Abstract
Organic anion and cation transporting proteins (OATs, OATPs, and OCTs), as well as the Multidrug and Toxin Extrusion (MATE) transporters of the Solute Carrier (SLC) family are playing a pivotal role in the discovery and development of new drugs due to their involvement in drug disposition, drug-drug interactions, adverse drug effects and related toxicity. Computational methods to understand and predict clinically relevant transporter interactions can provide useful guidance at early stages in drug discovery and design, especially if they include contemporary data science approaches. In this review, we summarize the current state-of-the-art of computational approaches for exploring ligand interactions and selectivity for these drug (uptake) transporters. The computational methods discussed here by highlighting interesting examples from the current literature are ranging from semiautomatic data mining and integration, to ligand-based methods (such as quantitative structure-activity relationships, and combinatorial pharmacophore modeling), and finally structure-based methods (such as comparative modeling, molecular docking, and molecular dynamics simulations). We are focusing on promising computational techniques such as fold-recognition methods, proteochemometric modeling or techniques for enhanced sampling of protein conformations used in the context of these ADMET-relevant SLC transporters with a special focus on methods useful for studying ligand selectivity.
Keywords: SLC transporters, Uptake transporters, Molecular modeling, Drug-drug interactions, Selectivity, Conformational sampling, Fold-recognition, Open data
Graphical abstract
1. Introduction: Clinically Relevant Transporters of the SLC Family and Implications in Drug Discovery
Assessing a compounds' transporter pharmacology is an established paradigm in drug discovery and development, and efforts to document clinically relevant interactions with transporters are systematically undertaken since at least a decade as demonstrated by the White Paper from the International Transporter Consortium from 2010 [111]. Such transporters broadly cover members of the ATP-binding cassette (ABC) transporter and the solute carrier (SLC) transporter superfamilies. Reviews covering the broader topic of modeling approaches on the SLC transporters are available from Colas et al. and Schlessinger et al. [20,102,103]. In this review, we are focusing on members of the SLC family which were identified to have major implications in the pharmacokinetics of drugs. Mostly these transporters are uptake transporters, only MATE1 and MATE2K - the Multidrug and Toxin Extrusion transporters (MATEs; belonging to SLC47A subfamily) - are representatives of efflux pumps that transfer substances out of cells (mainly expressed in kidney and liver cells). The other discussed transporters herein are uptake transporters belonging to the families of Organic Anion Transporting Polypeptides (OATPs; SLCO), Organic Anion Transporters (OATs; SLC22A), Organic Cation Transporters (OCTs; SLC22A), and Organic Carnitine Transporters (OCTNs; SLC22A).
The tissue expression of the different SLC members discussed in this review is quite heterogeneous and was already described extensively by others (summarized in Table 1) [84,97]. However, predominant organs of expression include liver, kidney, and to a lesser extent intestine. Pharmacological barriers in these organs are crucial for absorption, metabolism, distribution, and excretion of drugs and any impairment in the function of one of these transporters can therefore lead to adverse effects or toxicity by altered pharmacokinetics of the drug or food ingredient administered. For instance, treatment with the immunosuppressant cyclosporine can result in statin-induced myopathy when administered at the same time since statins are known to be substrates for OATP1B1. Inhibition of OATP1B1 by cyclosporine therefore leads to increased plasma concentration of different statins [86].
Table 1.
Tissue expression profiles of clinically relevant SLC transporters: predominant organs/tissues of expression are written in bold.
| Human Transporter | Tissues of predominant expression | Reference |
|---|---|---|
| OCT1 | Liver (sinusoidal membrane) | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| OCT2 | Kidney (basolateral membrane) | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| OCT3 | Placenta, testis, brain, lung, intestine etc. | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| OCTN1 | Kidney (brush-border membrane), skeletal muscle, etc. | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| OCTN2 | Kidney (brush-border membrane), liver, heart, etc. | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| MATE1 | Kidney (brush-border membrane), liver (canalicular membrane), muscle, etc. | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| MATE2-K | Kidney (brush-border membrane) | Motohashi, H., & Inui, K. I. [84]. AAPS J, 15(2), 581–588. |
| OAT1 | Kidney, skeletal muscle, brain and placenta | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OAT2 | Liver, kidney | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OAT3 | Kidney, brain | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OATP1A2 | Brain, small intestine, kidney, testes, lung, Liver (cholangeocytes) | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OATP1B1 | Liver (basolateral membrane) | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OATP1B3 | Liver (basolateral membrane) | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
| OATP2B1 | Liver (basolateral membrane), intestine, placenta, heart, mammary gland, brain | Roth et al. [97]. Br J Pharmacol, 165(5), 1260–1287. |
OATP1B1 and OATP1B3 are exclusively expressed in hepatocytes. Due to their high sequence similarity (~80%) they transport common substrates and are inhibited by common inhibitors. Nevertheless, there are some compounds (e.g. pitavastatin for OATP1B1 [41], cholecystokinin octapeptide CCK-8 for OATP1B3 [48]) which are selectively binding to only one of the two transporters with high affinity.
OATP2B1 is ubiquitously expressed, with highest expression levels at basolateral membranes of hepatocytes [97]. Due to its more distant relationship to OATP1B1 and OATP1B3 (~30% sequence identity to both), also substrate and inhibitor profiles are overlapping to a lesser extent [55,113].
Expression of OATP1A2 in liver cells has been observed in cholangiocytes, possibly accounting for the re-uptake of xenobiotics from bile [67]. In addition, common expression of OATP1A2 and OATP2B1 at the luminal membrane of intestinal absorptive cells is potentially implicated in drug-food interactions [28]. Specifically, it has been found that components of fruit juices (especially naringin) cause the inhibition of OATP1A2, which in turn affects oral bioavailability of fexofenadine (substrate of OATP1A2) [7].
OAT1, and OAT3 are expressed at the basolateral membrane of kidney cells, pointing to their concerted interplay in the uptake of substrates and related drug-drug interactions (DDIs) [16,29,44]. For example, it has been found that probenecid inhibits methotrexate transport by OAT1 and OAT3 [88]. OCT2 is expressed at the luminal membrane of kidney cells [37] and its organ of highest expression is liver (rather than kidney).
OCTN1 and OCTN2 are highly expressed in kidney as well [76,124]. OCTN1 and OCTN2 are associated with several pathologies, such as inflammatory bowel disease, primary carnitine deficiency, diabetes, neurological disorders, and cancer [94]. Interestingly, in case of unresectable gastrointestinal stromal tumors treated with imatinib therapy, polymorphisms in OCTN1 and OCTN2 are associated with a prolonged time to progression in GIST patients receiving imatinib therapy [4]. In a different study, OCTN1-mediated uptake of cytarabine into tumor cells in a cohort of acute myeloid leukemia (AML) patients could be related to reduction in the development of resistance to chemotherapy [27].
Human MATE proteins have been shown to be widely distributed across different body tissues, including liver, skeletal muscles, testis, and kidney. Efflux- and uptake-activity of human MATEs have been observed to be pH-dependent [66]. Nevertheless, in renal cells MATE1 serves as an efflux pump, mediating the transport of the substances from kidney into urine. The OCT2-MATE1 interplay in uptaking and effluxing compounds in the kidney shows that also little related transporters can be conjointly involved in clinically relevant DDIs. MATE2-K is the human kidney-specific MATE2 active splice variant of MATE2 [84]. In addition, the ubiquitously expressed variant of MATE, MATE2-B, has also been detected. Nevertheless, this variant probably does not have any functional activity [76].
For being able to assess a compounds' risk to interact with any of these ADMET-relevant transporters, having predictive in silico models for all of them at hand would be useful at an early stage in the drug discovery pipeline. As will be discussed in the following sections, such efforts are complicated by limitations in data availability in the domain of ADMET-relevant SLC transporters. Other challenges include the overlapping substrate or inhibitor specificities across these transporters which are arising from the promiscuous nature of these proteins [111], as well as from the substrate/inhibitor promiscuity.
In this review article, data science and computational modeling approaches for unravelling ligand-transporter interactions for the important class of clinically relevant SLC transporters are discussed. In chapter 2, a general overview on important ligand- and structure-based modeling methods which are traditionally being used in computer-aided drug discovery is provided. Next, challenges arising from data sparseness on both the ligand and protein side in the field of clinically relevant SLC transporters are discussed (chapter 3). Chapter 4 is providing more details about different ligand-based (LB) and structure-based (SB) modeling methods that have been used in the context of clinically relevant SLC transporters and discusses insights that were delivered by the different studies. The emphasis is further put on the combination of LB and SB methods to provide a more comprehensive picture of ligand recognition. In chapter 5, studies and different methods providing insights into transporter selectivity are highlighted. Finally, the challenge of including information on transporter flexibility into the SB-modeling procedure is discussed (chapter 6) and some recent developments in the field are highlighted (such as the use of elastic network models for conformational sampling). In conclusion, a computational workflow for studying ligand interactions with clinically relevant SCL transporters is proposed, interconnecting ligand data integration, data curation and analysis, as well as LB and SB modeling techniques in order to come up with binding mode hypotheses.
2. Molecular Modeling Approaches at a Glance
Computational approaches have become a standard paradigm in area of preclinical drug discovery [78]. Molecular modeling is an interdisciplinary field that incorporates theoretical concepts and efficient computational algorithms to study chemical phenomena [65]. The main idea is to use approximative mathematical models while being able to predict behaviour of chemical systems as closely as possible [129]. In the field of computer-aided drug design (CADD), molecular modeling methods are generally divided into two categories: (1) Ligand-based (LB) and (2) Structure-based (SB) modeling. LB modeling is also known as ‘indirect drug design’, since in the modeling approach protein information is not taken into account. On the other hand, SB molecular modeling techniques enable to study protein-ligand complexes at the molecular level. SB drug design is also termed ‘direct drug design’ since in these cases 3D information of the target protein or a structural model built on basis of (phylogenetically or structurally) a related protein template is included into the modeling process.
In terms of LB modeling, a system of interest is represented mostly as a statistical model incorporating knowledge on the associated compounds (substrate or inhibitor data) possessing experimentally determined activities against a particular target. Compound series can be used to derive an abstract representation of important molecular features being crucial for a binding event. This approach is known as LB pharmacophore modeling and it is especially useful to overcome difficulties in aligning and searching for structurally little related compounds which persist similar in terms of chemical features (so-called “scaffold-hopping” concept) [45,128]. Another widely used approach is Quantitative Structure-Activity Relationship (QSAR) which is based on correlating physico-chemical properties (or other representations of the chemical structure, such as molecular fingerprints of compounds) to their biological activity [23]. QSAR modeling and related qualitative approaches (such as binary classification models) have currently become especially popular due to the observed increase of the deployment of machine learning and deep learning algorithms in CADD. Especially classification models (models being trained to distinguish compounds into e.g. ‘active’ and ‘inactive’ class) can subsequently be used for virtual screening of chemical databases with the aim to detect new (potentially active) compounds. Qualitative models can also aid in the interpretation of ligand profiles which might trigger a compound's activity against a particular target [113]. Beyond the conventional SAR techniques, 3D-QSAR modeling provides a natural extension to the classical QSAR formalism by superimposing 3D ligand structures to retrieve 3D-based ligand features [116].
SB molecular modeling approaches are significantly hampered by the limited number of experimentally resolved structures for membrane transporters. So far (updated on January 2019), the number of the available crystal structures for membrane proteins in Protein Data Bank (PDB) did not exceed ~3,5% of all deposited entries. Such a small fraction of resolved membrane proteins is primarily caused by problems when overexpressing membrane proteins in bacteria [101], as well as by the obstacles accompanying crystallization of membrane proteins [89]. These include, for example, finding optimal conditions for crystallization [87], as well as difficulties to account for a membrane-like environment which in turn is being crucial for crystallizing the native form of membrane proteins. However, substantial progress has been made by e.g. improving protein engineering to increase the stability of a protein of interest [110]. The latter issue is commonly solved by using micelle detergents to solubilize membrane proteins for purification purposes [106]. Except for X-ray crystallization, solid-state NMR techniques can be used to resolve structure of membrane proteins [63]. In this methodology, membrane nanodiscs have successfully been applied as membrane-mimetics [98]. If the crystal, NMR, or recently also Cryo-EM structure of a respective target is not available, 3D structural models can be built “from scratch” upon basic thermodynamic principles [39]. These methods are known as ab initio (de novo) structural predictions and are rather rarely used, mainly due to the challenges to efficiently sample the whole conformational space at a feasible time scale [51]. In most of the cases, 3D structural models are created on basis of sequence-homologous proteins with known structure (“homology modeling” [52]), or more evolutionary distant proteins with conserved fold (“fold-recognition methods” or “protein threading” [75]). Fitting of a target sequence onto the 3D coordinates of a sequentially- or structurally- related protein (“template”) is further accompanied by a global geometry optimization to satisfy structural restraints imposed by internal coordinates, as well as local optimization to remove steric clashes and/or reduce the noise caused by poorly modeled side chain rotamers [62,99,119]. This procedure is commonly applicable for the transmembrane core of membrane proteins. However, it might not be sufficient for modeling intra- and extra-cellular domains, which are mostly consisting of large loop regions [32]. Loop modeling is a non-trivial step in protein structure prediction, since the loops are intrinsically disordered regions, often requiring enhanced conformational sampling [33]. In addition, loops are usually indeterminate parts in the crystallization process due to their high conformational flexibility and thus low electron density in X-ray diffraction patterns. Homology modeling is subsequently limited by incomplete sequence alignment because of missing loop regions. Modeling of extremely short segments (<3 amino acids) can be satisfied by geometrical constraints of their bond lengths and angles. Template-based loop construction by using a database of known structural fragments (not necessarily with identical sequence) is a common modeling approach for medium-size loops (~10 amino acids). In general, energy minimization combined with MD and simulated annealing is advisable to refine modeled loops. For longer loops (~25 amino acids), de novo coarse grained modeling methods (employing e.g. united residue models) have successfully been applied [49].
Furthermore, molecular dynamics (MD) simulations with enhanced sampling techniques can additionally be integrated to the 3D model building procedure especially to refine low-confidence regions, such as flexible loops (as discussed above) and less structurally-conserved parts of the protein [34]. When performing MD simulations on membrane proteins, one should also account for the substantial role of phospholipid membranes which spatially restrain a protein's 3D structure [82]. The computationally less demanding approach is to treat membrane environment as a mean-field continuum model which replaces explicit protein-lipid interactions by effective interactions being a priori included in force-field parameters of a membrane-protein system [31]. These approaches have become particularly useful for the simulations of large time scale events, such as protein folding, albeit for the cost of lacking all-atom representation of a simulated system. The lack of high-resolution accuracy by using implicit membrane models can be corrected by the explicit representation of lipid bilayers in the simulation box. However, running MD simulations with explicit lipid bilayers might be unfeasible for biologically relevant time scales.
Quality assessment of 3D protein models is required to distinguish the native protein structure from the physically non-relevant states [40]. For this purpose, several scoring functions, including statistics-based, knowledge-based, physics-based, or their combinations, have been developed. To give an example, ProQM is a statistics-(learning-) based method using Support Vector Machines (SVM) models which are trained on the known structures to predict correct structural features of membrane proteins, such as membrane topology or conserved structural motifs [95]. Another example is C-score which estimates the confidence of target-template alignments based on the fold-recognition methods [127]. TM-score function is a metric of 3D similarity between two proteins when performing structural alignment [131].
The next step in SB modeling is to apply docking algorithms to iteratively search for preferred orientations of a ligand molecule relatively to the protein binding site(s) [68]. Subsequently, protein-ligand complexes can be ranked on basis of scoring functions (knowledge-based [85], physics/force field-based [35], machine-learning [2], and/or empirical [12]), estimating the likelihood of all possible binding poses with respect to the energetically (un)favorable intermolecular interactions. Molecular docking can be performed by using different strategies, ranging from rigid docking, where the protein structure is kept fixed and only the ligand's conformational space is explored [9], to induced-fit docking, where the protein local backbone movements are allowed to adjust the proper ligand-protein binding pose [117]. More sophisticated and computationally demanding methods are treating the whole protein structure as flexible [123]. Docking screens are typically evaluated on basis of score accuracy (by comparing the predicted binding affinities to the experimental ones, if available [8]), enrichment factor (by checking if the docking screens are able to discriminate between known actives and known inactives/decoys [46]), or on basis of prospective validation (by measuring e.g. IC50 values [47]). Traditional docking approaches can be complemented by more accurate free energy calculations not only to estimate absolute free energy of binding [24], but also to study ligand selectivity profiles [3]. Approximations of binding free energies are given by methods like MM/GBSA (Molecular Mechanics/Generalized Born Surface Area), as originally described by Kollman et al. [59]. However, free energy perturbation (FEP) provides better estimates of free energy of ligand binding, and it offers the possibility to directly evaluate the impact of mutated functional groups of the ligand from the energetic point of view [118]. On the protein side, biochemical mutational studies can be informed by docking exercises and vice versa as demonstrated for the interleukin 8-gene [22].
Following the same strategy as in case of LB pharmacophores, SB pharmacophore models can be created upon projection of the important pharmacophoric features of a target-ligand complex to an abstract representation [107]. In the recent past, new techniques combining LB and SB approaches have become popular. For example, proteochemometric modeling (PCM) can outperform traditional QSAR modeling by simultaneous evaluation of the similarity of ligands and targets [115]. The great advantage here is the possibility to extrapolate the activities of known ligands against known targets to novel targets without knowing their 3D structure.
3. Data Sparseness as a Major Challenge in Computer-aided Drug Discovery & Implications to the Exploration of Uptake Transporters
Chapter2 provides on overview of main computational techniques which are traditionally being used in the drug discovery pipeline. When it comes to investigations on clinically relevant SLC transporters, however, the direct application of above-mentioned methods is far from being trivial. The main obstacle hampering computational studies is caused by data sparseness on both ligand and protein levels in the domain of uptake transporters. In addition, the promiscuous nature of uptake transporters being able to bind both endogenous compounds and xenobiotics [56], considerable transporter flexibility which accompanies translocation processes (such as “rocker-switch” mechanism as proposed for Major Facilitator Superfamily members) [120], as well as the likely existence of multiple binding sites [43,70,74,125], turns all modeling efforts into even more challenging tasks.
From a ligand's perspective, modeling studies are strongly impeded by the inconsistent and mostly insufficient number of high-quality bioactivity data which is spread over different data sources in the open domain. For LB studies on uptake transporters, single-point percentage inhibition data has often become the only source for QSAR modeling [1,14,19,55,58,60,122]. Such models trained on single-point inhibition data can achieve high accuracies, particularly if the measurements were retrieved following the same experimental protocols, as demonstrated by e.g. the classification models by Karlgren et al. [55] (with accuracies between 73% and 92% for the different models). However, even in the case of almost identical assay protocols, other parameters such as the substrate concentration might to a significant extent change the final value of the read out (e.g. percentage inhibition value). In case of the studies by Ahlin et al. [1] and Chen et al. [19] which both measured reuptake inhibition for OCT1 inhibitors by using the same probe substrate (4–4-dimethylaminostyryl-N-methylpyridinium, abbreviated as ASP+), a different substrate concentration (2 μM in Chen et al. and 1 μM in Ahlin et al., respectively) as well as inhibitor concentration (20 μM in Chen et al. and 50 μM in Ahlin et al.) led to a percentage of around 14% of all overlapping compounds (measured in both papers) being differently classified by the different studies (by using a cutoff of 50%). Obviously, the study by Chen et al. was much more rigorous in assigning the label “inhibitor”, since many conflicting annotations with Ahlin et al. turned out to be rather false negatives (data not shown).
In most of these cases, the activity cut-off for binary classification into actives and inactives was set to 50% [1,19,55,60,122]. In some studies, however, the authors applied more stringent activity criteria for setting a treshold by removing weak actives from the data set, e.g. in the study by van de Steeg et al. [114]. Here, the activity cut-off for OATP1B1 inhibitors was defined as ≥ 60%, while OATP1B1 non-inhibitors were defined as ≤ 40%. It concludes that all “grey zone” data points (the weak actives) in the range (40;60) were excluded from the dataset. On one hand, removal of weak actives can be beneficial to reduce the noise caused by the variations in the experimental measurements [114]. On the other hand, complete exclusion of weak actives might decrease the applicability domain of such models, e.g. when attempting to predict the activity of compounds which are structurally closely related to those from the “grey zone”. For establishing LB pharmacophore models for MATE1 [126] and OCT2 inhibitors [125], classification of inhibition/substrate data into binary classes was done via manual literature searches with the aim to extract recommended activity thresholds proposed by the authors of primary literature sources. For deciding upon the cutoffs for percentage inhibition data in order to generate predictive classification models for OATP1B1, OATP1B3, and OATP2B1 inhibition from diverse data source we have chosen the same strategy [113].
Structurally and pharmacologically distinct compounds can also be partitioned into discrete clusters based on their molecular features, as applied for e.g. the classification of OCT2 inhibitors/non-inhibitors [58]. After the clustering step, different activity cut-offs (inhibitory effect ≥ 50% or ≥75%) were probed to examine which clusters contained the highest fraction of the inhibitors based on the applied cut-off. Specifically, by applying the more stringent cut-off (≥ 75%) the inhibition patterns of identified clusters became more pronounced. Distinct OCT2 clusters were subsequently used for deriving several independent SAR analyses to explain complementary inhibitory mechanisms of OCT2 inhibitors.
Full dose-response curve data has been exploited to much lesser extent. Examples include IC50 measurements for LB studies on MATE1, MATE2-K [5], and OCT2 [122], Km values for OATP1B1 substrate pharmacophore models [17], as well as Ki values for pharmacophore modeling of OCT2 stereoselective binding [79].
If the amount of bioactivity data is not sufficient for model development, direct usage of categorical annotations, such as “substrate”, “non-substrate”, “inhibitor”, or “non-inhibitor”, can be applied. For this purpose, Drugbank (containing a collection of marketed or FDA-approved drugs [121]), or Metrabase (primarily containing substrates for OCT1, OATP1A2, OATP1B1, OATP1B3, and OATP2B1 [72]) can serve as rich, open sources for LB modeling. For example, substrate annotations from Metrabase were retrieved for QSAR and PCM modeling to predict OCT1, OATP1A2, OATP1B1, OATP1B3, and OATP2B1 substrates [104]. It has to be pointed out, that using manual activity annotations for binary classification modeling seems to be quite error-prone since it is not clear how data curators decided upon assignment of activity annotations in certain cases. As demonstrated in our current LB studies on hepatic OATPs [113], an extensive comparison of Metrabase annotations for OATP1B1, OATP1B3, and OATP2B1 (non-)substrates and (non-)inhibitors with numerical bioactivity measurements from CHEMBL revealed activity misclassifications for Metrabase data up to 74%. On the other hand, categorical annotations can still be utilized in developing accurate computational models when e.g. performing selective fusion of more independent classifiers and therefore increasing the confidence in the model's predictability [104].
With increasing efforts of making bioactivity data publicly available to the scientific community, new challenges are arising. Nowadays, it is no longer only access to data but the proper usage of data which can provide a competitive advantage in drug discovery. Finally, filtering out high-quality data is essential as well as making use of the possibilities to interconnect different types of data, including data originating from diverse sources. In the light of those efforts, data analytics platforms for creating automated data workflows, such as the Konstanz Information Miner (KNIME [11]) or Pipeline Pilot [108], have become particularly useful. Notably, integrative data mining (i.e., data fusion from different sources) can enrich existing data sets by not only the size in enumerated compounds, but also by obtaining novel scaffolds which can lead to an expansion of the available chemical space as demonstrated recently for hepatic OATPs [113]. Moreover, merging data from multiple independent bioactivity measurements (Km, IC50, EC50, Ki, percentage inhibition data) for a single compound can significantly increase the confidence of bioactivity data. When it comes to QSAR modeling, it is usually not recommended to mix compound data originating from different bioactivity end-points [53,61]. On the other hand, binary classification (e.g. separating substrates from non-substrates) should be independent from a certain assay or experimental protocol used [80]. Another benefit when considering multiple bioactivity measurements is the ability to rationally decide upon activity thresholds for binary label assignment (e.g. active/inactive) by studying the distribution of bioactivities within the data set [113].
As recently demonstrated [113], above mentioned pipelining tools are quite handy for semi-automatically fetching ligand data from different open data sources, such as CHEMBL [10], UCSF-FDA Transportal [83], IUPHAR [90], and Drugbank [121] and Metrabase [72]. It has to be emphasized that ligand data originating from different sources might be highly inconsistent with respect to their structural format used. To overcome this issue, applying a standardization protocol, as e.g. by Atkinson (available at https://wwwdev.ebi.ac.uk/chembl/extra/francis/standardiser/), has proven to be useful for mapping data from different sources.
From Table 2 and Table 3 it becomes clear that the most comprehensive data set is currently available for OATP1B1 and OATP1B3, with CHEMBL being detected as the most prominent source (1993 and 1972 unique compounds, respectively). This trend is reflected by the high number of LB modeling studies with a special focus on OATP1B1: QSAR/classification models [6,54,55,60,104,105,113,114], PCM [14,104], and LB pharmacophore model [17].
Table 2.
Number of unique compounds/bioactivities from open domain databases. In case of Metrabase and IUPHAR, only numerical bioactivity values have been extracted from the data bases (categorical annotations were discarded).
| Transporter | UNIPROT ID | CHEMBL | Metrabase | IUPHAR | Transportal | Total number of unique compounds |
|---|---|---|---|---|---|---|
| OCT1 | O15245 | 290/437 | 230/607 | 1/1 | 44/86 | 307 |
| OCT2 | O15244 | 126/221 | No entries | 1/1 | 67/120 | 144 |
| OCT3 | O75751 | 37/44 | No entries | 1/1 | 28/44 | 54 |
| OCTN1 | Q9H015 | 26/33 | No entries | No entries | 11/20 | 26 |
| OCTN2 | O76082 | 67/96 | No entries | No entries | 6/10 | 68 |
| MATE1 | Q96FL8 | 70/139 | No entries | 3/4 | 31/55 | 86 |
| MATE2-K | Q86VL8 | 44/55 | No entries | 3/4 | 23/46 | 60 |
| OAT1 | Q4U2R8 | 111/205 | No entries | 1/1 | 74/132 | 123 |
| OAT2 | Q9Y694 | 32/39 | No entries | No entries | 32/39 | 50 |
| OAT3 | Q8TCC7 | 113/180 | No entries | No entries | 68/102 | 131 |
| OATP1B1 | Q9Y6L6 | 1993/2566 | 307/752 | 1/1 | 61/139 | 2052 |
| OATP1B3 | Q9NPD5 | 1972/2469 | 249/408 | 3/3 | 45/95 | 2015 |
| OATP1A2 | P46721 | 70/100 | 54/96 | 2/2 | 17/24 | 75 |
| OATP2B1 | O94956 | 252/392 | 232/461 | 0/0 | 21/46 | 294 |
Table 3.
Number of unique compounds with categorigal activity annotations from the open domain.
| Transporter | UNIPROT ID | Metrabase | Drugbank | Total number of unique compounds |
|---|---|---|---|---|
| OCT1 | O15245 | 504 | 69 | 470 |
| OCT2 | O15244 | No entries | 56 | 56 |
| OCT3 | O75751 | No entries | 29 | 29 |
| OCTN1 | Q9H015 | No entries | 27 | 27 |
| OCTN2 | O76082 | No entries | 54 | 54 |
| MATE1 | Q96FL8 | No entries | 6 | 6 |
| MATE2-K | Q86VL8 | No entries | 1 | 1 |
| OAT1 | Q4U2R8 | No entries | 111 | 111 |
| OAT2 | Q9Y694 | No entries | 36 | 36 |
| OAT3 | Q8TCC7 | No entries | 77 | 77 |
| OATP1B1 | Q9Y6L6 | 375 | 75 | 385 |
| OATP1B3 | Q9NPD5 | 338 | 44 | 345 |
| OATP1A2 | P46721 | 111 | 61 | 107 |
| OATP2B1 | O94956 | 352 | 33 | 338 |
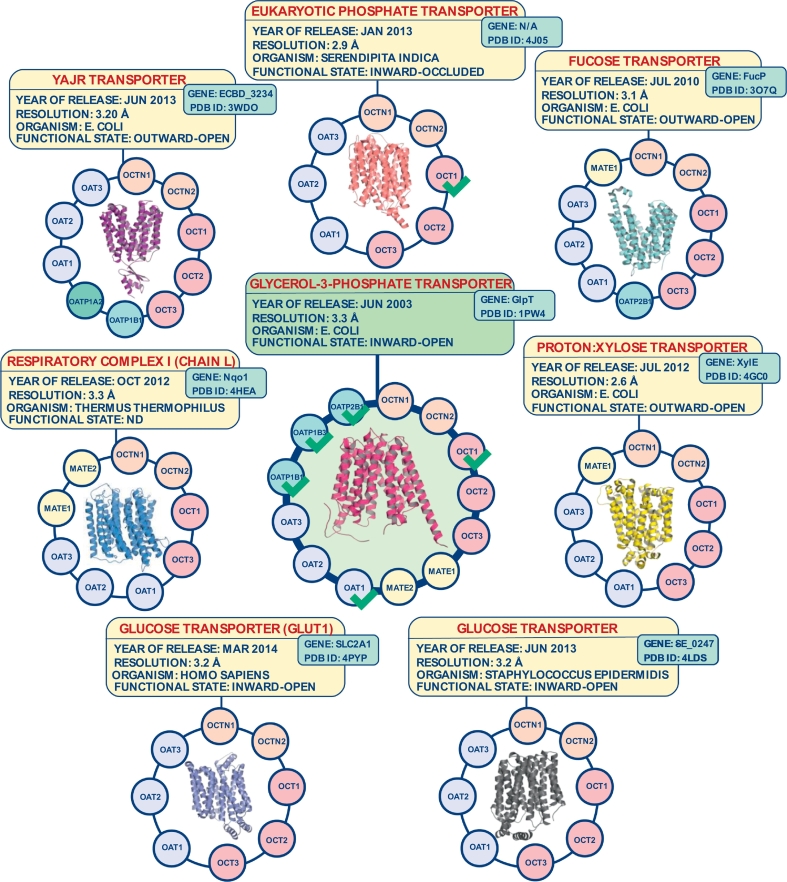
In addition to this quite restricted availability of compound bioactivity data for uptake transporters which limits over all the applicability domain of respective LB models, SB modeling efforts are strongly impeded by the lack of crystal structures in this domain. Only MATE1 bacterial templates are available (NorM from Vibrio cholera21, pdb id: 3mkt; pfMATE from Pyrococcus furiosus, pdb id: 3vvn). Both of the available crystal structures share approximately 24% identity with human MATE1. Except for this case, there are no available homologs for the other clinically relevant SLC transporters. These drawbacks can be diminished by the use of fold-recognition methods to search for structurally related analogues with conserved fold. The rationale behind is that a protein's secondary structure should have been conserved to a higher extent during evolution than its amino acid sequence [52]. An overview of available crystal structures which were predicted as potentially useful templates for SB modeling of ADMET relevant SLC transporters is provided in Fig. 1. Predictions were performed by using pGenThreader prediction server [71]. It appears interesting that all predicted templates (except for chain L from respiratory complex I) belong to the Major Facilitator Superfamily (MFS) [92]. All protein structures are built up of twelve transmembrane helices. It is interesting to note that sequence similarity among discussed transporters and the detected template structures is generally rather low (10–25%), which obviously slowed down the generation of comparative models on basis of these templates compared to other protein families (numbers of published studies per year including comparative modeling for clinically relevant SLC transporters is depicted in Fig. 2). As visible from Fig. 1, different templates are reflecting different functional states of the transporters (inward-open, occluded, outward-open). Out of these templates, Glycerol-3-Phosphate transporter (pdb id: 1pw4) was the most abundantly used template, specifically for building computational models for OATP1B1 [69], OATP1B3 [36,73,77], OATP2B1 [57,77], OAT1 [93,112], and OCT1 [13]. The popularity of Glycerol-3-Phosphate transporter as a structural template for this class of transporters can mainly be attributed to the fact that it appeared as the first available template in 2003.
Fig. 1.
Predicted structural templates for ADMET relevant SLC transporters by using pGenThreader (p < 0.001). Templates already used in structure-based modeling for a respective uptake transporter are indicated by a green check mark. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
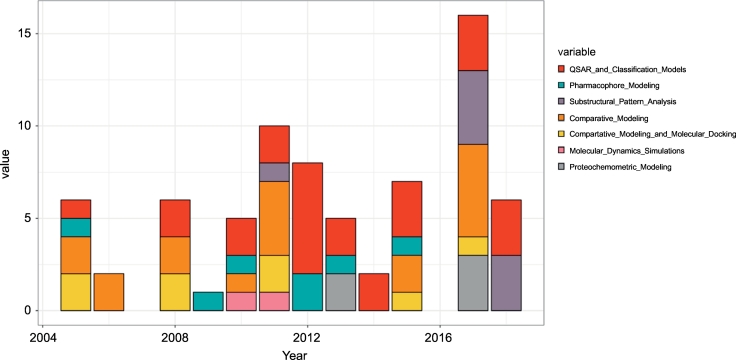
Fig. 2.
Time evolution of different computer-based methods used in published in silico studies on clinically relevant SLC transporters. abscissae....year; ordinate.....numbers of published studies.
4. Linking Ligand- and Structure-Based Modeling Methods for Studying Uptake Transporters
Natural increase in available ligand bioactivity data and structural templates also led to an increase of published computational studies on clinically relevant SLC transporters over the years. Fig. 2 shows the time evolution of different computational methods that have been used in the context of uptake transporter modeling. In general, we can observe an increase in in silico studies over the years for both LB and SB approaches. Both approaches have been used early on and we can observe peaks in the emergence of QSAR/classification models and new homology models in 2011/12. In 2017, one publication reported the establishment of new comparative models for OCT1, OCT2, OCT3, OCTN1, and OCTN2, which is also visible as a peak from Fig. 2. Interestingly, MD simulations were integrated only twice into the process of clinically relevant SLC transporter modeling, namely for OAT1 and MATE1 (in 2011/12). With the recent increase in available comparative models for uptake transporters, a revival of MD-based methods can be expected especially for performing enhanced conformational sampling of distinct transporter states in the future (see also chapter 5). Naturally, studies including PCM modeling appeared later in literature, since they incorporate information from the ligand and protein side. Thus, for such a technique to be effective it requires a minimum amount of ligand data and ideally knowledge on potential protein binding sites.
Also, from a static perspective, LB and SB methods have been used to approximately the same extend (e.g. 26 studies on QSAR/classification modeling; 26 studies including comparative modeling). If a fair amount of compound bioactivity data for a particular transporter is available (at least a few hundred unique compounds), LB approaches can deliver quicker and more comprehensive insights into important molecular features driving compound affinity towards a particular target than SB approaches. On the other hand, traditional SAR-based methods do not account for the hypothetical presence of distinct binding sites. Such drawbacks can be diminished by sub-structural pattern analyses such as studying pharmacological profiles of enriched scaffolds in the data set. The hereby retrieved congeneric SAR series are likely to interact with the same binding pocket and differences in pharmacological activities are likely triggered by subtle modifications at the side chain level [113]. Thus, such compound series are useful collections for subsequent structure-based docking studies.
In addition, LB pharmacophore models can be useful in detecting important pharmacophoric ligand features which can complement SB docking studies into comparative models. An interesting pharmacophore-based modeling approach where in vitro and in silico (‘IVIS’) methods are combined has been adopted by Diao et al. to map pharmacophoric features of human OCTN2 inhibitors [25,26]. This methodology has subsequently been used also by Astorga et al. to study the inhibitory profiles of human MATE1 [5]. Specifically, the IVIS procedure aims to iteratively build 3D-pharmacophore models, further used for database screening and subsequent experimental testing of new hits. Afterwards, high-affinity detected compounds are used to rebuild initial pharmacophore hypotheses to perform another round of database screening, and so on. For human MATE1 inhibitors, a common-features pharmacophore has initially been developed by merging pharmacophoric features of both high- and low-affinity MATE1 inhibitors [25]. The aim of mixing high- and low-affinity compounds for building a single pharmacophore model is to detect the minimal essential properties which are crucial for the effective interaction with MATE1. The iterative procedure has finally led to a pharmacophore with two hydrophobic features, one H-bond acceptor and one ionizable (cationic) feature. It is noteworthy to mention that the generation of quantitative pharmacophores has been strongly dependent on the probe substrates used in the in vitro measurements. Specifically, when using 4-4-dimethylaminostyryl-N-methyl-pyridinium instead of 1-methyl-4-phenylpyridinium, the final pharmacophore model was composed of three hydrophobic features, two H-bond acceptors, and three excluded volumes, spatially arranged in strikingly different configuration from the original one. These findings suggest that human MATEs might contain multiple substrate (and potentially inhibitor) binding sites. Therefore, for developing a complete computational model of MATE inhibitory mechanisms it would be required to build up multiple pharmacophore models per distinct binding site. For OCTN2 inhibitors, the common pharmacophore model revealed three hydrophobic features and one positive ionizable feature [26]. This IVIS-based pharmacophore hypothesis was partially confirmed by the upcoming study on OCTN2, showing that two hydrophobic features, one H-bond donor, as well as one positively ionizable feature are likely driving OCTN2 inhibitory activity [25].
As already introduced in chapter 3, SB modeling is complicated by the lack of native family member templates. Exclusively human and rabbit MATE1 structural models have been constructed on basis of sequence-homologous templates retrieved by the BLAST algorithm [126,130]. The first structural model for human MATE1 has been built upon the sequence similarity with NorM crystal structure from Vibrio cholerae (pdb id: 3mkt, 35.6% sequence similarity) [130]. After structural model generation, molecular dynamics (MD) simulation has been performed to test the stability of generated human/rabbit MATE1 structural model. In parallel, MD simulations have been performed for the NorM template structure to see if the conformational dynamics of Norm crystal and derived homology models remains conserved. A 50 ns long production MD confirmed overall stability. Interestingly, several helices in the human MATE1 homology model (for example, TM6 and TM9) reoriented and assumed opposite tilt angles when compared to the template. Obviously, a partial closing of the translocation pore was happening for the homology model. This interesting use case highlights the fact that each transporter ortholog might possess its internal dynamics, which cannot always easily be captured by the template structure.
Other reported SB studies rely on the structural similarity with MFS members. For details describing studies for a respective uptake transporter, see Table 4. In the recent past, a multiscale approach for 3D structural modeling of seven human OCTs (OCT1, OCT2, OCT3, OCTN1, OCTN2, OCT6, and FLIPT1) has been published. This study [21]. This study [144] introduces a comprehensive modeling pipeline for tertiary structure prediction, starting from the comparative sequence alignment, and fold-recognition 3D model building combined with ab-initio modeling performed via I-TASSER [145]). In addition, post-translation modifications of functionally relevant structural motifs (e.g. phosphorylation, ubiquitination, and/or glycosylation sites) were predicted via bioinformatic tools (PhosphoSitePlus available at http://www.phosphosite.org/ and NetNGlyc 1.0 server available at http://www.cbs.dtu.dk/services/NetNGlyc/). An integrative approach for 3D structure prediction is followed by the comprehensive evaluation of the modeled structures based on different metrics, including sequence identity between the target and template, query coverage, and consensus Z-score of the top threading programs. Structural analysis of generated models has revealed that the 3D structural models generated here share structural similarity with the human glucose transporter GLUT3 (pdb id: 5c65). Visual inspection of the obtained models further implies 2-pseudofold symmetry, as well as the hypothesis about two distinct functional states (inward- and outward-open). These observations were fully supported by the structural superposition of 3D generated models of OCTs with the GLUT3 transporter. This use case again demonstrates that phylogenetically unrelated transporters (since OCTs belong to SLC22A and GLUTs belong to SLC2A subfamily) can share the same fold.
Table 4.
List of available ligand- and structure-based molecular modeling studies on uptake transporters discussed in this review.
| Paper | Transporter | Method | Description | Results |
|---|---|---|---|---|
| Tanihara et al.[132] | MATE1 | Binary classification modeling | Identification of key features for inhibitory mechanism | Cationic charge is crucial for MATE1 inhibition. |
| Diao et al. [25] | MATE1 | Bayesian machine learning modeling | Identification of key features for inhibitory mechanism | Six-membered rings including nitrogen are important for MATE1 inhibition. |
| Astorga et al. [5] | MATE1 | IVIS pharmacophore modeling | Iterative identification of new MATE1 inhibitors by using pharmacophore-based virtual screening | Two hydrophobic features, H-bond acceptor and cationic feature have occurred in final pharmacophore model for MATE1. |
| Zhang et al. [130] | MATE1 | Structural model building, molecular dynamics simulation | Topology of human MATE1 transporter, stability of constructed structural model | Human MATE1 transporter consists of 12 TM which have a functional role; 13th TM is not required for the transport. |
| Wittwer et al. [122] | MATE1 | Binary classification modeling | Identification of key features for inhibitory mechanism | Cationic charge, molecular weight, and lipophilicity are important features for MATE1 inhibition. |
| Xu et al. [126] | MATE1 | Combinatorial pharmacophore modeling | Studying multiple inhibitory mechanisms of MATE1 inhibitors | Four different binding sites (two competitive, one non-competitive and one mixed inhibition binding site) were identified for MATE1. |
| Xu et al. [126] | MATE1 | Structural model building, molecular docking | Elucidate the evidence of multiple binding sites from combinatorial pharmacophore model | Four different binding sites (two competitive, one non-competitive and one mixed inhibition binding site) were identified for MATE1. |
| Astorga et al. [5] | MATE2-K | IVIS pharmacophore modeling | Iterative identification of new MATE2-K inhibitors by using pharmacophore-based virtual screening | Two hydrophobic features, H-bond acceptor and cationic feature have occurred in final pharmacophore model. |
| Perry et al. [93] | OAT1 | Structural model building | Identification of critical residues important for OAT1 transport function | Importance of aromatic amino acid at at position 230 has been discovered. |
| Truong et al. [133] | OAT1, OAT3 | QSAR modeling | Comparison of interactions of antiviral drugs with OAT1 versus OAT3 | Number of H-bond donors (alcohols and amides) and total polar surface area have triggered a preferred inhibitory activity towards OAT1. |
| Tsigelny et al. [112] | OAT1 | Molecular dynamics simulation | Investigations of dynamics events accompanying OAT1 transport | Tilting mechanism of two hemi-domains is crucial for the initialization of transport process. |
| Soars et al. [134] | OAT1, OAT3 | QSAR modeling | Comparison of inhibitor features between OAT1 and OAT3 | OAT1 and OAT3 inhibitors have statistically significant inhibitory profiles. |
| Bednarczyk et al. [135] | OCT1 | LB pharmacophore modeling | Identification of important pharmacophoric features for OCT1 inhibition | Three hydrophobic and one positive ionizable feature are important features for OCT1 inhibition. |
| Moaddel et al. [79] | OCT1 | LB pharmacophore modeling | Studying stereroselective recognition of OCT1 transporters | One positive ionizable feature, one hydrophobic, and two H-bond acceptor features are important for OCT1 inhibition. |
| Ahlin et al. [1] | OCT1 | QSAR modeling | Identification of molecular features being important for inhibitory activity | H-bond donors, lipophilicity, cationic charge positively correlate with OCT1 inhibition. |
| Badolo et al. [6] | OCT1 | QSAR modeling | Identification of molecular features being important for inhibitory activity | Topological polar surface area negatively correlates with OCT1 inhibition. |
| Shaikh et al. [104] | OCT1, OATP1B1, OATP1B3, OATP2B1 | QSAR/PCM modeling, substructural analysis | Identification of important molecular features and structural fragments for substrate activity against reported transporters | Developed models were used for prediction of substrate propensity for blood-brain barrier transporters. |
| Dakal et al. [22] | OCT1, OCT2, OCT3, OCTN1, OCTN2 | Structural model building | Multiscale structural models construction for OCT transporters | Constructed structural models for OCTs share close structural similarity with GLUT3 transporter (pdb id: 5c65). |
| Chen et al. [19] | OCT1 | Structural model building, molecular docking | Identification of critical residues important for OCT1 activity; virtual screening for sake of detecting new OCT1 inhibitors | D474 is important for ligand binding; detection of two distinct binding sites in translocation channel. |
| Boxberger et al. [13] | OCT1 | Structural model building, molecular docking | Identification of critical residues important for OCT1 activity | Identification of three distinct binding sites based on the presence of critical residues (W218, Y222, T226, I443, I447, Q475). |
| Kido et al. [58] | OCT2 | QSAR modeling | Identiffication of molecular determinants for OCT2 inhibitors | Suggestion of multiple binding sites for OCT2 transporter. |
| Suhre et al. [136] | OCT2 | 2D-QSAR modeling, Comparative Molecular Field Analysis (CoMFA) | Identification of molecular determinants of OCT2 substrates and inhibitors | Hydrophobicity, steric factor, and number of rotatable bonds were identified as important features for OCT2 inhibition. |
| Wittwer et al. [122] | OCT2 | QSAR modeling | Identification of molecular determinants of OCT2 inhibitors | Occurrence of both zwitterionic and basic functional groups is important for OCT2 inhibition. |
| Xu et al. [125] | OCT2 | Combinatorial pharmacophore modeling | Studying multiple inhibitory mechanism of OCT2 inhibitors | Four distinct pharmacophore hypotheses, corresponding to the competitive inhibition (one hypothesis), non-competitive inhibition by occlusion (two hypotheses), and one mixed inhibition pattern, have been identified for OCT2 inhibitors. |
| Diao et al. [26] | OCTN2 | IVIS pharmacophore modeling | Iterative identification of new OCTN2 inhibitors by using pharmacophore-based virtual screening | Three hydrophobic and one positive ionizable feature are important for OCTN2 inhibition. |
| Diao et al. [25] | OCTN2 | IVIS pharmacophore modeling, Bayesian modeling | Iterative identification of new OCTN2 inhibitors by using pharmacophore-based virtual screening | Two hydrophobic features, one H-bond donor, and positive ionizable feature are important for OCTN2 inhibitors; aromatic and tertiary-amine groups have also been detected via Bayesian modeling. |
| Mandery et al. [73] | OATP1A2, OATP1B3, OATP2B1 | Structural models construction | Comparison of structural determinants for ligand activity among OATP family | K361 and K399 are highly conserved residues across OATP family; K361 is pointing towards the translocation pore; variable loop located within a translocation pore differs in terms of crucial residues for respective targets (R58 and S62 in OATP1B3, Q58 and P62 in OATP1A2, and S64 in OATP2B1). |
| Chang [17] | OATP1B1 | LB pharmacophore modeling | Detection of pharmacophoric features for OATP1B1 substrates | Two H-bond acceptors and two or three hydrophobic features are important for OATP1B1 substrates. |
| Badolo et al. [6] | OATP1B1, OATP1B3 | QSAR modeling | Identification of molecular features being important for OATP1B1/1B3 inhibition | Lipophilicity, polarity, lower base pKa, higher number of H-bond acceptors, and higher molecular weight correlate with OATP1B inhibition. |
| Soars et al. [105] | OATP1B1 | QSAR modeling | Identification of molecular features being important for OATP1B1 inhibition | Low number of aromatic bonds (<7), lipophilicity, and hydrogen-bonding potential are important for OATP1B1 inhibition. |
| Karlgren et al. [54] | OATP1B1 | QSAR modeling | Virtual screening for detecting new OATP1B1 inhibitors | Lipophilicity, larger molecular weight, larger polar surface area |
| Karlgren et al. [55] | OATP1B1, OATP1B3, OATP2B1 | QSAR modeling | Comparison of molecular determinants for ligand activity among hepatic OATPs | Lipophilicity and polar surface area are general features for OATP inhibition; OATP2B1 inhibitors are less dependent on polarity than OATP1B1/1B3 inhibitors. |
| Van de Steeg et al. [114] | OATP1B1 | Bayesian modeling | Identification of molecular features being important for OATP1B1 inhibition | Conjugated-bond systems, (hetero)cycles with acceptor/donor atoms inside or outside the rings, molecular weight, molecular surface area, lipophilicity, number of rings, number of rotatable bonds, number of H-bond acceptors are important for OATP1B1 inhibition. |
| Bruyn et al. [14] | OATP1B1, OATP1B3 | PCM modeling | Comparison of molecular determinants for ligand activity for OATP1B1 and OATP1B3 transporter | Lipophilicity, absence of cationic charge, number of ringbonds, presence of an anionic functional group, molecular volume, and substantial number of H-bond acceptors are important for general OATP1B inhibition; low number of aromatic bonds correlates with OATP1B1 inhibition, whereas higher lipophilicity and moderate number of H-bond donors corresponds with OATP1B3 inhibition. |
| Kotsampasakou et al. [60] | OATP1B1, OATP1B3 | QSAR modeling | Comparison of molecular determinants for ligand activity for OATP1B1 and OATP1B3 transporters; virtual screening to search for new OATP1B ligands | Number of H-bond donors and acceptors, LogP, molecular refractivity, topological surface area, molecular weight, number of rotatable bonds, topological radius, topological diameter, topological shape, global topological charge index, have been used to develop models for OATP1B1 and OATP1B3. |
| Türkova et al. [113] | OATP1B1, OATP1B3, OATP2B1 | Substructural analysis, QSAR modeling | Comparison of molecular determinants for ligand activity among hepatic OATPs | Lipophilicity, molecular weight, number of atoms, molecular refractivity, and flexibility are important features for general OATP inhibition; OATP2B1 inhibitors tend to be more planar than OATP1B1/1B3 inhibitors. |
| Li et al. [69] | OATP1B1 | Structural model construction, molecular docking | Exploring the importance of selected amino acids from TM2 on the uptake of Estrone-3-sulphate | D70 and F73 are involved in the interaction with substrates; two distinct binding sites (low- and high- affinity site) for Estrone-3-sulphate have been identified. |
| Hong et al. [137] | OATP1B1 | Structural model construction, molecular docking | Exploring the importance of selected amino acids from TM11 on the uptake of prototypic substrates | Importance of negative charge at position 596 for OATP1B1 uptake. |
| Glaeser et al., [36] | OATP1B3 | Structural model construction,molecular docking | Identification of important amino acids on OATP1B3 transport function | Importance of positive charge at position 41, importance of R580 residue on OATP1B3 transport. |
| Meier-Abt et al. [77] | OATP1B3, OATP2B1 | Structural model construction, molecular docking | Comparison of important amino acids on OATP1B3 and OATP2B1 transport function | R181 might contribute to the OATP1B substrate specificity, while H579 is hypothesized to be crucial for OATP2B family; conservation of H-bonds patterns, as well as helix-breaking residues (proline and glycine patterns), have also been detected. |
| Gui and Hagenbuch [38] | OATP1B1, OATP1B3 | Structural model construction, molecular docking | Comparison of important amino acids on OATP1B1 and OATP2B1 transport function | TM10 is pronounced to drive the differences between OATP1B1 and OATP1B3. |
| Khuri et al. [57] | OATP2B1 | Structural model construction, molecular docking, QSAR modeling | Identification of molecular features being important for OATP2B1 inhibition; virtual screening for new OATP2B1 inhibitors | OATP2B1 inhibitors are lipophilic. |
Two very promising ways of integrating ligand and protein information are so-called combinatorial pharmacophores as well as PCM modeling approaches. Combinatorial pharmacophore modeling was first reported for OCT2 inhibitors in 2013 [125]. In general, this approach represents a multi-step combinatorial scheme to generate a set of diverse LB pharmacophore models including the available information about their binding modes: First, generated pharmacophore models with identical pharmacophoric features in a close spatial arrangement are grouped in order to reduce the large pool of potential hypotheses and a combinatorial approach is employed to test all possible combinations of different pharmacophore hypotheses. The main idea behind the combinatorial pharmacophores is to study how different pharmacophoric patterns are corresponding to (potential) multiple binding mode hypotheses of uptake transporters. For this purpose, different sub-categories of reference inhibitors are being used - (1) competitive inhibitors (i.e., binding to orthosteric binding site), (2) occluding inhibitors (i.e., noncompetitive inhibitors, occluding the substrate binding site and locking the conformation transformation of the tarnsporter), and (3) allosteric inhibitors (i.e., modulating the transporter's function by binding to the different – allosteric – binding site). A use case on MATE1-OCT2 selectivity profiling is presented in chapter 5.
Proteochemometric modeling (PCM) is conceptualized as an advanced extension to the conventional QSAR-based modeling by simultaneous considerations of the similarity between multiple ligands and multiple targets [115]. PCM modeling can thus be categorized as a method at the interface between ligand- and structure-based modeling. The two-dimensional structural sequence information can be integrated into the PCM model either as a whole amino acid sequence, or the pre-selection of key residues (e.g. those occurring in the binding pocket and/or other conserved residues) can be performed. ([64,93]; [138]). Protein sequences can then be reduced to a more abstract representation by calculating the Z-scales which are corresponding to the principal components of multi-property matrices combining different physico-chemical properties, such as lipophilicity, volume, and polarity for respective residues [64,115]. In case of uptake transporters discussed herein, PCM modeling was used for the investigations of structural determinants between OATP1B1 and OATP1B3 [14], as well as in a recent study by Shaikh et al. for investigating transporter substrates of OCT1, OATP1A2, OATP1B1, OATP1B3, and OATP2B1 [104].
5. Selectivity Profiling: Linking Knowledge of Related Uptake Transporters
Since uptake transporters are often co-expressed at pharmacological barriers and generally transport a wide variety of pharmaceutical agents, it is of medical interest to increase the understanding of the interplay of such related transporters. A prominent example are hepatic OATPs – OATP1B1, OATP1B3, and OATP2B1 – which are responsible for e.g. bile acids uptake (such as taurocholic acid), but also pharmaceuticals, hormones etc. into hepatocytes ([139], [140]). It is only insufficiently understood to date, how ligand activity and selectivity towards one of the three transporters is achieved. Such knowledge could not only pave the way for functional studies on these transporters (by the use of truly selective tool compounds), but would also increase our knowledge on critical compound/drug properties associated with the onset of clinically relevant drug-drug interactions.
On principle, studies on determining factors for selectivity can include knowledge from the ligand side (QSAR/classification modeling, pharmacophore modeling), the protein side (comparative modeling, molecular docking, virtual screening, MD simulations), or both (PCM modeling or combinations of the latter approaches).
In case of OATP1B1, OATP1B3, and OATP2B1, comparative QSAR modeling has been performed by Karlgren et al. already in 2012 [55] which identified important molecular features for general OATP inhibition (vs. non-inhibition): higher lipophilicity, molecular weight and polarity. Just recently our group identified additional features discriminating hepatic OATP inhibitors from non-inhibitors: higher polarizability, molecular refractivity (corresponding to the distribution of charge over a molecule's surface), and flexibility (expressed as a higher number of rotatable bonds) [146]. Development of in silico models for individual OATP transporters by Karlgren et al. [55] has identified certain differences between the OATP1B and OATP2B subfamily. In contrast to OATP1B1 and OATP1B3, OATP2B1 inhibitory activity has been negatively correlated with nonpolar- and total- surface area, proposing that OATP2B1 inhibitors might be less dependent on polarity than OATP1B1 and OATP1B3 inhibitors. In addition, in our current study we could highlight additional properties to be responsible for OATP1B1 and OATP1B3 versus OATP2B1 inhibition (the latter seem to be more planar, whereas OATP1B members tend to possess a large number of amide bonds) [113].
Another way to explore ligand (and potentially selectivity) profiles is an enrichment analysis in substructures among actives of one target of interest vs the other(s). Again, for hepatic OATPs, this methodology led to a list of enriched scaffolds possessing a certain activity profile (i.e.,OATP1B1 selective inhibition, OATP1B1/OATP1B3 dual inhibition, OATP1B1/OATP1B3/OATP2B1 pan-inhibition). As an outcome, e.g. the pravastatin-like scaffold showed a preferential inhibitory activity for OATP1B1 (over OATP1B3 and OATP2B1) and the cyclosporine-like scaffold accounted for OATP1B1/OATP1B3 dual inhibition. Interestingly, the steroidal scaffold has been found to be enriched in the actives of all three hepatic OATPs. Here depending on the side-chain variations, preferred activity towards one of the targets might be achieved [113].
Pharmacophore modeling is also interesting for studying ligand selectivity across different species. To give an example, human- and rabbit-OCT2 pharmacophore models indicate that despite the similarity of most of the pharmacophoric features (reflected by 83% sequence identity of these two OCT2 variants), there is a difference in the spatial arrangement of hydrogen bonding features [109].
Finally, even ligand profiles and selectivity among uptake transporters of different families might be of interest, in particular if they are commonly expressed at the same pharmacological barrier. A way to tackle this is comparing pharmacophore hypotheses generated for the two targets of interest, like in the case of OCT2 and MATE1, which both are playing a significant role in renal disposition and toxicity (König et al. [141). It has been shown that charge distribution was one of the important factors, favoring the inhibitory activity of one transporter with respect to the other. Specifically, OCT2 inhibitors comprise both zwitterionic and basic functional groups, whereas MATE1 inhibitors are less enriched with basic groups and do not necessarily contain zwitterionic groups [122].
An even more comprehensive understanding of OCT2-MATE1 selectivity profiling was delivered by a combinatorial pharmacophore-based approach [125,126], as introduced in chapter 4. Since MATE1 and OCT2 are commonly expressed in the kidney, it is interesting to learn about their interplay and selectivity switches to better understand transporter-mediated drug distribution and drug elimination processes (König et al., 2011). A combinatorial pharmacophore model approach developed for both OCT2 [125] and MATE1 [126] can therefore reveal which features are shared and which ones are unique for just one of these two transporters. The latter can give hints for selectivity switches at the ligand level. For OCT2, combinatorial pharmacophore modeling has revealed four distinct pharmacophoric hypotheses [125]. An aromatic feature was included in all four hypotheses, suggesting the essential role of pi-pi interactions in the OCT2-ligand recognition. In addition, a cationic charge has appeared in three out of four pharmacophoric hypotheses, which corresponds to previous findings [122]. Xu et al. also compared the molecular weights for the inhibitors matching different pharmacophoric features which provided additional insights into the constitution and/or size of distinctive binding site(s) within the transporter. Following the same strategy as for OCT2, the authors studied multiple inhibitory mechanisms of MATE1 ligands in a follow-up paper [126]. The model reveals significant importance of aromatic rings, as well as hydrophobicity to induce MATE1 inhibition. When compared to the combinatorial model for OCT2 inhibition, it becomes obvious that one of the hypotheses was the same in both transporters, thus proposing one common binding mode hypothesis which can accommodate a substantial number of dual MATE1 and OCT2 inhibitors.
In the future more sophisticated methods, such as multi-label classification might come into play, depending on the availability of compound data with consistent bioactivity measurements for targets under study. Such methods were recently used for studying selectivity profiles of ABC transporters [81].
In addition to the above discussed LB approaches for studying selectivity, the molecular basis for selectivity is delivered by the protein structure, and more specifically by subtle differences in residues interacting with the ligand during binding and transport. To give an illustrative example, attempts to understand selectivity among hepatic OATPs at a molecular level are discussed. Transmembrane regions for both OATP1B3 and OATP2B1 were built on basis of templates from the MFS family, namely glycerol-3-phosphate (pdb id: 1pw4) and lactose permease (pdb id: 1pv6) from Escherichia coli [77]. The model quality has subsequently been validated via docking of the cardiac glycoside digoxin into the putative translocation channel. Based on the 3D models and multiple sequence alignment across OATP family members, the analyses suggest that the pore-facing residue R181 might contribute to the substrate specificity of OATP1B transporters, as this residue is fully conserved across the OATP1B family. In analogy, H579 is hypothesized to be crucial for binding of ligands to members of the OATP2B family and it is found at a spatially adjacent position to R181 [77]. Another SB modeling effort for understanding commonalities and differences of the more closely related hepatic transporters OATP1B1 and OATP1B3 (~80% sequence identity) led to the construction of a series of chimeric proteins between OATP1B3 and 1B1 [38]. The aim here was the determination of structural domains and/or residues responsible for substrate selectivity of OATP1B3, specifically for CCK-8. Homology modeling and molecular docking led to binding mode hypotheses which were further validated experimentally. When replacing TM10 in OATP1B3 with TM10 of OATP1B1 a dramatically reduced degree of CCK-8 transport was observed, indicating that TM10 is indeed crucial for recognition and/or translocation of CCK-8. Using site-directed mutagenesis, key residues for substrate binding namely, Y537, S545, and T550 in TM10 were identified [38].
Using ligand and protein information in conjunction for selectivity profiling can be conducted by using PCM modeling. This technique outperforms conventional QSAR models and can be used to virtually screen for selective compounds that are solely active on a single member of a subfamily of targets [115]. In the field of clinically relevant SLC transporters, PCM modeling was first undertaken for investigating chemical features favoring OATP1B1 and OATP1B3 inhibition [14]. Performing multiple sequence alignment for OATP1B1, OATP1B3, OATP2B1, and OATP1A2 aided in identifying most conserved regions. Further, critical protein residues were prioritized on basis of SB modeling studies previously done for hepatic OATPs [73,77]. This step again demonstrates the usefulness of combining different computational approaches. PCM models were developed for OATP1B inhibition (2-class classification model, i.e., ‘OATP1B inhibitor’ or ‘OATP1B non-inhibitor’), and individual OATP1B1/1B3 inhibition (4-class classification model, i.e., ‘OATP1B dual inhibitor’, ‘OATP1B dual non-inhibitor’, ‘OATP1B1 selective inhibitor’, or ‘OATP1B3 selective inhibitor’). When looking at protein properties, only limited conclusions could be deduced from this study. The limited interpretability of target information is caused by the fact that only two proteins were included into the PCM modeling (van Westen et al., 2012). As a future perspective, the authors suggest to apply the PCM procedure to more OATP proteins, as there are bioactivity measurements for 22 OATP isoforms available in CHEMBL. In conclusion, the developed 2- and 4-class classification models united the important molecular features reported from previous computational studies [6,17,55,105], but also provided new information about OATP1B1/1B3 inhibition (e.g. high number of ring bonds).
Pharmacological profiles can also be investigated for a whole group of simultaneously expressed transporters located at the same pharmacological barrier. For example, Shaikh et al. combined QSAR and PCM modeling to perform an extensive exploration of substrate interactions for 13 clinically relevant efflux and uptake transporters, including OCT1, OATP1B1, OATP1B3, OATP2B1, and OATP1A2 [104]. The motivation behind such an extensive study is to predict transport across major pharmacological barriers as a whole by a sequence of computational models. Thus, consensus models by applying various machine learning techniques were finally constructed and the developed models were used to predict the substrate propensity of compounds for blood-brain barrier (BBB) transport.
6. Accounting for Transporter Flexibility
Since substrate translocation via transporters requires the protein to adapt different conformational states, protein flexibility has to be taken into account also when performing SB modeling [30]. So far, most of the docking studies on uptake transporters were performed by considering only a single transporter conformation (e.g., OAT1 [93], OCT1 [13], OATP1B1 [69], OATP1B3 [38]). However, building a structural model on basis of a single conformation can inherently bias subsequent docking screens since e.g. some ligands might not fit into a narrow binding pocket of a particular conformation. Khuri et al. attempted to capture OATP2B1 transporter dynamics by building multiple OATP2B1 comparative models based on seven templates considering different conformational states: inward-open, outward-open, and occluded [57]. In general, multiple comparative models can together provide a more comprehensive representation of a ligand binding event [100]. However, combining drastically different conformational states (such as outward-open with inward) is risky, since allosteric modulations rather happen upon subtle changes in protein conformation and slight rotameric variations of side chains [96].
An orthogonal approach to the selection of multiple template structures for sampling protein conformations is the generation of template protein ensembles by using MD simulations. For lactose permease (pdb code: 1pv6), a representative of the Major Facilitator Superfamily which to a high degree relates to uptake transporters by secondary structure, both all-atom [42,91] and coarse-grained [50] simulations have been performed for elucidating ligand binding and even transport events. However, direct usage of MD simulations to mimic the translocation process of template-based comparative models is only rarely used since the majority of available template structures cover only transmembrane regions. For example, available templates for OATP1B1 cover <400 amino acids out of 691 in total. Running MD simulations with an incomplete target structure could lead to artifacts. Furthermore, the limited time scale for all-atom simulations (hundreds of nanoseconds to a few microseconds), as well as the choice of including or omitting the lipid bilayer into the simulated system, can heavily affect the correctness and interpretability of the MD simulations. Contemporary simulation techniques, such as stochastic Monte Carlo, or MD simulations with enhanced sampling (e.g. by applying replica-exchange methods) can be used for structural refinement of extra- and intracellular domains.
To date, enhanced sampling techniques have been used only once for building a complete structural model for any of the discussed uptake transporters. As shown in a MD simulation study on OAT1 [112], modeling of the complete OAT1 transporter structure was divided into two stages: First, only the transmembrane region was modeled based on a template with high secondary structure similarity (glycerol-3-phosphate transporter; pdb code: 1pw4). At the second step, the extracellular domain was iteratively sampled by simulated annealing, while the transmembrane region was kept restrained. Results of this study indicate that the structural refinement by applying enhanced sampling methods could significantly improve existing structural models for uptake transporters which in turn would enhance the understanding of functional aspects of the transport mechanisms.
Only recently, reduced representation methods, such as normal mode analysis by applying elastic network models, can be used to overcome shortcomings arising from the high computational demands of conventional MD simulations ([[142], [143]]). For example, elastic network models have been used for the structurally-related fucose transporter (pdb code: 3o7q) to study its molecular basis for allosteric modulations [18]. Furthermore, by building comparative models in different functional states (inward- and outward-open conformation), elastic network models were capable to reproduce the whole translocation pathway of this transporter.
For multiple ensemble docking, normal mode simulations have been shown to be particularly useful to e.g. detect a biologically relevant conformation of dopamine D3 receptor, which has subsequently been prospectively validated by the existing dopamine D3 crystal structure [15]. One might argue that the generation of multiple conformations for a template structure might inherently bias the construction of structural models, since they can possess its internal dynamics which probably cannot be completely captured by the template. However, as shown and discussed in case of the human MATE1 homology model [130], the overall conformational stability between the template and the derived MATE1 homology models remained unchanged.
As a conclusion, the use of normal mode simulations in structure-based modeling studies can potentially improve conformational sampling when modeling uptake transporters. This in turn can lead to more accurate docking poses with the aim to better understand ligand-protein binding events and potentially selectivity switches.
7. Summary, Conclusions & Future Perspectives
ADMET-relatedSLC transporters are proteins of emerging interest in the framework of preclinical drug design. As demonstrated herein - by collecting available ligand and protein information from the open domain – data sparseness resulted in quite limited understanding of these transporter to date. Other factors complicating effective exploration of this class of proteins is their promiscuous nature, with potentially multiple binding sites, as well as overlapping substrate- and inhibitor profiles.
As demonstrated by discussed examples of molecular modeling and data analysis herein, new emerging technologies are on the rise also for these targets being particularly hard to unlock. Especially, data integration techniques and data analysis can lead to useful hypothesis about interesting SAR series at the beginning of an in silico study. Further, combining LB and SB methods seems to be an effective strategy, especially when it comes to selectivity profiling (like in the case of PCM modeling), or the exploration of knowledge about multiple binding sites (like in the case of combinatorial pharmacophores). In general, inclusion of in vitro experiments is a must especially for SB methods, e.g. to test the established binding mode hypotheses. In return, those in vitro measurements will lead to an increase in data points for a particular target, which can further be explored by statistical methods (such as machine learning approaches). For SB approaches, it would be interesting to include more systematically conformational sampling of protein conformations and multiple template structures into the comparative modeling step. It will be interesting to then compare results to those from docking into a single static template.
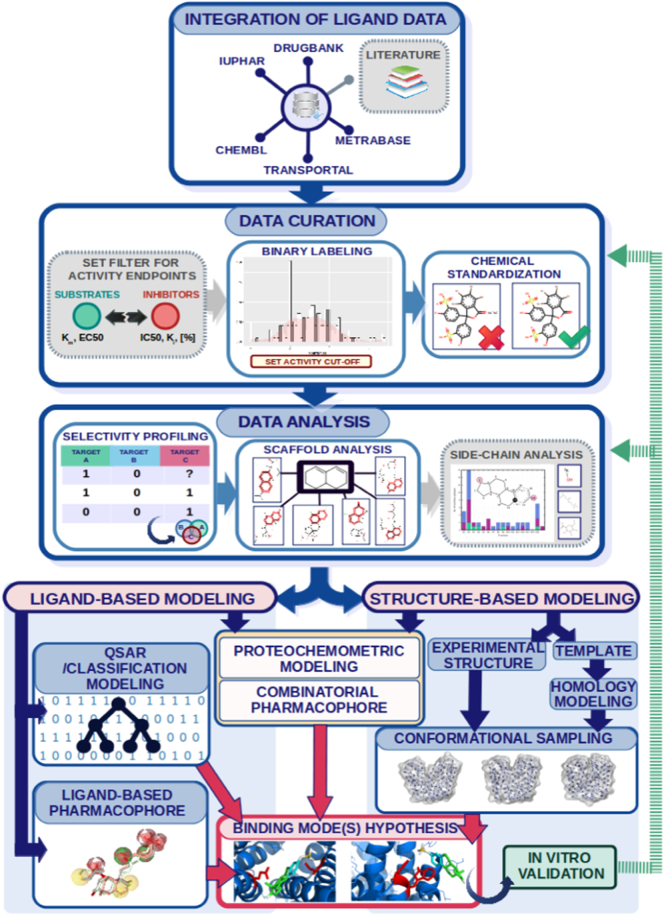
We are proposing a general workflow for in silico modeling of clinically relevant SLC transporters (see Fig. 3) which makes use of all available molecular modeling approaches and combines them with timely data science approaches (as far as the available data allows the different methods).
Fig. 3.
Proposed computational workflow for studying ligand interactions with ADMET-relevant SLC transporters. Results from in vitro validation can be inputted to the stages of data curation and data analysis and subsequently be used for a new iteration of modeling.
With the aim to develop safer medicine, it is of extraordinary importance to increase our understanding of the molecular basis of respective transporter-ligand interactions on an individual level (transporter by transporter), but also from a global point of view (how they act in concert). During evolution, transporters were optimized to help metabolizing chemical matter that we were exposed to. Obviously, they found a way to transport a wide variety of chemically distinct compounds. Understanding the interplay of clinically relevant transporters in transporting chemical matter, as well as the mechanisms underlying transporter selectivity, can help to unravel potential drug-drug or drug-food interactions which will finally lead to safer drugs in the future.
Acknowledgements
We acknowledge financial support provided from the Austrian Science Fund (FWF), grant no. P 29712.
References
- 1.Ahlin G., Karlsson J., Pedersen J.M., Gustavsson L., Larsson R., Matsson P. Structural requirements for drug inhibition of the liver specific human organic cation transport protein 1. J Med Chem. 2008;51:5932–5942. doi: 10.1021/jm8003152. [DOI] [PubMed] [Google Scholar]
- 2.Ain Q.U., Aleksandrova A., Roessler F.D., Ballester P.J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdisc Rev. 2015;5:405–424. doi: 10.1002/wcms.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldeghi M., Heifetz A., Bodkin M.J., Knapp S., Biggin P.C. Predictions of ligand selectivity from absolute binding free energy calculations. J Am Chem Soc. 2017;139:946–957. doi: 10.1021/jacs.6b11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angelini S., Pantaleo M.A., Ravegnini G., Zenesini C., Cavrini G., Nannini M. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol Res. 2013;68(1):1–6. doi: 10.1016/j.phrs.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Astorga B., Ekins S., Morales M., Wright S.H. Molecular determinants of ligand selectivity for the human multidrug and toxin extruder proteins MATE1 and MATE2-K. J Pharmacol Exp Ther. 2012;341:743–755. doi: 10.1124/jpet.112.191577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badolo L., Rasmussen L.M., Hansen H.R., Sveigaard C. Screening of OATP1B1/3 and OCT1 inhibitors in cryopreserved hepatocytes in suspension. Eur J Pharm Sci. 2010;40:282–288. doi: 10.1016/j.ejps.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Bailey D.G., Dresser G.K., Leake B.F., Kim R.B. Naringin is a major and selective clinical inhibitor of organic anion-transporting polypeptide 1A2 (OATP1A2) in grapefruit juice. Clin Pharmacol Ther. 2007;81:495–502. doi: 10.1038/sj.clpt.6100104. [DOI] [PubMed] [Google Scholar]
- 8.Ballante F. Protein-ligand docking in drug design: Performance assessment andbinding-pose selection. In: Mavromoustakos T., Kellici T.F., editors. Rational drug design: methods and protocols. Methods in Molecular Biology. Springer New York; New York, NY: 2018. pp. 67–88. [DOI] [PubMed] [Google Scholar]
- 9.Baumgartner M.P., Camacho C.J. Choosing the optimal rigid receptor for docking and scoring in the CSAR 2013/2014 experiment. J Chem Inf Model. 2016;56:1004–1012. doi: 10.1021/acs.jcim.5b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T. KNIME - the Konstanz information miner: version 2.0 and beyond. SIGKDD Explor Newsl. 2009;11:26–31. [Google Scholar]
- 12.Böhm H.-J. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J Comput Aided Mol Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 13.Boxberger K.H., Hagenbuch B., Lampe J.N. Ligand-dependent modulation of hOCT1 transport reveals discrete ligand binding sites within the substrate translocation channel. Biochem Pharmacol. 2018;156:371–384. doi: 10.1016/j.bcp.2018.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruyn T.D., Westen G.J.P.V., IJzerman A.P., Stieger B., Witte de P., Augustijns P.F. Structure-based identification of OATP1B1/3 inhibitors. Mol Pharmacol Mol. 2013 doi: 10.1124/mol.112.084152. 112.084152. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson J., Coleman R.G., Setola V., Irwin J.J., Fan H., Schlessinger A. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat Chem Biol. 2011;7:769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha S.H., Sekine T., Fukushima J.I., Kanai Y., Kobayashi Y., Goya T. Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol Pharmacol. 2001;59:1277–1286. doi: 10.1124/mol.59.5.1277. [DOI] [PubMed] [Google Scholar]
- 17.Chang C. Comparative pharmacophore modeling of organic anion transporting polypeptides: a meta-analysis of rat Oatp1a1 and human OATP1B1. J Pharmacol Exp Ther. 2005;314:533–541. doi: 10.1124/jpet.104.082370. [DOI] [PubMed] [Google Scholar]
- 18.Chang S., Li K., Hu J., Jiao X., Tian X. Allosteric and transport behavior analyses of a fucose transporter with network models. Soft Matter. 2011;7:4661–4671. [Google Scholar]
- 19.Chen E.C., Khuri N., Liang X., Stecula A., Chien H.-C., Yee S.W. Discovery of competitive and noncompetitive ligands of the organic cation transporter 1 (OCT1; SLC22A1) J Med Chem. 2017;60:2685–2696. doi: 10.1021/acs.jmedchem.6b01317. [DOI] [PubMed] [Google Scholar]
- 20.Colas C., Ung P.M.-U., Schlessinger A. SLC transporters: structure, function, and drug discovery. Medchemcomm. 2016;7:1069–1081. doi: 10.1039/C6MD00005C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dakal T.C., Kumar R., Ramotar D. Structural modeling of human organic cation transporters. Comput Biol Chem. 2017;68:153–163. doi: 10.1016/j.compbiolchem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Dakal T.C., Kala D., Dhiman G., Yadav V., Krokhotin A., Dokholyan N.V. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms in IL8 gene. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-06575-4. 6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dastmalchi S., Hamzeh-Mivehroud M., Sokouti B., Hamzeh-Mivehroud M., Sokouti B. CRC Press; 2018. Quantitative structure – activity relationship : a practical approach. [Google Scholar]
- 24.Deng N., Flynn W.F., Xia J., Vijayan R.S.K., Zhang B., He P. Large scale free energy calculations for blind predictions of protein–ligand binding: the D3R grand challenge 2015. J Comput Aided Mol Des. 2016;30:743–751. doi: 10.1007/s10822-016-9952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diao L., Ekins S., Polli J.E. Quantitative structure activity relationship for inhibition of human organic cation/carnitine transporter. Mol Pharm. 2010;7:2120–2131. doi: 10.1021/mp100226q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diao L., Ekins S., Polli J.E. Novel inhibitors of human organic cation/carnitine transporter (hOCTN2) via computational modeling and in vitro testing. Pharm Res. 2009;26:1890–1900. doi: 10.1007/s11095-009-9905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drenberg C.D., Gibson A.A., Pounds S.B., Shi L., Rhinehart D.P., Li L. OCTN1 is a high-affinity carrier of nucleoside analogs. Cancer Res. 2017;77(8):2102–2111. doi: 10.1158/0008-5472.CAN-16-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dresser G.K., Bailey D.G., Leake B.F., Schwarz U.I., Dawson P.A., Freeman D.J. Fruit juices inhibit organic anion transporting polypeptide-mediated drug uptake to decrease the oral availability of fexofenadine. Clin Pharmacol Ther. 2002;71:11–20. doi: 10.1067/mcp.2002.121152. [DOI] [PubMed] [Google Scholar]
- 29.Enomoto A., Takeda M., Shimoda M., Narikawa S., Kobayashi Yukari, Kobayashi Yasuna. Interaction of human organic anion transporters 2 and 4 with organic anion transport inhibitors. J Pharmacol Exp Ther. 2002;301:797–802. doi: 10.1124/jpet.301.3.797. [DOI] [PubMed] [Google Scholar]
- 30.Erickson J.A., Jalaie M., Robertson D.H., Lewis R.A., Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47:45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- 31.Feig M. Implicit membrane models for membrane protein simulation. Methods Mol Biol. 2008;443:181–196. doi: 10.1007/978-1-59745-177-2_10. [DOI] [PubMed] [Google Scholar]
- 32.Fiser A., Do R.K., Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiser A., Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 34.Flohil J.A., Vriend G., Berendsen H.J.C. Completion and refinement of 3-D homology models with restricted molecular dynamics: application to targets 47, 58, and 111 in the CASP modeling competition and posterior analysis. Proteins. 2002;48:593–604. doi: 10.1002/prot.10105. [DOI] [PubMed] [Google Scholar]
- 35.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discovery. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glaeser H., Mandery K., Sticht H., Fromm M.F., König J. Relevance of conserved lysine and arginine residues in transmembrane helices for the transport activity of organic anion transporting polypeptide 1B3. Br J Pharmacol. 2010;159:698–708. doi: 10.1111/j.1476-5381.2009.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorboulev V., Ulzheimer J.C., Akhoundova A., Ulzheimer-Teuber I., Karbach U., Quester S. Cloning and characterization of two human polyspecific organic cation transporters. DNA Cell Biol. 1997;16:871–881. doi: 10.1089/dna.1997.16.871. [DOI] [PubMed] [Google Scholar]
- 38.Gui C., Hagenbuch B. Amino acid residues in transmembrane domain 10 of organic anion transporting polypeptide 1B3 are critical for cholecystokinin Octapeptide transport †. Biochemistry. 2008;47:9090–9097. doi: 10.1021/bi8008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardin C., Pogorelov T.V., Luthey-Schulten Z. Ab initio protein structure prediction. Curr Opin Struct Biol. 2002;12:176–181. doi: 10.1016/s0959-440x(02)00306-8. [DOI] [PubMed] [Google Scholar]
- 40.Heim A.J., Li Z. Developing a high-quality scoring function for membrane protein structures based on specific inter-residue interactions. J Comput Aided Mol Des. 2012;26:301–309. doi: 10.1007/s10822-012-9556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirano M., Maeda K., Shitara Y., Sugiyama Y. Contribution of OATP2 (OATP1B1) and OATP8 (OATP1B3) to the hepatic uptake of pitavastatin in humans. J Pharmacol Exp Ther. 2004;311:139–146. doi: 10.1124/jpet.104.068056. [DOI] [PubMed] [Google Scholar]
- 42.Holyoake J., Sansom M.S.P. Conformational change in an MFS protein: MD simulations of LacY. Structure. 2007;15:873–884. doi: 10.1016/j.str.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Hoshino Y., Fujita D., Nakanishi T., Tamai I. Molecular localization and characterization of multiple binding sites of organic anion transporting polypeptide 2B1 (OATP2B1) as the mechanism for substrate and modulator dependent drug–drug interaction. Med Chem Commun. 2016;7:1775–1782. [Google Scholar]
- 44.Hosoyamada M., Sekine T., Kanai Y., Endou H. Molecular cloning and functional expression of a multispecific organic anion transporter from human kidney. Am J Physiol. 1999;276:F122–F128. doi: 10.1152/ajprenal.1999.276.1.F122. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y., Stumpfe D., Bajorath J. Recent advances in scaffold hopping: miniperspective. J Med Chem. 2017;60:1238–1246. doi: 10.1021/acs.jmedchem.6b01437. [DOI] [PubMed] [Google Scholar]
- 46.Huang N., Shoichet B.K., Irwin J.J. Benchmarking sets for molecular docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irwin J.J. Community benchmarks for virtual screening. J Comput Aided Mol Des. 2008;22:193–199. doi: 10.1007/s10822-008-9189-4. [DOI] [PubMed] [Google Scholar]
- 48.Ismair M.G., Stieger B., Cattori V., Hagenbuch B., Fried M., Meier P.J. Hepatic uptake of cholecystokinin octapeptide by organic anion-transporting polypeptides OATP4 and OATP8 of rat and human liver. Gastroenterology. 2001;121:1185–1190. doi: 10.1053/gast.2001.28704. [DOI] [PubMed] [Google Scholar]
- 49.Jamroz M., Kolinski A. Modeling of loops in proteins: a multi-method approach. BMC Struct Biol. 2010;10:5. doi: 10.1186/1472-6807-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jewel Y., Dutta P., Liu J. Exploration of conformational changes in lactose permease upon sugar binding and proton transfer through coarse-grained simulations. Proteins. 2017;85:1856–1865. doi: 10.1002/prot.25340. [DOI] [PubMed] [Google Scholar]
- 51.Jothi A. Principles, challenges and advances in ab initio protein structure prediction. Protein Pept Lett. 2012;19:1194–1204. doi: 10.2174/092986612803217015. [DOI] [PubMed] [Google Scholar]
- 52.Kaczanowski S., Zielenkiewicz P. Why similar protein sequences encode similar three-dimensional structures? Theor Chem Acc. 2010;125:643–650. [Google Scholar]
- 53.Kalliokoski T., Kramer C., Vulpetti A., Gedeck P. Comparability of mixed IC50 data – a statistical analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karlgren M., Ahlin G., Bergström C.A.S., Svensson R., Palm J., Artursson P. in vitro and in silico strategies to identify OATP1B1 inhibitors and predict clinical drug–drug interactions. Pharm Res. 2012;29:411–426. doi: 10.1007/s11095-011-0564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karlgren M., Vildhede A., Norinder U., Wisniewski J.R., Kimoto E., Lai Y. Classification of inhibitors of hepatic organic anion transporting polypeptides (OATPs): influence of protein expression on drug–drug interactions. J Med Chem. 2012;55:4740–4763. doi: 10.1021/jm300212s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kell D.B., Dobson P.D., Bilsland E., Oliver S.G. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Discov Today. 2013;18:218–239. doi: 10.1016/j.drudis.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Khuri N., Zur A.A., Wittwer M.B., Lin L., Yee S.W., Sali A. Computational discovery and experimental validation of inhibitors of the human intestinal transporter OATP2B1. J Chem Inf Model. 2017;57:1402–1413. doi: 10.1021/acs.jcim.6b00720. [DOI] [PubMed] [Google Scholar]
- 58.Kido Y., Matsson P., Giacomini K.M. Profiling of a prescription drug library for potential renal drug–drug interactions mediated by the organic cation transporter 2. J Med Chem. 2011;54:4548–4558. doi: 10.1021/jm2001629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 60.Kotsampasakou E., Brenner S., Jäger W., Ecker G.F. Identification of novel inhibitors of organic anion transporting polypeptides 1B1 and 1B3 (OATP1B1 and OATP1B3) using a consensus vote of six classification models. Mol Pharm. 2015;12:4395–4404. doi: 10.1021/acs.molpharmaceut.5b00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kramer C., Kalliokoski T., Gedeck P., Vulpetti A. The experimental uncertainty of heterogeneous public K(i) data. J Med Chem. 2012;55:5165–5173. doi: 10.1021/jm300131x. [DOI] [PubMed] [Google Scholar]
- 62.Krieger E., Joo K., Lee Jinwoo, Lee Jooyoung, Raman S., Thompson J. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77(Suppl. 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ladizhansky V. Applications of solid-state NMR to membrane proteins. Biochimica et Biophysica Acta (BBA) - proteins and proteomics. Biophys Canada. 2017;1865:1577–1586. doi: 10.1016/j.bbapap.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Lapinsh M., Prusis P., Lundstedt T., Wikberg J.E.S. Proteochemometrics modeling of the interaction of amine G-protein coupled receptors with a diverse set of ligands. Mol Pharmacol. 2002;61:1465–1475. doi: 10.1124/mol.61.6.1465. [DOI] [PubMed] [Google Scholar]
- 65.Leach A. 2nd. ed. Pearson; Harlow, England ; New York: 2001. Molecular modelling: principles and applications. [Google Scholar]
- 66.Lechner C., Ishiguro N., Fukuhara A., Shimizu H., Ohtsu N., Takatani M. Impact of experimental conditions on the evaluation of interactions between multidrug and toxin extrusion proteins and candidate drugs. Drug Metab Dispos. 2016;44:1381–1389. doi: 10.1124/dmd.115.068163. [DOI] [PubMed] [Google Scholar]
- 67.Lee W., Glaeser H., Smith L.H., Roberts R.L., Moeckel G.W., Gervasini G. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280:9610–9617. doi: 10.1074/jbc.M411092200. [DOI] [PubMed] [Google Scholar]
- 68.Lengauer T., Rarey M. Computational methods for biomolecular docking. Curr Opin Struct Biol. 1996;6:402–406. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 69.Li N., Hong W., Huang H., Lu H., Lin G., Hong M. Identification of amino acids essential for Estrone-3-sulfate transport within transmembrane domain 2 of organic anion transporting polypeptide 1B1. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X., Huang J., Sun Y., Zhan K., Zhang Z., Hong M. Identification of multiple binding sites for substrate transport in bovine organic anion transporting polypeptide 1a2. Drug Metab Dispos. 2013;41:602–607. doi: 10.1124/dmd.112.047910. [DOI] [PubMed] [Google Scholar]
- 71.Lobley A., Sadowski M.I., Jones D.T. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]
- 72.Mak L., Marcus D., Howlett A., Yarova G., Duchateau G., Klaffke W. Metrabase: a cheminformatics and bioinformatics database for small molecule transporter data analysis and (Q)SAR modeling. J Chem. 2015;7:31. doi: 10.1186/s13321-015-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mandery K., Sticht H., Bujok K., Schmidt I., Fahrmayr C., Balk B. Functional and structural relevance of conserved positively charged lysine residues in organic anion transporting polypeptide 1B3. Mol Pharmacol. 2011;80:400–406. doi: 10.1124/mol.111.071282. [DOI] [PubMed] [Google Scholar]
- 74.Martínez-Guerrero L.J., Wright S.H. Substrate-dependent inhibition of human MATE1 by cationic ionic liquids. J Pharmacol Exp Ther. 2013;346:495–503. doi: 10.1124/jpet.113.204206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Šali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 76.Masuda S., Terada T., Yonezawa A., Tanihara Y., Kishimoto K., Katsura T. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol. 2006;17:2127–2135. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 77.Meier-Abt F., Mokrab Y., Mizuguchi K. Organic anion transporting polypeptides of the OATP/SLCO superfamily: identification of new members in nonmammalian species, comparative modeling and a potential transport mode. J Membr Biol. 2006;208:213–227. doi: 10.1007/s00232-005-7004-x. [DOI] [PubMed] [Google Scholar]
- 78.Meyer E.F., Swanson S.M., Williams J.A. Molecular modelling and drug design. Pharmacol Ther. 2000;85:113–121. doi: 10.1016/s0163-7258(99)00069-8. [DOI] [PubMed] [Google Scholar]
- 79.Moaddel R., Ravichandran S., Bighi F., Yamaguchi R., Wainer I.W. Pharmacophore modelling of stereoselective binding to the human organic cation transporter (hOCT1) Br J Pharmacol. 2007;151:1305–1314. doi: 10.1038/sj.bjp.0707341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montanari F., Ecker G.F. BCRP inhibition: from data collection to ligand-based modeling. Mol Inf. 2014;33:322–331. doi: 10.1002/minf.201400012. [DOI] [PubMed] [Google Scholar]
- 81.Montanari F., Zdrazil B., Digles D., Ecker G.F. Selectivity profiling of BCRP versus P-gp inhibition: from automated collection of polypharmacology data to multi-label learning. J Chem. 2016;8 doi: 10.1186/s13321-016-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mori T., Miyashita N., Im W., Feig M., Sugita Y. Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms. Biochim Biophys Acta (BBA) - Biomembr. 2016;1858:1635–1651. doi: 10.1016/j.bbamem.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morrissey K.M., Wen C.C., Johns S.J., Zhang L., Huang S.-M., Giacomini K.M. The UCSF-FDA TransPortal: a public drug transporter database. Clin Pharmacol Ther. 2012;92:545–546. doi: 10.1038/clpt.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Motohashi H., Inui K. Organic cation transporter OCTs (SLC22) and MATEs (SLC47) in the human kidney. AAPS J. 2013;15:581–588. doi: 10.1208/s12248-013-9465-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muegge I. PMF scoring revisited. J Med Chem. 2006;49:5895–5902. doi: 10.1021/jm050038s. [DOI] [PubMed] [Google Scholar]
- 86.Neuvonen P.J., Niemi M., Backman J.T. Drug interactions with lipid-lowering drugs: mechanisms and clinical relevance. Clin Pharmacol Ther. 2006;80:565–581. doi: 10.1016/j.clpt.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Newstead S., Ferrandon S., Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nozaki Y., Kusuhara H., Kondo T., Iwaki M., Shiroyanagi Y., Nakayama H. Species difference in the inhibitory effect of nonsteroidal anti-inflammatory drugs on the uptake of methotrexate by human kidney slices. J Pharmacol Exp Ther. 2007;322:1162–1170. doi: 10.1124/jpet.107.121491. [DOI] [PubMed] [Google Scholar]
- 89.Parker J.L., Newstead S. Membrane protein crystallisation: current trends and future perspectives. Adv Exp Med Biol. 2016;922:61–72. doi: 10.1007/978-3-319-35072-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pawson A.J., Sharman J.L., Benson H.E., Faccenda E., Alexander S.P.H., Buneman O.P. The IUPHAR/BPS guide to pharmacology: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014;42:D1098–D1106. doi: 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pendse P.Y., Brooks B.R., Klauda J.B. Probing the periplasmic-open state of lactose permease in response to sugar binding and proton translocation. J Mol Biol. 2010;404:506–521. doi: 10.1016/j.jmb.2010.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perland E., Fredriksson R. Classification Systems of Secondary Active Transporters. Trends Pharmacol Sci. 2017;38:305–315. doi: 10.1016/j.tips.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 93.Perry J.L., Dembla-Rajpal N., Hall L.A., Pritchard J.B. A three-dimensional model of human organic anion transporter 1: aromatic amino acids required for substrate transport. J Biol Chem. 2006;281:38071–38079. doi: 10.1074/jbc.M608834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pochini L., Scalise M., Galluccio M., Indiveri C. OCTN cation transporters in health and disease: role as drug targets and assay development. J Biomol Screen. 2013;18:851–867. doi: 10.1177/1087057113493006. [DOI] [PubMed] [Google Scholar]
- 95.Ray A., Lindahl E., Wallner B. Model quality assessment for membrane proteins. Bioinformatics. 2010;26:3067–3074. doi: 10.1093/bioinformatics/btq581. [DOI] [PubMed] [Google Scholar]
- 96.Rodgers T.L., Townsend P.D., Burnell D., Jones M.L., Richards S.A., McLeish T.C.B. Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roth M., Obaidat A., Hagenbuch B. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies: OATPs, OATs and OCTs. Br J Pharmacol. 2012;165:1260–1287. doi: 10.1111/j.1476-5381.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rouck J., Krapf J., Roy J., Huff H., Das A. Recent advances in nanodisc technology for membrane proteins studies (2012–2017) FEBS Lett. 2017;591:2057–2088. doi: 10.1002/1873-3468.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 100.Schafferhans A., Klebe G. Docking ligands onto binding site representations derived from proteins built by homology modelling11Edited by J. Thornton. J Mol Biol. 2001;307:407–427. doi: 10.1006/jmbi.2000.4453. [DOI] [PubMed] [Google Scholar]
- 101.Schlegel S., Klepsch M., Gialama D., Wickström D., Slotboom D.J., De Gier J. Revolutionizing membrane protein overexpression in bacteria. J Microbial Biotechnol. 2010;3:403–411. doi: 10.1111/j.1751-7915.2009.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schlessinger A., Khuri N., Giacomini K.M., Sali A. Molecular modeling and ligand docking for solute carrier (SLC) transporters. Curr Top Med Chem. 2013;13:843–856. doi: 10.2174/1568026611313070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schlessinger A., Welch M.A., van Vlijmen H., Korzekwa K., Swaan P.W., Matsson P. Molecular modeling of drug-transporter interactions-an international transporter consortium perspective. Clin Pharmacol Ther. 2018;104:818–835. doi: 10.1002/cpt.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shaikh N., Sharma M., Garg P. Selective fusion of heterogeneous classifiers for predicting substrates of membrane transporters. J Chem Inf Model. 2017;57:594–607. doi: 10.1021/acs.jcim.6b00508. [DOI] [PubMed] [Google Scholar]
- 105.Soars M.G., Barton P., Ismair M., Jupp R., Riley R.J. The development, characterization, and application of an OATP1B1 inhibition assay in drug discovery. Drug Metab Dispos. 2012;40:1641–1648. doi: 10.1124/dmd.111.042382. [DOI] [PubMed] [Google Scholar]
- 106.Sonoda Y., Newstead S., Hu N.-J., Alguel Y., Nji E., Beis K. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19:17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steindl T.M., Schuster D., Wolber G., Laggner C., Langer T. High-throughputstructure-based pharmacophore modelling as a basis for successful parallel virtual screening. J Comput Aided Mol Des. 2006;20:703–715. doi: 10.1007/s10822-006-9066-y. [DOI] [PubMed] [Google Scholar]
- 108.Stevenson J.M., Mulready P.D. Pipeline pilot 2.1 by Scitegic, 9665 Chesapeake drive, suite 401, San Diego, CA 92123-1365. J Am Chem Soc. 2003;125:1437–1438. www.scitegic.com See web site for pricing information. [Google Scholar]
- 109.Suhre W.M., Ekins S., Chang C., Swaan P.W., Wright S.H. Molecular determinants of substrate/inhibitor binding to the human and rabbit renal organic cation transporters hOCT2 and rbOCT2. Mol Pharmacol. 2005;67:1067–1077. doi: 10.1124/mol.104.004713. [DOI] [PubMed] [Google Scholar]
- 110.Tate C.G., Schertler G.F.X. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 111.The International Transporter Consortium, Giacomini K.M., Huang S.-M., Tweedie D.J., Benet L.Z., Brouwer K.L.R. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsigelny I.F., Kovalskyy D., Kouznetsova V.L., Balinskyi O., Sharikov Y., Bhatnagar V. Conformational changes of the multispecific transporter organic anion transporter 1 (OAT1/SLC22A6) suggests a molecular mechanism for initial stages of drug and metabolite transport. Cell Biochem Biophys. 2011;61:251–259. doi: 10.1007/s12013-011-9191-7. [DOI] [PubMed] [Google Scholar]
- 113.Türková A., Jain S., Zdrazil B. Integrative data mining, scaffold analysis, and sequential binary classification models for exploring ligand profiles of hepatic organic anion transporting polypeptides. J Chem Inf Model. 2018 doi: 10.1021/acs.jcim.8b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van de Steeg E., Venhorst J., Jansen H.T., Nooijen I.H.G., DeGroot J., Wortelboer H.M. Generation of Bayesian prediction models for OATP-mediated drug–drug interactions based on inhibition screen of OATP1B1, OATP1B1∗15 and OATP1B3. Eur J Pharm Sci. 2015;70:29–36. doi: 10.1016/j.ejps.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 115.van Westen G.J.P., Wegner J.K., IJzerman A.P., van Vlijmen H.W.T., Bender A. Proteochemometric modeling as a tool to design selective compounds and for extrapolating to novel targets. Med Chem Commun. 2011;2:16–30. [Google Scholar]
- 116.Verma J., Khedkar V.M., Coutinho E.C. 3D-QSAR in drug design—a review. Curr Top Med Chem. 2010;10:95–115. doi: 10.2174/156802610790232260. [DOI] [PubMed] [Google Scholar]
- 117.Wei B.Q., Weaver L.H., Ferrari A.M., Matthews B.W., Shoichet B.K. Testing a flexible-receptor docking algorithm in a model binding site. J Mol Biol. 2004;337:1161–1182. doi: 10.1016/j.jmb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 118.Williams-Noonan B.J., Yuriev E., Chalmers D.K. Free energy methods in drug design: prospects of “alchemical perturbation” in medicinal chemistry. J Med Chem. 2018;61:638–649. doi: 10.1021/acs.jmedchem.7b00681. [DOI] [PubMed] [Google Scholar]
- 119.Wilson C., Gregoret L.M., Agard D.A. Modeling side-chain conformation for homologous proteins using an energy-based rotamer search. J Mol Biol. 1993;229:996–1006. doi: 10.1006/jmbi.1993.1100. [DOI] [PubMed] [Google Scholar]
- 120.Wisedchaisri G., Park M.-S., Iadanza M.G., Zheng H., Gonen T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat Commun. 2014;5:4521. doi: 10.1038/ncomms5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wittwer M.B., Zur A.A., Khuri N., Kido Y., Kosaka A., Zhang X. Discovery of potent, selective multidrug and toxin extrusion transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug profiling and computational modeling. J Med Chem. 2013;56:781–795. doi: 10.1021/jm301302s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wong C.F. Flexible receptor docking for drug discovery. Expert Opin Drug Discovery. 2015;10:1189–1200. doi: 10.1517/17460441.2015.1078308. [DOI] [PubMed] [Google Scholar]
- 124.Wu X., Prasad P.D., Leibach F.H., Ganapathy V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem Biophys Res Commun. 1998;246:589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 125.Xu Y., Liu X., Li S., Zhou N., Gong L., Luo C. Combinatorial pharmacophore modeling of organic cation transporter 2 (OCT2) inhibitors: insights into multiple inhibitory mechanisms. Mol Pharm. 2013;10:4611–4619. doi: 10.1021/mp400423g. [DOI] [PubMed] [Google Scholar]
- 126.Xu Y., Liu X., Wang Y., Zhou N., Peng J., Gong L. Combinatorial pharmacophore modeling of multidrug and toxin extrusion transporter 1 inhibitors: a theoretical perspective for understanding multiple inhibitory mechanisms. Sci Rep. 2015;5:13684. doi: 10.1038/srep13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang J., Zhang Y. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics. 2015;52:5.8.1–5.815. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang S.-Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 129.Young D. John Wiley & Sons; 2004. Computational chemistry: A practical guide for applying techniques to real world problems. [Google Scholar]
- 130.Zhang X., He X., Baker J., Tama F., Chang G., Wright S.H. Twelve transmembrane helices form the functional core of mammalian multidrug and toxin extruder 1 (MATE1) J Biol Chem. 2012 doi: 10.1074/jbc.M112.386979. jbc.M112.386979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Y., Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 132.Tanihara Y., Masuda S., Sato T., Katsura T., Ogawa O., Inui K.-I. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem. Pharmacol. 2007;74:359–371. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 133.Truong D.M., Kaler G., Khandelwal A., Swaan P.W., Nigam S.K. Multi-level Analysis of Organic Anion Transporters 1, 3, and 6 Reveals Major Differences in Structural Determinants of Antiviral Discrimination. J. Biol. Chem. 2008;283:8654–8663. doi: 10.1074/jbc.M708615200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Soars M.G., Barton P., Elkin L.L., Mosure K.W., Sproston J.L., Riley R.J. Application of an in vitro OAT assay in drug design and optimization of renal clearance. Xenobiotica. 2014;44:657–665. doi: 10.3109/00498254.2013.879625. [DOI] [PubMed] [Google Scholar]
- 135.Bednarczyk D., Ekins S., Wikel J.H., Wright S.H. Influence of molecular structure on substrate binding to the human organic cation transporter, hOCT1. Mol. Pharmacol. 2003;63:489–498. doi: 10.1124/mol.63.3.489. [DOI] [PubMed] [Google Scholar]
- 136.Suhre W.M., Ekins S., Chang C., Swaan P.W., Wright S.H. Molecular determinants of substrate/inhibitor binding to the human and rabbit renal organic cation transporters hOCT2 and rbOCT2. Mol. Pharmacol. 2005;67:1067–1077. doi: 10.1124/mol.104.004713. [DOI] [PubMed] [Google Scholar]
- 137.Hong W., Wu Z., Fang Z., Huang J., Huang H., Hong M. Amino Acid Residues in the Putative Transmembrane Domain 11 of Human Organic Anion Transporting Polypeptide 1B1 Dictate Transporter Substrate Binding, Stability, and Trafficking. Molecular Pharmaceutics. 2015;12:4270–4276. doi: 10.1021/acs.molpharmaceut.5b00466. [DOI] [PubMed] [Google Scholar]
- 138.van Westen G.J.P., Hendriks A., Wegner J.K., Ijzerman A.P., van Vlijmen H.W.T., Bender A. Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kalliokoski A., Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stieger B., Hagenbuch B. Organic Anion Transporting Polypeptides. Curr Top Membr. 2014;73:205–232. doi: 10.1016/B978-0-12-800223-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.König J., Zolk O., Singer K., Hoffmann C., Fromm M.F. Double-transfected MDCK cells expressing human OCT1/MATE1 or OCT2/MATE1: determinants of uptake and transcellular translocation of organic cations. Br. J. Pharmacol. 2011;163:546–555. doi: 10.1111/j.1476-5381.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bahar I. On the functional significance of soft modes predicted by coarse-grained models for membrane proteins. Journal of General Physiology. 2010;135:563–573. doi: 10.1085/jgp.200910368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Isin B., Tirupula K.C., Oltvai Z.N., Klein-Seetharaman J., Bahar I. Identification of Motions in Membrane Proteins by Elastic Network Models and Their Experimental Validation. Methods Mol Biol. 2012;914:285–317. doi: 10.1007/978-1-62703-023-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dakal T.C., Kumar R., Ramotar D. Structural modeling of human organic cation transporters. Comput Biol Chem. 2017;68:153–163. doi: 10.1016/j.compbiolchem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 145.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Türková A., Jain S., Zdrazil B. Integrative Data Mining, Scaffold Analysis, and Sequential Binary Classification Models for Exploring Ligand Profiles of Hepatic Organic Anion Transporting Polypeptides. J Chem Inf Model. 2018 doi: 10.1021/acs.jcim.8b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]