Abstract
Phytomics or metabolomics is analysis of large-scale primary and secondary metabolites of plant extracts and provides very meaningful data to monitor or evaluate cellular function or systems biology. The activity of plant extracts depends on the synergistic/antagonistic effect of different metabolites rather than single active metabolites. Matrix metalloproteinases (MMPs) have an active role in the formation of many diseases. To our knowledge, there is no study on the correlation between the phytomics and MMP inhibitory activity of Achillea millefolium, Achillea filipendulina (Asteraceae), Mentha piperita, and Salvia officinalis (Lamiaceae), (AAMS). Therefore, this study aimed to correlate the metabolomics profiling of AAMS extracts to identify the metabolites responsible for the MMP inhibitory activity based on phytomics data. The AAMS extracts showed a significant MMP inhibitory effect (57.73–92.73%) at different concentrations (25–500 μg/mL). In order to identify the metabolites responsible for such activities in the extract, the metabolomic profiling of the plants was investigated using gas chromatography-mass spectrometry (GC-MS). After deconvolution and aligning of the chromatograms, 284 metabolites were detected, of which 149 were annotated using retention index libraries. Multivariate analyses results indicated that A. millefolium and A. filipendulina showed similar metabolomic profiles, while M. piperita and S. officinalis differed both from each other and from Achillea species. The correlation analysis was applied to evaluate the correlation between metabolomic levels and MMP inhibitory activities, and 96 metabolites had a negative correlation (r ≤ −0.70) and 55 had a highly positive correlation (r ≥ 0.70) with MMP inhibitory activity. This is the first study which revealed that phytomics, plant metabolomics, can be used for activity evaluation and a single metabolite may not be responsible for a specific activity. In conclusion, phytomics can be a more useful tool for the evaluation of the activities than investigating a single metabolite. This new perspective can also provide a better understanding of plant metabolomics and can be easily employed for future research on plant activity.
Keywords: Metabolomics, Phytomics, Herbal extracts, Matrix metalloproteinases, GC-MS
1. Introduction
Plant metabolomics and bioactivity correlation are relatively new approaches. Metabolomic formation is achieved after certain steps starting with DNA analysis (Verpoorte et al., 2008). The metabolomes of medicinal plants are particularly valuable for the evidence-based development of new phytotherapeutics because medicinal plants are still considered an important source of new chemicals with potential therapeutic effects (Mahdi et al., 2013, Elwy and Tabl, 2012, Elwy and Ghada, 2013). Today, the search for new pharmacologically active agents obtained by screening natural sources, such as plant extracts has led to the discovery of many clinically useful drugs for the treatment of many diseases (Kamatou et al., 2005, Kamatou et al., 2006, Kamatou et al., 2008, Elwy and Tabl, 2012). Natural organic compounds in plants are produced by primary and secondary metabolites. In the primary metabolic pathway, fewer products occur than in the secondary pathway, and primary metabolites found in all plants have essential and usually evident metabolic roles. All secondary metabolites derive from primary metabolites and are not essential for the survival of plants (Dennis, 1987, Dennis et al., 1997, Stitt, 1998). Although there are numerous publications about the potential biological activity of plant secondary metabolites, the contribution of primary metabolites to such activity has not yet been investigated.
Identification of low amounts of active phytochemical components in medicinal plants is difficult, and pharmacological activity observed in such plants may appear synergistic or antagonistic (Williamson, 2001).
MMPs are involved in the etiology of many diseases and medical conditions, such as metastasis, rheumatoid arthritis, osteoarthritis, periodontal diseases, inflammatory diseases, atherosclerosis, diabetes, Parkinson's disease pathology, and skin aging. During aging, increased MMP-1, MMP-8, and MMP-13 from the collagenase family break down collagen and elastin, leading to degenerative changes of the extracellular matrix, and accelerate wrinkle formation, and sagging and aging of the skin (Bailey, 2001, Gosline et al., 2002, Heim et al., 2006, Nema et al., 2013) MMP-1 and MMP-8 are also known to cause osteoarthritis (Fuchs et al., 2004). The physiological activity process of MMPs is controlled by endogenous tissue inhibitor metalloproteinases (TIMPs) (Öncel, 2012). Sustaining physiological events in an organism requires a constant balance between MMP activity and specific endogenous TIMPs. Stopping the activity of MMP enzymes means fighting against skin aging and osteoarthritis by preventing collagen destruction. If sufficient TIMPs cannot be secreted in response to increased MMP activity, then MMP inhibitory supplements need to be provided. Nature is a unique important source for the development of new active products.
Today, some plants, including A. millefolium, A. filipendulina (Giorgi, 2005), M. piperita (Nair, 2001), and S. officinalis, (Daniela, 1993) are included in combination products used against skin aging. Furthermore, A. millefolium is prescribed in osteoarthritis (Akram, 2013), and M. piperita has very good analgesic and anti-inflammatory properties (Dara and Belamkar, 2014).
This study, for the first time in the literature, brings a new perspective to complex biological systems by investigating the whole metabolomic pool, rather than a single active compound in terms of activities. For this purpose, we correlated the metabolomic profiles of AAMS methanolic extracts with their MMP-1, MMP-8 and MMP-13 inhibitory activities. Correlation analyses were also performed to determine the highly positively and negatively correlated metabolites.
2. Methods
2.1. Plant material
Aerial parts of Achillea millefolium (TBÇ-A-002), Achillea filipendulina (TBÇ-A-001) (Asteraceae), Mentha piperita L. (TBÇ-M-002), and Salvia officinalis L. (TBÇ-S-001) (Lamiaceae) were purchased during the flowering period from the cultural areas of Selçuk University, Faculty of Agriculture, Department of Medicinal Plants. The plant specimens were deposited in the medicinal and aromatic plants herbarium of the same university.
2.2. Chemicals and reagents
All solvents used were of analytical grade. MMP inhibitor screening assay kits (MMP-13 ab139450, MMP-8 ab139452, MMP-1 ab139443) are purchased from ABCAM. N-Methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) were purchased from Thermo Fisher. Ultrapure water (Millipore Direct-Q, Meck KGaA) was used for all reagents.
2.3. Sample preparations
2.3.1. Extraction of plant materials
All plant materials were weighed approximately 2 g, extracted with 25 mL methanol in a reflux cooler at 30 °C for 25 min, and filtered. Then, the residue was extracted with 25 mL methanol for 30 min and filtered again. The filtrates were combined, evaporated to dryness, and lyophilized.
2.3.2. Sample preparation for MMP inhibitory activity
2 mg lyophilized plant material was dissolved in 4 mL methanol (500 μg/mL). The dilutions (250 μg/mL, 100 μg/mL, 50 μg/mL, 25 μg/mL, and 10 μg/mL) were prepared from 500 μg/mL stock solution using methanol as a diluent.
2.3.3. Reference solution preparation for MMP inhibitory activity
2 mg reference compound was dissolved in 4 mL methanol (500 μg/mL). The dilutions (50 μg/mL, 25 μg/mL, 10 μg/mL, 5 μg/mL, and 2 μg/mL) were prepared from 500 μg/mL stock solution using methanol as a diluent.
2.3.4. Sample preparation for GC-MS
1 mg lyophilized plant material was dissolved in 10 mL methanol (100 μg/mL). 200 μL of this solution was evaporated to dryness in a vacuum dryer concentrator. The residuals were methoxyaminated using 20 μL of 20 mg/mL methoxyamine in pyridine and derivatized using 80 μL MSTFA with 1% TMCS. After derivatization, the samples were transferred into GC-MS vials with 200 μL silanized inserts.
2.3.5. MMP-1, MMP-8 and MMP-13 inhibitory screening assays
MMP inhibitor screening was performed according to the instructions of the kit manufacturer. The assay kit consisted of a microplate, MMP enzyme (30.6 U/µL of MMP-1, 9.2 U/µL of MMP-8, and 3.45 U/µL of MMP-13), MMP inhibitor (1.3 mM NNGH in DMSO), MMP substrate (25 mM in DMSO) and colorimetric assay buffer (obtained from ABCAM). The MMP substrate and MMP inhibitor were brought to room temperature from 80 °C. The MMP inhibitor was diluted at 1/200 in assay buffer and warmed up to the reaction temperature of 37 °C. The MMP substrate was diluted at 1/25 in assay buffer to the required total volume (10 µL per well). The MMP enzyme was diluted (1/40 for MMP-1, 1/100 for MMP-8, and 1/50 for MMP-13) in assay buffer to the required total volume (20 µL per well) and warmed up to the reaction temperature of 37 °C shortly before assay. The assay buffer was pipetted into each well. The microplate was allowed to equilibrate to the assay temperature. 20 µL of the diluted MMP enzyme was added to the control, MMP inhibitor and test inhibitor wells. 20 µL of the diluted MMP inhibitor was only added to the MMP inhibitor wells; then the desired volume of test inhibitor was added to appropriate wells. The plate was incubated for 30–60 min at the reaction temperature to allow inhibitor/enzyme interaction. The reaction was started by the addition of 10 µL of the diluted MMP substrate. The continuous absorbance of the wells was measured at A412nm using a microplate reader, and data analysis was performed.
2.4. Metabolomic analysis
This analysis was performed using GC-MS as previously described (Nemutlu et al., 2015). Briefly, metabolomic profiling was undertaken using GC-MS (Shimadzu GC-MS-QP2010 Ultra) with a DB-5MS stationary phase column (30 m + 10 m duraguard × 0.25 mm i.d. and 0.25-µm film thickness). Once the analysis was completed, the complex chromatograms were deconvoluted using AMDIS, and the retention time was corrected and data matrixes were created using SpectConnect software. The correlation analysis between each of metabolites in plant extract and MMPs activities was performed using Microsoft Excel. The pathway enrichment analysis in KEGG database was performed using MetaboAnalyst software.
3. Results
3.1. MMP-1, MMP-8 and MMP-13 inhibitory effects of A. millefolium, A. filipendulina, M. piperita and S. officinalis methanolic extracts
The AAMS extracts showed a significant MMP inhibitory effect at different concentrations. These inhibition percentages were close to the positive control. Among the AAMS extracts, M. piperita showed the highest inhibitory effect on MMPs compared to the other extracts. The inhibitory activity of the extracts on MMP-1, MMP-8 and MMP-13 are presented in Table 1, Table 2, Table 3, respectively.
Table 1.
Inhibition percentages of the methanolic extracts of AAMS on the MMP-1 enzyme.
| Medicinal plants | 25 μg/mL | 50 μg/mL | 100 μg/mL | 250 μg/mL | 500 μg/mL |
|---|---|---|---|---|---|
| A. millefolium | 67.24 | 70.32 | 73.20 | 77.62 | 81.27 |
| A. filipendulina | 57.73 | 63.98 | 66.95 | 71.47 | 78.29 |
| M. piperita | 60.04 | 68.88 | 72.24 | 75.31 | 84.44 |
| S. officinalis | 60.13 | 65.90 | 70.99 | 75.79 | 70.51 |
*The MMP-1 inhibitory activity of 1.3 μM NNGH (positive control) was 90.8%.
Table 2.
Inhibition percentages of the methanolic extracts of AAMS on the MMP-8 enzyme.
| Medicinal plants | 25 μg/mL | 50 μg/mL | 100 μg/mL | 250 μg/mL | 500 μg/mL |
|---|---|---|---|---|---|
| A. millefolium | 71.98 | 73.34 | 72.96 | 74.47 | 75.30 |
| A. filipendulina | 70.32 | 71.90 | 73.19 | 72.89 | 62.99 |
| M. piperita | 70.62 | 73.04 | 76.21 | 78.25 | 80.36 |
| S. officinalis | 70.85 | 73.19 | 77.79 | 80.51 | 80.06 |
*The MMP-8 inhibitory activity of 1.3 μM NNGH (positive control) was 93.88%.
Table 3.
Inhibition percentages of the methanolic extracts of AAMS on the MMP-13 enzyme.
| Medicinal plants | 25 μg/mL | 50 μg/mL | 100 μg/mL | 250 μg/mL | 500 μg/mL |
|---|---|---|---|---|---|
| A. millefolium | 80.99 | 76.39 | 77.90 | 83.37 | 89.20 |
| A. filipendulina | 77.83 | 76.10 | 77.32 | 82.00 | 84.31 |
| M. piperita | 82.72 | 73.94 | 90.64 | 92.73 | 91.72 |
| S. officinalis | 78.83 | 86.47 | 85.53 | 89.27 | 89.63 |
*The MMP-1 inhibitory activity of 1.3 μM NNGH (positive control) was 91.94%.
3.2. Metabolomic data analysis
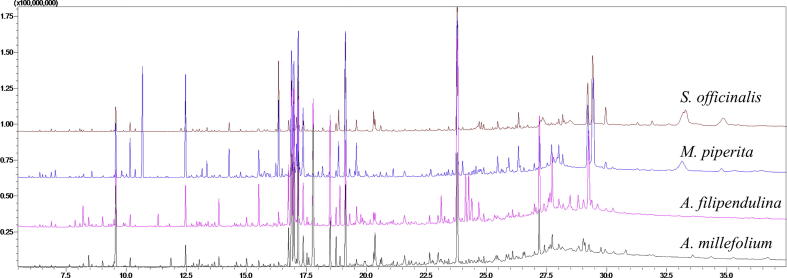
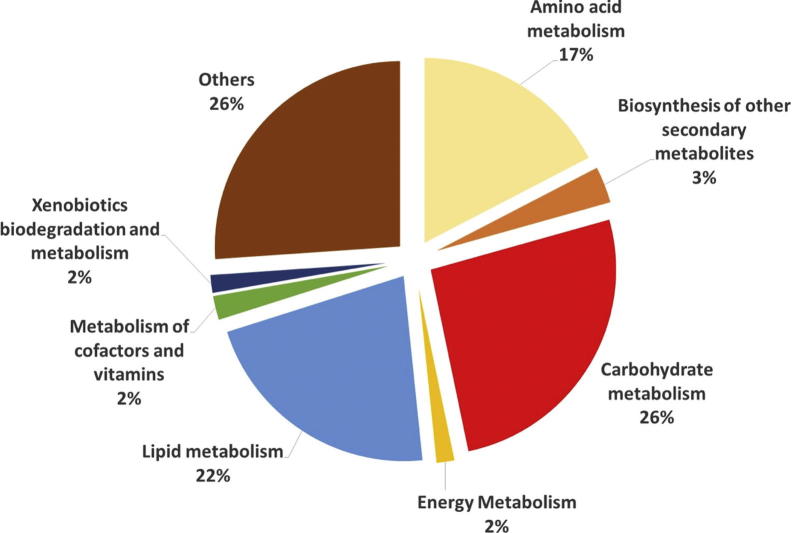
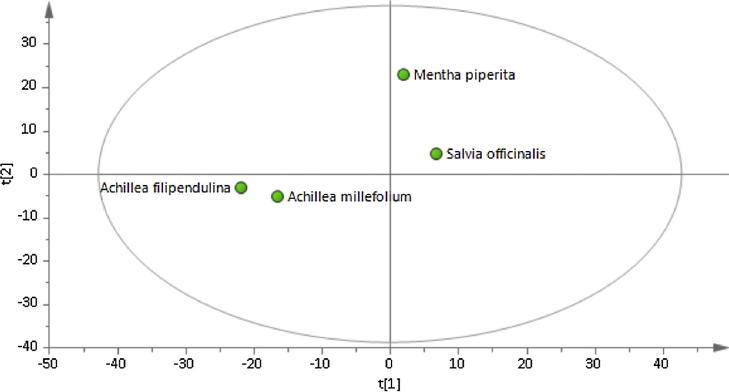
In order to identify the metabolites responsible for the MMP inhibitory activities in the extracts, the metabolomic profiling of the plants were investigated using GC-MS. Representative GC-MS chromatograms were presented in the Fig. 1. After deconvolution and alignment of the chromatograms, 284 metabolites were detected, of which 149 were annotated using retention index libraries. The identified metabolites were belonging to metabolic pathways of biosynthesis of other secondary metabolites and metabolism of amino acid, carbohydrate, energy, lipid, cofactors/vitamins and xenobiotics biodegradation (Fig. 2). The results of the multivariate analyses showed that Asteraceae family members, A. millefolium and A. filipendulina, exhibited similar metabolomic profiles, while Lamiaceae members M. piperita and S. officinalis differed both from each other and from Achillea species (Fig. 3).
Fig. 1.
Representative GC-MS chromatograms of the plants.
Fig. 2.
The metabolomic pathway distribution of the annotated metabolites.
Fig. 3.
Principal component score plot of AAMS.
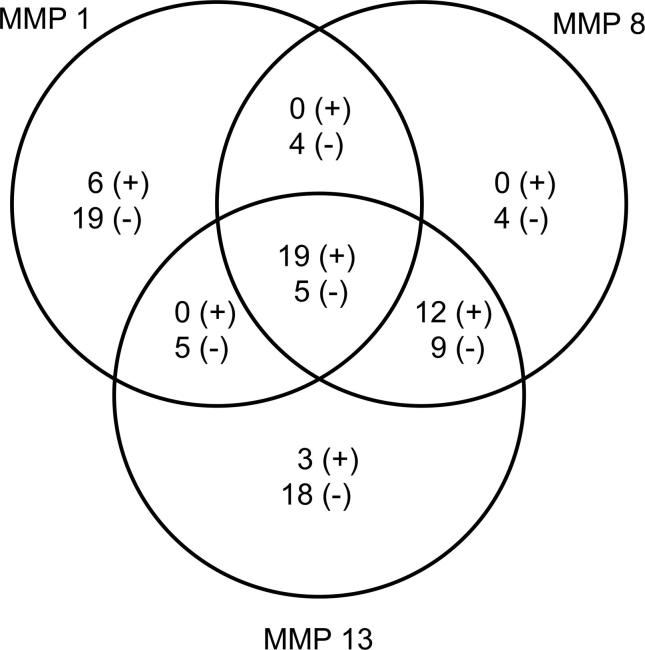
The correlation analysis was applied to evaluate the correlation between metabolomic levels and MMP inhibitory activities. According to the results, 96 metabolites had a negative correlation (r ≤ −0.70) and 55 metabolites had a very high positive correlation (r ≥ 0.70) with MMP inhibitory activity (Fig. 4).
Fig. 4.
Metabolites showing positive and/or negative interaction with MMP-1,8,13. Metabolites showing positive interactions- with MMP1: octadecanoic acid, D-pinitol, gly-pro, palmitic a, N-methylglutamic a, hexadecanol, with MMP8: melibiose, citramalic a, gulose, pentadecanoic a, xylitol, tyrosine, epsilon-caprolactam, glucaric a, methyl-β-D-galactopyranoside, trans-ferulic a, ribose, tyramine, with MMP13: tartaric a., 2,4-bishydroxybutanoic a., galactonic a., tryptophane, glyceric a, malonic a, maleic a, ribonic a, β-alanine, 3-hydroxy-3-methylglutaric a, myo-inositol, ferulic a, 2-hydroxyglutaric a, N-acetyl-D-mannosamine, raffinose, succinic a, monomethylphosphate, alanine, L-(+) lactic a, with MMP1-8: none, with MMP1-13: none, with MMP8-13: 2,3-Dihydroxybutanedioic a, 2,4,6-tri-tert.-butylbenzenethiol, cis-aconitic a, citric a, digalactosylglycerol, DL-3,4-dihydroxyphenylglycol, fumaric a, galactinol, gentiobiose, glycerol, L-alanine, L-proline, myo-inositol-1-phosphate, threonic a, ursolic a, with MMP1-8-13: 2-O-glycerol-β-D-galactopyranoside, benzyl thiocyanate, hydroxylamine Metabolites showing negative interactions- with MMP1: 1-ethylglucopyranoside, 4-isopropylbenzoic a, α-D-glc-(1,2)- β-D-Fru, α-tocopherol, cis-aconitic a, D-allose, DL-3,4-dihydroxyphenyl glycol, gluconic acid lactone, L-norleucine, L-serine, L-threonine, malic a, maltotriose, melibiose, p-cymene, pyruvic a, raffinose, sucrose, trehalose, with MMP8: capric a, D-threitol, glutaric acid, glycine, mannitol, octadecanoic a, phosphoric a, salicylic acid glucopyranoside, squalene, with MMP13: 2-ketoisocaproic acid galactosylglycerol, myristic a, oleic a, porphine, with MMP1-8: 1-monooctadecanoylglycerol, kaempferol, lactic a, triacontanoic a, with MMP1-13: D-sphingosine, monomethylphosphate, oxalic a, shikimic a, xylitol, with MMP8-13: 2-hydroxypyridine, 2-pyrrolidinone, 2,4,6-tri-tert.-butylbenzenethiol, 4-aminobutyric a., 4-hydroxy-3-methoxybenzoic a., 4-hydroxybenzoic a, allo-inositol, α-ketoglutaric a, arachidic a, benzoic a, β-sitosterol, citraconic a, D-mannitol, D-pinitol, docosanol, dodecanoic a, dotriacontanol, eicosanoic a, eicosanol, ethanolamine, glucopyranose, hesperetin, hexacosanol, hexadecanoic a, hydroquinone, L-alanine, L-proline, O-acetylsalicylic a, palatinitol, palmitic a, phytol, piceatannol, threitol, triacontanol, urea, vanillin, with MMP1-8-13: 1-benzylglucopyranoside, 1,3-bisethynylbenzene, 3,4-dihydroxybenzoic a, 4-trans-caffeoylquinic a, 5-trans-caffeoylquinic a, allose, behenic a, chlorogenic a, fructose, octacosanol, pipecolic a, proline, pyroglutamic a, quinic a, stearic a, tetracosanol, valine, octadecanol.
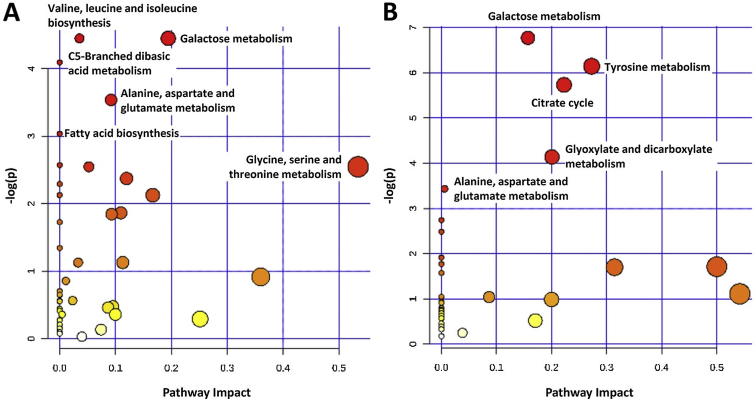
The pathway analysis was done using the library of metabolic pathways for Arabidopsis thaliana assembled from the KEGG database (Xia and Wishart, 2010). This library is the only one that sufficiently close to our samples (Chong et al., 2018). The pathway enrichment analyses were performed using positively and negatively correlated metabolites in order to identify the most relevant pathways involved in the study (Fig. 5). The significantly altered pathways (p < 0.05) were marked on the Fig. 5 The metabolomic pathways of valine, leucine and isoleucine biosynthesis; galactose metabolism; C5-branched dibasic acid metabolism; alanine, aspartate and glutamate metabolism; fatty acid biosynthesis; and glycine, serine and threonine metabolism were negative correlated with MMPs activities. While galactose metabolism; tyrosine metabolism; citrate cycle; glyoxylate and dicarboxylate metabolism; and alanine, aspartate and glutamate metabolism were positive correlated.
Fig. 5.
Pathway analysis on the negatively (A) and positively (B) correlated metabolites. The pathways significantly altered (p < 0.05) were labeled.
When the contents of the four plant extracts are examined, the compounds contained in all were found to have been formed by the shikimic acid biosynthetic pathway. Table 4 presents the correlation between MMP inhibitory activity and the shikimic acid biosynthetic pathway members, namely quinic acid, shikimic acid, phenylalanine, and their final products kaempferol and hydroquinone.
Table 4.
The correlation between MMP inhibitory activities and metabolites of the shikimic acid pathway.
| Metabolites | Correlation coefficient |
||
|---|---|---|---|
| MMP-1 | MMP-8 | MMP-13 | |
| Quinic acid | −0.92 | −0.90 | −0.92 |
| Shikimic acid | −0.71 | −0.32 | −0.71 |
| Phenylalanine | 0.62 | −0.11 | −0.41 |
| Kaempferol | −0.92 | −0.71 | 0.63 |
| Hydroquinone | −0.68 | −0.93 | −0.81 |
4. Discussion
New approaches that offer insight into the mode of action of plants and natural products are developing day by day. Studies published in recent years show that the interest in metabolomics has increased. Instead of focusing on specific individual component(s) or groups of compound(s), metabolomics studies are now based on a holistic approach in which the whole population of chemical components is taken into consideration (Ulrich-Merzenich et al., 2007). However, in addition to the new opportunities offered by these novel approaches, they have new or unexpected effects that need to be explored (Verpoorte et al., 2005). Therefore, the present study aimed to determine the correlation between comprehensive chemical fingerprints and bioactivity. We compared the metabolomic profiles of A. millefolium, A. filipendulina, M. piperita and S. officinalis to identify the metabolites with stimulating or inhibiting activities for MMPs.
In some cases, it is necessary to focus on a single active ingredient, but it is very well known that the activity of an extract cannot be attributed to a single compound or a single class of compounds since stimulating or inhibiting effects may lead to synergism or antagonism between the compounds (Ulrich-Merzenich et al., 2007, Verpoorte et al., 2005). From this point of view, in the current study, MMP inhibitory effects of the extracts obtained from AAMS were investigated.
When the overall results were examined comparatively, it was determined that the investigated plant extracts had a significant MMP inhibitory effect at five concentrations. M. piperita at the 500 μg/mL concentration had the highest inhibitory effect on MMP-1 (84.44%), MMP-8 (80.46%) and MMP-13 (91.02%), similar to the positive control NNGH (90%). S. officinalis was one of the weakest inhibitors of MMP-1, while it had the second most inhibiting activity for MMP-8 (80.06%) and MMP-13 (89.63%) after M. piperita (Table 1, Table 2, Table 3).
A. millefolium and A. filipendulina had a similar inhibitory effect on all three MMPs. The MMP inhibitory activities of AAMS were around 70% at the 100 μg/mL concentration. Although they have different metabolomic profiles, they all strongly inhibited the activities of MMPs. In order to identify the metabolites responsible for the MMP inhibitory activities in the extracts, the metabolomic profiling of the plants was investigated using GC-MS.
After deconvolution and alignment of the chromatograms, 284 metabolites were detected, and 149 of these metabolites were annotated using retention index libraries. Firstly, the obtained data from the GC/MS analysis of AAMS were subjected to a principal component analysis (PCA). The differences or similarities between the metabolomic profiles of the samples were visualized using a scatterplot (Fig. 3). As expected, the Asteraceae family members, A. millefolium and A. filipendulina, had similar metabolomic profiles, whereas the Lamiaceae family members, M. piperita and S. officinalis differed both from each other and from Achillea species. The scatterplot revealed that the differences and similarities in the metabolomic profiles can be used for classification and can also explain similar activities of different species.
A correlation analysis was applied to evaluate the correlation between metabolomic levels and MMP inhibitory activities to identify the metabolites responsible for these activities in the four investigated plants based on the synergist and antagonistic effect between the metabolites, instead of a single metabolite. We found that 96 metabolites had a negative correlation (r ≤ −0.70) and 55 had a highly positive correlation with MMP inhibitory activity (r ≥ 0.70) (Fig. 5).
The pathway analysis was done using the library of metabolic pathways for Arabidopsis thaliana assembled from the KEGG database. There is no currently available pathway database for Asteraceae and Lamiaceae families. Therefore the closest and available plant metabolic pathway (Arabidopsis thaliana) was selected in order to highlight metabolomic pathways correlated with activities. According to pathway analysis, amino acid (valine, leucine and isoleucine biosynthesis; and glycine, serine and threonine metabolism) and lipid (fatty acid biosynthesis) metabolism were negatively correlated with MMPs activities while energy metabolism (citrate cycle and glyoxylate and dicarboxylate metabolism) were positively correlated. There are also common pathways (galactose metabolism; and alanine, aspartate and glutamate metabolism) showed both positive and negative correlations with MMPs activities.
The results of correlation studies showed that MMP inhibitory activities were positively associated with phytochemicals, including 2-O-glycerol β-D-galactopyranoside, benzyl thiocyanate, and hydroxylamine, which were common in the AAMS materials. On the other hand, valine, stearic acid, pipecolic acid, allose, behenic acid, quinic acid, fructose, tetracosanol, octadecanol, 1-benzylglucopyranoside, 1,3-bisethynylbenzene, octacosanol, 3,4-dihydroxybenzoic acid, 4-trans-caffeoylquinic acid, 5-trans-caffeoyl quinic acid, chlorogenic acid, proline, and pyroglutamic acid decreased the inhibitory effect on MMP-1, MMP-8, and MMP-13.
Interestingly, the members of the shikimic acid pathway (quinic acid, shikimic acid, phenylalanine, cinnamic acid, and the final products kaempferol and hydroquinone) identified in our metabolomic analysis had a negative correlation with MMP activities except for phenylalanine, which was positively correlated with MMP-1. Considering that kaempferol’s interaction was −0.92, the MMP inhibitory effect of the extract seems to have been reduced. Although a high correlation was expected between phenolic compounds and the MMP inhibitory effect, our results did not confirm this for kaempferol and hydroquinone (Table 4).
The amino acids detected in the extracts were L-serine, L-threonine, L-norleucine, L-valine, β-alanine, L-tyrosine, L-tryptophan and L-alanine. The first four of these amino acids had a negative interaction with MMP-1, but were not significantly correlated with MMP-8 or MMP-13. While alanine, β-alanine and L-tryptophan had a positive correlation with MMP-13, tyrosine was positively correlated only with MMP-8.
Some sugars, such as melibiose, gulose and gentiobiose had a positive correlation with MMP-8, and raffinose and gentiobiose with MMP-13.
2-O-glycerol β-D-galactopyranoside, benzyl thiocyanate and hydroxylamine, were positively correlated with MMPs. β-alanine and L-tryptophan had a positive correlation with MMP-13, and tyrosine and some sugars with MMP-8. Phenolic compounds are known to have many biological effects. Although they were also expected to produce significant results in this study, it was interesting to obtain findings indicating the inefficacy of kaempferol and hydroquinone in particular.
5. Conclusion
This is the first study demonstrating that metabolomic profiling can be used for evaluation of activities in plants. The reason for the detection of the whole metabolite pool is that it takes into consideration the synergistic and antagonistic interaction between the metabolites, instead of investigating a single metabolite. This new perspective can be used to explain the activities of different plants with similar activities. The activity of a plant may not arise from a single metabolite, and the synergistic and antagonistic effects must be considered. As it is known, nature is changeable, and therefore when considering the effect of a molecule, its synergistic interaction with other molecules should also be considered, and metabolomic profiling and correlation analysis present as the best approach to achieve this.
Conflict of interest
No conflict to disclose.
Acknowledgement
This study was supported by a financial grant from Hacettepe University Scientific Research Projects (Project No: THD-2016-9171).
Footnotes
Peer review under responsibility of King Saud University.
References
- Akram M. The minireview on Achillea millefolium Linn. J. Membr. Biol. 2013;9:661–663. doi: 10.1007/s00232-013-9588-x. [DOI] [PubMed] [Google Scholar]
- Bailey A.J. Molecular mechanisms of ageing in connective tissues. Mech Ageing Dev. 2001;122:735–755. doi: 10.1016/s0047-6374(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucl. Acids Res. 2018;26:2342–2344. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniela T. Salvia officinalis l. I. Botanic characteristics, composition, use and cultivation. Ceskoslovenska Farmacie. 1993;42(3):111–116. [PubMed] [Google Scholar]
- Dara S.K.A., Belamkar S. A review on anti-inflammatory and analgesic activity of herbal origin. Asian J. Pharm. Res. Develop. 2014;2(3):45–53. [Google Scholar]
- Dennis D.T. Chapman and Hall; New York: 1987. Energy Utilisation in Plants. [Google Scholar]
- Dennis D.T., Huang Y., Negm F.B. Glycolysis, the pentose phosphate pathway and anaerobic respiration. In: Dennis D.T., Layzell D.B., Lefebvre D.D., Turpin D.H., editors. Plant Metabolism. Longman Singapore Publishers; Singapore: 1997. pp. 105–124. [Google Scholar]
- Elwy A.M., Tabl G. Anti-inflammatory and immune regulatory effects of Salvia officinalis extract on ova-induced asthma in mice. Life Sci. J. 2012;9(25):191–196. [Google Scholar]
- Elwy A.M., Ghada Tabl. Anti-Inflammatory and immune regulatory effects of Salvia officinalis extract on OVA-induced asthma in mice. Life Sci. J. 2013;10(1):1874–1878. [Google Scholar]
- Fuchs S., Skwara A., Bloch M., Dankbar B. Differential induction and regulation of matrix metalloproteinases in osteoarthritic tissue and fluid synovial fibroblasts. Osteoarthritis Cartilage. 2004;12(5):409–418. doi: 10.1016/j.joca.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Giorgi A., Bononi M., Tateo F., Cocucci M. Yarrow (Achillea millefolium L.) growth at different altitudes in Central Italian Alps: biomass yield, oil content and quality. J. Herbs, Spices Medicinal Plants. 2005:47–58. [Google Scholar]
- Gosline J., Lillie M., Carrington E., Guerette P., Ortlepp C., Savage K. Elastic proteins: biological roles and mechanical properties. Philos Trans: Biol Sci. 2002;357(1418):121–132. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A.J., Matthews W.G., Koob T.J. Determination of the elastic modulus of native collagen fibrils via radial indentation. Appl. Phys. Lett. 2006;89(18):181902–181903. [Google Scholar]
- Kamatou G.P.P., Viljoen A.M., Gono-Bwalya A.B., Van Zyl R.L., Van Vuuren S.F., Lourens A.C.U., Baser K.H.C., Demirci B., Lindsey K.L., Van Staden J., Steenkamp P. The in vitro pharmacological activities and a chemical investigation of three South African Salvia species. J. Ethnopharmcol. 2005;102:382–390. doi: 10.1016/j.jep.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Kamatou G.P.P., Van Zyl R.L., Van Vuuren S.F., Viljoen A.M., Figueiredo A.C., Pedro L.G., Barroso J.G., Tilney P.M. Chemical composition, leaf trichome types and biological activities of the essential oils of four related Salvia species indigenous to southern Africa. J. Essent. Oil Res. 2006;18:72–79. [Google Scholar]
- Kamatou G.P.P., Makunga N.P., Ramogola W.P.N., Viljoen A.M. South African Salvia species: a review of biological activities and phytochemistry. J. Ethnopharm. 2008;119:664–672. doi: 10.1016/j.jep.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Mahdi J., Al-Musayeib N., Mahdi E., Pepper C. Pharmacological importance of simple phenolic compounds on inflammatıon, cell proliferation and apoptosis with a special reference to beta-d-salicin and hydroxybenzoic acid. Eur. J. Inflamm. 2013;11(2):327–336. [Google Scholar]
- Nair B. Final report on the safety assessment of Mentha Piperita (Peppermint) Oil, Mentha Piperita (Peppermint) Leaf Extract, Mentha Piperita (Peppermint) Leaf, and Mentha Piperita (Peppermint) Leaf Water. Int. J. Toxicol. 2001;20(Suppl 3):61–73. [PubMed] [Google Scholar]
- Nema N.K., Maity N., Sarkar B.K., Mukherjee P.K. Matrix metalloproteinase, hyaluronidase and elastase inhibitory potential of standardized extract of Centella asiatica. Pharm. Biol. 2013;51(9):1182–1187. doi: 10.3109/13880209.2013.782505. [DOI] [PubMed] [Google Scholar]
- Nemutlu E., Zhang S., Yi-Zhou Xu., Terzic A., Zhong L., Dzeja P.D., Cha Y.M. Cardiac resynchronization therapy induces adaptive metabolic transitions in the metabolomic profile of heart failure. J. Card. Fail. 2015;21(6):460–469. doi: 10.1016/j.cardfail.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öncel M. Matriks Metalloproteinazlar ve Kanser. Eur. J. Basic Med. Sci. 2012;2(3):91–100. [Google Scholar]
- Stitt M. Pyrophosphate as an alternative energy donor in the cytosol of plant cells: an enigmatic alternative to ATP. Bot. Acta. 1998;111:167–175. [Google Scholar]
- Ulrich-Merzenich G., Zeitler H., Jobst D., Panek D., Vetter H., Wagner H. Application of the “-Omic” technologies in phytomedicine. Phytomedicine. 2007;70:70–82. doi: 10.1016/j.phymed.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Verpoorte R., Choi Y.H., Kim H.K. Ethnopharmacology and systems biology: a perfect holistic match. J. Ethnopharmacol. 2005;100:53–56. doi: 10.1016/j.jep.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Verpoorte R., Choi Y.H., Mustafa N.R., Kim H.K. Metabolomics: back to basics. Phytochem. Rev. 2008;7(3):525–537. [Google Scholar]
- Williamson E.M. Synergy and other interactions in phytomedicines. Phytomedicine. 2001;8:401–409. doi: 10.1078/0944-7113-00060. [DOI] [PubMed] [Google Scholar]
- Xia J., Wishart D.S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]