy of Child and Adolescent Psychiatry 55, 784–791, doi: 10.1016/j.jaac.2016.06.010 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Leivonen S et al. Prenatal Maternal Smoking and Tourette Syndrome: A Nationwide Register Study. Child psychiatry and human development 47, 75–82, doi: 10.1007/s10578-015-0545-z (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wehby GL, Prater K, McCarthy AM, Castilla EE & Murray JC The Impact of Maternal Smoking during Pregnancy on Early Child Neurodevelopment. Journal of human capital 5, 207–254, doi: 10.1086/660885 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polanska K, Jurewicz J & Hanke W Smoking and alcohol drinking during pregnancy as the risk factors for poor child neurodevelopment - A review of epidemiological studies. International journal of occupational medicine and environmental health 28, 419–443, doi: 10.13075/ijomeh.1896.00424 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Jung Y, Lee AM, McKee SA & Picciotto MR Maternal smoking and autism spectrum disorder: meta-analysis with population smoking metrics as moderators. Scientific reports 7, 4315, doi: 10.1038/s41598-017-04413-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joubert BR et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environmental health perspectives 120, 1425–1431, doi: 10.1289/ehp.1205412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joubert BR et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am J Hum Genet 98, 680–696, doi: 10.1016/j.ajhg.2016.02.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawashima A et al. Effects of maternal smoking on the placental expression of genes related to angiogenesis and apoptosis during the first trimester. PloS one 9, e106140, doi: 10.1371/journal.pone.0106140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterton Z et al. In utero exposure to maternal smoking is associated with DNA methylation alterations and reduced neuronal content in the developing fetal brain. Epigenetics & chromatin 10, 4, doi: 10.1186/s13072-017-0111-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betancur C, Sakurai T & Buxbaum JD The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends in neurosciences 32, 402–412, doi: 10.1016/j.tins.2009.04.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai NP et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151, 1581–1594, doi: 10.1016/j.cell.2012.11.040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S, Frisen J & Barbacid M Aberrant axonal projections in mice lacking EphA8 (Eek) tyrosine protein kinase receptors. The EMBO journal 16, 3106–3114, doi: 10.1093/emboj/16.11.3106 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu SN, Kollu R & Banerjee-Basu S AutDB: a gene reference resource for autism research. Nucleic Acids Res 37, D832–836, doi: 10.1093/nar/gkn835 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birnbaum R, Jaffe AE, Hyde TM, Kleinman JE & Weinberger DR Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am J Psychiatry 171, 758–767, doi: 10.1176/appi.ajp.2014.13111452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427, doi: 10.1038/nature20612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe AE et al. Developmental And Genetic Regulation Of The Human Cortex Transcriptome In Schizophrenia. bioRxiv, doi: 10.1101/124321 (2017). [DOI] [PMC free article] [PubMed]
- 33.Jaffe AE et al. qSVA framework for RNA quality correction in differential expression analysis. Proc Natl Acad Sci U S A 114, 7130–7135, doi: 10.1073/pnas.1617384114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanwood GD & Levitt P Drug exposure early in life: functional repercussions of changing neuropharmacology during sensitive periods of brain development. Current opinion in pharmacology 4, 65–71, doi: 10.1016/j.coph.2003.09.003 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Rice D & Barone S Jr. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environmental health perspectives 108 Suppl 3, 511–533 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodier PM Developing brain as a target of toxicity. Environmental health perspectives 103 Suppl 6, 73–76 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Mamun A et al. Does maternal smoking during pregnancy predict the smoking patterns of young adult offspring? A birth cohort study. Tob Control 15, 452–457, doi: 10.1136/tc.2006.016790 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandel DB, Wu P & Davies M Maternal smoking during pregnancy and smoking by adolescent daughters. Am J Public Health 84, 1407–1413 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieb R, Schreier A, Pfister H & Wittchen HU Maternal smoking and smoking in adolescents: a prospective community study of adolescents and their mothers. Eur Addict Res 9, 120–130, doi: 10.1159/000070980 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Demyanenko GP et al. Neural cell adhesion molecule NrCAM regulates Semaphorin 3F-induced dendritic spine remodeling. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 11274–11287, doi: 10.1523/JNEUROSCI.1774-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzli D et al. A direct interaction of axonin-1 with NgCAM-related cell adhesion molecule (NrCAM) results in guidance, but not growth of commissural axons. The Journal of cell biology 149, 951–968 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakurai T The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Molecular and cellular neurosciences 49, 351–363, doi: 10.1016/j.mcn.2011.12.002 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Sakurai T et al. Association analysis of the NrCAM gene in autism and in subsets of families with severe obsessive-compulsive or self-stimulatory behaviors. Psychiatr Genet 16, 251–257, doi: 10.1097/01.ypg.0000242196.81891.c9 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Moy SS, Nonneman RJ, Young NB, Demyanenko GP & Maness PF Impaired sociability and cognitive function in Nrcam-null mice. Behavioural brain research 205, 123–131, doi: 10.1016/j.bbr.2009.06.021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marui T et al. Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. The international journal of neuropsychopharmacology 12, 1–10, doi: 10.1017/S1461145708009127 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Barbeau D, Liang JJ, Robitalille Y, Quirion R & Srivastava LK Decreased expression of the embryonic form of the neural cell adhesion molecule in schizophrenic brains. Proc Natl Acad Sci U S A 92, 2785–2789 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennaman LH & Maness PF NCAM in neuropsychiatric and neurodegenerative disorders. Advances in experimental medicine and biology 663, 299–317, doi: 10.1007/978-1-4419-1170-4_19 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Poltorak M et al. Disturbances in cell recognition molecules (N-CAM and L1 antigen) in the CSF of patients with schizophrenia. Experimental neurology 131, 266–272 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Padula AE et al. KCNN Genes that Encode Small-Conductance Ca2+-Activated K+ Channels Influence Alcohol and Drug Addiction. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 40, 1928–1939, doi: 10.1038/npp.2015.42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cadet JL et al. Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol Psychiatry, doi: 10.1038/mp.2016.48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talhout R et al. Hazardous compounds in tobacco smoke. International journal of environmental research and public health 8, 613–628, doi: 10.3390/ijerph8020613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaffe AE et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nature neuroscience 19, 40–47, doi: 10.1038/nn.4181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ursini G et al. Placental gene expression mediates the interaction between obstetrical history and genetic risk for schizophrenia. bioRxiv, doi: 10.1101/147207 (2017). [DOI]
- 54.Quinn PD et al. Association Between Maternal Smoking During Pregnancy and Severe Mental Illness in Offspring. JAMA Psychiatry 74, 589–596, doi: 10.1001/jamapsychiatry.2017.0456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Onofrio BM, Van Hulle CA, Goodnight JA, Rathouz PJ & Lahey BB Is maternal smoking during pregnancy a causal environmental risk factor for adolescent antisocial behavior? Testing etiological theories and assumptions. Psychological medicine 42, 1535–1545, doi: 10.1017/S0033291711002443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D’Onofrio BM et al. Smoking during pregnancy and offspring externalizing problems: an exploration of genetic and environmental confounds. Dev Psychopathol 20, 139–164, doi: 10.1017/S0954579408000072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cecil CA et al. DNA methylation and substance-use risk: a prospective, genome-wide study spanning gestation to adolescence. Translational psychiatry 6, e976, doi: 10.1038/tp.2016.247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collado-Torres L et al. Flexible expressed region analysis for RNA-seq with derfinder. Nucleic Acids Res 45, e9, doi: 10.1093/nar/gkw852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883, doi: 10.1093/bioinformatics/bts034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gene Ontology C Gene Ontology Consortium: going forward. Nucleic Acids Res 43, D1049–1056, doi: 10.1093/nar/gku1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Wang LG, Yan GR & He QY DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609, doi: 10.1093/bioinformatics/btu684 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Fabregat A et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 44, D481–487, doi: 10.1093/nar/gkv1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milacic M et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers 4, 1180–1211, doi: 10.3390/cancers4041180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G, Wang LG, Han Y & He QY clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : a journal of integrative biology 16, 284–287, doi: 10.1089/omi.2011.0118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452, doi: 10.1093/nar/gku1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.