Abstract
Objective:
Cytokine expression is tightly regulated post-transcriptionally but high levels of IL-6 in osteoarthritis (OA) indicate disruption of regulatory mechanisms. ZCCHC6 enzyme is implicated in post-transcriptional regulation of inflammatory cytokine expression but its role in OA pathogenesis is unknown. Here we studied whether ZCCHC6 directs the expression of IL-6 and influence OA pathogenesis in vivo.
Methods:
Human and mouse chondrocytes were stimulated with recombinant IL-1β. We knocked down the expression of ZCCHC6 in human chondrocytes by siRNAs. IL-6 transcript stability was determined by Actinomycin-D chase and 3’-uridylation of miRNAs was determined by deep sequencing. Zcchc6−/− mice were produced by gene targeting. OA was surgically induced in the knee joints of mice and the disease severity was scored using a semi-quantitative scoring system.
Results:
ZCCHC6 was markedly upregulated in the damaged cartilage from human OA patients and from wild type mice with surgically-induced OA. Overexpression of ZCCHC6 induced the expression of IL-6 and its knockdown reduced the IL-6 transcript stability and IL-1β-induced expression in chondrocytes. Reintroduction of Zcchc6 in Zcchc6−/− chondrocytes rescued the IL-1β-induced IL-6 expression. Knockdown of ZCCHC6 reduced the population of miR-26b with 3’-uridylation by 60%. Zcchc6−/− mice with surgically-induced OA produced low levels of IL-6 and showed reduced cartilage damage and synovitis in the joints.
Conclusions:
ZCCHC6 enhance IL-6 expression in chondrocytes through transcript stabilization and by uridylating miR-26b which abrogates repression of IL-6. Inhibition of IL-6 expression and significantly reduced OA severity in Zcchc6−/− mice identify ZCCHC6 as a novel therapeutic target to inhibit disease pathogenesis.
Introduction
Osteoarthritis (OA) is the most common form of arthritis that affects diarthrodial joints and causes pain and disability (1). OA pathology is now being recognized as driven by a pro-inflammatory component as high level of IL-6 and other cytokines are present in the synovial fluid of OA patients and also seen in animal models of OA (2–5). A prospective population study on a cohort of British women showed a correlation of higher BMI and elevated serum levels of IL-6 with development of radiographic knee OA (6). IL-6 has also been shown to act as a crucial mediator of MMP13 expression in HIF-2α-induced experimental OA in mice (7) and affects the anabolic processes in cartilage such as downregulation of type II collagen expression (8). IL-6 has been reported to increase the severity of OA and the inhibition of IL-6/Stat3 signaling slows the progression of experimental OA (9, 10). However, regulation of IL-6 expression in OA is still not fully understood.
Gene expression of cytokines is a tightly regulated process to control tissue inflammation. The cytokine gene expression is regulated by various factors at transcriptional, post-transcriptional and translational levels (11) and miRNAs have been found to play a critical role in the regulation of cytokine gene expression (12). Recent work has shown the presence of non-templated uridine residues added by terminal uridylyltransferases (TUTases) on pre-microRNAs, miRNAs and mRNAs (13–16). ZCCHC6 (zinc-finger, CCHC domain-containing protein 6 or TUT-7) is a non-canonical poly(A)-polymerase and a member of the nucleotidyltransferase superfamily that has been shown to uridylate the 3’-end of miRNAs and modulate the biogenesis and stability of mRNAs and miRNAs (17, 18). It was found that knockdown of ZCCHC11 expression in A549 cells suppressed the expression of IL-6 by altering the 3’-uridylation of miRNAs involved in the regulation of IL-6 expression (19). ZCCHC6, a homologue of ZCCHC11, has recently been shown to be involved in the regulation of inflammatory cytokines in mouse macrophages (20) but its role in chondrocytes or in OA pathogenesis has not been investigated.
In this study, we used human OA cartilage and a mouse model of surgically induced OA to investigate the expression of ZCCHC6 and whether ZCCHC6 regulates IL-6 expression in chondrocytes and affects OA pathogenesis in vivo. Here, we report that ZCCHC6 expression was highly upregulated in human and mouse OA cartilage and chondrocytes and was also modulated by IL-1β in vitro. Human chondrocytes transfected with ZCCHC6 targeting siRNAs expressed low levels of IL-6 while its overexpression enhanced the expression of IL-6. ZCCHC6 modulated both the stability and expression of IL-6 mRNAs in chondrocytes. ZCCHC6 knockdown had no effect on the overall expression level of miRNAs but decreased the population of 3’-uridylated miRNA miR-26b which targets IL-6 mRNA in human chondrocytes. Zcchc6−/− mice with surgically induced OA expressed low levels of IL-6 in the joints and showed reduced synovitis, significantly less cartilage damage and loss of proteoglycans compared to the wild type littermates. Taken together, our gain-of-function and loss-of-function studies clearly demonstrated that ZCCHC6 acts as a novel regulator of IL-6 expression and OA pathogenesis in a mouse model of OA.
Materials and Methods
Chondrocytes preparation and culture.
Studies for the use of discarded, deidentified human cartilage samples were approved as non-human subject study under 45 CFR, Exemption 4, by the NEOMED and SUMMA Health IRB. Chondrocytes from OA cartilage were prepared by the enzymatic digestion and cultured as described previously (21). Chondrocytes from mouse joints were prepared using femoral condyles and tibial plateaus as described (22). Chondrocyte’s cultures when 80% confluent were serum starved overnight and then stimulated with IL-1β [5 ng/ml; human IL-1β (Cat# 201-LB-025) or mouse IL-1β (Cat# 401-ML-005), R & D Systems, St Paul, MN, USA] for indicated time points. Culture supernatants were centrifuged at 20,000xg for 5 minutes to remove debris and was used for ELISA and cytokine Multiplex assays. Chondrocyte phenotype was analyzed using a PCR array for chondrocyte-specific genes (Cat #GK064, ScienCell™).
Experimental OA and Histological analyses of mouse knee joints.
All animal studies were approved by the Institutional Animal Use and Care Committee of the Northeast Ohio Medical University. Zcchc6+/− mice were generated by Gene Targeting (SIGTR ES cell line AE0325, Mutant Mouse Regional Resource Center, University of California, Davis) and Zcchc6−/− mice were produced by Zcchc6+/− × Zcchc6+/− and then by Zcchc6−/− × Zcchc6−/− breeding. Experimental OA was induced in the right knee of 12 weeks old male Zcchc6−/− and WT littermates (n=10/group) by surgical destabilization of medial meniscus (DMM) as described (23). Sham surgery was performed on the left knee, which served as control. Mice were sacrificed 8-weeks post-surgery and the joints were used for the histological assessment of the severity of the disease and scored using the semi quantitative OARSI scoring system (24). Synovitis was determined by Safranin-O and Haematoxylin staining and degree of synovial inflammation was scored from 0–3 as previously described (25).
Knockdown and overexpression of ZCCHC6.
Human OA chondrocytes were transfected with 100 nmoles of scrambled or ZCCHC6 targeting siRNAs (OnTarget Plus, SMARTPOOL siRNAs, Dharmacon) or ZCCHC6 expression plasmid [generously provided by Dr. Chris Norbury, Sir William Dunn School of Pathology, University of Oxford, Oxford, UK (26)] using the Lonza 4D Nucleofector System and Amaxa P3 Primary Cell 4D Nucleofection kit and were cultured as above. Gene expression analyses were performed 48 hours post-transfection.
Total RNA isolation from the cartilage and chondrocytes and gene expression analysis by TaqMan assays.
Human cartilage samples were stained with India ink and cartilage from the damaged and undamaged areas was resected and frozen in liquid nitrogen and pulverized to powder using a Spex freezer mill (Spex 6770) which was then used to extract DNA free total RNA using miRNeasy kit (#217004, Qiagen). Total RNA from human or mouse chondrocytes was also prepared as above. Integrity of RNA preparations was determined using TapeStation 4200 (Agilent Technologies, Santa Clara, CA, USA) and 1-μg of total RNA was used to synthesize cDNA (Applied Biosystems, cDNA synthesis kit, #4368814) and the mRNA expression was determined by TaqMan assays.
mRNA Decay Experiment.
The stability of IL-6 mRNA transcripts in human OA chondrocytes with knockdown of ZCCHC6 expression or in Zcchc6−/− and Zcchc6+/+ mouse chondrocytes was determined by Actinomycin D chase experiments as described previously (27).
RNA Immunoprecipitation (RIP) Assay.
The RIP assay was performed as described earlier (27). Briefly, Chondrocytes (5×106 cells per 100mm dish, total of 10 dishes) were stimulated with IL-1β for 6 hours, harvested and washed with ice cold PBS followed by lysis in Polysomes lysis buffer (100 mM KCl, 5 mM MgCL2, 10 mM HEPES, 0.5% NP40, 1 mM DTT, 100 units/ml RNasin, 400 μM VRC and protease inhibitor cocktail). The lysate was incubated with anti-ZCCHC6 antibody or rabbit IgG as control for overnight at 4°C. The antibody-lysate mix was incubated with Protein A agarose beads for pull-down and RNA was prepared from the immunoprecipitate using Trizol-Chloroform method followed by TaqMan assay for IL-6 mRNA quantitation.
Western Immunoblotting.
The protein expression was determined by Western blotting as described (28). Briefly, chondrocytes were lysed in RIPA buffer containing complete protease inhibitor cocktail (Roche, Indianapolis, IN, # 11697498001) and the protein concentration was estimated by Bradford dye assay. The lysates were resolved by SDS-PAGE and transferred to PVDF membrane (Bio-Rad, # 1704156). Membranes were probed with primary polyclonal antibodies specific for ZCCHC6 (validated by siRNA mediated knockdown of ZCCHC6) (Santa Cruz Biotechnology, #sc-137947) or a monoclonal anti-β-Actin antibody (Santa Cruz Biotechnology; sc-47778) followed by appropriate HRP conjugated secondary antibodies (Cell Signaling Technologies). Images were acquired using the Syngene Pxi-Imaging System (Syngene, Frederick, MD) and the band intensities were quantified by ImageJ software.
IL-6 ELISA.
IL-6 concentration in the culture supernatants was determined using a human IL-6 ELISA kit according to manufacturer’s instructions (R & D Systems, Cat# D6050). Plates were read using a Synergy H1 Hybrid Plate Reader (Biotek Instruments, VT). The sensitivity of the assay was 0.7 pg IL-6/ml.
Cytokine Multiplex Immunoassay:
Levels of the secreted cytokines and chemokines in the culture supernatants were determined using the Procarta Plex® 65-plex immunoassay (Thermo Fischer Scientific; Cat No: EPX650-10065-901) for human chondrocytes and 36-plex immunoassay (Thermo Fischer Scientific; Cat No: EPX360-26092-901) for mouse chondrocytes. Human chondrocytes with knockdown of ZCCHC6 expression or chondrocytes isolated from Zcchc6−/− mice and WT littermates were stimulated with IL-1β for 6 hrs and 50μl aliquots of the supernatants in duplicate were assayed for the secreted molecules as per manufacturer’s instructions using Luminex 200 System and analyzed by ProcartaPlex Analyst 1.0 (Luminex Corporation).
Cytokine PCR Array Analyses.
Human OA chondrocytes with knockdown of ZCCHC6 expression were stimulated with IL-1β for 6 hours and total RNA was extracted for cDNA synthesis as described above. Gene expression was analyzed using a panel of 88 genes involved in immune cell response, inflammation, cellular signaling and 8 housekeeping genes as normalization control (Realtimeprimers.com, #HAIIR-I).
Immunohistochemistry (IHC).
Full thickness human cartilage pieces were taken from the tibial plateau and were fixed in 4% paraformaldehyde for 48 hrs. Samples were dehydrated, embedded in paraffin and 5 μM thick serial sections were cut for IHC. To analyze the expression of ZCCHC6, sections were probed with anti-ZCCHC6 antibody as primary and appropriate HRP tagged secondary antibody. The sections were developed with DAB substrate kit (Pierce, #34002) and images were captured by Olympus VS120 Scanning microscope.
Deep Sequencing of Small RNAs and miRNA 3’ end Uridylation Analyses.
Total RNA from control and ZCCHC6 depleted chondrocytes from three different donors were prepared using miRNeasy kit as above. RNA samples with RNA integrity number≥8 were used for deep sequencing. The small RNA library was prepared using TruSeq small RNA Library Prep Kit (Illumina, cat # RS-200–0012) per manufacturer’s instructions and the sequencing was performed on Illumina MiSeq system using the MiSeq reagent kit v2. The miRNA 3’-uridylation analysis was performed by Chimira (29).
Transfection of human OA chondrocytes with wild type and modified miR-26a/26b mimetics.
Standard miR-26a and miR-26b oligos and oligos with addition of 1 or 2 non-template Uridines were purchased from IDT (San Diego, CA, USA). The oligo transfected, and mock transfected human OA chondrocytes were treated with IL-1β for 6 hours and IL-6 mRNA expression was determined by TaqMan assay.
Statistical analyses.
All the statistical analyses were performed using GraphPad Prism software (Version 7.04). Statistical significance between two groups was calculated using two tailed student t-test or and in more than two groups using one-way analysis of variance (ANOVA) followed by Tukey’s test for post hoc analysis. P values ≤0.05 were considered significant.
Results
ZCCHC6 is upregulated in human and mouse OA cartilage and chondrocytes treated with IL-1β.
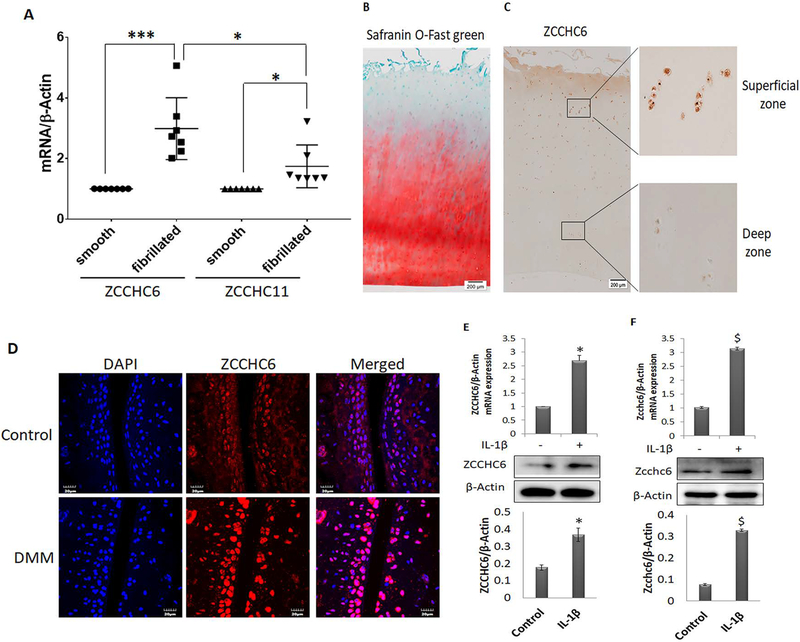
To explore the role of TUTases in OA pathogenesis, we first analyzed the expression profile of ZCCHC6 (TUT7) and ZCCHC11 (TUT4) in human OA cartilage. Our data showed that the expression levels of both ZCCHC6 and ZCCHC11 were high in the damaged cartilage compared with the undamaged cartilage of the same patient (Figure 1A). Importantly, the expression of ZCCHC6 was significantly higher than that of ZCCHC11 (p=0.02) (Figure1A). Consistent with the increased mRNA expression, immunohistochemistry showed increased number of ZCCHC6 positive chondrocytes in the damaged OA cartilage (Figure 1B, C). Similarly, the number of Zcchc6 positive chondrocytes were significantly increased in the cartilage of mice with surgically induced OA (Figure 1D). Additionally, stimulation of both human and mouse chondrocytes with IL-1β caused significant upregulation of the ZCCHC6 mRNA and protein expression (Figure 1 E and F respectively).
Figure1. ZCCHC6 is highly expressed in the damaged areas of human OA cartilage.
(A) Total RNA was prepared from damaged and undamaged areas of OA cartilage (n=7) and the expression of ZCCHC6 and ZCCHC11 were determined by TaqMan assays. The data was normalized with β-Actin and is represented as fold change relative to the expression level in the smooth cartilage. (B) Human OA cartilage (n=5) sections were stained with Safranin O-Fast Green. Representative image shows the superficial zone with excessive loss of proteoglycans compared to deep zone. (C) ZCCHC6 protein expression in human OA cartilage was determined by IHC. The superficial damaged area showed increased expression of ZCCHC6 protein in human OA cartilage in comparison to deep zone. (D) OA was induced in the knee joints of male C57BL/6 mice by DMM surgery and expression of Zcchc6 was visualized by immunofluorescence staining. The staining showed enhanced expression of Zcchc6 in mouse knee joints with DMM surgery compared to control. (E) IL-1β treatment increased the expression of ZCCHC6 in human OA chondrocytes and (F) in mouse chondrocytes at mRNA and protein levels. The top bar graphs represent mRNA quantification and bottom bar graphs represent quantification of Western blots from at least three independent experiments (*p<0.05 vs human chondrocytes control and $p<0.05 vs mouse chondrocytes control).
ZCCHC6 regulates the expression of IL-6 in chondrocytes under pathological conditions.
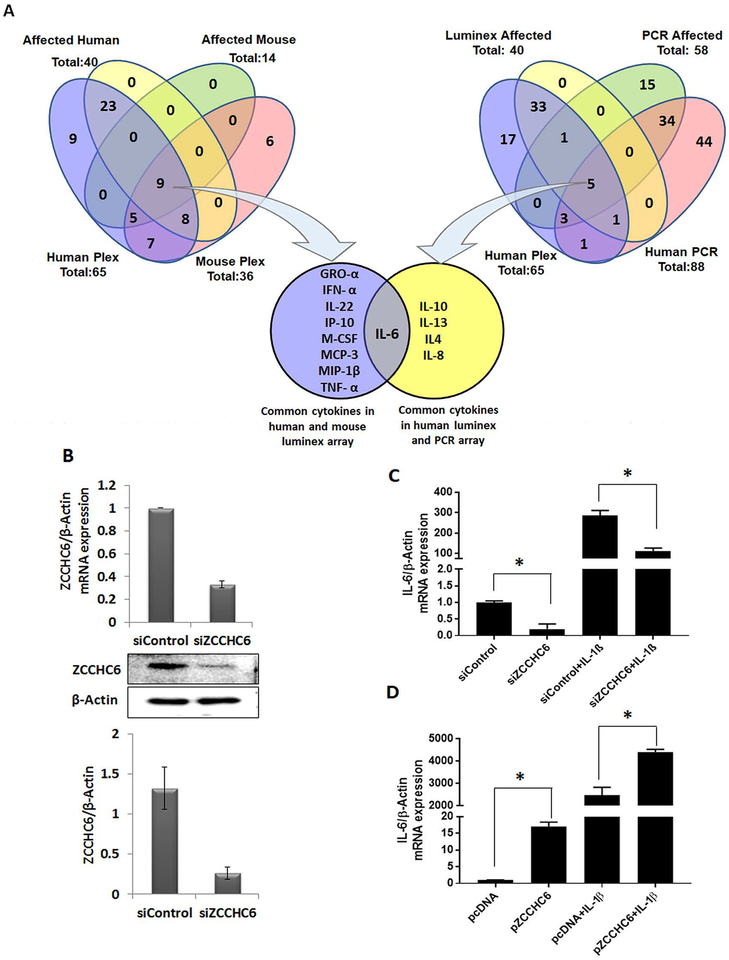
To delineate the target(s) of ZCCHC6 in chondrocytes, a systematic and comprehensive analysis was performed to define the common cytokine(s)/chemokine(s) affected by ZCCHC6 depletion in both human and mouse chondrocytes. The Venn diagram analysis (Figure 2A) of the data derived from cytokine PCR array and multiplex assay (Supplementary Table 1) from human and mouse chondrocytes indicated that IL-6 is the major cytokine regulated by ZCCHC6. Based on the above analysis and published reports on the role of IL-6 in OA pathogenesis (7, 8), we chose to study the regulation of IL-6 by ZCCHC6 in human and mouse chondrocytes by using loss of function and gain of function approaches. Chondrocytes with siRNA mediated knockdown of ZCCHC6 (Figure 2B) significantly suppressed the expression of constitutive and IL-1β induced IL-6 mRNA and protein expression (Figure 2C and Supplementary Figure 1). Furthermore, human OA chondrocytes with overexpression of ZCCHC6 (Supplementary Figure 2) showed significantly increased constitutive expression of IL-6, which was further increased several folds upon stimulation with IL-1β (Figure 2D). To determine if the observed effect of ZCCHC6 knockdown and overexpression is specific to ZCCHC6 or other TUTases also regulate IL-6 expression, we knockdown the expression of TUT-1 and TUT-2 using siRNAs in human OA chondrocytes and determined the expression of IL-6 mRNA. Interestingly knockdown of TUT-1 and TUT-2 expression did not suppress the expression of IL-6 (Supplementary Figure 3) demonstrating that the observed effect in chondrocytes with knockdown of ZCCHC6 expression was ZCCHC6 specific. Of importance is our finding that knockdown of ZCCHC6 did not alter the expression of ZCCHC11 (Supplementary Figure 4). Taken together, these data demonstrated that ZCCHC6 plays an essential role in the regulation of IL-6 expression in OA chondrocytes under pathological conditions.
Figure 2. ZCCHC6 regulates IL-6 expression in chondrocytes.
(A) Cytokine multiplex assay and qPCR were performed with the culture supernatant of ZCCHC6 depleted human chondrocytes and Zcchc6−/− mouse chondrocytes and total RNA prepared from ZCCHC6 depleted human chondrocytes and their respective controls. The cytokine expression from all the three different experiments were compared and the Venn diagram showed that IL-6 was the common cytokine downregulated in both human and mouse chondrocytes. (Human plex = cytokine multiplex assay of human samples with a total number of cytokines=65 and number of affected cytokines was 40. Mouse plex = cytokine multiplex assay of mouse samples with a total number of cytokines = 36 and number of affected cytokines was 14. Human PCR = cytokine PCR array of human samples with a total number of targets = 88 and number of affected targets was 58). (B) siRNA mediated knockdown of ZCCHC6 at mRNA and protein level was confirmed by qPCR and Western blotting respectively. (C) Human OA chondrocytes with siRNA mediated knockdown of ZCCHC6 expressed low levels of constitutive and IL-1β induced compared to controls (*p<0.05). (D) Overexpression of ZCCHC6 in human OA chondrocytes induced the constitutive and IL-1β induced expression of IL-6 (*p<0.05).
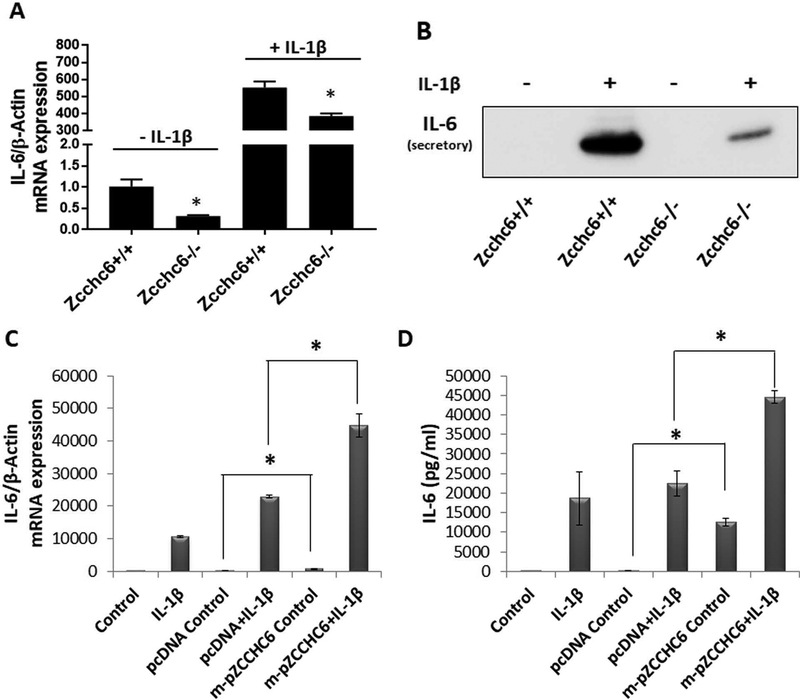
To determine whether ZCCHC6 also regulate the expression of IL-6 in mouse chondrocytes, we prepared articular cartilage chondrocytes from Zcchc6−/− mice and their corresponding WT (Zcchc6+/+) littermates and the expression of IL-6 was determined as described above. Similar to the data obtained with human chondrocytes, expression of IL-6 in Zcchc6−/− mouse chondrocyte was significantly low in comparison to the levels detected in Zcchc6+/+ mouse chondrocytes (Figure 3A). Stimulation of mouse chondrocytes with IL-1β resulted in significant increase in the expression of IL-6 mRNA and protein in both Zcchc6+/+ and Zcchc6−/− chondrocytes, however the IL-6 expression in Zcchc6−/− chondrocytes was significantly low compared to the levels in Zcchc6+/+ chondrocytes (Figure 3A, 3B). To confirm the above results that the observed effect on IL-6 expression was due to depletion of Zcchc6, mouse chondrocytes prepared from Zcchc6−/− mice were transfected with the Zcchc6 expression plasmid and the expression of IL-6 was determined. Restoration of Zcchc6 expression in Zcchc6−/− chondrocytes not only rescued but also increased the constitutive and IL-1β induced expression of IL-6 (Figure 3C and 3D respectively) indicating that Zcchc6 is an important regulator of IL-6 expression in chondrocytes.
Figure 3: Zcchc6−/− mouse chondrocytes expressed low levels of IL-6 upon stimulation with IL-1β.
(A) Mouse chondrocytes were prepared from the knee joints of Zcchc6−/− and Zcchc6+/+ littermates and were either untreated or treated with IL-1β. The expression of IL-6 mRNA was downregulated in Zcchc6−/− mouse chondrocytes compared to Zcchc6+/+ chondrocytes in the presence and absence of IL-1β (*p<0.05). (B) IL-6 protein expression in the culture supernatant was determined by Western blotting. (C) and (D) The Zcchc6−/− mouse chondrocytes were transfected with Zcchc6 overexpression plasmid followed by stimulation with IL-1β. Zcchc6 expression restored the constitutive and induced IL-6 expression in Zcchc6−/− mouse chondrocytes. pcDNA was used as control. (*p<0.05). The experiments were repeated at least three times with mouse chondrocytes.
ZCCHC6 binds IL-6 mRNA transcripts in chondrocytes.
ZCCHC6 is a zinc finger containing protein which are known to bind to RNA (30) but the binding of ZCCHC6 to IL-6 mRNA has not been reported. We performed RNA immunoprecipitation assay and found significant enrichment of IL-6 mRNA in the anti-ZCCHC6 antibody pulldown fractions compared to control IgG pull down fractions (Figure 4A). We further confirmed the binding of ZCCHC6 protein with IL-6 mRNA by co-localization studies using immunofluorescence (IF) staining of ZCCHC6 protein and FISH of IL-6 mRNA and found that the IL-6 mRNA colocalized with the ZCCH6 protein in human chondrocytes (Supplementary Figure 5).
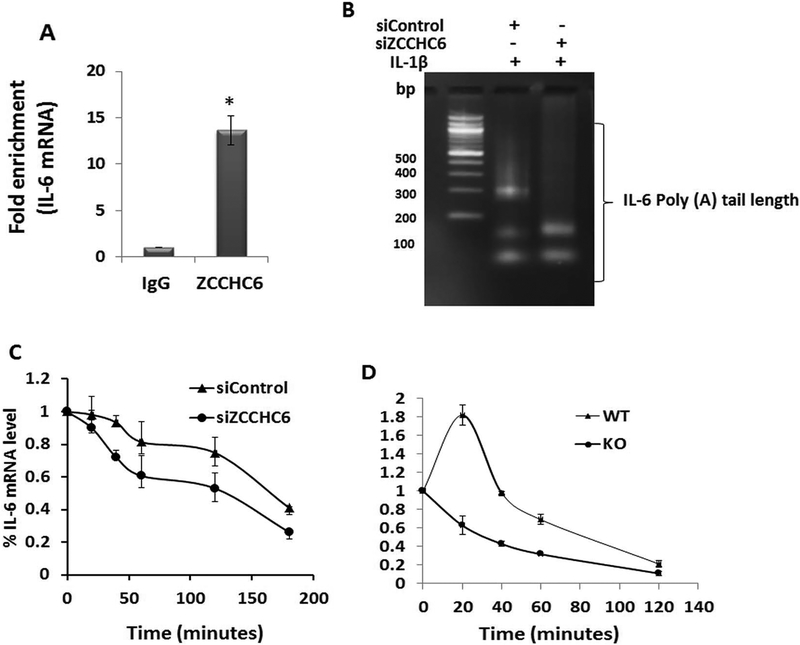
Figure 4: ZCCHC6 binds to IL-6 mRNA and regulates its stability.
(A) Human OA chondrocytes were stimulated with IL-1β for 6 hrs and harvested for RIP assay using anti-ZCCHC6 antibody. IgG was used as negative control. RIP showed significant enrichment of IL-6 mRNA. (*p<0.05) (B) poly(A) tail length of IL-6 mRNA was reduced in ZCCHC6 depleted human OA chondrocytes. (C) Human chondrocytes depleted of ZCCHC6 showed significantly (p<0.05) reduced half-life of IL-6 mRNA (t1/2 in siControl = 157.74±5.33 minutes and t1/2 in siZCCHC6= 83.18±10.29 minutes). (D) Zcchc6−/− mouse chondrocytes showed significantly (p<0.05) reduced half-life of IL-6 mRNA. (t1/2 in Zcchc6+/+ = 55.37±5.4 minutes and t1/2 in Zcchc6−/− = 28.68±1.5 minutes). The experiments were repeated at least three times with OA and mouse chondrocytes.
ZCCHC6 plays an essential role in maintaining the poly(A) tail lengths and stability of IL-6 mRNA transcripts in chondrocytes.
Poly(A) tail length of mammalian mRNA functions as an important determinant of mRNA stability and translation (31). Since ZCCHC6 is a non-canonical poly(A) polymerase and our data demonstrated its binding with IL-6 mRNA (Figure 4A), we investigated whether ZCCHC6 plays a role in maintaining the poly(A) tail length of IL-6 mRNA in human chondrocytes using the loss of function approach. In human chondrocytes with knockdown of ZCCHC6 expression, the poly(A) tail length of IL-6 mRNA was decreased (Figure 4B). This suggested that in the absence of ZCCHC6, IL-6 mRNA may not be stable and may have shorter half-life. To test this, human OA chondrocytes with siRNA mediated knockdown of ZCCHC6 were stimulated with IL-1β followed by Actinomycin D chase experiment. The half-life (t1/2) of IL-6 mRNA in control chondrocytes was found to be 157.74±5.33 minutes and was reduced to 83.18±10.29 minutes in chondrocytes with siRNA mediated knockdown of ZCCHC6 (Figure 4C). In agreement with these findings, chondrocytes prepared from Zcchc6−/− mice and stimulated with IL-1β also showed almost 50% reduction (t1/2 28.68±1.5 min in Zcchc6−/− chondrocytes vs t1/2 55.37±5.4 min in wild type chondrocytes) in the half-life of IL-6 mRNA (Figure 4D). In summary, absence of ZCCHC6 negatively affects the poly(A) tail length and the stability of IL-6 mRNA in chondrocytes suggesting that maintaining the poly(A) tail length is a step through which ZCCHC6 regulates IL-6 expression.
ZCCHC6 knockdown reduces the 3’-uridylation of IL-6 targeting miRNAs.
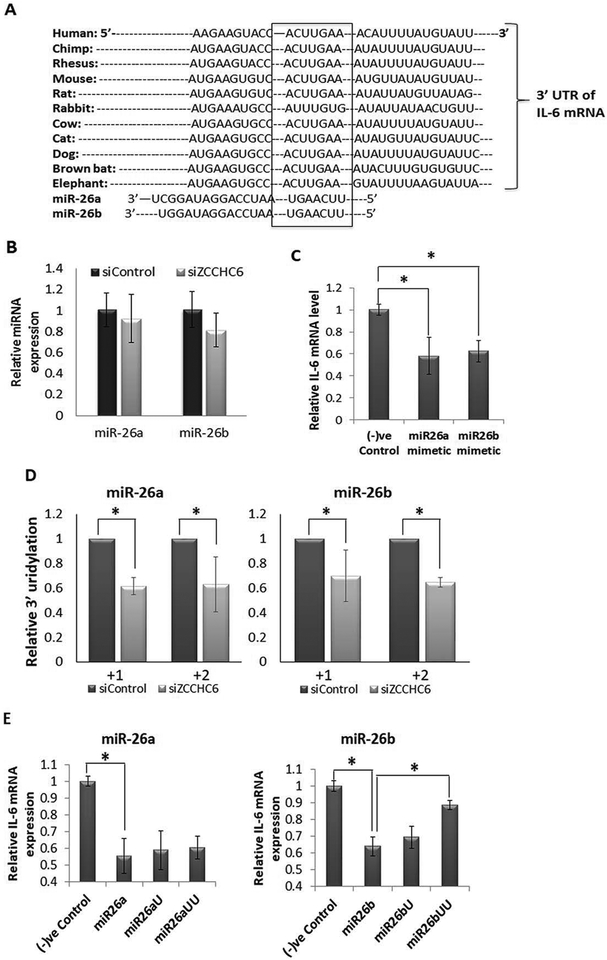
Previous studies have shown that the closely homologous enzyme ZCCHC11 regulates the expression of IL-6 by 3’-uridylation of miRNAs (19). Since ZCCHC6 has been shown to uridylate RNAs (31), we determined whether ZCCHC6 plays a role in maintaining high levels of IL-6 expression through the inactivation of miRNAs that repress IL-6 expression via non-template additions of uridines at the 3’-ends. We analyzed the 3’-uridylation of miRNAs at global level in ZCCHC6 depleted chondrocytes by deep sequencing. Data analyses using the online available miRNA modification analysis tool, Chimira (29) showed that ZCCHC6 depletion in human OA chondrocytes resulted in the global reduction of 3’-mono and di-uridylation of miRNAs (Supplementary Figure 6). Since our aim was to investigate the regulation of IL-6 expression, we chose miR-26a and miR-26b that are known to repress IL-6 expression (19) by binding to the highly conserved “seed sequence” present in the 3’-UTR of IL-6 mRNA (Figure 5A). We first determined the expression of miR-26a and miR-26b miRNAs in human chondrocytes with depleted expression of ZCCHC6 and found that the expression levels of miR-26a and miR-26b were not affected (Figure 5B). Further, overexpression of miR-26a and miR-26b suppressed the IL-1β induced expression of IL-6 in human OA chondrocytes (Figure 5C). We next analyzed the deep sequencing data to investigate the 3’-uridylation of miR-26a and miR-26b miRNAs and discovered that the 3’-uridylation of miR-26a/26b miRNAs at +1 and +2 positions was significantly decreased in the ZCCHC6 depleted chondrocytes (Figure 5D). To test whether 3’-uridylation of miR-26a and miR-26b abrogate their suppressive effects in chondrocytes, we used miR-26a and miR-26b mimetics with additional “U” and “UU” at their 3’-end and determined their ability to suppress the expression of IL-1β induced expression of IL-6 in human chondrocytes. Interestingly, we observed that addition of uridines at the 3′ end of miR-26b, but not of miR-26a, abrogated the suppression of IL-1β induced expression of IL-6 in human OA chondrocytes (Figure 5E). Taken together, this suggests that the increased expression of IL-6 may be due to the inactivation of miR-26b by 3’-uridylation due to increased expression and activity of ZCCHC6 in chondrocytes under pathological conditions.
Figure 5: IL-6 mRNA expression is regulated by miR26a/b in chondrocytes.
(A) Targetscan prediction showed a conserved site for miR26a/b family of miRNAs in the 3’-UTR of IL-6 mRNA. (B) The expression level of miR26a or miR26b was not altered in human chondrocytes with knockdown of ZCCHC6. (C) Expression of miR26a and miR26b suppressed the IL-1β induced expression of IL-6 in human OA chondrocytes (*p<0.05). (D) 3’-uridylation analyses of miR26a and miR26b by deep sequencing showed decreased uridylation at +1 and +2 position. (E) Human OA chondrocytes were transfected with miR26a or miR26b mimetics and miR26a and miR26b mimetics with additional mono- or di-Uridines at the 3’-ends followed by IL-1β treatment for 6 hrs and total RNA was prepared for qPCR analysis of IL-6 mRNA. miR26b mimetics with di-uridine at the 3’-end failed to suppress the IL-1β-induced expression of IL-6 mRNA (*p<0.05). (–)ve control represent scrambled oligo. The experiments were repeated with chondrocytes from at least three OA donors.
Genetic inactivation of Zcchc6 abrogates OA pathogenesis:
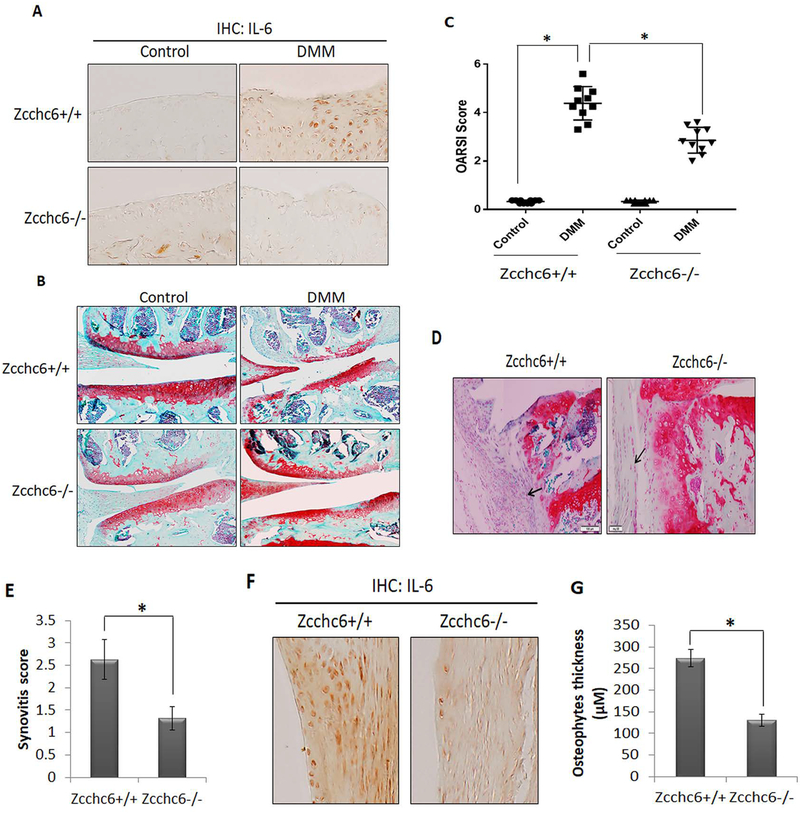
Zcchc6−/− mice were used to investigate the role of Zcchc6 in regulating IL-6 expression and OA pathogenesis in vivo. Zcchc6−/− mice were born in Mendelian ratios without any overt abnormalities. Since female mice are resistant to the development of OA (32), we performed DMM surgery on 12 weeks old Zcchc6+/+ and Zcchc6−/− male mice and assessed the effect of Zcchc6 inactivation on the severity of OA 8 weeks post-DMM. We used serial sections harvested at 80 μM intervals of frontally embedded knee joints for histological scoring and intervening sections were used to analyze the expression of IL-6 by immunohistochemistry. Our results showed that the Zcchc6−/− mice, compared to wild type littermates, had low levels of IL-6 in the joints with DMM-induced OA (Figure 6A). Additionally, the cartilage erosion was significantly reduced in Zcchc6−/− mice compared to wild type littermates (Figure 6B). OARSI scoring further confirmed a significant reduction in cartilage damage in Zcchc6−/− mice (Figure 6C). We also found that Zcchc6−/− mice developed reduced synovitis (Figure 6D and 6E) and showed decreased expression of IL-6 in the synovium (Figure 6F). There was also a significant reduction in the osteophytes thickness (P<0.05, Figure 6G). Overall, these results support the conclusion that Zcchc6 plays a critical role in the disease pathogenesis through upregulated expression of IL-6 in mice with DMM-induced OA.
Figure 6: Zcchc6 inactivation reduced the severity of experimental osteoarthritis in mice.
(A) 12 weeks old Zcchc6−/− or Zcchc6+/+ C57BL/6 male mice (n=10, each) were subjected to DMM surgery on the right knee (the left knee was used as control). Mice were sacrificed after 8-week post-DMM surgery and joints were collected and processed for histological analysis. IHC with anti-IL-6 antibody showed decreased expression of IL-6 protein in the Zcchc6−/− mice joints subjected to DMM compared to Zcchc6+/+. (B) Loss of proteoglycans in the knee joints with surgically-induced OA was analyzed by Safranin O/Fast Green staining. (C) Osteoarthritis scores were determined as per OARSI guidelines. (*p<0.005). Zcchc6 deletion showed protective effect on cartilage degeneration in mice with surgically-induced OA. (D and E) Synovium from Zcchc6−/− or Zcchc6+/+ littermates with surgically-induced OA were analyzed by staining with Safranin O-Hematoxylin. Synovial thickness and synovitis score were significantly reduced in Zcchc6−/− mice (*p< 0.005). (F) IHC of IL-6 in Zcchc6−/−and Zcchc6+/+ mice joints subjected to DMM showed reduced expression of IL-6 in synovium. (G) Osteophyte thickness was significantly reduced in Zcchc6−/− mice joints subjected to DMM surgery compared to Zcchc6+/+ joints (*p<0.005).
Discussion
A hallmark of OA is the progressive and irreversible cartilage degradation due to the enhanced production of inflammatory cytokines by activated chondrocytes, such as IL-6, that amplifies the inflammatory effect in the affected joints and induces the expression of several cartilage extracellular matrix degrading enzymes. The overall cytokine repertoire of OA synovial fluid was shown to be primarily dependent on the extent of OA disease (5). It is of importance to note that our data show that human chondrocyte cultures derived from pathologic cartilage samples produced a cytokine repertoire that included elevated levels of IL-6 (Supplementary Table 1) mirroring that of OA synovial fluid (2). Highly elevated levels of IL-6 were also detected in knee cartilage from OA patients (3) and was found to positively correlate with Kellgren-Lawrence (K-L) scores (4).
Cytokines are central mediators of tissue inflammation and their expression is tightly regulated at various levels, including transcriptional, post-transcriptional and translational levels. We have shown previously that various transcription factors including NFκB and AP1 regulate the expression of IL-6 at transcriptional level in human chondrocytes (33, 34). Post-transcriptional regulation by RNA binding proteins and miRNAs have been shown to play a vital role in controlling the expression of cytokines by modulating mRNA stability and translation (12). ZCCHC6 and its homologue, ZCCHC11 have been reported to uridylate and alter the stability of mRNAs in HeLa cells (18). It was also found that ZCCHC11 knockdown in A549 cells downregulated the expression of IL-6 (19). However; the involvement of ZCCHC6 or any other TUTase in regulating IL-6 gene expression in chondrocytes and its role in OA pathogenesis has not been reported.
In the present study, we used loss of function and gain of function approaches to investigate the potential role of ZCCHC6 in the post-transcriptional regulation of IL-6 expression. Our data showed that ZCCHC6 downregulation in human OA chondrocytes significantly reduced the expression of IL-6 mRNA and protein. This is similar to the effect of ZCCHC11 depletion in A549 cells (19). Here we show that similar to human OA chondrocytes, the expression of IL-6 in Zcchc6 deficient mouse chondrocytes was also downregulated in both in vitro and in vivo studies (Figure 3). In a recent study, in contrast to results reported here, it was shown that Zcchc6 deficient mouse macrophages showed increased expression of IL-6 (20) indicating a cell type and context specific regulation of IL-6 expression by Zcchc6 in vivo.
The poly(A) tail length of eukaryotic mRNA is a critical determinant of transcript stability and translational efficiency (35). We found that IL-6 mRNA had shorter poly(A) tail length in chondrocytes depleted of ZCCHC6 expression indicating that ZCCHC6 functions in maintaining IL-6 transcript stability and translation. This is supported by our findings that overexpression of ZCCHC6 in human chondrocytes upregulated the expression of IL-6 mRNA and protein. This has not been demonstrated in any cell type before. Thus this activity of ZCCHC6 in maintaining the poly(A)tail lengths of IL-6 mRNA in chondrocytes mimics that of ZCCHC11 in immune cells (19). Since ZCCHC11 expression was not altered in chondrocytes with knockdown of ZCCHC6, and knockdown of TUT-1 and TUT-2 did not affect the IL-6 mRNA expression indicates that the downregulation of IL-6 expression was specific to ZCCHC6 knockdown in chondrocytes. We postulate that these enzymes may have cell specific nonredundant functions as well.
Cytokine expression is tightly regulated by miRNAs (12). ZCCHC6 and ZCCHC11, both have been shown to regulate the expression of various cytokines by altering the biogenesis and stability of miRNAs (17). ZCCHC6 has been found to selectively uridylate miRNAs and modulate their stability and mRNAs repressive activity (36, 37). Interestingly, we found that ZCCHC6 knockdown in human OA chondrocytes decreased the 3’-uridylation of miRNAs with significant reduction in the population of 3’-uridylated miR-26b. Furthermore, we also show here that miR-26b mimetic with additional “UU” at the 3’-end did not suppress the expression of IL-6 mRNA indicating that the miR-26b with two uridine at the 3’-end found in our RNA-Seq analyses represents the population of miR-26b incapable of suppressing IL-6 expression and identifies a mechanism through which increased ZCCHC6 activity constitute to increased IL-6 expression in chondrocytes under pathological conditions.
Our results are consistent with another study, wherein it was also found that 3’-uridylation of miR-26b abrogated its IL-6 mRNA repressive activity in A549 cells (19). As IL-6 plays an important role in the pathogenesis of OA, we also investigated whether inhibition of IL-6 may prevent cartilage damage in OA. Indeed, we found that the expression of IL-6 and damage to the cartilage was significantly reduced in the Zcchc6−/− mouse with DMM-induced OA. These results strongly support the hypothesis that Zcchc6 contributes to OA pathogenesis by facilitating the increased expression of IL-6 in chondrocytes. However; our data of IL-6 regulation does not rule out the possibility of the regulation of other OA related genes by ZCCHC6 as well and will be reported in future studies.
In summary, the present study is the first in vivo analysis of the effects of ZCCHC6 on IL-6 expression in articular cartilage chondrocytes and its role in OA pathogenesis. We demonstrated that genetic inactivation of Zcchc6 reduced IL-6 expression and cartilage damage in a mouse model of experimental OA. Also, we found that the expression pattern of ZCCHC6 and IL-6 was similar in human OA cartilage and the mouse OA cartilage. Taken together, these data demonstrate a previously unknown function of ZCCHC6 in regulating IL-6 expression in chondrocytes and in OA pathogenesis and identifies a potential therapeutic target for the management of OA.
Supplementary Material
Acknowledgements
This work was supported in part by USPHS/National Institutes of Health grants (RO1-AT-005520; RO1-AT-007373; RO1-AR- 067056) and funds from the Northeast Ohio Medical University to TMH.
References
- 1.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nature reviews Rheumatology. 2016;12(7):412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malemud CJ. Biologic basis of osteoarthritis: state of the evidence. Current opinion in rheumatology. 2015;27(3):289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moktar NM, Yusof HM, Yahaya NH, Muhamad R, Das S. The transcript level of interleukin-6 in the cartilage of idiopathic osteoarthritis of knee. La Clinica terapeutica. 2010;161(1):25–8. [PubMed] [Google Scholar]
- 4.Orita S, Koshi T, Mitsuka T, Miyagi M, Inoue G, Arai G, et al. Associations between proinflammatory cytokines in the synovial fluid and radiographic grading and pain-related scores in 47 consecutive patients with osteoarthritis of the knee. BMC musculoskeletal disorders. 2011;12:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchida AI, Beekhuizen M, t Hart MC, Radstake TR, Dhert WJ, Saris DB, et al. Cytokine profiles in the joint depend on pathology, but are different between synovial fluid, cartilage tissue and cultured chondrocytes. Arthritis research & therapy. 2014;16(5):441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livshits G, Zhai G, Hart DJ, Kato BS, Wang H, Williams FM, et al. Interleukin-6 is a significant predictor of radiographic knee osteoarthritis: The Chingford Study. Arthritis and rheumatism. 2009;60(7):2037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu JH, Yang S, Shin Y, Rhee J, Chun CH, Chun JS. Interleukin-6 plays an essential role in hypoxia-inducible factor 2alpha-induced experimental osteoarthritic cartilage destruction in mice. Arthritis and rheumatism. 2011;63(9):2732–43. [DOI] [PubMed] [Google Scholar]
- 8.Poree B, Kypriotou M, Chadjichristos C, Beauchef G, Renard E, Legendre F, et al. Interleukin-6 (IL-6) and/or soluble IL-6 receptor down-regulation of human type II collagen gene expression in articular chondrocytes requires a decrease of Sp1.Sp3 ratio and of the binding activity of both factors to the COL2A1 promoter. The Journal of biological chemistry. 2008;283(8):4850–65. [DOI] [PubMed] [Google Scholar]
- 9.Latourte A, Cherifi C, Maillet J, Ea HK, Bouaziz W, Funck-Brentano T, et al. Systemic inhibition of IL-6/Stat3 signalling protects against experimental osteoarthritis. Annals of the rheumatic diseases. 2017;76(4):748–55. [DOI] [PubMed] [Google Scholar]
- 10.Nasi S, So A, Combes C, Daudon M, Busso N. Interleukin-6 and chondrocyte mineralisation act in tandem to promote experimental osteoarthritis. Annals of the rheumatic diseases. 2016;75(7):1372–9. [DOI] [PubMed] [Google Scholar]
- 11.Kovarik P, Ebner F, Sedlyarov V. Posttranscriptional regulation of cytokine expression. Cytokine. 2017;89:21–6. [DOI] [PubMed] [Google Scholar]
- 12.Palanisamy V, Jakymiw A, Van Tubergen EA, D’Silva NJ, Kirkwood KL. Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. Journal of dental research. 2012;91(7):651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5’ to 3’ and 3’ to 5’. Genes & development. 2008;22(1):50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rissland OS, Norbury CJ. Decapping is preceded by 3’ uridylation in a novel pathway of bulk mRNA turnover. Nature structural & molecular biology. 2009;16(6):616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306(5698):997. [DOI] [PubMed] [Google Scholar]
- 16.Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). Rna. 2012;18(10):1875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim B, Ha M, Loeff L, Chang H, Simanshu DK, Li S, et al. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. The EMBO journal. 2015;34(13):1801–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159(6):1365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nature cell biology. 2009;11(9):1157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlowski E, Wasserman GA, Morgan M, O’Carroll D, Ramirez NP, Gummuluru S, et al. The RNA uridyltransferase Zcchc6 is expressed in macrophages and impacts innate immune responses. PloS one. 2017;12(6):e0179797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansari MY, Khan NM, Ahmad I, Haqqi TM. Parkin clearance of dysfunctional mitochondria regulates ROS levels and increases survival of human chondrocytes. Osteoarthritis and cartilage. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nature protocols. 2008;3(8):1253–60. [DOI] [PubMed] [Google Scholar]
- 23.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis and cartilage. 2007;15(9):1061–9. [DOI] [PubMed] [Google Scholar]
- 24.Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis and cartilage. 2010;18 Suppl 3:S17–23. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Jeon J, Shin M, Won Y, Lee M, Kwak JS, et al. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell. 2014;156(4):730–43. [DOI] [PubMed] [Google Scholar]
- 26.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Molecular and cellular biology. 2007;27(10):3612–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansari MY, Haqqi TM. Interleukin-1beta induced Stress Granules Sequester COX-2 mRNA and Regulates its Stability and Translation in Human OA Chondrocytes. Scientific reports. 2016;6:27611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari MY, Khan NM, Haqqi TM. A standardized extract of Butea monosperma (Lam.) flowers suppresses the IL-1beta-induced expression of IL-6 and matrix-metalloproteases by activating autophagy in human osteoarthritis chondrocytes. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;96:198–207. [DOI] [PubMed] [Google Scholar]
- 29.Vitsios DM, Enright AJ. Chimira: analysis of small RNA sequencing data and microRNA modifications. Bioinformatics. 2015;31(20):3365–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown RS. Zinc finger proteins: getting a grip on RNA. Current opinion in structural biology. 2005;15(1):94–8. [DOI] [PubMed] [Google Scholar]
- 31.Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nature reviews Molecular cell biology. 2013;14(10):643–53. [DOI] [PubMed] [Google Scholar]
- 32.Ma HL, Blanchet TJ, Peluso D, Hopkins B, Morris EA, Glasson SS. Osteoarthritis severity is sex dependent in a surgical mouse model. Osteoarthritis and cartilage. 2007;15(6):695–700. [DOI] [PubMed] [Google Scholar]
- 33.Haseeb A, Ansari MY, Haqqi TM. Harpagoside suppresses IL-6 expression in primary human osteoarthritis chondrocytes. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2017;35(2):311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasheed Z, Akhtar N, Haqqi TM. Advanced glycation end products induce the expression of interleukin-6 and interleukin-8 by receptor for advanced glycation end product-mediated activation of mitogen-activated protein kinases and nuclear factor-kappaB in human osteoarthritis chondrocytes. Rheumatology. 2011;50(5):838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Critical reviews in biochemistry and molecular biology. 2004;39(4):197–216. [DOI] [PubMed] [Google Scholar]
- 36.Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, et al. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic acids research. 2014;42(18):11777–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, et al. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS genetics. 2012;8(11):e1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.