Abstract
Members of the RAS proto-oncogene superfamily are indispensable molecular switches that play critical roles in cell proliferation, differentiation, and cell survival. Recent studies have attempted to prevent the interaction of RAS/GTP with RAS guanine nucleotide exchange factors (GEFs), impair RAS-effector interactions, and suppress RAS localization to prevent oncogenic signalling. The present study aimed to investigate the effect of the natural triterpenoic acid inhibitor glycyrrhetinic acid, which is isolated from the roots of Glycyrrhiza plant species, on RAS stability. We found that glycyrrhetinic acid may bind to the P-loop of RAS and alter its stability. Based on our biochemical tests and structural analysis results, glycyrrhetinic acid induced a conformational change in RAS. Meanwhile, glycyrrhetinic acid abolishes the function of RAS by interfering with the effector protein RAF kinase activation and RAS/MAPK signalling.
Abbreviations: CD, circular dichroism; DTT, d,l-dithiothreitol; FTIs, farnesyltransferase inhibitors; FTS, fluorescence-based thermal shift; GA, glycyrrhetinic acid; GAPs, GTP hydrolysis by GTPase-activating proteins; GEFs, guanine nucleotide exchange factors; HOBt, hydroxybenzotrizole; Kobe, Kobe0065; N3-tag, 3-azido-7-hydroxycoumarin; NH2-MMs, Fe3O4 amino magnetic microspheres; RAS, GTPases RAS; SPR, surface plasmon resonance; Sulfo-SADP, sodium1-((3-((4-azidophenyl)disulfanyl)propanoyl)oxy)-2,5-dioxopyrrolidine-3-sulfonate; Tip, tipifarnib
KEY WORDS: Glycyrrhetinic acid, RAS, Allosteric inhibitor, RAS/MAPK signalling
Graphical abstract
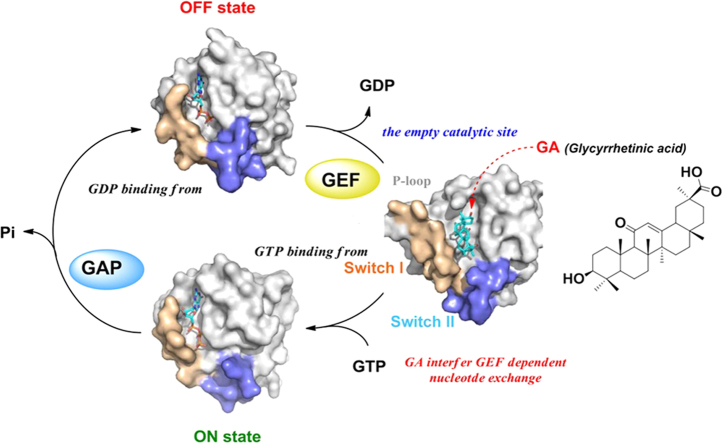
Glycyrrhetinic acid binds to the conserved P-loop region of RAS protein, induces a conformational change, and alters its stability. Meanwhile, glycyrrhetinic acid abolishes the function of RAS by interfering with the effector protein RAF kinase activation and RAS/MAPK signalling.
1. Introduction
RAS GTPases (RAS) are signal transduction pathway regulators that control cell proliferation, survival, differentiation, and apoptosis1., 2., 3.. The RAS protein is an intracellular guanine nucleotide-binding protein (G protein) that comprises two domains: The G domain and the hypervariable region. The G domain binds guanine nucleotides and comprises three dynamic structural motifs: switch I (residues 30–38), switch II (residues 60–76), and the phosphate-binding (P)-loop (residues 10–17). The switch I and II motifs constitute the nucleotide-binding site and interfaces for downstream effectors4., 5.. The P-loop regulates vital interactions with the phosphate groups of nucleotides6., 7., 8.. Normally, RAS activity is regulated by a switch between the GTP-bound state (ON) and the GDP-bound state (OFF) following GTP hydrolysis by GTPase-activating proteins (GAPs)9 or acceleration of GDP release and GTP binding by guanine nucleotide exchange factors (GEFs, such as SOS)10. Consequently, small-molecule inhibitors selectively maintain the OFF conformation to inhibit RAS signalling by disrupting the binding of either effectors11.
RAS mutations have been reported in over 30% of solid tumours and in approximately 20% of common myeloid malignancies, particularly in pancreatic, lung, and colon cancers12. These mutations are often tumorigenic and inhibit the GTPase activity of RAS, resulting in the accumulation of activated RAS. In particular, RAS mutations have been shown to inhibit the interaction of RAS with GAPs, thereby decelerating the rate of hydrolysis and accelerating the activation of GTP-bound effectors such as PI3K13, RAF14, and RalGDS15, thereby affecting the cytoskeletal organization, transcriptional regulation, or membrane trafficking pathways in the cell16.
RAS inhibitors are difficult to establish due to the lack of well-defined surface pockets that facilitate drug binding17., 18.. Recent studies have identified certain small molecules that directly inhibit and disrupt crucial functions of RAS. For example, andrographolide and its derivative (SRJ23) inhibit GDP–GTP exchange and abrogate the interactions of oncogenic RAS with its effectors19. Some mutant-specific inhibitors form covalent bond with the thiol of the G12C cysteine residue in the P-loop, which constitutively activate RAS through locked GTP binding20. The study by Gentile et al.21 utilized disulphide tethering of a non-natural cysteine (K-RAS (M72C)) to identify a new switch-II pocket (S-IIP) binding ligand (2C07) that engages the active GTP state. FarnesyltransfeRASe inhibitors (FTIs) target the CAAX motif in RAS and prevent membrane binding of RAS22., 23.. Despite previous efforts, anti-RAS therapies have not been implemented in the clinical setting.
Glycyrrhetinic acid (GA), a triterpenoic acid derived from the roots of the Glycyrrhiza plant, has widespread clinical applications due to its anti-allergic24, hepatoprotective, anti-inflammatory, antimicrobial, and antitumour effects25., 26.. From in our previous study, GA exerts a synergistic anti-asthmatic effects via a β2-adrenergic receptor-mediated pathway27. In the present study, chemical biology strategies were primarily used to identify potential target proteins of GA. The RAS protein was evaluated as a potential target of GA using fluorescence-based thermal drift (FTS) and surface plasmon resonance (SPR) analyses. In addition, negative-staining electron microscopy and SPR analyses of the interaction between GA and the wild-type or mutant H-RAS protein revealed that GA binds to the P-loop of RAS, triggers a conformational change, impairs RAS interaction with its effector proteins, and disrupts oncogenic RAS function.
2. Materials and methods
2.1. Reagents, cell culture, SDS-PAGE, and Western blotting
A549 (human adenocarcinoma cells) and HepG2 (hepatocellular carcinoma cells) cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). Cell lines were tested for murine pathogens and mycoplasma and found to be negative. A549 and HepG2 cell lines were maintained in Dulbecco׳s modified Eagle׳s medium (DMEM) supplemented with 10% (v/v) foetal bovine serum and 100 unit/mL penicillin and incubated at 37 °C in a 5% CO2 atmosphere. Western blot and SDS-PAGE analyses are consistent with the literature28 using the antibodies anti-RAS (#3965), anti-P38 (#9212), anti-phospho-P38 (#9211), anti-ERK1/2 (#9102), anti-phospho-ERK1/2 (#4370), anti-GAPDH (#2118), and a goat anti-rabbit IgG (#7074) secondary antibody, which were purchased from Cell Signaling Technology (Beverly, MA, USA). C-RAF (#3965), phospho-c-RAF (S338, #9427, S259, ab173539), phospho-B-RAF (S729, ab124794, T401, ab68215), and Alexa Fluor 594-conjugated goat anti-rabbit IgG (ab150084) were purchased from Abcam (Cambridge, UK). The HRAS-c-RAF inhibitor Kobe0065 (Kobe) and farnesyltransferase (FTase) inhibitor tipifarnib (Tip) were purchased from Selleck (Houston, Texas, USA). Kobe is a RAS family small GTPase inhibitor that inhibits the interaction of RAS/GTP with multiple effectors, including RAF, PI3K, and RalGDS and SOS29. Tip can inhibit the prenylation of the CAAX tail motif of FTase, which allows RAS to bind to the membrane30. Glycyrrhetinic acid (purity>98.5%, as determined by HPLC) and propargylamine were purchased from J&K Chemical (Beijing, China).
2.2. Enrichment of target proteins in cells
A549 cells were cultured with 10 µmol/L alkynyl-GA probe for 6 h. After three washes with precooled PBS, 500 µL of lysis buffer (Solarbio, Beijing, China) was added and incubated on ice. Cell lysates were then treated with GA-modified functionalised magnetic microspheres (probe 1) and incubated with a catalyst (2.0 mmol/L sodium ascorbic acid and 1.0 mmol/L CuSO4 in precooled PBS) overnight at 4 °C. Afterwards, probe 1 was separated with magnets and washed by PBS. Enriched probe 1 was then treated with d,l-dithiothreitol (DTT) (100 mmol/L) to release the captured protein targets. Then SDS-PAGE and Western blotting were performed in accordance with a previously reported method28.
2.3. Co-localization of target proteins and GA
A549 cells were cultured in flasks with 1 µmol/L alkynyl-GA probe. After washing, fixation with 4% paraformaldehyde, and blocking with 10% goat serum, anti-RAS antibodies (1:1000) were added and incubated overnight at 4 °C. Afterwards, Alexa Fluor 594-conjugated secondary antibodies (1:1000) were added to the cells and incubated for 1 h at room temperature. The N3-tag substrate (10 µmol/L) was then added to the cells for the probe 2 click reaction with the aforementioned catalyst system. After an adequate number of washes, fluorescence images were obtained with a confocal microscope (Leica TCS SP8, Japan): Alexa Fluor594: λEx: 594 and λEe: 617 nm; probe 2: λEx: 488 and λEe: 520 nm. DAPI (1:1000) was used to stain the nucleus and images were captured at λEx: 405 and λEe: 430 nm.
2.4. Evaluation of RAS activation
A New East Assay Kit (NewEast Biosciences, Wuhan, China) was used for RAS detection according to the manufacturer׳s instructions. We appended a brief explain in the Supporting Information.
2.5. Fluorescence-based thermal shift (FTS) assay
The recombinant H-RAS protein, GA and Tip were diluted in their corresponding incubation buffers. The H-RAS protein was complexed with each compound at a 1:10 ratio (protein concentration, 10 μmol/L) and the Protein Thermal Shift Dye KitTM (1:6000) in a total volume of 20 μL in a 96-well plate. Scanning (37–95 at 0.03 °C/s) was conducted by a real-time PCR machine (LightCycler®96, Roche, Switzerland). Data collection and organization are consistent with the literature31.
2.6. Surface plasmon resonance (SPR) analysis of H-RAS
SPR experiments of the interaction of H-RAS with GA were performed by a Biacore T200 optical biosensor (GE Healthcare, Pittsburgh, PA, USA). Immobilization of recombinant wild-type or mutant H-RAS protein (50 μg/mL) was carried out in sodium acetate buffer (pH 5.5). During each binding cycle, the GA solution (1.9 to 250 μmol/L) was injected at a flow rate of 30 μL/min for 1 min and the dissociation was monitored for 300 s. Data collection and organization are consistent with the literature31.
2.7. Negative stain electron microscopy
Briefly, the uranyl formate staining process is consistent with the literature32. Images were obtained from a Gatan 1400 TEM operated at 120 kV (Talos F200C, Waltham, MA, USA), using a 4 × 4 k CCD camera with a nominal magnification of 30 k, corresponding to 3.71 Å/pixel. Defocus value ranged between 1 and 2 μm.
2.8. Circular dichroism (CD) spectroscopy
The purified H-RAS protein was dialyzed against sodium phosphate buffer (pH 7.4). CD spectral analysis was performed at 20 °C using a MOS-450 spectropolarimeter (Bio-Logic, France) equipped with a Peltier unit for temperature control.
2.9. Migration and invasion assays
A549 cells were treated with various concentrations of GA (20 or 40 μmol/L) or Tip (10 µmol/L) and cultured for 24 h in 24-well plates in the upper wells of transwell migration inserts and transwell BD BioCoatTM MatrigelTM invasion chambers (pore size: 8 μm, BD Biosciences, Franklin Lake, New Jersey, USA). Cells that had migrated and invaded the lower surface were stained with crystal violet and then enumerated manually.
2.10. Cell cycle analysis
The A549 cells were incubated with certain concentrations of GA, kobe and Tip for 24 h. Then, the cells were harvested and fixed in ice-cold 70% (v/v) ethanol overnight at −20 °C. The cell pellet was resuspended in PBS and stained with a mixture of RNase (10 μg/mL) and PI (25 mg/mL) in sodium citrate containing 0.5% Triton X-100 for 30 min in the dark. Fluorescence was measured using a flow cytometer (BD LSRFortessa, Franklin Lake, New Jersey, USA).
2.11. Xenografts in nude mice
Six-week-old female BALB/c nude mice (specific pathogen-free [SPF]) were purchased from the Laboratory Animal Experimental Center of Academy of Military Medical Sciences (No. 1601563; Beijing, China). Studies using experimental animals were performed according to protocols approved by Nankai University Ethics Committee on Pre-Clinical Studies. A549 cells (1 × 107) were subcutaneously (s.c.) injected into the right flanks of the mice. The mice were randomly assigned to 4 groups (n = 6 mice per group): two groups were intragastrically (i.g.) administered GA (50 or 100 mg/kg) daily for 40 days, and one group was i.g. administered Kobe (80 mg/kg) daily. Tumour volumes (V) were calculated using previously described methods33.
2.12. H&E and IHC staining
Tumour tissue sections were deparaffinised, rehydrated, and boiled in sodium citrate for 15 min for antigen retrieval. After quenching endogenous peroxidases, sections were incubated with an anti-P-ERK1/2 antibody (1:500) overnight at 4 °C. Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:1000) was applied to the sections and incubated at 37 °C for 45 min. Fluorescence images were generated with a confocal microscope (Leica TCS SP8).
2.13. Statistical analysis
The results were reported as means ± S.D. (standard deviations). For single comparisons, significant differences between the means were determined using Student׳s t-test. A P-value less than 0.05 was regarded as indicate significant differences. All data were processed using the GraphPad Prism statistical software, version 5.01 (La Jolla, CA, USA).
3. Results
3.1. In silico docking and target fishing of GA probe
To screen for potential targets of GA, a virtual docking process was adopted using PhamMapper (http://59.78.96.61/phammapper). The first 40 candidate targets of GA are listed in Supporting Information Table S1. Next, we studied the candidate targets by putting them into String 10.0 (http://www.string-db.org/) and found that RAS was involved in the MAPK signalling pathway and constitutes the RAS/RAF/MEK/ERK axis (Supporting Information Fig. S1). Then, the candidate targets were selected and assessed used AutoDock and Molecular Operating Environment software (Supporting Information Fig. S2). The binding energies of GA targeting MAP2K1, PKCα, MAPK14, MAPK10, H-RAS and MAPK1 were found to be –9.93, –8.77, –8.82, –7.93, –7.47 and –7.36 kcal/mol, respectively.
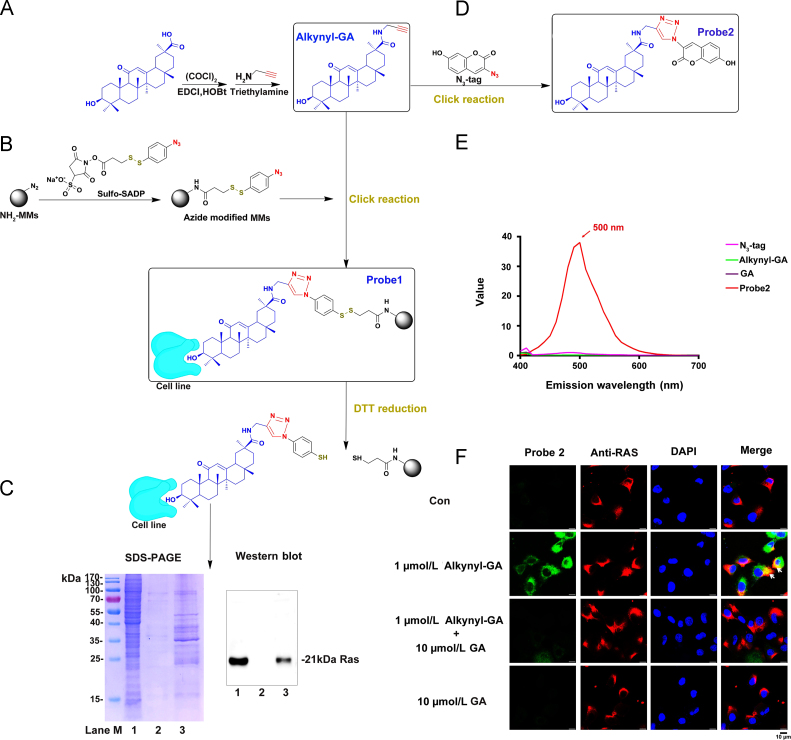
To validate the results of this in silico analysis, alkynyl-GA was synthesized according to the scheme shown in Fig. 1A. To identify the potential targets, the GA-modified functionalized MMs (probe 1) were used to capture protein targets in A549 cells. The captured proteins were released by DTT reduction (Fig. 1B). To assess collection efficiency, SDS-PAGE and Western blotting were then performed. Levels of the RAS protein, which was presented as an approximately 21 kDa band, were significantly increased in the probe 1-enriched microspheres compared with the non-alkynyl-GA-modified microspheres in the control group (Fig. 1C, right and left panel). Meanwhile, we performed capture capacity assays for other targets. The result is shown that MAPK1, MAPK14 and PKCα were also captured in the same system as the RAS protein. However, JNK and MEK1 did not yield clear positive results (Supporting Information Fig. S3). Since MAPK1, MAPK14 and PKCα are located downstream of the RAS signalling pathway. Therefore, the goal of GA is still focused on the RAS protein. Next, tracking probe 2 was synthesized (Fig. 1D). As expected, the click product probe 2 showed strong fluorescence, while the three substrates (alkynyl-GA, GA and N3-tag) showed almost no fluorescence (Fig. 1E). Under identical conditions, analysis of co-localization of alkynyl-GA probe and the RAS protein expression by confocal microscopy on A549 cells. The GA (10 µmol/L) treatment group showed little fluorescence. But obviously alkynyl-GA (1 µmol/L) fluorescence was observed in the cytoplasm. In addition, the specific fluorescence was competitively ablated by a 10 µmol/L GA treatment (Fig. 1F). This phenomenon indicated the specificity of the cytochemical staining for GA. The routes used by synthesize alkynyl-GA, probe 1 and probe 2 are shown in the supplementary data (Supporting Information Figs. S4–S8).
Figure 1.
GA targets RAS and co-localizes with RAS in A549 cells. (A) Synthesis of alkynyl-modified GA (alkynyl-GA). (B) Synthesis of GA-modified functionalized MMs (probe 1) and the process for capturing and releasing the target protein. (C) Lane 1 shows A549 lysate as a loading control, Lane 2 shows the lysate captured by the azide-modified MMs as a negative control, and Lane 3 shows the lysate captured by probe 1. Markers indicate the molecular weight. The concentrations of all protein samples were adjusted to equal amounts before capture and were adjusted to the same volume after capture for SDS-PAGE and Western blot analyses. (D) Synthesis of fluorescent click product (probe 2). (E) Fluorescence intensity of the click product (probe 2) compared with alkynyl-GA, GA and N3-tag. (F) Analysis of the co-localization of alkynyl-GA and the RAS protein using fluorescence confocal microscopy. The GA (10 µmol/L) treatment group showed little fluorescence. But obvious alkynyl-GA (1 µmol/L) fluorescence was observed in the cytoplasm (green). The specific fluorescence was competitively ablated by a 10 µmol/L GA treatment. Alexa Fluor594 staining for RAS (red) was found in the cytoplasm and membrane and partially co-localized with alkynyl-GA (yellow), as indicated by the arrows, scale bar 10 µm.
3.2. GA binds to RAS and changes its conformational state
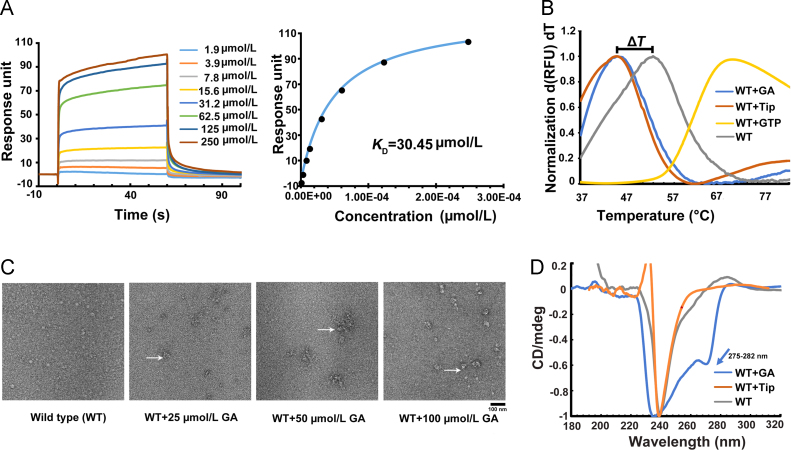
To further evaluate the feasibility of GA combining with RAS as a ligand, adopting the conventional SPR tests to verify the binding of GA to RAS targets. Depending on SPR results, GA bound to the immobilized H-RAS protein and the binding affinities of GA was 30.45 μmol/L (Fig. 2A). Then, the FTS assay was conducted to assess H-RAS thermal stability upon GA binding. The principle of FTS assay measurement is that ligand binding changes the thermal stability of proteins34. Purified H-RAS protein was subjected to FTS in the presence and absence of GA (Fig. 2B). Strikingly, GA caused a 6–8 °C decrease in the protein melting temperature (ΔT), which was strongly suggesting that considerable conformational changes occurred in H-RAS upon GA binding and similar to the FTIs tipifarnib (Tip) control group. In contrast, treatment with the positive control GTP substantially increased H-RAS thermal stability and caused 15 °C increase in ΔT. Furthermore, the leftward shift always indicates substantial instability31. In addition, a negative stain electron microscopy experiments were performed to verify the binding of GA to H-RAS target. Electron microscopic observation of wild-type H-RAS protein was homogenously distributed. However, the H-RAS protein formed many irregular aggregates after incubation with GA, and these irregular aggregates existed in a dose-dependent manner (Fig. 2C). To confirm the sensitive conformation alterations induced by hydrophobic amino acid residues, circular dichroism (CD) spectroscopy was performed (Fig. 2D). After charging with GA, the CD spectra exhibited a slight change at approximately 275–282 nm. The region represented the specific absorbance of tyrosine residues35. The result indicated that GA displayed the greatest impact on the stability of H-RAS. The H-RAS protein expression and purification process are shown in the supplemental data (Supporting Information Fig. S9).
Figure 2.
GA binds to RAS and changes its conformational state. (A) SPR analysis of interactions between GA and H-RAS. A Biacore CM5 chip was used to capture H-RAS. Measures of GA/RAS association and dissociation were performed in the presence of various GA concentrations ranging from 1.9 to 250 μmol/L. After passage over the surface of the CM5 chip, an apparent KD of 30.45 μmol/L was obtained. (B) The thermal stability of the H-RAS protein incubated with GA was assessed using the thermal shift assay. (C) Negative stain electron microscopy analysis of changes in wild-type H-RAS morphology before and after treatment with 25, 50 and 100 µmol/L GA. Arrows indicate irregular oligomers of the RAS protein, scale bar: 100nm. (D) The conformational state of the H-RAS protein (40 µmol/L) was analysed by CD after treatment with 400 µmol/L GA or 400 µmol/L Tip.
3.3. GA targets the P-loop region of H-RAS
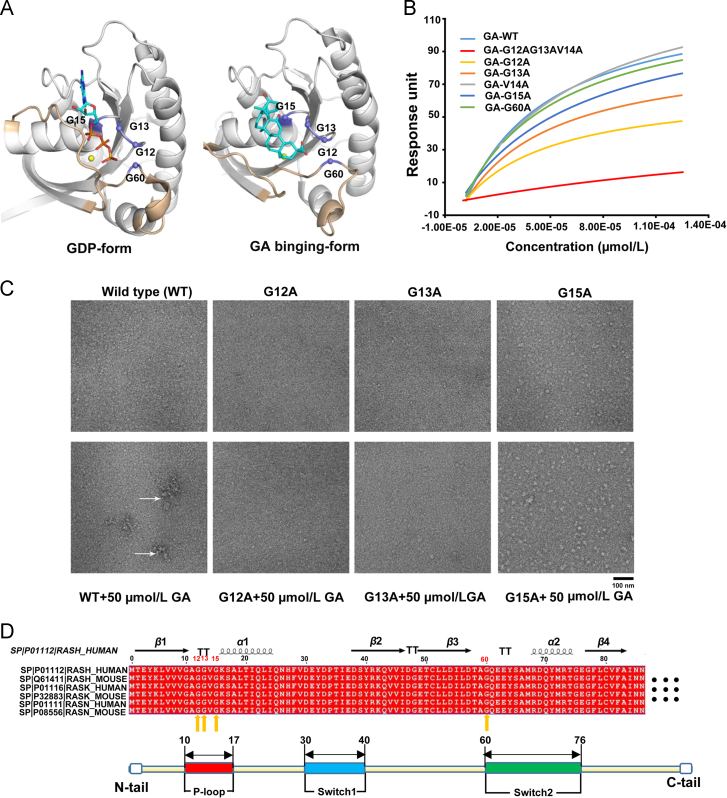
For predicting the position of GA might bind in the RAS-GTP binding pocket, multiple molecular dynamics simulations of RAS (PDB ID codes 4EFL, 3×1W, and 2CL7) complexed with GA were conducted (Supporting Information Fig. S10). The predicted ligand binding site involves two distinct Pockets: Pocket 1, which includes the effector P-loop (residues 12–15), and Pocket 2, which includes switch II (residue 60). The P-loop is very important for binding nucleotides mainly binds to the β-phosphate of GDP and the β,γ-phosphates of GTP36., 37.. The predicted ligand binding site involves residues G12, G13 and G15 that possibly make hydrogen bonds with GA (Fig. 3A, right panel), which is identical to the GDP pocket (G13, G15, G16 and G17) (Fig. 3A, left panel). To verify the reliability of predicted amino acid sites, we prepared H-RAS mutants, which harboured alanine substitution at G12, G13, V14, G15 and G60, to examine ligand binding capacity using SPR approach38. Usually, alanine replaced proteins are used to identify residues important for protein stability, function and shape39. Compared to the wild-type protein, a mutation at G12, G13 and G15 resulted in nearly 60%, 50% and 40% loss of binding of GA, respectively. And multipoint mutations at G12/G13/V14 resulted in nearly lose binding of GA (Fig. 3B and Supporting Information Fig. S11). Furthermore, the negative stain electron microscopy of wild-type and mutant H-RAS with GA was carried respectively. As showed in Fig. 3C, the binding of GA strongly induced irregular aggregates of the wild-type H-RAS, and the irregular aggregate state was observed reduction for three mutants G12A, G13A and G15A. Taken together, these results suggested that GA interacts with the nucleotides binding region P-loop of H-RAS through specific residues and triggers H-RAS irregular aggregates. P-loop sequence is highly conserved across not only the H-RAS but also the whole RAS family (H-RAS, K-RAS and N-RAS) (Fig. 3D). These key basic residues (G12, G13, and G15) are reported to play essential roles in GTP-hydrolysis40. It is speculated that GA targets the P-loop and might affect all RAS family proteins normal functions.
Figure 3.
GA targets the P-loop region of RAS. (A) Molecular docking analysis of GDP and GA. A detailed docking model for H-RAS showed that carbonyl and hydroxide radicals in the GA structure form hydrogen bonds with Gly12, Gly13, Gly15, and Gly60. GDP was also docked into the P-loop pocket. (B) SPR response of GA incubated with different H-RAS mutant proteins. (C) Negative stain electron microscopy analysis of changes in wild-type and mutant H-RAS before and after treatment with 50 µmol/L GA. Arrows indicate the irregular oligomers of the RAS protein, scale bar: 100 nm. (D) Comparison of the main sequence of human and mouse members of the RAS family.
3.4. GA inhibits the action of RAS and affects the function of downstream effectors
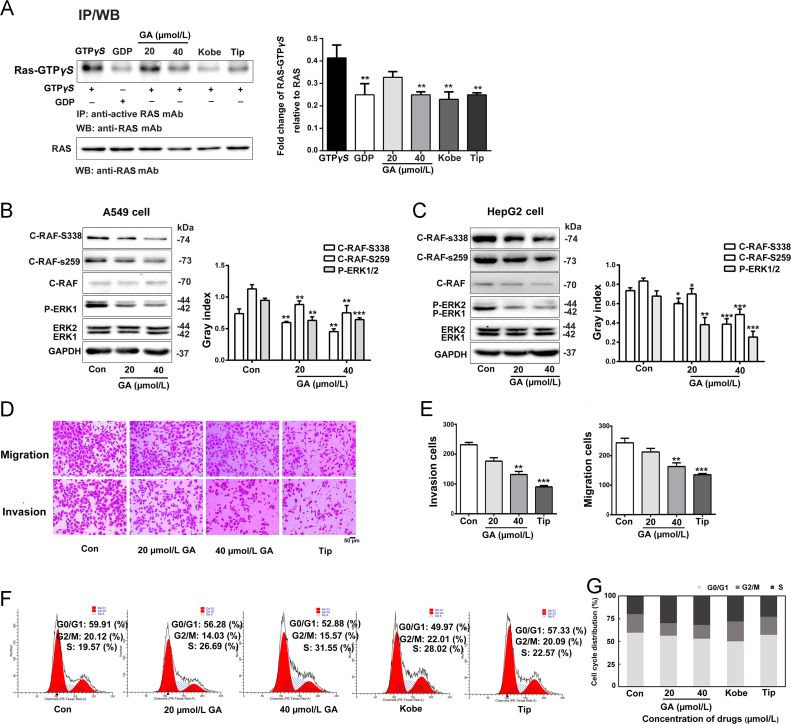
Normally, GTP binds to activate RAS and is inactivated when GTP is hydrolyzed to GDP. To further confirm the inhibitory effect of GA on RAS protein, a RAS activation assay kit was used to measure active RAS-GTP levels based on the use of a configuration-specific anti-RAS-GTPγS monoclonal antibody, then the bound active RAS was pulled down by protein A/G agarose. The precipitated active RAS was detected by immunoblot analysis using anti-RAS antibody. As showed in Fig. 4A, 40 μmol/L GA significantly inhibited intermolecular interactions of activation on A549 cells. RAS regulates proliferation by activating the mitogen-activated protein (MAP) kinase (ERK) cascade41., 42.. The first step in RAS-dependent activation of ERK signalling is RAS binding to members of the RAF family43., 44.. RAF kinase is a key RAS effector protein and is phosphorylated by RAS at residues S338 and S25945. Therefore, phosphorylated c-RAF and ERK1/2 were tested in A549 cells and HepG2 cells. c-RAF and ERK1/2 expression exhibited no evident changes, but the level of phosphorylation c-RAF at both Ser338 and Ser259 sites and the level of phosphorylation ERK1/2 were decreased following GA treatment in a dose-dependent manner (Fig. 4B and C).
Figure 4.
GA inhibits GTP binding and the activation of downstream effectors. (A) GA dose-dependently inhibits GTP binding and the activation of downstream effectors in vitro. (B) and (C) Western blots showing the levels of activated downstream effectors, c-RAF phosphorylated at S338 and S259 and phosphorylated ERK1/2 levels, in A549 cells and HepG2 cells after GA treatment. GAPDH expression was used as an internal control for normalization. The assay was performed three times, Error bars indicate means ± S.D. (*P < 0.05, **P < 0.01, ***P < 0.001 compared to the control). (D) and (E) A549 cells were cultured with 20 or 40 µmol/L GA. Migration (upper panel) and invasion (lower panel) were investigated using Transwell and Matrigel assays (n = 5), scale bar: 50 µm. (F) and (G) PI staining was performed to investigate the effect of GA on the cell cycle. Cells were treated with GA (20 or 40 µmol/L) for 24 h and then stained with PI. GA induced S phase arrest in A549 cells. Histogram shows the percentage of cells in G0/G1, G2/M and S Phase (n = 3).
3.5. GA decreases cell migration, invasion, and inhibits cell cycle progression
To assess the function of GA on the chemotactic motility of cancer cells in vitro, the effects of GA on cell migration and invasion were examined. Compared with the untreated control, the population of migrating cells decreased by 23.74% and 43.29% and the population of invasive cells decreased by 12.76% and 33.33% following treatment with 20 and 40 µmol/L GA, respectively. Consistent results were observed when A549 cells were treated with the RAS inhibitor Tip (Fig. 4D and E). Previous results show that RAS is active between G0 and S phases of the cell cycle46., 47.. Previous results show that RAS activity is required for both cell cycle initiation and subsequent G1 progression through the activation of various RAS effector pathways48., 49.. In order to explore whether the cell cycle arrest contributed to GA-induced proliferation inhibition, we further analysed the cell cycle distribution, and we found that GA induced S phase arrest in A549 cells (Fig. 4F and G).
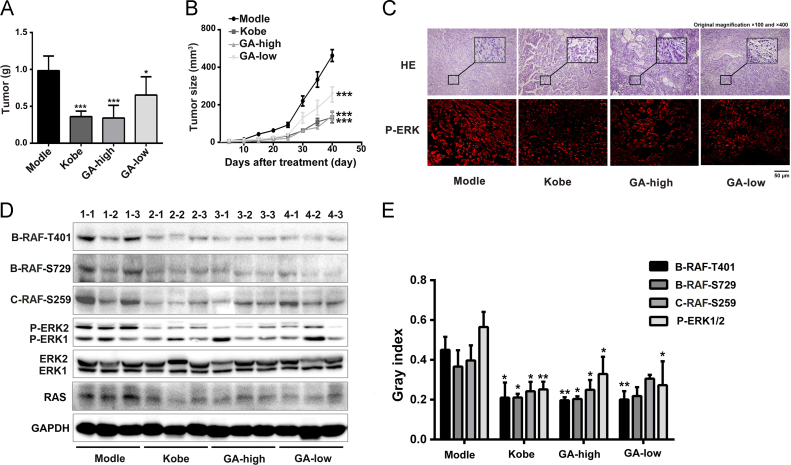
3.6. GA inhibits tumour growth in the mouse xenograft model
The antitumour activity of GA was assessed using a xenograft of A549 cells in nude mice. The tumour size of the model group displayed a significant increase compared with the GA-treated mice (Fig. 5A and B). A high dose of GA inhibited tumour growth by 50%–60%, similar to animals treated with Kobe29. For the histological analysis, tumour tissue sections were stained with haematoxylin & eosin (H&E). Compared with the model group, the high-dose GA and Kobe groups exhibited an obviously ameliorated severity of necrosis and pyknosis in the nuclei of tumour cells. Immunohistochemistry (IHC) of the tumour tissue revealed a substantial decrease in ERK1/2 phosphorylation of ERK1/2 in response to the GA treatment (Fig. 5C). Furthermore, we tested the expression of phospho-c-RAF, phospho-B-RAF and phospho-ERK1/2 in the tumour tissues from both the GA- and Kobe-treated groups. The level of phosphorylation of c-RAF at Ser259, B-RAF at both Ser729 and Thr401, ERK1/2 was decreased following GA treatment high-dose group. (Fig. 5D and E).
Figure 5.
Treatment with GA inhibited the growth of A549 cells in vivo. (A) and (B) The GA treatment suppressed the formation of tumours by A549 cells in vivo. Mice were treated with 0.9% NaCl (model), GA (high dose, 100 mg/kg; low dose, 50 mg/kg) or Kobe (80 mg/kg). (A) Tumour weights were monitored. (B) Growth curves of the xenograft tumours. Values are presented as means ± S.D. (n = 6). (C) Images of H&E (original magnification × 100 and × 400), and IHC staining of the tumour tissues show the levels of P-ERK1/2, scale bar: 50 µm. (D) and (E) Levels of the phosphorylated c-RAF-S259, B-RAF-S729, B-RAF-T401 and ERK1/2 proteins in the tumour tissues were analysed by Western blotting. The data are reported as means ± S.D. (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 compared to the model.
4. Discussion
Here, our study demonstrates that GA acts as a ligand targeting the P-loop of the RAS protein. They are identified by chemical biology strategies and biophysical techniques. P-loop is a conserved sequence motif (GxxxxGKS/T) found in the superfamily of P-loop nucleoside triphosphate hydrolases (NTPases)50. In generally, all GTPases are involved in signal transduction and regulation belongs to P-loop NTPases51. The P-loop maintains the GTP in an appropriate configuration for nucleophilic attack by a water molecule interacting with the switch I and II regions52. GDP–GTP exchange is mainly regulated by GEFs, which assist in displacing the bound GDP and replacing it with GTP, and by GAPs, which are responsible for catalysing GTP hydrolysis9. Conformational changes in the P-loop may be a common strategy contributing to GEF-stimulated nucleotide exchange in G-proteins. Thus, Structural disruption of the P-loop is likely the essential mechanism underlying the sharply reduced catalytic function of GEFs for GDP release and GDP–GTP exchange53, which leads to changes in affinity and kinetics of interaction with effectors.
According to us results of in silico docking and biophysical techniques results, GA is predicted to bind P-loop and switch II of RAS which are composed of certain basic residues (G12, G13, V14, G15 and G60). Replacement of G12, G13 and G15 with alanine leaded to obvious loss of binding ability of GA, and these residues involved in GA-induced irregular oligomers. Studies have demonstrated that the switch I and II domains bind to γ-phosphate via the backbone NH groups of the invariant T35 and G60 residues, which may be referred to as the loading spring mechanism8. Release of the γ-phosphate after GTP hydrolysis allows the switch regions to relax into a different conformation. We hypothesized that GA binds to the P-loop region of the RAS protein, and release of γ-phosphate after normal GTP hydrolysis allows the two switch regions to relax into a GDP-specific conformation that is disturbed. A possible explanation for this phenomenon is that the GA exhausts the pool of GTP-RAS over time by promoting the intrinsic hydrolysis of bound GTP and simultaneously preventing the reactivation of GTP via GEF-catalysed GDP–GTP exchange. According to the results of FTS assay, the thermal stability of RAS protein was reduced by GA as it caused a 6–8 °C decrease in ΔTm. In contrast to competitive inhibitors, GA exhibits the characteristics of an allosteric inhibitor. At present, the inhibitor SCH54292 was designed to directly block RAS by competing with nucleotides and binding to RAS, which revealed a new binding site, Lys101, under switch II54. A series of small-molecule compounds were administered to inhibit intrinsic nucleotide exchange, similar to the RAS neutralizing antibody Y13–259, which binds to amino acid residues in switch II (Glu-63, Ser-65, Ala-66, Met-67, and Arg-73)55. A natural product, andrographolide, and its derivatives bind to switch II (Tyr 64 and Tyr 71) and impair its interaction with RAS, thereby preventing GEF-assisted nucleotide exchange19.
In cell signaling networks, the involvement of H-RAS, N-RAS or K-RAS in the RAS-RAF-MAPK pathway has shown to be critical for controlling proliferation, differentiation and survival56., 57.. Based on our intracellular enzyme activity and effector protein assay results, the activity of RAS decreased following the addition of 40 μmol/L GA. The level of phosphorylation c-RAF at both Ser338 and Ser259 sites decreased following GA treatment in a dose-dependent manner. We performed in vitro and in vivo studies to determine the effects of GA on tumor cell migration, invasion, and cell cycle progression using A549 cells and nude mice. GA successfully reduced tumor cell migration and invasiveness and inhibited cell cycle progression, thereby inhibiting the RAS/MAPK signalling pathway and associated downstream effectors RAF and ERK. The above results are consistent with previous reports that GA-treated tumor cells exhibit down-regulation of H-RAS protein, which affects cell cycle arrest and cell growth inhibition58.
Naturally occurring triterpenoid saponin and its derivatives, such as, ursolic acid59, including GA60 have displayed a broad spectrum of anticancer activity and low toxicity. However, the identification of its precise anti-tumour targets is still challenging61. In contrast to competitive inhibitors, GA exhibits the characteristics of an allosteric inhibitor in our study. GA exerts an inhibitory effect by changing the conformation of the RAS protein. GA may be an effective and comparatively safe compound for the design and development of RAS-related antineoplastic drugs.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Grant Nos. 81430095, 81673616, and 81473403); and International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 81761168039).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.apsb.2018.11.002.
Contributor Information
Yuanyuan Hou, Email: houyy@nankai.edu.cn.
Gang Bai, Email: gangbai@nankai.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Cox A.D., Der C.J. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnoub A.E., Weinberg R.A. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stieglitz B., Bee C., Schwarz D., Yildiz O., Moshnikova A., Khokhlatchev A. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 2008;27:1995–2005. doi: 10.1038/emboj.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmuller L., Lautwein A., Schmitz F. The Ras--RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 6.Harrison R.A., Lu J., Carrasco M., Hunter J., Manandhar A., Gondi S. Structural dynamics in Ras and related proteins upon uucleotide switching. J Mol Biol. 2016;428:4723–4735. doi: 10.1016/j.jmb.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller M.P., Goody R.S. Review: Ras GTPases and myosin: qualitative conservation and quantitative diversification in signal and energy transduction. Biopolymers. 2016;105:422–430. doi: 10.1002/bip.22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 9.McCormick F. Ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 10.Bonfini L., Karlovich C.A., Dasgupta C., Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 11.Spiegel J., Cromm P.M., Zimmermann G., Grossmann T.N., Waldmann H. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 12.Schubbert S., Shannon K., Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 13.Castellano E., Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351 Pt 2:289–305. [PMC free article] [PubMed] [Google Scholar]
- 15.Ferro E., Trabalzini L. RalGDS family members couple Ras to Ral signalling and that׳s not all. Cell Signal. 2010;22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Roberts P.J., Der C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 17.Wang W., Fang G., Rudolph J. Ras inhibition via direct Ras binding—is there a path forward? Bioorg Med Chem Lett. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]
- 18.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 19.Hocker H.J., Cho K.J., Chen C.Y., Rambahal N., Sagineedu S.R., Shaari K. Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc Natl Acad Sci U S A. 2013;110:10201–10206. doi: 10.1073/pnas.1300016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter J.C., Gurbani D., Ficarro S.B., Carrasco M.A., Lim S.M., Choi H.G. In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc Natl Acad Sci U S A. 2014;111:8895–8900. doi: 10.1073/pnas.1404639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentile D.R., Rathinaswamy M.K., Jenkins M.L., Moss S.M., Siempelkamp B.D., Renslo A.R. Ras binder induces a modified switch-II pocket in GTP and GDP states. Cell Chem Biol. 2017;24: doi: 10.1016/j.chembiol.2017.08.025. 1455-66. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox A.D., Der C.J., Philips M.R. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 24.Akasaka Y., Yoshida T., Tsukahara M., Hatta A., Inoue H. Glycyrrhetinic acid prevents cutaneous scratching behavior in mice elicited by substance P or PAR-2 agonist. Eur J Pharmacol. 2011;670:175–179. doi: 10.1016/j.ejphar.2011.08.043. [DOI] [PubMed] [Google Scholar]
- 25.Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2015;5:310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csuk R., Schwarz S., Siewert B., Kluge R., Strohl D. Conversions at C-30 of glycyrrhetinic acid and their impact on antitumor activity. Arch Pharm. 2012;345:223–230. doi: 10.1002/ardp.201100046. [DOI] [PubMed] [Google Scholar]
- 27.Shi Q., Hou Y., Hou J., Pan P., Liu Z., Jiang M. Glycyrrhetic acid synergistically enhances β2-adrenergic receptor-Gs signaling by changing the location of Galphas in lipid rafts. PLoS One. 2012;7:e44921. doi: 10.1371/journal.pone.0044921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang R., Cui Q., Sun J., Duan X., Ma X., Wang W. PDK1/Akt/PDE4D axis identified as a target for asthma remedy synergistic with β2 AR agonists by a natural agent arctigenin. Allergy. 2015;70:1622–1632. doi: 10.1111/all.12763. [DOI] [PubMed] [Google Scholar]
- 29.Shima F., Yoshikawa Y., Ye M., Araki M., Matsumoto S., Liao J. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci U S A. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparano J.A., Moulder S., Kazi A., Coppola D., Negassa A., Vahdat L. Phase II trial of tipifarnib plus neoadjuvant doxorubicin–cyclophosphamide in patients with clinical stage IIB-IIIC breast cancer. Clin Cancer Res. 2009;15:2942–2948. doi: 10.1158/1078-0432.CCR-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu X., Wang Z., Li L., Dong S., Li Z., Jiang Z. Novel chemical ligands to Ebola virus and Marburg virus nucleoproteins identified by combining affinity mass spectrometry and metabolomics approaches. Sci Rep. 2016;6:29680. doi: 10.1038/srep29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P.H., Unger V., He X. Structure of full-length human PDGFRβ bound to its activating ligand PDGF-B as determined by negative-stain electron microscopy. J Mol Biol. 2015;427:3921–3934. doi: 10.1016/j.jmb.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei Y., Yang Q., Zhang Y., Zhao T., Liu X., Zhong J. Plumbagin restrains hepatocellular carcinoma angiogenesis by suppressing the migration and invasion of tumor-derived vascular endothelial cells. Oncotarget. 2017;8:15230–15241. doi: 10.18632/oncotarget.14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo M.C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Kelly S.M., Price N.C. The application of circular dichroism to studies of protein folding and unfolding. Biochim Biophys Acta. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 36.Wittinghofer A., Vetter I.R. Structure–function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita K., Sadanami K., Kidera A., Go N. Structural motif of phosphate-binding site common to various protein superfamilies: all-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 1999;12:11–14. doi: 10.1093/protein/12.1.11. [DOI] [PubMed] [Google Scholar]
- 38.Xiao J., Xing F., Liu Y., Lv Y., Wang X., Ling M.T. Garlic-derived compound S-allylmercaptocysteine inhibits hepatocarcinogenesis through targeting LRP6/Wnt pathway. Acta Pharm Sin B. 2018;8:575–586. doi: 10.1016/j.apsb.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatellier J., Mazza A., Brousseau R., Vernet T. Codon-based combinatorial alanine scanning site-directed mutagenesis: design, implementation, and polymerase chain reaction screening. Anal Biochem. 1995;229:282–290. doi: 10.1006/abio.1995.1414. [DOI] [PubMed] [Google Scholar]
- 40.Singh H., Longo D.L., Chabner B.A. Improving prospects for targeting RAS. J Clin Oncol. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 41.Hou Y., Nie Y., Cheng B., Tao J., Ma X., Jiang M. Qingfei Xiaoyan Wan, a traditional Chinese medicine formula, ameliorates Pseudomonas aeruginosa-induced acute lung inflammation by regulation of PI3K/AKT and Ras/MAPK pathways. Acta Pharm Sin B. 2016;6:212–221. doi: 10.1016/j.apsb.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F., Yang X., Geng M., Huang M. Targeting ERK, an Achilles׳ Heel of the MAPK pathway, in cancer therapy. Acta Pharm Sin B. 2018;8:552–562. doi: 10.1016/j.apsb.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moodie S.A., Willumsen B.M., Weber M.J., Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X.F., Settleman J., Kyriakis J.M., Takeuchi-Suzuki E., Elledge S.J., Marshall M.S. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe M., Miyajima N., Igarashi M., Endo Y., Watanabe N., Sugano S. Sodium phenylacetate inhibits the Ras/MAPK signaling pathway to induce reduction of the c-Raf-1 protein in human and canine breast cancer cells. Breast Cancer Res Treat. 2009;118:281–291. doi: 10.1007/s10549-008-0215-y. [DOI] [PubMed] [Google Scholar]
- 46.Feig L.A., Cooper G.M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobrowolski S., Harter M., Stacey D.W. Cellular ras activity is required for passage through multiple points of the G0/G1 phase in BALB/c 3T3 cells. Mol Cell Biol. 1994;14:5441–5449. doi: 10.1128/mcb.14.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones S.M., Kazlauskas A. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat Cell Biol. 2001;3:165–172. doi: 10.1038/35055073. [DOI] [PubMed] [Google Scholar]
- 49.Kumar A., Marques M., Carrera A.C. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S phase entry. Mol Cell Biol. 2006;26:9116–9125. doi: 10.1128/MCB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson D., Madera M., Vogel C., Chothia C., Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 52.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercier E., Girodat D., Wieden H.J. A conserved P-loop anchor limits the structural dynamics that mediate nucleotide dissociation in EF-Tu. Sci Rep. 2015;5:7677. doi: 10.1038/srep07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganguly A.K., Pramanik B.N., Huang E.C., Liberles S., Heimark L., Liu Y.H. Detection and structural characterization of ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg Med Chem. 1997;5:817–820. doi: 10.1016/s0968-0896(97)00021-7. [DOI] [PubMed] [Google Scholar]
- 55.Hattori S., Clanton D.J., Satoh T., Nakamura S., Kaziro Y., Kawakita M. Neutralizing monoclonal antibody against ras oncogene product p21 which impairs guanine nucleotide exchange. Mol Cell Biol. 1987;7:1999–2002. doi: 10.1128/mcb.7.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Der C.J., Krontiris T.G., Cooper G.M. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982;79:3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parada L.F., Tabin C.J., Shih C., Weinberg R.A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 58.Yu T., Yamaguchi H., Noshita T., Kidachi Y., Umetsu H., Ryoyama K. Selective cytotoxicity of glycyrrhetinic acid against tumorigenic r/m HM-SFME-1 cells: potential involvement of H-Ras downregulation. Toxicol Lett. 2010;192:425–430. doi: 10.1016/j.toxlet.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 59.Mendes V.I.S., Bartholomeusz G.A., Ayres M., Gandhi V., Salvador J.A. Synthesis and cytotoxic activity of novel A-ring cleaved ursolic acid derivatives in human non-small cell lung cancer cells. Eur J Med Chem. 2016;123:317–331. doi: 10.1016/j.ejmech.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Z.H., Zhang L.L., Li T., Lu J.H., Ma D.L., Leung C.H. Glycyrrhetinic acid induces cytoprotective autophagy via the inositol-requiring enzyme 1a-c-Jun N-terminal kinase cascade in non-small cell lung cancer cells. Oncotarget. 2015;6:43911–43926. doi: 10.18632/oncotarget.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi L.W., Wang C.Z., Yuan C.S. Isolation and analysis of ginseng: advances and challenges. Nat Prod Rep. 2011;28:467–495. doi: 10.1039/c0np00057d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material