Abstract
The present study aimed to describe the emergence of carbapenem-resistant Pseudomonas aeruginosa isolated from clinical Lebanese patients. The resistance of these isolates is due to the presence of the plasmid-encoded blaVIM-2 gene. We provide its first description in Lebanon, as well as a description of disruption of the oprD gene by mutations.
Keywords: Carbapenem resistance, oprDgene, Pseudomonas aeruginosa, VIM-2
Pseudomonas aeruginosa is an important pathogen that is the main cause of acute nosocomial infections, especially in immunocompromised patients [1]. The resistance of P. aeruginosa to carbapenem is becoming a major global threat and is exacerbated by the excessive use of carbapenem [2], [3]. This resistance is mainly due to the alteration or loss of the outer membrane porin protein (oprD), to the increased expression of the efflux pumps and the production of carbapenemase, mainly Verona integron-encoded metallo-β-lactamase (VIM) and imipenem (IMP) [2], [3].
In this study, we report the emergence of carbapenem-resistant P. aeruginosa isolated from rectal swabs of 23 intensive care unit patients treated with carbapenem for more than 1 week between October 2016 and February 2017 from Saint-George Hospital in Lebanon. Carbapenem-resistant organisms were screened using agar plates with Ertapenem (2 μg/mL). Four carbapenem-resistant P. aeruginosa were isolated and identified by MALDI-TOF MS. Antimicrobial susceptibility testing was performed on Müller-Hinton agar using the disc diffusion method, and Etest was performed to determine the MIC of IMP, as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (https://www.sfm-microbiologie.org/wp-content/uploads/2019/01/CASFM2019_V1.0.pdf). The phenotypic detection of carbapenemase was confirmed using the Carba NP test [4]. The carbapenemase encoding genes were screened by real-time PCR and standard PCR, and were then sequenced. Molecular characterization of the oprD gene was performed using PCR amplification and sequencing [5]. Analysis of the sequenced oprD gene was compared against the reference strain P. aeruginosa PA01 using Multalin alignment software (http://multalin.toulouse.inra.fr/multalin/). Multilocus sequence typing (MLST) was performed to determine the genetic relationship among the clinical isolate as described on the Institute Pasteur's MLST website (https://pubmlst.org/paeruginosa/).
The results indicated that P. aeruginosa isolates were resistant to all antibiotics tested except to colistin and fosfomycin, with MICs for IMP >32 μg/mL. All isolates harboured the blaVIM-2 gene, except P. aeruginosa (PA-4) (Table 1). In addition, all isolates had mutations in the oprD gene (Fig. 1). MLST analysis revealed that three P. aeruginosa (PA-3, PA-6, PA-16), and one P. aeruginosa (PA-4) isolates harboured sequence types (ST) 357 and ST233 respectively (Table 1). Conjugal transfer between carbapenemase-producing P. aeruginosa and Escherichia coli (J35) succeeded, to yield E. coli transconjugants harbouring a ∼45 kb plasmid, except for the clone ST233, suggesting that these metallo-β-lactamase blaVIM-2 were plasmid encoded for ST357 and chromosomally encoded for ST233.
Table 1.
Phenotypic and genotypic features of carbapenem-resistant clinical isolates
| Pseudomonas aeruginosa strain | Source | Antibiotic resistance profile | IMP MIC (μg/mL) | Carba NP test | VIM-2 | ST |
|---|---|---|---|---|---|---|
| PA-3 | Rectal swab | TIC, TCC, TZP, CAZ, FEP, IMP, ERT, AK, TOB, CIP, F, DO, SXT, R | >32 | + | + | 357 |
| PA-16 | Rectal swab | TIC, TCC, TZP, CAZ, FEP, IMP, ERT, AK, TOB, CIP, F, DO, SXT, R | >32 | + | + | 357 |
| PA-6 | Rectal swab | TIC, TCC, TZP, CAZ, FEP, IMP, ERT, AK, TOB, CIP, F, DO, SXT, R | >32 | + | + | 357 |
| PA-4 | Rectal swab | TIC, TCC, TZP, CAZ, FEP, IMP, ERT, AK, TOB, CIP, F, DO, SXT,R | >32 | + | − | 233 |
AK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; DO, doxycycline; ERT, ertapenem; F, nitrofurantoin; FEP, cefepime; FF, fosfomycin; IPM, imipenem; R, rifampicin; ST, sequence, type; SXT, trimethoprim/sulfamethoxazole; TCC, ticarcillin/clavulanic, acid; TIC, ticarcillin; TOB, tobramycin; TZP, piperacillin/tazobactam.
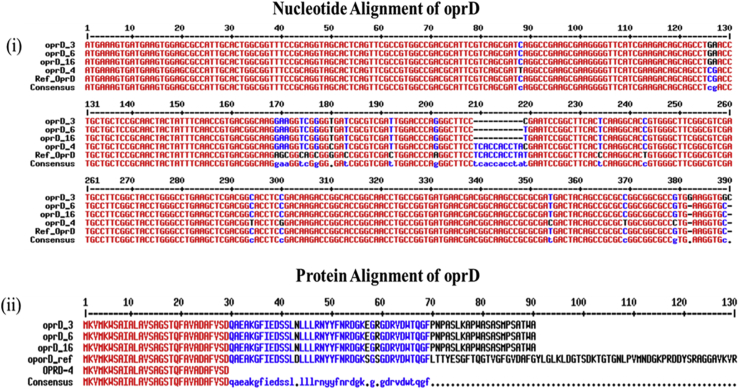
Fig. 1.
Genetic representation of oprD gene. (a) Nucleotide alignment of oprD gene and (b) protein alignment of oprD gene. For Pseudomonas aeruginosa PA-3, PA-16 and PA-6, large deletion of ten nucleotides from position 209 to 218 led to premature stop codon TGA in oprD, resulting in truncated polypeptide made of 90 aa residues. For P. aeruginosa PA-4, C to T substitution in nucleotide position 88 led to premature stop codon TAG in oprD, resulting in truncated polypeptide made of 29 aa residues.
Here we describe the emergence of carbapenem-resistant P. aeruginosa in Saint-Georges Hospital due to the presence of blaVIM-2 gene and mutations of the oprD gene. These results are in concordance with those previously reported in Lebanon, where Al Bayssari et al. [3], [6] have reported the emergence of VIM-2–producing P. aeruginosa in humans and animals. Other studies have also shown the spread of VIM-2–producing P. aeruginosa in different Lebanese hospitals [7], [8]. In addition, Christophy et al. [9] revealed the presence of the blaVIM gene in P. stutzeri collected from cancer patients in North Lebanon. However, none of those studies has revealed a plasmidic location of the blaVIM-2 gene. Our study also demonstrated that mutations leading to premature stop codon resulting in a defective protein oprD were the main cause of P. aeruginosa's resistance to carbapenem, as described above [3], [10]. The main finding in our study was the emergence of P. aeruginosa harbouring the VIM-2 plasmid, which has never been detected before in Lebanon, where all detected isolates had the chromosomic blaVIM-2 gene, or the studies did not specify the genetic location of the blaVIM gene [3]. MLST analysis showed that the three P. aeruginosa isolates harbouring the plasmid-encoded blaVIM-2 gene belonged to the ST357 clone, which has been found in different countries of central Europe [11]. However, P. aeruginosa ST233, which has the chromosomic blaVIM-2 gene, has already been described in Lebanon as well as in various countries in the Mediterranean basin [3], [12].
To conclude, our study is the first to report the detection of the plasmid-encoded blaVIM-2 gene in Lebanon. This finding poses a serious public health problem because the plasmid containing this β-lactamase is a major source of dissemination of this enzyme. An urgent strategy must be implemented to control the spread of these resistant microorganisms in hospitalized patients.
Acknowledgements
The authors thank CookieTrad for English-language editorial work. Supported in part by the Lebanese Council for Research and the French government under the ‘Investissements d'avenir’ programme managed by the Agence Nationale de la Recherche (Méditerranée Infection 10-IAHU-03).
Conflict of interest
None declared.
References
- 1.Bellés A., Bueno J., Rojo-Bezares B., Torres C., Javier Castillo F., Sáenz Y. Characterisation of VIM-2–producing Pseudomonas aeruginosa isolates from lower tract respiratory infections in a Spanish hospital. Eur J Clin Microbiol Infect Dis. 2018;37:1847–1856. doi: 10.1007/s10096-018-3318-3. [DOI] [PubMed] [Google Scholar]
- 2.Bouaziz A., Loucif L., Ayachi A., Guehaz K., Bendjama E., Rolain J.M. Migratory white stork (Ciconia ciconia): a potential vector of the OXA-48–producing Escherichia coli ST38 clone in Algeria. Microb Drug Resist. 2018;24:461–468. doi: 10.1089/mdr.2017.0174. [DOI] [PubMed] [Google Scholar]
- 3.Al Bayssari C., Diene S.M., Loucif L., Gupta S.K., Dabboussi F., Mallat H. Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob Agents Chemother. 2014;58:4966–4970. doi: 10.1128/AAC.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakour S., Olaitan A.O., Ammari H., Touati A., Saoudi S., Saoudi K. Emergence of colistin-and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report. Microb Drug Resist. 2015;21:279–285. doi: 10.1089/mdr.2014.0214. [DOI] [PubMed] [Google Scholar]
- 5.Diene S.M., L’homme T., Bellulo S., Stremler N., Dubus J.-C., Mely L. ISPa46, a novel insertion sequence in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from a cystic fibrosis patient in Marseille, France. Int J Antimicrob Agents. 2013;42:268–271. doi: 10.1016/j.ijantimicag.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Al Bayssari C., Dabboussi F., Hamze M., Rolain J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother. 2015;70:950–951. doi: 10.1093/jac/dku469. [DOI] [PubMed] [Google Scholar]
- 7.Hammoudi Halat D., Moubareck C.A., Sarkis D.K. Heterogeneity of carbapenem resistance mechanisms among Gram-negative pathogens in Lebanon: results of the first cross-sectional countrywide study. Microb Drug Resist. 2017;23:733–743. doi: 10.1089/mdr.2016.0077. [DOI] [PubMed] [Google Scholar]
- 8.Hammoudi D, Moubareck CA, Kanso A, Nordmann P, Sarkis DK. Surveillance of carbapenem non-susceptible Gram-negative strains and characterization of carbapenemases of classes A, B, and D in a Lebanese hospital. J Med Liban n.d.;63:66–73. [DOI] [PubMed]
- 9.Christophy R., Osman M., Mallat H., Achkar M., Ziedeh A., Moukaddem W. Prevalence, antibiotic susceptibility and characterization of antibiotic-resistant genes among carbapenem-resistant Gram-negative bacilli and yeast in intestinal flora of cancer patients in North Lebanon. J Infect Public Health. 2017;10:716–720. doi: 10.1016/j.jiph.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.Y., Ko K.S. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012;40:168–172. doi: 10.1016/j.ijantimicag.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Hrabak J., Cervena D., Izdebski R., Duljasz W., Gniadkowski M., Fridrichova M. Regional spread of Pseudomonas aeruginosa ST357 producing IMP-7 metallo-lactamase in Central Europe. J Clin Microbiol. 2011;49:474–475. doi: 10.1128/JCM.00684-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellouk F.Z., Bakour S., Meradji S., Al-Bayssari C., Bentakouk M.C., Zouyed F. First detection of VIM-4–producing Pseudomonas aeruginosa and OXA-48–producing Klebsiella pneumoniae in northeastern (Annaba, Skikda) Algeria. Microb Drug Resist. 2017;23:335–344. doi: 10.1089/mdr.2016.0032. [DOI] [PubMed] [Google Scholar]