Abstract
P2X purinergic receptors are ATP-driven ionic channels expressed as trimers and showing various functions. A subtype, the P2X4 receptor present on microglial cells is highly involved in neuropathic pain. In this study, in order to prepare antibodies recognizing the native structure of rat P2X4 (rP2X4) receptor, we immunized mice with rP2X4’s head domain (rHD, Gln111–Val167), which possesses an intact structure stabilized by S-S bond formation (Igawa and Abe et al. FEBS Lett. 2015), as an antigen. We generated five monoclonal antibodies with the ability to recognize the native structure of its head domain, stabilized by S-S bond formation. Site-directed mutagenesis revealed that Asn127 and Asp131 of the rHD, in which combination of these amino acid residues is only conserved in P2X4 receptor among P2X family, were closely involved in the interaction between rHD and these antibodies. We also demonstrated the antibodies obtained here could detect rP2X4 receptor expressed in 1321N1 human astrocytoma cells.
Electronic supplementary material
The online version of this article (10.1007/s11302-019-09646-5) contains supplementary material, which is available to authorized users.
Keywords: P2X4 receptor, Neuropathic pain, Monoclonal antibody, FSEC
Introduction
More than 20 million patients suffer from neuropathic pain worldwide. P2X receptors are ATP-activated trimeric ion channels expressed displaying various functions. In particular, the subtype P2X4 receptor is involved in neuropathic pain [1–3]. Specifically, during neuropathic pain, the P2X4 receptor present in mammalian microglial cells is activated; ATP then binds to the P2X4 receptor, after which Ca2+ ions flow into microglial cells [4]. The activation of microglial cells leads to the release of brain-derived neurotrophic factor (BDNF), which in turn suppresses the expression of the anion efflux pump KCC2 and increases the chloride ion concentration in nerve cells [5]. This effect causes a depolarization of nerve cells by abnormal inhibitory signals, resulting in neuropathic pain such as allodynia, a pain evoked by innocuous stimuli. The P2X4 receptor in activated microglia plays a key role in neuropathic allodynia, as suggested by the findings that a P2X4-specific inhibitor suppresses pain and that pain is suppressed in P2X4-knockout mice [6, 7]. Furthermore, the expression level of the P2X4 receptor on microglial cells has been strongly correlated with the quantitative level of neuropathic pain [4]. Thus, P2X4 receptor is one of targets involved in neuropathic pain.
In the crystal structure of P2X4 receptor, the extracellular domain adopts a dolphin-shaped structure comprising an extracellular head, a body, and a fluke domain [8]. These domains interact with each other in the characteristic P2X4 trimer. When ATP binds, the movement of these domains modulates the conformational change of the trimer structure and subsequently gives rise to the opening and closing of pores causing Ca2+ influx [8, 9]. Therefore, the extracellular domain is a target for the regulation of P2X4 receptor function.
Previously, in order to prepare an anti-rat P2X4 receptor monoclonal antibody, we expressed its extracellular domain (Asp58–Ile333) in Escherichia coli, then refolded the domain and used it as an antigen for immunization [10]. The antibody generated was reactive in Western blotting for immunoprecipitation of the P2X4 receptor on transfected 1321N1 human astrocytoma cells. This antibody can now be obtained from Wako. However, this established antibody cannot be employed for immunostaining on P2X4 receptor–expressing cells, because it is unable to recognize the native structure of the P2X4 receptor on the cell surface (data not shown).
The head domain is small, having three of the five disulfide bonds present in the P2X4 receptor. Recently, we expressed the head domain of rat P2X4 (rat head domain of P2X4 receptor (rHD), Gln111–Val167) in E. coli and showed an intact structure with three correctly formed S-S bonds [11]. In this study, we developed an anti-rHD monoclonal antibody using the head domain of the rat P2X4 (rHD, Gln111–Val167) as an antigen for immunization. As a result, we obtained five monoclonal antibodies recognizing the conformational epitope on the head domain. Moreover, we demonstrated the evidence that the antibody obtained here could detect rP2X4 receptor expressed on 1321N1 human astrocytoma cells.
Materials and methods
Preparation of the rP2X4 head domain and its KLH conjugation
The head domain of rP2X4 (rHD) was prepared according to a previous report [11]. To prepare rHD conjugated with KLH, 0.5 mg/ml KLH after dialysis in PBS buffer and 1 mg/ml rHD were incubated with 0.2% glutaraldehyde at 4 °C for 1 h. After incubation, 200 mmol/l glycine was added to the mixture to stop the reaction.
Point mutations of rHD were incorporated into the protein expression plasmid pET-22b(+) by the megaprimer method with the primers given in Table 1. Mutagenesis were confirmed by DNA sequencing. These mutants were expressed and purified by the methodology outlined for wild-type protein.
Table 1.
List of oligonucleotide primers employed in this study
| 5′ | CGCAAGCTTCATATGCACCACCACCACCACCACATGCAAACACAAAGTACCTGTCCAG |
| 3′ | CGCCGAATTCTCACACCGGGCACCATGCAGCC |
| K122A | CAAATGCTGGTCGCATCAGGAATCTC |
| S124T | CTGAATTACAAATAGTGGTCTTATCAG |
| I125V | CTGAATTACACACGCTGGTCTTATC |
| N127K | CGGCGTCTGATTTACAAATGCTG |
| D131S | CCAGGAGTGCAGCTGGCGTCTG |
| P151A | GAAGATGTGTTGCTTTCAATGAGTCTG |
| E154G | GTTCCTTTCAATGGGTCTGTGAAGACC |
| C116A | CGCAAGCTTCATATGCACCACCACCACCACCACATGCAAACACAAAGTACCGCTCCAG |
| C126A | GCGTCTGAATTAGCAATGCTGGTC |
| C132A | GCCAGGAGTGGCGTCGGCGTC |
| C149A | GACTGGAAGAGCTGTTCCTTTCAATG |
| C159A | GTGAAGACCGCTGAGGTGGCTG |
| C165A | CGCCGAATTCTCACACCGGGGCCCATGCAGCC |
Preparation of anti-rP2X4 monoclonal antibody
Seven-week-old female BALB/c mice and C57BL/6 mice were obtained from KBT Oriental (Saga, Japan). Seven-week-old female MRT mice (MRL/MpJJmsSlc-lpr/lpr: autoimmune disease mice) were obtained from Japan SLC (Shizuoka, Japan). Two mice from each strain were injected i.p. with 0.1 ml saline emulsified 1:1 in 0.1% (v/v) Complete Freund’s adjuvant (CFA) containing rHD was conjugated with Keyhole limpet hemocyanin (KLH) (50 μg/mouse) on day 0. Each mouse received i.p. booster immunization with the same antigen in 0.1% (v/v) incomplete Freund’s adjuvant (IFA) on day 14 followed by immunizations with the same antigen in saline every 2 weeks. The splenocytes were isolated and fused with a HAT-sensitive mouse myeloma cell line, SP2/0-Ag14, by the polyethylene glycol (PEG) method [12]. Hybridomas producing a monoclonal antibody against rHD were cloned by the limited dilution method [13]. Established hybridomas were cultured in 15% FCS eRDF medium. The produced monoclonal antibodies were purified by COSMOGEL® Ig-Accept ProteinA (Nakalai Tesque, Kyoto, Japan).
ELISA
A 96-well immunoplate (NUNC MaxiSorp; Thermo Scientific, Waltham, MA, USA) was incubated and adsorbed by 100 μl of 1 μmol/l rHD in 15 mmol/l Na2CO2 and 35 mmol/l NaHCO3 buffer (pH 9.6) for 1 h. The plate was treated with 300 μl phosphate-buffered saline (PBS) containing 5% skim milk (Nakalai Tesque) and 0.005% Tween 20 for 1 h to reduce nonspecific adsorption. The plate was washed with PBS containing 0.005% Tween 20 (T-PBS) and reacted with 100 μl of the prepared monoclonal antibody for 1 h. The plate was then washed with T-PBS three times and incubated with 100 μl of 1000-fold diluted alkaline phosphatase–conjugated goat anti-mouse IgG (Abcam, Cambridge, UK) for 1 h. After the plate was washed three times with T-PBS, 100 μl substrate solution, consisting of 0.3 mg/ml ABTS (Wako Pure Chemical Industries, Tokyo) in 60 mmol/l citrate and 40 mmol/l sodium citrate buffer containing 0.003% (v/v) hydrogen peroxide, was added to each well. The plate was then incubated for 30 min. Absorbance at 405 nm was measured with a Multiskan FC plate reader (Thermo Scientific). All reactions were conducted at 37 °C.
Dot blot and Western blot
SDS-treated rHD samples were prepared by incubation in 2% SDS aqueous solution at 100 °C for 3 min. Ten microliters of protein solution (10 μg/ml) was directly added to methanol-treated PVDF membrane. For Western blotting, protein samples were subjected to 12% (w/v) SDS-polyacrylamide gel electrophoresis. Separated proteins were blotted onto a PVDF membrane (Millipore, Billerica, MA, USA). Proteins were detected using anti-rat P2X4 monoclonal antibody and an alkaline phosphatase–conjugated goat anti-mouse IgG (Bio-Rad, Hercules, CA, USA) as a secondary antibody. Staining was performed using SIGMAFAST BCIP/NBT tablet solution (Sigma-Aldrich, St. Louis, MO, USA) in 100 mmol/l Tris-HCl (pH 9.5) containing 100 mmol/l NaCl and 5 mmol/l MgCl2.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) analyses were performed using MicroCal Auto-iTC200 (GE Healthcare, Little Chalfont, UK). Rat HD (60 μmol/l) was injected (1 × 0.4 μl and 18 × 2 μl) into 3 μmol/l antibody solution in PBS buffer at 25 °C. The data were fitted to a single-site binding model using the Origin software.
Surface plasmon resonance
The kinetic parameters for interactions between antibody and rHD mutants were measured by SPR using a Biacore X100 instrument (GE Healthcare). The rat HD was immobilized on a CM chip by amine coupling chemistry. Serial dilutions of rHD, respectively, in 10 mM HEPES buffer (pH 7.4) containing 150 mmol/l NaCl, 0.05 mmol/l EDTA, and 0.005% (v/v) Tween 20 were injected into the protein-immobilized and blank channels (for reference subtraction) for 100 s at a flow rate of 30 μl/min, followed by 100 s of buffer to monitor antibody dissociation. The chip surface was regenerated with 10 mmol/l glycine buffer (pH 3) prior to injection of new analyte. Kinetic parameters were calculated by fitting to a 1:1 (Langmuir) binding model.
Fluorescence-detection size-exclusion chromatography
GFP-fused rat P2X4 expressed in 1321N1 cells [14] was prepared in 50 mmol/l Tris-HCl (pH 7.5) containing 1% (w/v) dodecylmaltoside (DDM), 0.2% (w/v) cholesteryl hemisuccinate (CHS), and 200 mmol/l NaCl and allowed to stand on ice for 1 h. After centrifugation (14,000g × 10 min), the supernatants were applied to a fluorescence-detection size-exclusion chromatography (FSEC) analytical system [15]. FSEC analysis was performed using a HPLC system equipped with a Superdex 200 5/150 GL (GE) column equilibrated with 20 mmol/l Tris-HCl (pH 7.5) containing 150 mmol/l NaCl, 0.03% (w/v) DDM, and 0.006% (w/v) CHS. The sample was injected and the GFP fluorescence was detected at an emission wavelength of 525 nm with excitation at 490 nm.
Immunocytochemistry on rP2X4-expressing cells
Rat P2X4-expressing 1321N1 cells were seeded at 5000 cells/well in clear-bottom 96-well plates coated with poly-d-lysine and grown overnight at 37 °C and 5% CO2 incubator. The antibodies were prepared at the indicated concentrations and then pre-incubated at 37 °C for 20 min. After washing with BSS, the cells were incubated with the pre-incubated antibody at 37 °C for 30 min and fixed with 4% formaldehyde for 20 min at room temperature. After washing four times with PBS, the cells were blocked for 15 min at room temperature in blocking buffer (PBS containing 3% normal goat serum, Life Technologies, Waltham, MA, USA). The secondary antibody Alexa488 anti-mouse IgG (1:1000) was added and incubated for 1 h at room temperature. After washing three times with PBS, 200-fold diluted Hoechst was added and incubated for 15 min at room temperature for nucleus staining. After three washes with PBS, microscopy was carried out on an IN Cell Analyzer 2000 (GE Healthcare) at room temperature.
Measurement of ATP-induced Ca2+ influx
Measurement of Ca2+ imaging in 96-well plates was performed using the Functional Drug Screening System 7000EX (FDSS7000EX, Hamamatsu, Japan). 1321N1 astrocytoma cells stably expressing rP2X4 receptor were cultured for 24 h at 37 °C on 96-well plates (2.0 × 104 cells/well). Cells were loaded with 2.5 μmol/l Fura-2-AM containing 0.02% (v/v) pluronic (Molecular Probes) in a balanced salt solution (BSS; 150 mmol/l NaCl, 5 mmol/l KCl, 1.8 mmol/l CaCl2, 1.2 mmol/l MgCl2, 10 mmol/l d-glucose, and 25 mmol/l HEPES, pH 7.4) at 37 °C for 30 min and then washed with BSS. After pretreatment with reagent or antibody for 10 min, cells were stimulated with 10 μmol/l ATP (adenosine 5′-triphosphate disodium salt).
Results
Generation of novel anti-rat P2X4 head domain monoclonal antibodies
The folded rHD was expressed in Escherichia coli and purified as described in our previous study [11]. The purified rHD was then conjugated with KLH. The resultant conjugated protein was used for immunization of mice. In this study, we used the MRL mouse strain, which is an autoimmune disease model mouse, to boost immunogenicity, because the primary structure of rat and mouse P2X4 is very similar to each other (identity 95%). In fact, the antibody titer was higher in immunized MRT mice than in BALB/c mice or C57BL/6 mice (data not shown). About 2 × 108 splenocytes were produced and fused with SP2/0-Ag14 HAT-sensitive mouse myeloma cells according to a previously reported procedure [16]. After HAT-selection incubation, we carried out screening by using direct ELISA and single-cell cloning by the limited dilution method [17]. Thirty-eight hybridomas producing monoclonal antibodies reactive to rHD were obtained by direct ELISA screening.
Screening of antibodies recognizing the native rHD
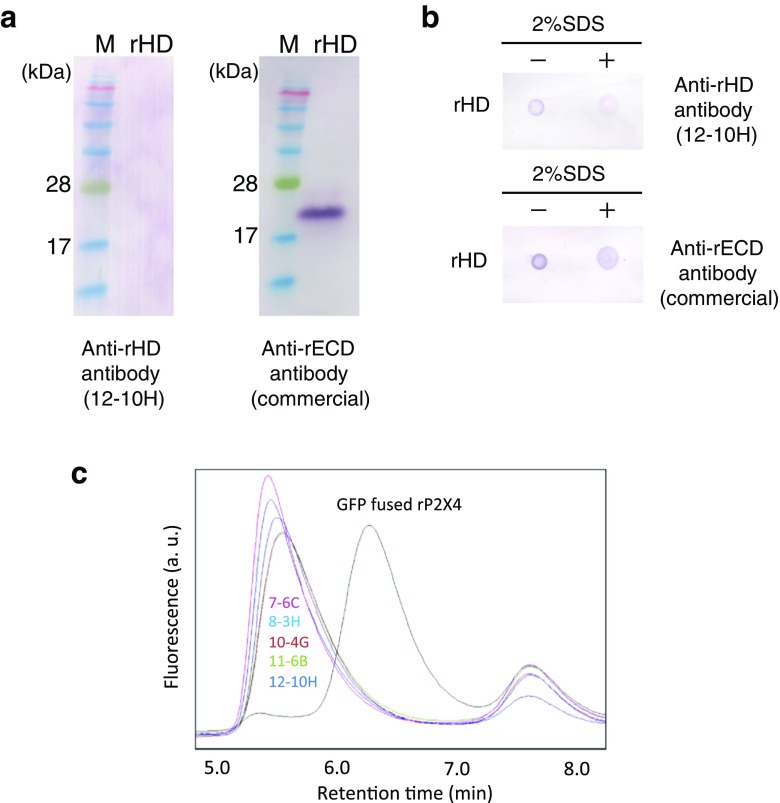
To examine whether the obtained monoclonal antibodies recognize the native conformation of rHD, we carried out Western blotting after SDS-PAGE under non-reducing conditions. Using Western blotting, we obtained five hybridomas producing monoclonal antibodies (7-6C, 8-3H, 10-4G, 11-6B, and 12-10H) that did not react with the denatured rHD (data not shown). A typical result using an antibody obtained here (12-10H) and our previously established monoclonal antibody [10] is shown in Fig. 1a. In dot blotting, these antibodies did not react with SDS-denatured rHD but did react with the native conformation of rHD (Fig. 1b). Unlike these five antibodies, our previously established monoclonal antibody only reacted with SDS-denatured rHD.
Fig. 1.
Screening of monoclonal antibodies. a Western blotting for rHD using anti-rHD antibody 12-10H (left) and anti-rECD antibody (right). b Dot blot of rHD in the presence (right) or absence (left) of 2% SDS using anti-rHD antibody 12-10H (upper) and anti-rECD antibody (lower). c GFP-fused rat P2X4 in the absence (black) or the presence of 7-6C (magenta), 8-3H (cyan), 10-4G (brown), 11-6B (green), and 12-10H (indigo) was monitored by fluorescence-detection size-exclusion chromatography (FSEC). FSEC was performed with the Superdex 200 5/150 GL at flow rate 0.5 ml/min. The fluorescence was detected at 525 nm with excitation at 490 nm
To verify if these antibodies recognized the full-length rat P2X4, we performed fluorescence-detection size-exclusion chromatography (FSEC) using the GFP-fused rat P2X4 expressed in 1321N1 cells after solubilization by detergents. As shown in Fig. 1c, the fluorescence peak of GFP-fused rat P2X4 in the absence of antibody was found the at an elution time at 6.3 min. In contrast, the mixtures of GFP-fused rat P2X4 with antibodies (7-6C, 8-3H, 10-4G, 11-6B, and 12-10H) was eluted earlier (~ 5.5 min), suggesting the formation of a complex between rat P2X4 and antibody. These data indicate that we obtained five antibodies that detect the native conformation of rHD even at the low concentrations of the antigen and antibodies employed.
Analysis of affinity between antibodies and rHD by using ITC
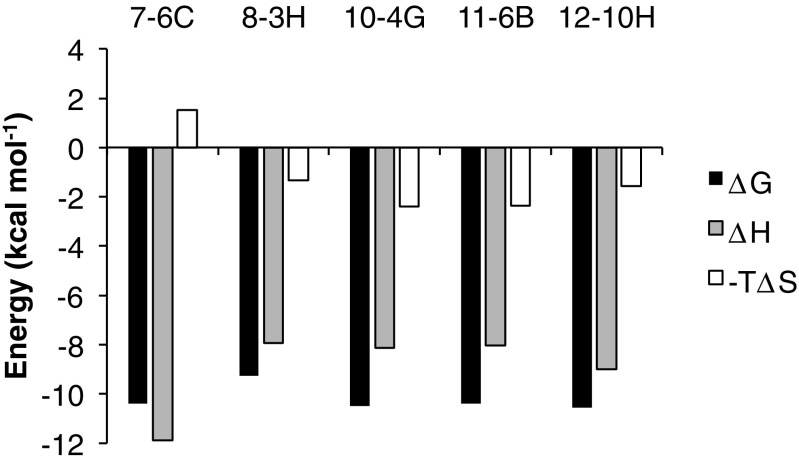
We examined the affinity between the established anti-P2X4 antibodies and rHD using ITC. Rat HD (rHD) in a titration syringe was added to antibody solution in a sample cell. In each titration, we observed an exothermic reaction during the rHD titration. The calorimetric parameters suggested that the dissociation constants of the complex between rHD and each antibody were around 20 nmol/l, except in the case of 8-3H which showed a value of 160 nmol/l (Table 2). From the titration results, we determined the thermodynamic parameters of each reaction (Fig. 2), suggesting that the interaction between rHD and each antibody is favored by enthalpy-driven interactions such as hydrogen bonds and van der Waals interactions.
Table 2.
Thermodynamics parameters of the interaction between anti-rHD antibodies and rHD
| Antibody | n | KD (nM) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|
| 7-6C | 2.37 ± 0.04 | 24 ± 11 | − 11.9 ± 0.3 | 1.5 ± 0.5 |
| 8-3H | 2.41 ± 0.06 | 164 ± 48 | − 7.9 ± 0.3 | − 1.3 ± 0.5 |
| 10-4G | 2.27 ± 0.03 | 20 ± 10 | − 8.1 ± 0.2 | − 2.4 ± 0.6 |
| 11-6B | 2.07 ± 0.02 | 24 ± 7 | − 8.0 ± 0.1 | − 2.4 ± 0.3 |
| 12-10H | 2.21 ± 0.04 | 18 ± 10 | − 9.0 ± 0.3 | − 1.6 ± 0.6 |
n indicates the number of binding sites in antibody
Fig. 2.
Thermodynamics of binding of each monoclonal antibody to rHD at 25 °C. ΔG (black bar), ΔH (dotted bar), and −TΔS (white bar) are shown
Determination of the epitope in rHD
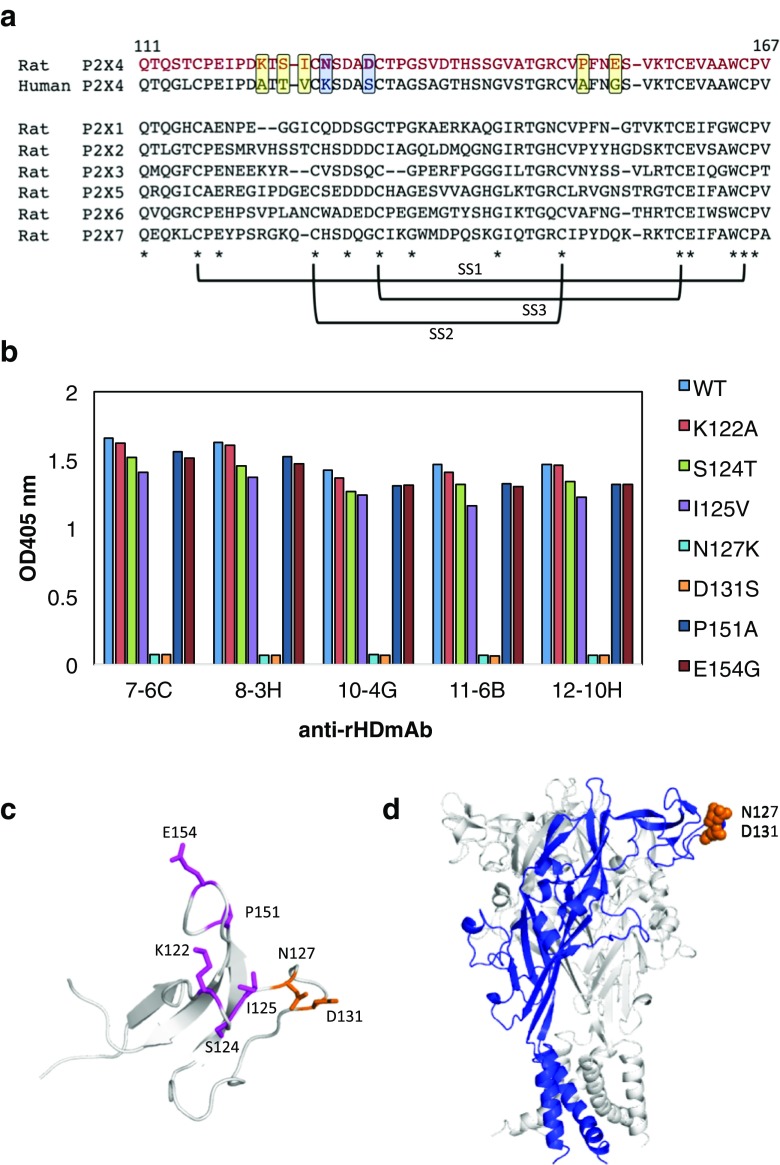
To further characterize these antibodies, we identified the epitope of rHD. Our previously established monoclonal antibody reacted with the extracellular domain of rat P2X4 but not with that of human P2X4 [10]. Similarly, a typical established antibody hardly recognize with human P2X4 receptor (unpublished results). From the alignment between rHD and human HD, we observed seven residues located on the protein surface that are not present in human HD (Fig. 3a). Therefore, we prepared seven rHD mutants where each residue was substituted to the corresponding residue of human HD. Using ELISA, we show that mutants N127K and D131S bind significantly less to the antibodies than the wild-type rHD or the other mutants (Fig. 3b). To confirm these results for antibody 12-10H, we used surface plasmon resonance (SPR). The antibody 12-10H was immobilized on the chip, and the association and dissociation curves of wild-type rHD and mutants were obtained in order to calculate the dissociation constants (Table 3). The dissociation constants of the complex between the wild-type rHD and antibody 12-10H were 57 nmol/l, which is threefold weaker than that determined by ITC, possibly because of the effect by the immobilization of the antibody. No binding was observed for mutants N127K or D131S. The dissociation constants of the other mutants were not very different from that of the wild-type rHD consistent with the ELISA data. These results indicated that Asn127 and Asp131 are key elements of the epitope of rHD for the binding of these antibodies (Fig. 3c, d). Combination of these residues is not conserved among rat P2X1~P2X7 (Fig. 3a), suggesting that these antibodies may specifically bind to the head domain of rP2X4.
Fig. 3.
Epitope of rP2X4 for anti-rHD antibody. a Sequence alignment of the head domain of human P2X4 and rat P2X1~P2X7. The mutation sites were enclosed in squares. Conserved amino acids are indicated by asterisks and three S-S bond formations are represented. b The results of ELISA for rHD mutants (K122A, S124T, I125V, N127K, D131S, P151A, and E154G) using each monoclonal antibody as a primary antibody and alkaline phosphatase–conjugated anti-mouse IgG as a secondary antibody. c Mutation sites (stick model) were mapped on the structure of rHD. The epitope is shown in blue. d The epitope is indicated on the homology model of the rP2X4 trimer. One monomer in the rP2X4 trimer is shown in blue
Table 3.
Kinetic parameters of the interaction between immobilized anti-rHD antibodies and rHD mutants
| Antibody | rHD | ka (1/Ms) | kd (1/s) | KD (M) |
|---|---|---|---|---|
| 12-10H | Wild type | 1.8e5 | 1.0e−2 | 5.7e−8 |
| K122A | 1.6e5 | 8.8e−3 | 5.4e−8 | |
| S124T | 7.1e4 | 8.6e−3 | 1.2e−7 | |
| I125V | 9.8e4 | 1.9e−2 | 1.7e−7 | |
| N127K | N. D. | N. D. | N. D. | |
| D131S | N. D. | N. D. | N. D. | |
| P151A | 1.8e5 | 1.1e−2 | 5.9e−8 | |
| E154G | 1.1e5 | 1.9e−2 | 1.8e−7 | |
| 8-3H | Wild type | 2.0e5 | 2.1e−2 | 1.0e−7 |
| K122A | 2.2e5 | 1.6e−2 | 7.1e−8 | |
| S124T | 2.2e5 | 3.6e−2 | 1.6e−7 | |
| I125V | 8.9e4 | 1.8e−2 | 2.0e−7 | |
| N127K | N. D. | N. D. | N. D. | |
| D131S | N. D. | N. D. | N. D. | |
| P151A | 1.9e5 | 2.6e−2 | 1.3e−7 | |
| E154G | 1.0e5 | 4.2e−2 | 4.1e−7 |
N. D., not detected
Contribution of S-S bond formation to antibody recognition
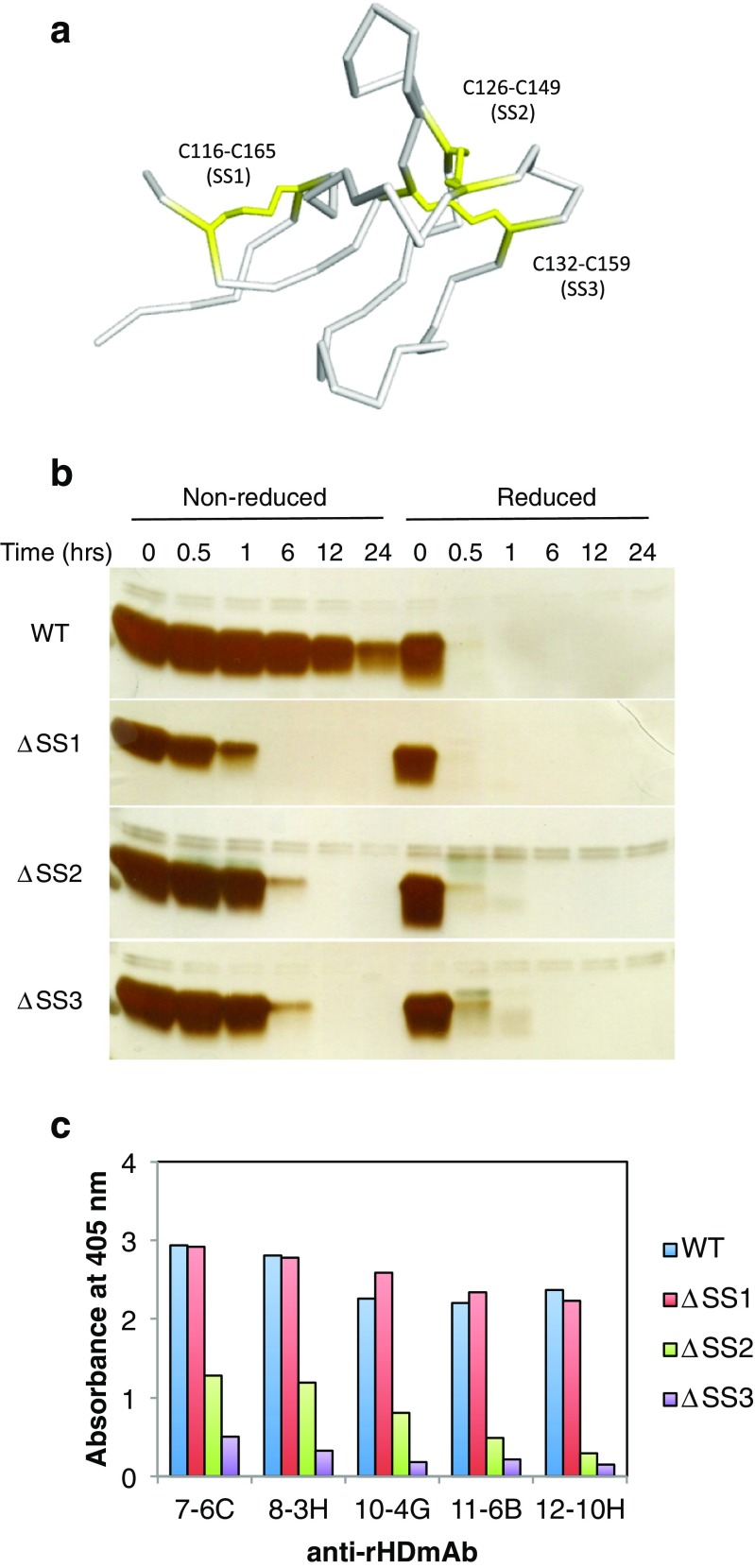
The head domain possesses three of the five disulfide bonds present in the full-length P2X4 receptor. To evaluate the individual contribution of each S-S bond in the recognition of rHD by the antibodies, we prepared three S-S bond deletion mutants, where two individual cysteines were substituted to alanines. The cysteine combinations were Cys116/Cys165, Cys126/Cys149, and Cys132/Cys159 that we termed ΔSS1, ΔSS2, and ΔSS3, respectively (Fig. 4a). These cysteine mutants were more sensitive to a protease, cathepsin B, than wild-type rHD, as it can be seen from the time course in Fig. 4b. However, given that the action of the protease is not immediate, some three-dimensional structure must remain in all deletion mutants. In contrast, under reducing conditions, these mutants as well as wild type were completely digested by cathepsin B before the first time point, suggesting that S-S bonds are necessary to keep a properly folded structure (Fig. 4b).
Fig. 4.
Disulfide bond contribution of anti-rHD antibody binding to rHD. a Disulfide bonds of rHD are depicted on the structure of rHD. Cysteines are shown in yellow. b Protease sensitivity of S-S bond deletion mutants of rHD, ΔSS1 (C116A/C165A), ΔSS2 (C126A/C149A), and ΔSS3 (C132A/C159A), and wild-type rHD. Proteins (25 μM) were treated with 100 μg/ml cathepsin B for the indicated times at 25 °C. Under reducing conditions, the sample was incubated with 52 mmol/l 2-mercaptoethanol for 40 °C and 90 min before digestion. The presence of protein in the gels was detected with silver staining. c The results of ELISA for S-S bond deletion rHD mutants, ΔSS1 (C116A/C165A), ΔSS2 (C126A/C149A), and ΔSS3 (C132A/C159A), using each monoclonal antibody
In addition, we carried out ELISA assay using these Cys-deletion mutants with the monoclonal antibodies (Fig. 4c). The results indicate that ΔSS2 and ΔSS3 did not bind strongly to the monoclonal antibodies, suggesting that the S-S bond formation in Cys126/Cys149 and Cys132/Cys159 is involved in the antibody binding. Whereas the deletion of the S-S bond between Cys116 and Cys165 hardly influences binding, the S-S bonds in Cys126/Cys149 and Cys132/Cys159 contribute to the retention of the epitope’s structure, i.e., native structure of the head domain. Collectively, the data indicated that these antibodies recognized the native structure of rat P2X4 receptor (rHD).
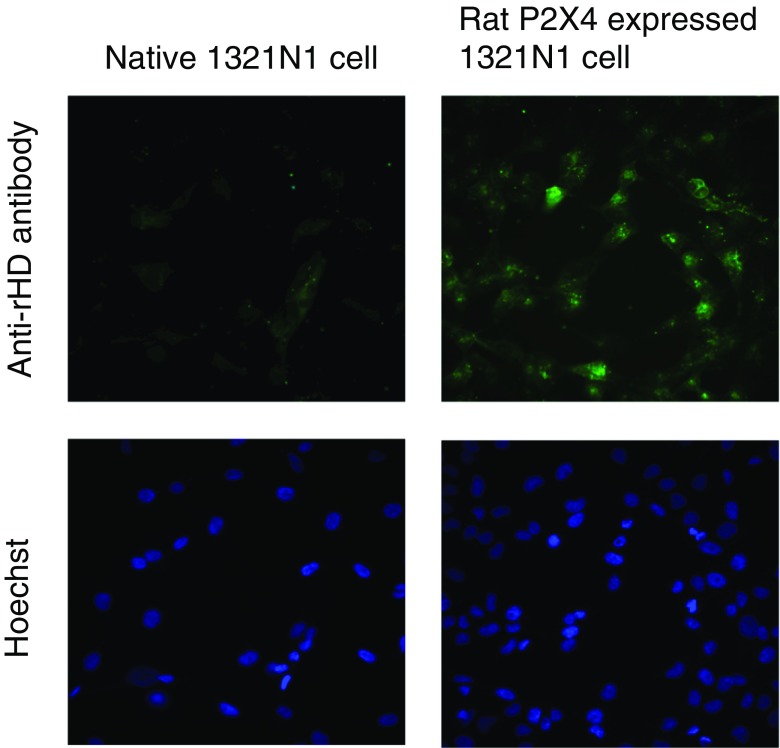
Imaging of rat P2X4 expressed on 1321N1 cells
In order to determine the suitability of the antibodies in immunocytochemistry, we cultured rat P2X4-transfected 1321N1 human astrocytoma cells overnight. The cultured cells were incubated with the established monoclonal antibody (12-10H) and then with a secondary antibody labeled with Alexa488. As shown in Fig. 5, the specific expression of rat P2X4 was more apparent in these cells than in the control cells (cells that had not been transfected with the P2X4 gene) by fluorescence microscopy. Therefore, the monoclonal antibody (12-10H) adequately recognized the native structure of rHD, allowing the detection of rat P2X4 expression on cells.
Fig. 5.
Detection of rP2X4 expressed on the cell by monoclonal antibody. Anti-rHD monoclonal antibody (12-10H, 10 μg/ml) and Alexa488-conjugated anti-mouse IgG were used for staining rP2X4 expressed on the 1321N1 cell (upper right). A native 1321N1 cell was also stained using both antibodies as a control (upper left). Hoechst was used to stain nuclei simultaneously (lower)
Discussion
In our previous report [15], we determined the solution structure of the head domain in rat P2X4 (Gln111-V167) by using NMR, showing the same backbone structure as that reported in zebrafish P2X4 receptor determined by X-ray crystallography [11]. In the present study, we employed the natively folded rHD as an antigen in an autoimmune disease model mouse strain, leading to the generation of 38 hybridomas producing anti-P2X4 monoclonal antibodies. From the results of Western blotting and SDS-treated dot blotting (Fig. 1), we narrowed down to five monoclonal antibodies that recognized the rHD epitope in the native protein. When we immunized mice with rHD lacking the disulfide bond of Cys116-Cys165 (ΔSS1) as the antigen, we could not obtain useful antibodies (data not shown). Therefore, since the disulfide bond (Cys116-Cys165) may contribute to the stability of head domain (Fig. 3b), we suggest that antigen stability is necessary to generate antibodies recognizing the native structure of rHD.
Additionally, we used FSEC for screening the binding of the antibodies to full-length rat P2X4. Using FSEC, the sensitivity is greatly enhanced and even when using minute quantities a clear response is observed by the fluorescence sensitivity of GFP-fused membrane proteins on the cell membrane [15, 17]. Small amounts of antibody from the supernatants of hybridoma cell culture are also an advantage of this approach. In this study, we clearly showed the shift of the position of the fluorescence elution peak upon complex formation between full-length rat P2X4 and the antibodies. Therefore, we have confirmed that the method of FSEC is a useful tool for the screening of the antibody binding to the GFP-fused membrane proteins.
To clarify the recognition mechanism, we found Asn127 and Asp131 as key residues of the epitope on rHD (Fig. 3c, d and Table 3). These results were consistent with that the disulfide bond deletion experiments (Fig. 3c), since both residues are located in the close proximity of the key disulfide bonds (Cys126-Cys149, Cys132-Cys159; Figs. 3c and 4a). Together with the result that the reduction of these three disulfide bonds in head domain was susceptible to a protease due to lack in its conformational stability (Fig. 4b), we may be able to generate these antibodies available in immunohistochemistry of native P2X4 receptor expressed on 1321N1 cells by using the antigen with retention of native conformation of rHD with three disulfide bonds.
Rituximab, which is still a blockbuster therapeutic antibody, binds to antigen with a dissociation constant of 4~5 nmol/l [18] and tocilizumab, which is also an antibody often used not only in Japan but also in Europe, binds to antigen with a dissociation constant of 2 nmol/l [19]. Although the affinity of the antibody (12-10H) and rHD (20 nmol/l, Table 2) from an analysis of ITC was weaker than that of rituximab, it was shown that the antibody had an affinity enough to be useful to image P2X4 receptor in immunohistochemistry (Fig. 5). As far as we know, there was no report on an antibody recognizing the native structure of rat P2X4 receptor.
Although the epitope is close to the ATP-binding site, these antibodies did not affect the influx of Ca2+ ion (Supplemental Fig. 1) and therefore cannot be used to inhibit rP2X4. From a FACS analysis, we confirmed that the monoclonal antibody (12-10H) reacted with preferentially bound to the rat P2X4 expressed on the primary cultured rat microglial cells (unpublished result). Therefore, as the antibodies obtained here did not affect P2X4 function, they will be useful for various applications such as live imaging of rat P2X4 receptors, which are highly expressed in neuropathic pain, in vivo in the near future.
Electronic supplementary material
(DOCX 107 kb)
Acknowledgements
We thank Dr. Jose Caaveiro of Kyshu University for critical readership of the manuscript. ITC measurements were supported by the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations
- CD
Circular dichroism
- CFA
Complete Freund’s adjuvant
- ECD
Extracellular domain of P2X4 receptor
- IFA
Incomplete Freund’s adjuvant
- KLH
Keyhole limpet hemocyanin
- PBS
Phosphate-buffered saline
- rHD
Rat head domain of P2X4 receptor
- TAPS
Trimethylammonium propyl sulfonate
Funding information
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to UT (23390155) and YA (18K06597) and by a grant from the Japan Foundation for Applied Enzymology.
Conflicts of interest
Tatsuhiro Igawa declares that he has no conflict of interest.
Shuhei Kishikawa declares that he has no conflict of interest.
Yoshito Abe declares that he has no conflict of interest.
Tomohiro Yamashita declares that he has no conflict of interest.
Saki Nagai declares that she has no conflict of interest.
Mitsunori Shiroishi declares that he has no conflict of interest.
Chinatsu Shinozaki declares that he has no conflict of interest.
Hiroyuki Tanaka declares that he has no conflict of interest.
Hidetoshi Tozaki-Saitoh declares that he has no conflict of interest.
Makoto Tsuda declares that he has no conflict of interest.
Kazuhide Inoue declares that he has no conflict of interest.
Tadashi Ueda declares that he has no conflict of interest.
Ethical approval
All animal experiments were conducted according to the relevant national and international guidelines in the Act on Welfare and Management of Animals (Ministry of Environment of Japan) and the Regulation of Laboratory Animals (Kyushu University), and under the protocols approved by the Institutional Animal Care and Use Committee review panels at Kyushu University.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tatsuhiro Igawa, Shuhei Kishikawa and Yoshito Abe contributed equally to this work.
Contributor Information
Kazuhide Inoue, Phone: +81-92-642-6566, Email: inoue@phar.kyushu-u.ac.jp.
Tadashi Ueda, Phone: +81-92-642-6662, Email: ueda@phar.kyushu-u.ac.jp.
References
- 1.Watkins LR, Maier SF. Glia: a novel drug discovery target for clinical pain. Nat Rev Drug Discov. 2003;2(12):973–985. doi: 10.1038/nrd1251. [DOI] [PubMed] [Google Scholar]
- 2.Watkins LR, Milligan ED, Maier SF. Spinal cord glia: new players in pain. Pain. 2001;93(3):201–205. doi: 10.1016/S0304-3959(01)00359-1. [DOI] [PubMed] [Google Scholar]
- 3.Watkins LR, Milligan ED, Maier SF. Glial activation: a driving force for pathological pain. Trends Neurosci. 2001;24(8):450–455. doi: 10.1016/S0166-2236(00)01854-3. [DOI] [PubMed] [Google Scholar]
- 4.Tsuda M, Shigemoto-Mogami Y, Koizumi S, Mizokoshi A, Kohsaka S, Salter MW, Inoue K. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature. 2003;424(6950):778–783. doi: 10.1038/nature01786. [DOI] [PubMed] [Google Scholar]
- 5.Coull JA, Beggs S, Boudreau D, Boivin D, Tsuda M, Inoue K, Gravel C, Salter MW, De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438(7070):1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- 6.Matsumura Y, Yamashita T, Sasaki A, Nakata E, Kohno K, Masuda T, Tozaki-Saitoh H, Imai T, Kuraishi Y, Tsuda M, Inoue K. A novel P2X4 receptor-selective antagonist produces anti-allodynic effect in a mouse model of herpetic pain. Sci Rep. 2016;6:32461. doi: 10.1038/srep32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagata K, Imai T, Yamashita T, Tsuda M, Tozaki-Saitoh H, Inoue K. Antidepressants inhibit P2X4 receptor function: a possible involvement in neuropathic pain relief. Mol Pain. 2009;5:20. doi: 10.1186/1744-8069-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460(7255):592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485(7397):207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igawa T, Higashi S, Abe Y, Ohkuri T, Tanaka H, Morimoto S, Yamashita T, Tsuda M, Inoue K, Ueda T. Preparation and characterization of a monoclonal antibody against the refolded and functional extracellular domain of rat P2X4 receptor. J Biochem. 2013;153(3):275–282. doi: 10.1093/jb/mvs143. [DOI] [PubMed] [Google Scholar]
- 11.Igawa T, Abe Y, Tsuda M, Inoue K, Ueda T. Solution structure of the rat P2X4 receptor head domain involved in inhibitory metal binding. FEBS Lett. 2015;589(6):680–686. doi: 10.1016/j.febslet.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 12.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73:3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 13.Goding JW. Antibody production by hybridomas. J Immunol Methods. 1980;39(4):285–308. doi: 10.1016/0022-1759(80)90230-6. [DOI] [PubMed] [Google Scholar]
- 14.Toyomitsu E, Tsuda M, Yamashita T, Tozaki-Saitoh H, Tanaka Y, Inoue K. CCL2 promotes P2X4 receptor trafficking to the cell surface of microglia. Purinergic Signal. 2012;8(2):301–310. doi: 10.1007/s11302-011-9288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiroishi M, Moriya M, Ueda T. Micro-scale and rapid expression screening of highly expressed and/or stable membrane protein variants in Saccharomyces cerevisiae. Protein Sci. 2016;25(10):1863–1872. doi: 10.1002/pro.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoyama Y, Fukada T, Murakami H. Production of monoclonal antibodies and ELISA for the baine and codeine. Cytotechnology. 1995;19(1):55–61. doi: 10.1007/BF00749755. [DOI] [PubMed] [Google Scholar]
- 17.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14(4):673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83(2):435–445. [PubMed] [Google Scholar]
- 19.Mihara M, Kasutani K, Okazaki M, Nakamura A, Kawai S, Sugimoto M, Matsumoto Y, Ohsugi Y. Tocilizumab inhibits signal transduction mediated by both mIL-6R and sIL-6R, but not by the receptors of other members of IL-6 cytokine family. Int Immunopharmacol. 2005;5(12):1731–1740. doi: 10.1016/j.intimp.2005.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 107 kb)