Abstract
Studies on TCP1-1 ring complex (TRiC) chaperonin have shown its indispensable role in folding cytosolic proteins in eukaryotes. In a psychrophilic organism, extreme cold temperature creates a low-energy environment that potentially causes protein denaturation with loss of activity. We hypothesized that TRiC may undergo evolution in terms of its structural molecular adaptation in order to facilitate protein folding in low-energy environment. To test this hypothesis, we isolated G. antarctica TRiC (GaTRiC) and found that the expression of GaTRiC mRNA in G. antarctica was consistently expressed at all temperatures indicating their importance in cell regulation. Moreover, we showed GaTRiC has the ability of a chaperonin whereby denatured luciferase can be folded to the functional stage in its presence. Structurally, three categories of residue substitutions were found in α, β, and δ subunits: (i) bulky/polar side chains to alanine or valine, (ii) charged residues to alanine, and (iii) isoleucine to valine that would be expected to increase intramolecular flexibility within the GaTRiC. The residue substitutions observed in the built structures possibly affect the hydrophobic, hydrogen bonds, and ionic and aromatic interactions which lead to an increase in structural flexibility. Our structural and functional analysis explains some possible structural features which may contribute to cold adaptation of the psychrophilic TRiC folding chamber.
Electronic supplementary material
The online version of this article (10.1007/s12192-019-00969-1) contains supplementary material, which is available to authorized users.
Keywords: Molecular chaperone, TRiC, TCP-1, CCT, Psychrophiles, Chaperonin, Flexibility
Introduction
Protein synthesis and protein folding are temperature cellular processes that severely restrict microbial growth in low-energy environments in the absence of specific adaptations to cold. The challenge in protein synthesis and folding in psychrophiles has been addressed via genomics and proteomics studies (Casanueva et al. 2010). Extreme temperature fluctuation causes cold shock and heat shock damage. Cold shock affects cells by hindering transcription and translation, affecting stabilization of secondary structures in nucleic acids, and by causing malfunction of cellular processes of metabolism and signal transduction (Phadtare 2004). Meanwhile, heat shock causes well-defined damage in protein folding and denaturation (Phadtare 2004). As for psychrophiles, their acclimation to cold in proteomic analysis has been well studied in several bacteria such as Psychrobacter arcticus, Shewanella livingstonensis, Methanococcoides burtonii, and Pseudoalteromonas haloplanktis where the upregulation and even downregulation of protein chaperones in psychrophilic bacteria indicate their significant contribution in cold-adaptive mechanisms (Bergholz et al. 2009; Campanaro et al. 2011; Goodchild et al. 2004; Kawamoto et al. 2007; Piette et al. 2011; Zheng et al. 2007). It has also been reported that quantitative proteomics analysis supports the hypothesis that psychrophilic microorganisms have a defined set of chaperones that functions at low temperature and another set that functions at higher growth temperatures (Piette et al. 2011). Interestingly, studies of cold and heat shock responses in psychrophilic yeast are still in their infancy. Studies on cold shock responses with adaptation mechanisms in E. coli and S. cerevisiae display several differences between these two model organisms (Phadtare and Inouye 2008; Al-Fageeh and Smales 2006; Phadtare et al. 1999; Jones and Inouye, 1994). However, the extent of similarities or differences of protein stress response between a mesophilic yeast and a psychrophilic yeast has not been demonstrated.
In this study, the availability of Glaciozyma antarctica genome has provided significant insights into the cold adaptation strategies acquired by this psychrophilic eukaryote. A study on this microbe has classified it as an obligate psychrophile based on its optimum temperature of 12 °C and able to tolerate high temperatures up to 20 °C. The G. antarctica genome (GenBank accession ASRT0000000) has identified a total of 7857 protein-coding genes where several genes were shown to be involved in cold adaptation (Firdaus-Raih et al. 2018). At least 89 genes may demonstrate chaperone activity (Yusof et al. 2015). Interestingly, our genome sequence analysis has identified several protein-coding genes that are known to be related to cold survival such as antifreeze proteins and fatty acid desaturases (Firdaus-Raih et al. 2018). In addition, gene expression analysis has revealed significant expression in G. antarctica related to antifreeze proteins, fatty acid desaturase enzymes, hsp70, hsp90, and hsp100 genes, neutral trehalase, and MnSOD genes (Boo et al. 2013). Characterization of chitinase from G. antarctica shows an optimum temperature of 15 °C and displays a high catalytic efficiency at extremely low temperatures compared to mesophilic chitinase (Ramli et al. 2012). Motivated by these intriguing observations, we have chosen to investigate the functional and structural properties of a mega-sized chaperonin obtained from the cold-living Antarctic yeast Glaciozyma antarctica.
Molecular chaperones are defined as groups of unrelated proteins that assist protein folding and allow the functional state of proteins by stabilizing unfolded proteins, unfolding them for translocation across membranes or for degradation, and assisting correct protein folding (Hartl et al. 2011). As molecular chaperones, these proteins interact with unfolded or partially folded protein subunits, stabilize non-native conformation and assist proper folding of protein subunits and do not interact with native proteins nor do they form part of the final folded structures (Hartl 1996; Hartl et al. 2011). Some chaperones are non-specific and interact with broad types of polypeptide chains while others are restricted to specific targets (Hartl and Hayer-Hartl 2002). Most chaperones often couple ATP binding or hydrolysis to the folding process. Besides that, many are essential for viability. Chaperone expression is often increased by cellular stress (Bukau et al. 2006; Hartl 1996). Many studies have found that molecular chaperones, particularly the heat shock proteins, are synthesized in the normal physiological state and increase in expression when exposed to temperature assaults both as a protective response and to promote cell survival (Fulda and Gorman, 2010, Hartl and Hayer-Hartl 2009; Jolly and Morimoto 2000; Hensen et al. 2013; Tsai et al. 2015). Extensive studies on molecular chaperones have proven that molecular chaperones are vital for the maintenance of correct protein folding in cells and act as the main defense against both physiological and stress conditions (Tomoyasu et al. 2001). In this context, we have chosen the cold-adapted eukaryotic Group II chaperonin TRiC from G. antarctica which plays an essential role in protein folding differently from the prokaryotic Group I chaperonin GroEL.
The chaperonin containing t-complex polypeptide-1 which is also known as TRiC plays a central role in cellular homeostasis by facilitating the folding of approximately 10% or more of newly synthesized proteins which include tubulins, actins, luciferin, Von Hippel-Lindau disease tumor suppressor (VHL), histone deacetylase 3 and other client proteins (Feldman et al. 1999; Frydman et al. 1994; Guenther et al. 2002; Valpuesta et al. 2002). TRiC chaperonin has a cylindrical-barrel-like shape consisting of eight different subunits (TRiCα, TRiCβ, TRiCγ, TRiCδ, TRiCε, TRiCζ, TRiCη, and TRiCθ) in a ring forming a 16-subunits toroid, back-to-back rings, where each ring contains a folding cage for client proteins (Leitner et al. 2012; Yébenes et al. 2011). The ATP binding, hydrolysis, and allosteric signaling from the intra- and inter-rings of the complex orchestrate sequestration of client proteins in a closed conformation followed by targeted protein release (Leitner et al. 2012; Yébenes et al. 2011). TRiC complex plays essential role for cellular protein homeostasis where the list of potential TRiC substrates has been expanded greatly by proteomic approaches (Spiess et al. 2004). Hence, cells metabolic systems could malfunction if TRiC complexes are structurally prone to be compromised by temperature fluctuations (Dekker et al. 2008; Thulasiraman et al. 1999; Yam et al. 2008).
With the availability of the G. antarctica genome, cDNA amplification of all eight TRiC subunits was subjected to sequence analysis. We hypothesized that G. antarctica has evolved in terms of structure and function of a TRiC chaperonin that plays an important role in maintaining protein folding activity at low temperature. In order to test this hypothesis, gene expression profiling of all eight subunits of the GaTRiC complex was done using quantitative real-time PCR (qPCR). The gene expression induction pattern of expression was analyzed at exposure temperatures of − 12 °C, 0 °C, 12 °C, and 20 °C. Subsequently, purification of native G. antarctica TRiC was done using affinity chromatography and gel filtration. Its functional properties were tested using denatured luciferase where renaturation of luciferase to the functional stage was done in different temperatures of 0 °C, 4 °C, 12 °C, and 30 °C. Its structural properties were compared with mesophilic counterparts from Saccharomyces cerevisiae and Bos taurus whose structures were available in the Protein Data Bank. The structural study was done at the sequence level and tertiary structures where residue substitutions that may contribute to the cold adaptation in TRiC were analyzed.
Materials and methods
Culturing G. antarctica and exposure to different heat and cold shock temperatures
The isolated G. antarctica culture was obtained courtesy of the School of Biological Sciences, Universiti Sains Malaysia. The cells were cultured in yeast peptone dextrose broth (10% (w/v) yeast extract, 20% (w/v) peptone, and 20% (w/v) dextrose) at 12 °C until until its OD600 reached approximately 0.6–0.8. Subsequently, the cultures were then exposed to different temperatures of − 12 °C, 0 °C, 12 °C, and 20 °C. The cells were harvested after 6 h exposure to each temperature.
RNA extraction
The following materials were treated overnight with DEPC 0.1% (v/v) and sent for autoclave: pestle and mortar, microcentrifuge tubes, pipette tips, and spatula. RNA extraction was done using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. The concentration and purity of total RNA were measured using Nanodrop spectrophotometer at 260 nm (ThermoScientific, Wilmington, DE, USA). The quality of RNA was checked by running total RNA on 1% (w/v) agarose gel at 120 V for 1 h. Total RNA was stored at − 80 °C.
Cloning and sequence analysis
cDNA amplification of all eight GaTRiC subunits was done and cloned into pGEMT-T easy vector (PROMEGA, Madison, WI, USA). Finally, the annealed product was transformed into E. coli DH5α cloning host. The positive transformants were identified using sequencing analysis. The isoelectric point was determined using ProtParam tool (Gasteiger et al. 2005). The sequence domain was analyzed using InterPro Scan (Quevillon et al. 2005) and Pfam (Sonnhammer et al. 1997). Sequence alignment was done using ClustalW (Thompson et al. 1994). The sequence of the GaTRiC subunits have been deposited in the GenBank™ database under these accession numbers (KU659477 TRiC_alpha; KU659478 TRiC_beta; TRiC_gamma KU659479; KU659480 TRiC_delta; KU659481 TRiC_epsilon; KU659482 TRiC_zeta; KU659483 TRiC_eta and KU659484 TRiC_theta).
Quantitative real-time PCR analysis
Complementary DNA was synthesized from 10 ng of total RNA from each sample using specific primers and QuantiFast SYBR® Green RT-PCR Kit (QIAGEN, Germantown, MD, USA) in a 20 μL reaction. The expression patterns of GaTRiC subunits were analyzed using Thermal cycler (Eppendorf, USA). Each reaction contained 12.5 μL of 1X Mastermix, 1 μM of primer GaTRiCαF-RT, 1 μM of primer GaTRiCαR-RT, 0.25 μL Quantifast Mix, and 2 μL of the template. A total of 9 sets of primers, which included the 8 TRiC subunits and 18 S, were used in the reactions (Table 1). Real-time cycler conditions were set as follows; reverse transcription step at 50 °C for 10 min for 1 cycle; initial activation step at 95 °C for 5 min; 40 cycles of denaturation step at 95 °C for 10 s, annealing at 60 °C for 30 s; 1 cycle of 95 °C for 30 s, 55 °C for 30 s. The set point temperature was increased after cycle 2 by 0.5 °C. A standard curve was constructed using tenfold serial dilutions (100 ng, 10 ng, 1 ng, 0.1 ng, and 0.01 ng) of RNA amplified with 18 S reference gene and GaTRiC subunit primers. The analysis was done in triplicate. A one-way analysis of variance (ANOVA) at a significance level of P < 0.05 was used to compare the expression levels at different temperatures. The GaTRiC subunits expression profiles were normalized with the 18 S according to Hashim et al. (2013) and Firdaus-Raih et al. (2018) in order to compensate for any variation in the amount of starting material among the samples. Melting curves analysis of the PCR reactions were done to analyze the specificity of each set of primers.
Table 1.
Primers used in real-time PCR analysis
| Primers | Sequence (5–3′) |
|---|---|
| GaTRiCαF-RT | GTACTTGACGGAGCAGCTTT |
| GaTRiCαR-RT | ATGCTCGCAGTCAAGACCA |
| GaTRiCβF-RT | CAAGAGCGAGCAGAGAATGTC |
| GaTRiCβR-RT | AGATCACGGTCACCAACGAC |
| GaTRiCγF-RT | ATGGAGAGACGGGCAAGGT |
| GaTRiCγR-RT | GAGGAGTGCAGAACATGGG |
| GaTRiCδF-RT | ATGGCTGCACCTACCACT |
| GaTRiCδR-RT | ATGGACAAGATGATCACGACC |
| GaTRiCεF-RT | ATCGATTGCGCTGGAAAGGG |
| GaTRiCεR-RT | GGTGGTGGAGACGAGGAGTA |
| GaTRiCζF-RT | GGTTGTGCAGGGAAGGAAC |
| GaTRiCζR-RT | GTAACCAAGCCTTCCAGATCAA |
| GaTRiCηF-RT | AGGCCGAATGCCCTACAT |
| GaTRiCηR-RT | GGTATGGACAAGCTCATCGTC |
| GaTRiCθF-RT | TCAGTAGGCTGTATGCTGCG |
| GaTRiCθR-RT | ATTCGCTACGGTACTGACGC |
Primers were named based on the TRiC subunits (subunit α, β, γ, δ, ε, ζ, η, and θ), F indicates forward primers, R is for reverse primers, and RT indicates real-time PCR
Preparation of cells, purification, and protein analysis
Harvested cells from G. antarctica culture were homogenized in buffer A containing 20 mM Tris (pH 8), 20 mM KCL, 1 mM EDTA, 1 mM DTT, 1 mM MgCl2, 0.5 mM PMSF, 1X protease inhibitor cocktail set (500 μM AEBSF hydrochloride, 150 nM Aprotinin, 1 μM E-64 protease inhibitor, 0.5 mM EDTA and 1 μM Leupeptin) (Merck Millipore, Germany) at 4 °C for 50 g cells per 50 mL buffer A. Cells were lysed using sonication and centrifuged at 10000 rpm for 30 min at 4 °C. Purification was done using anion exchange chromatography using buffer A and eluted using MgCl2 linear gradient. Eluted fractions were analyzed using immunoblotting for the presence of TRiC. Collected fractions containing G. antarctica TRiC were purified using gel filtration Superose 6 13/30 (GE Healthcare, USA) and eluted using buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 2 mM EDTA, 1 mM MgCl2, 1 mM KCl, 0.5 mM PMSF, and 0.5 mM DTT and 5% glycerol). The purified GaTRiC was analyzed for its purity using denatured gel electrophoresis. Western blot analysis was done using primary antibody which was the rat monoclonal anti-TRiC anti-mouse TCP-1a (Thermo Fisher Scientific, USA) with the ratio of 1:1000 and secondary antibody which was the goat anti-rat IgG2a (Thermo Fisher Scientific, USA) with the ratio of 1:10000 for an hour with agitation at room temperature followed by detection using Luminate Forte Western HRP (Merck, USA).
In vitro renaturation of denatured luciferase using native GaTRiC
Luciferase (Promega, USA) was diluted to 1 μg/μL with stability buffer (25 mM Tris pH 8.0, 8 mM MgSO4, 0.1 mM EDTA, 1% Triton X-100, 10% glycerol) heated at 41 °C for 10 min. Denatured luciferase was incubated with 10 μg/μL purified TRiC diluted in folding buffer (40 mM Tris pH 8.0, 200 mM NaCl, 10 mM MgCl2, 10% glycerol, 10% PEG 4000, 1 mM DTT, 0.1 mM Adenosine 5′-triphosphate (ATP)) and incubated at different temperatures of 30 °C, 16 °C, 4 °C, and 0 °C for 6 h respectively. Unheated luciferase incubated with and without purified GaTRiC protein were used as positive controls while heated luciferase incubated with BSA (10 μg/μL) without any incubation with GaTRiC was used as a negative control. Renaturation of luciferase was measured with the presence of 0.5 mM D-luciferin as substrate at 560 nm emission wave using SynergyHT (Research Instrument, USA) luminometer at 0, 1, 2, 3, 4, and 5 min. The ratio of luciferase to purified GaTRiC protein concentration for the assay was 1:10.
Modeling of GaTRiC tertiary structures
Three-dimensional structures of GaTRiC were modeled to the Saccharomyces cerevisiae TRiC (PDB: 4V81) using the program SWISS-MODEL. This software was used for comparative modeling of protein three-dimensional structures with related structures in PDB which were elucidated either using crystallography or nuclear magnetic resonance (NMR) methods (Schwede et al. 2003; Guex and Peitsch 1997). Structure quality was evaluated using PROCHECK (Laskowski et al. 1993), Verify3D (Eisenberg et al. 1997) and ANOLEA (Melo et al. 1997). PROCHECK was used to check for proper protein stereochemistry; symmetry and geometry checks; chirality, bond lengths and angles and torsion angles (Morris et al. 1992). Verify3D was used to analyze the compatibility of an atomic model (3D) with its own amino acid sequence (1D) (Kalman and Ben-Tal 2010; Eisenberg et al. 1997). ANOLEA was used to detect errors in the build protein models by performing energy calculations at the atomic level in protein structures (Melo and Feytmans 1998). Superimposed structure of GaTRiC and template and comparative analyses were done using UCSF CHIMERA (Pettersen et al. 2004).
Comparison of structural features with its mesophilic homologs
The structure of GaTRiC subunits was superimposed with the S. cerevisiae TRiC and the G. antarctica residues substitutions were analyzed using bioinformatics software in terms of hydrophobicity, ionic interaction and aromatic interaction that could affect the intra and inter-protein interaction. Using UCSF CHIMERA, analysis on residue substitutions was done by analyzing the ion pair interactions using a cut off distance of 4 Å while hydrophobic interaction within 5 Å (Bae and Phillips 2004, Perl 2000, Veille and Zeikus 2001). Analysis was also done on aromatic dimers associated with a possible role in folding and protein-protein interaction within 6 Å (Lanzarottie et al. 2011).
Results
Sequence analysis of GaTRiC subunits
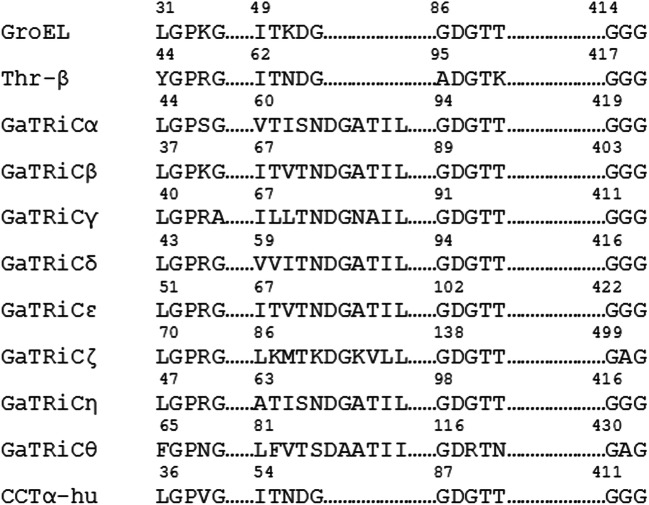
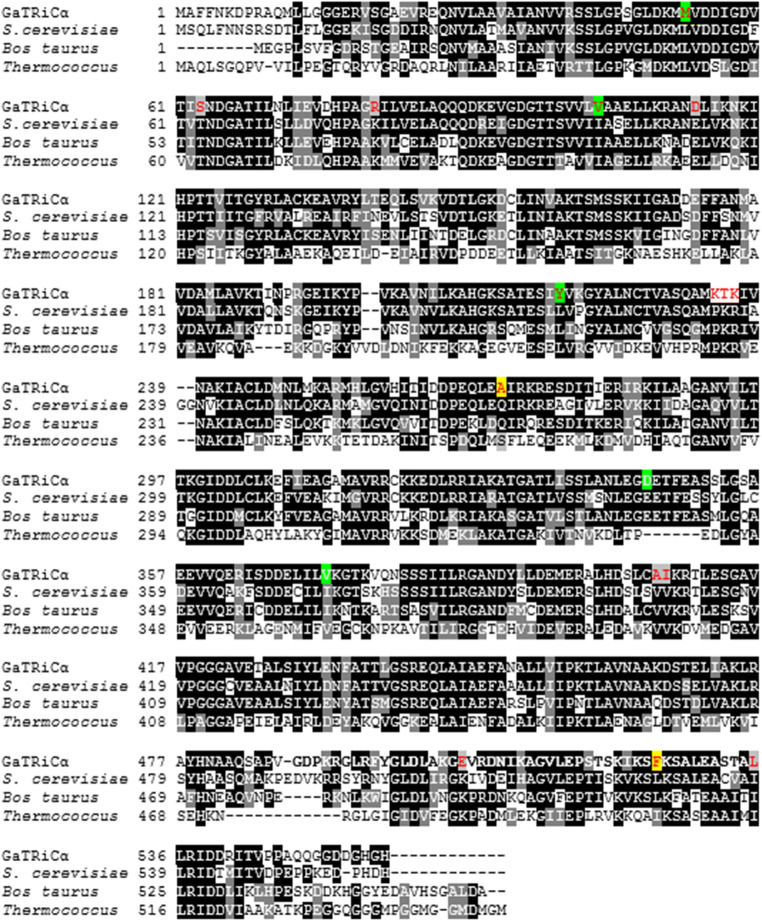
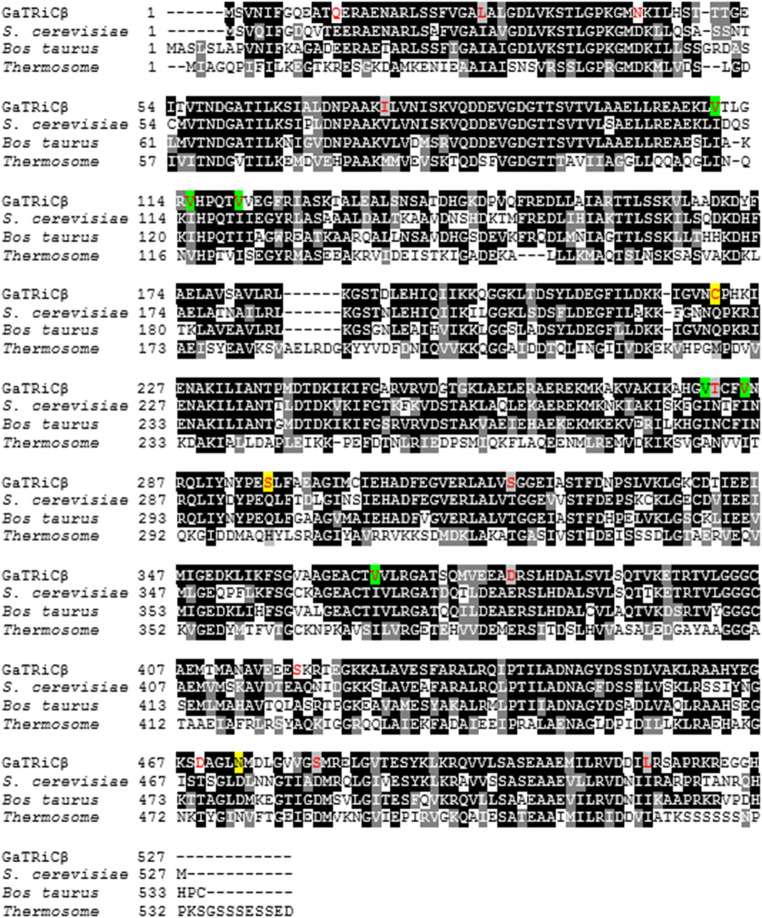
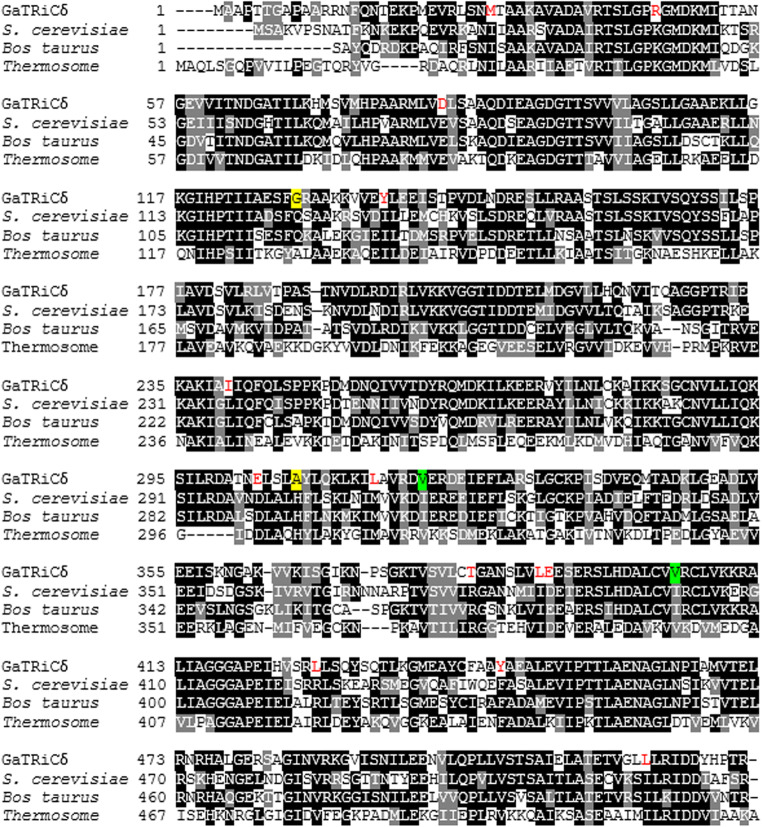
We isolated the gene encoding GaTRiC subunits and showed their relation to the mesophilic TRiC of S. cerevisiae and mammalian TRiC Bos taurus with which they shared high amino acid sequence identity (Table 2). Sequence alignment of all G. antarctica TRiC subunits, chaperonin of E. coli (GroEL), TRiCα-subunit from human (CCTα-human) and β-subunit from acidothermophilic archaeon Acidianus tengchongensis (Thr-β) showed a highly conserved ATP binding site in the equatorial domain (Fig. 1). The sequence alignment of Group I (GroEL) and Group II chaperonin (TRiC) showed that the equatorial domains in these chaperonins were conserved while the apical domains were found to have major sequence variations between all TRiC’s subunits. Multiple sequenced alignment analysis on the apical, intermediate and equatorial domains of G. antarctica TRiC α, β, and δ-subunits was done with TRiC templates from Saccharomyces cerevisiae (PDB ID: 4V81) and Bos taurus (PDB ID: 3IYG). Sequence alignment showed intriguing residue substitutions in the GaTRiCα (Fig. 2), GaTRiCβ (Fig. 3), and GaTRiCδ (Fig. 4). Interestingly, three categories of residue substitutions were found in these subunits—α, β, and δ: (i) bulky/polar side chains to alanine or valine, (ii) charged residues to alanine, and (iii) isoleucine to valine.
Table 2.
Amino acid sequence identity of G. antarctica TRiC with mesophilic TRiC from S. cerevisiae and mammalian TRiC of Bos taurus with E-value equals to 0
| G. antarctica TRiC | S. cerevisiae TRiC (%) | Bos taurus TRiC (%) |
|---|---|---|
| α | 71 | 68 |
| β | 69 | 71 |
| γ | 63 | 65 |
| δ | 65 | 66 |
| ε | 65 | 69 |
| ζ | 54 | 55 |
| η | 67 | 66 |
| θ | 54 | 53 |
Fig. 1.
Sequence alignment of all G. antarctica TRiC subunits, GroEL, CCTα-human, and Thr-β. Highly conserved residues were seen in the equatorial domain that plays important role in ATP hydrolysis. The GenBank accession number for these proteins are as following: GroEL from E. coli (AOX48130.1), CCTα-human (NP_110379.2) and β subunit from Acidianus tengchongensis (AAO47380.1). Starting from E. coli, yeast, archaea, and human, all of the chaperonin subunits have highly conserved residues where any changes were postulated to lead to severe effects (Dunn et al. 2001)
Fig. 2.
Sequence alignment between TRiCα from G. antarctica, S. cerevisiae, B. taurus and Thermococcus. The amino acid sequences of the TRiC α-subunit from G. antarctica was realigned relative to those of S. cerevisiae (PDB: 4V81_A), B. taurus (PDB: 4B2T_A) and Thermococcus (PDB: IA6D_A). Highly conserved residues are highlighted in black while partly conserved in gray. Residue substitutions of the GaTRiCα that differ with respect to the reference sequences are shown in red and significant changes in the tertiary structure interactions are highlighted in yellow for ionic interaction while green for hydrophobic interaction
Fig. 3.
Sequence alignment between TRiCβ from G. antarctica, S. cerevisiae, B. taurus, and Thermococcus The amino acid sequences of the TRiCβ from G. antarctica was realigned relative to those of S. cerevisiae (PDB: 4V81_B), B. taurus (PDB: 4B2T_B) and Thermococcus (PDB: IA6D_B). Highly conserved residues are highlighted in black while partly conserved in gray. Residue substitutions of the GaTRiCβ that differ with respect to the reference sequences are shown in red and significant changes in the tertiary structure interactions are highlighted in yellow for ionic interaction while green for hydrophobic interaction
Fig. 4.
Sequence alignment between TRiCδ from G. antarctica, S. cerevisiae, B. taurus, and Thermococcus. The amino acid sequences of the TRiC δ-subunit from G. antarctica was realigned relative to those of S. cerevisiae (PDB: 4V81_D), B. taurus (PDB: 4B2T_D) and Thermococcus (PDB: IA6D_D). Highly conserved residues are highlighted in black while partly conserved in gray. Residue substitutions of the GaTRiCδ that differ with respect to the reference sequences are shown in red and significant changes in the tertiary structure interactions are highlighted in yellow for ionic interaction while green for hydrophobic interaction
mRNA expression level of GaTRiC under different temperatures exposures
The RNA template was checked for genomic contamination using gel electrophoresis. The purity of the RNA was quantified using a spectrophotometer where a value of 1.9 to 2.0 indicated a high purity of RNA. The melting curve analysis showed single amplification product for 18 S and GaTRiC subunits. As for PCR efficiency, the efficiency of the primer sets used to amplify.
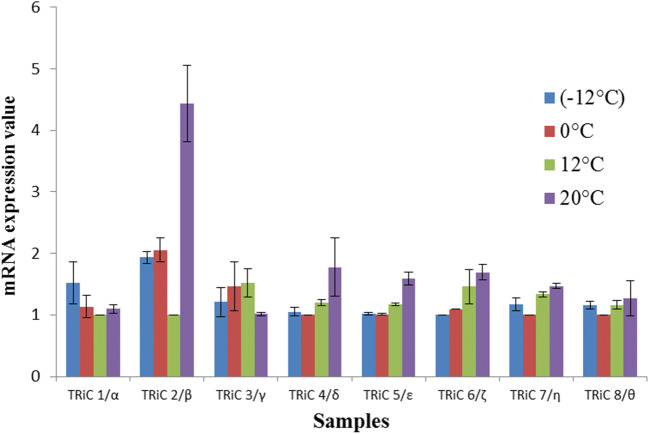
18 S and GaTRiC genes was acceptable as the slopes were within the range of − 3 to − 3.3. The R2-values fell within the range of 0.97 to 0.99 while the E-value for both primer sets was 110%. Thus, the template purity and PCR efficiency fulfilled the standard characteristics for accurate and reliable PCR data as stated in the general MIQE quantitative PCR guidelines (Bustin et al. 2009). The result of gene expression was presented in relative quantity to 18 S expression as the reference gene (Fig. 5). The expression of GaTRiC mRNA was measured at different temperatures to determine the induction pattern of GaTRiC subunits. The results showed that the expression of the GaTRiC subunits was constitutively expressed at all temperatures with the exception of GaTRiCβ at 20 °C where it reached 4.5-fold as compared to optimal growth at 12 °C. Analysis showed that the expression array of the G. antarctica TRiC was consistently expressed at all temperatures indicating their importance in cell regulation.
Fig. 5.
G. antarctica TRiC subunits mRNA expression at different temperatures analyzed using real-time PCR. The level of TRiC subunits was measured in cells exposed at the indicated temperatures for 6 h and normalized using 18 S level. The mRNA expression with the lowest expression for every subunit was set to 1 and other values were compared against this. Data were representative of three trials where the statistical significance was performed using ANOVA one-way test where P < 0.05
Purification and protein detection of GaTRiC
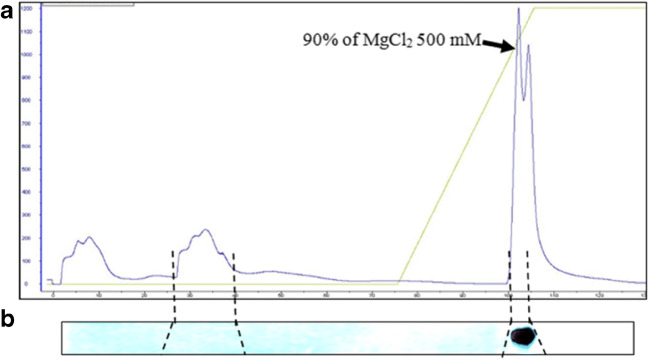
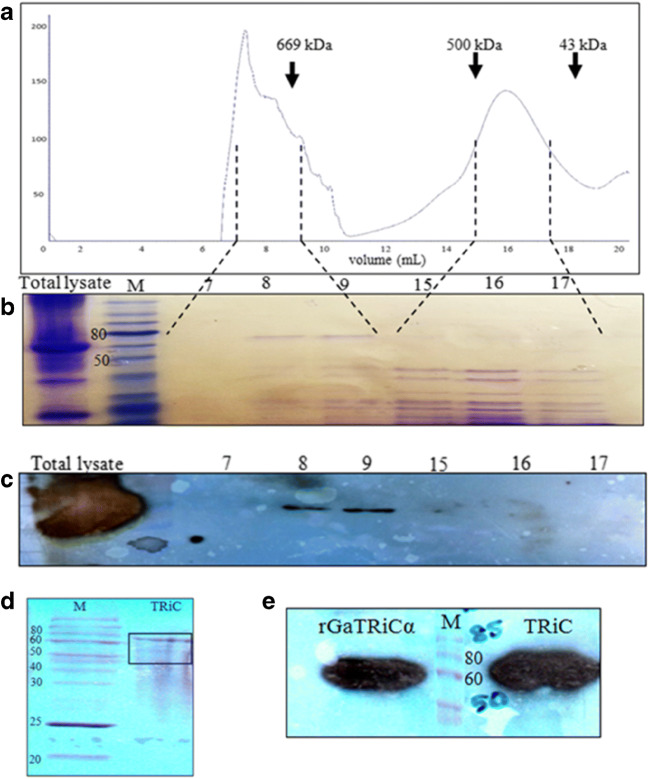
The first step for G. antarctica TRiC purification was using an anion exchange method. The G. antarctica lysate was loaded on an anion sepharose column and eluted by application of a linear MgCl2 gradient at 6 °C (Fig. 6a) and detected using anti-TRiCα immunoblot analysis (Fig. 6b). The fraction that contained GaTRiC native was eluted at 450 mM MgCl. Following chromatography on anion exchange, the final purification of G. antarctica TRiC was accomplished by using a gel filtration Superose 6 column at 4 °C. The elution profile of gel filtration using a Superose 6 column is shown in Fig. 7a. The fractions that contained GaTRiC complex were detected using SDS PAGE analysis (Fig. 7b) and immunoblot analysis of the purified GaTRiC was done using a monoclonal rat anti-mouse TCP-1α and a conjugated goat anti-rat IgG2a (Fig. 7c). The SDS-PAGE profile of the purified GaTRiC showed bands with the molecular weights anticipated for TRiC subunits between 55 and 70 kDa (Fig. 7d) and was detected using immunostaining by anti-TCP1α that recognized the presence of TRiC subunit-α (Fig. 7e). The gel cut of the GaTRiC complex was sent for protein sequencing and subunits from GaTRiC were identified.
Fig. 6.
Anion sepharose exchange chromatography of G. antarctica TRiC. Fractions containing GaTRiC were pooled and concentrated. Figure a) a chromatogram with the arrowhead showing the percentage of MgCl2 needed for TRiC elution and figure b) Immunoblot analysis of fractions eluted from the anion sepharose column using the anti-TRiCα antibodies
Fig. 7.
Size exclusion chromatography of G. antarctica TRiC. a A chromatogram of gel purification using Sepharose 6 with the arrowheads showing the positions of molecular size markers (thyroglobulin, 669 kDa; ferritin, 440 kDa and ovalbumin, 43 kDa). The numbers 0 mL to 20 mL in the chromatogram represented the eluted fractions. b A SDS-PAGE analysis of the fractions collected from the gel filtration column. The lane numbered as 7, 8, 9, 15, 16, and 17 were the chosen eluted fractions that correspond to the size of TRiC complex. c An immunoblot analysis of fractions from the Superose 6 column using anti-TRiCα antibodies. The lane numbered as 7, 8, 9, 15, 16, and 17 corresponding to the chosen eluted fractions. d Protein composition of fractions 7–9 from the Superose 6 column pooled and concentrated by ultrafiltration to 1 mg/mL. The bands in the gel were revealed by Coomassie blue staining. e Immunoblot analysis of purified G. antarctica TRiC complex using anti-TRiCα antibodies. Recombinant G. antarctica TRiCα was running together as a positive control
Folding activity of purified GaTRiC in luciferase folding assay
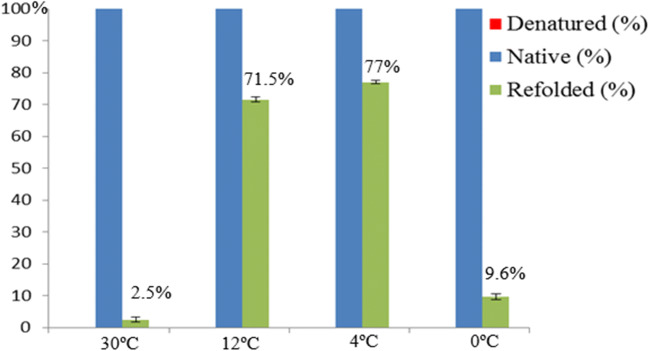
In order to determine whether GaTRiC possesses the ability to fold denatured proteins to the functional stage, a luciferase bioluminescence assay was done to determine the ATPase activity of the renatured luciferase. In this study, the denatured luciferase was incubated with purified GaTRiC at 0 °C, 4 °C, 12 °C, and 30 °C in order to determine the efficiency of folding activity at different temperatures. The refolding of denatured luciferase by GaTRiC was analyzed by assaying the luciferase activity (Fig. 8). Denatured luciferase lost almost 100% of its catalytic activity upon heating at 43 °C for 10 min. In this study, the incubation of denatured luciferase with GaTRiC for 6 h at different temperatures showed some recovery; incubation at 4 °C had the highest recovery of 77% followed by incubation at 12 °C with a recovery of 71.5%. However, incubation of denatured luciferase with GaTRiC at 0 °C and 30 °C showed low recoveries of 9.6% and 2.5%, respectively. This showed that GaTRiC can fold denatured luciferase to a functional state in cold temperatures as low as 0 °C. However, it performed its folding activity best between 4 °C and 12 °C. In terms of activity, GaTRiC was not very active at the extreme environment for G. antarctica growth of more than 30 °C and less than 0 °C.
Fig. 8.
Luciferase activity of denatured, native and refolded luciferase incubated with G. antarctica TRiC at different temperatures for 6 h. The activity of native luciferase which was not heat inactivated was 100% while the denatured luciferase was 0%. Renaturation of luciferase was recorded as highest at 4 °C followed by 12 °C, 0 °C, and 30 °C. The percent renaturation were 77%, 71.5%, 9.6%, and 2.5% respectively. Statistical significance was performed using the two-tailed t-test where P < 0.01
3D structure analysis of GaTRiC
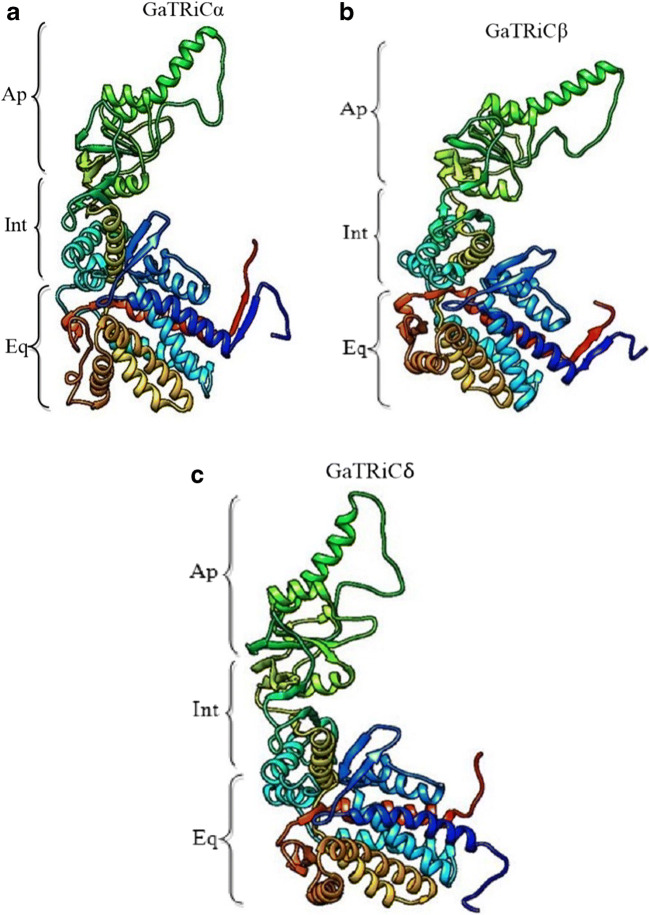
The GaTRiC model showed a common architecture of 3 domains which are the apical domain that is involved in substrate binding, the intermediate domain and the equatorial domain which carries ATPase activity (Fig. 9). A detailed summary of data collection and refinement statistics was previously published in Yusof et al. (2017). Overall, the PROCHECK analysis showed that the built model of GaTRiC fulfilled the Ramachandran plot with at least 99% in favored and allowed regions. Besides that, model verification using Verify3D showed that the constructed GaTRiC model had positive scores between 77.86% and 90.74%. The third evaluation was done by checking the energy calculation on a protein chain using ANOLEA. The results showed the low content of amino acids with high energy for all subunits which were lower than 30% except for subunit ζ and θ. The evaluations showed that all constructed models were acceptable for tertiary structure analysis except for subunit ζ and θ. The GaTRiC complex was built up by superimposing GaTRiC subunits and S. cerevisiae TRiC subunits where no pair exceeded 2 Å which reflected the high homology between both yeasts TRiC structures. For model analysis, the GaTRiCα, GaTRiCβ, and GaTRiCδ were selected as these models have the best quality for every evaluation analysis. Moreover, sequence analysis showed that these models contained residue substitutions that may reflect the interactions in their tertiary structures. The ion pair interactions were identified using a cut off distance of 4 Å. The analysis shows that the mesophilic S. cerevisiae TRiC has more ion pairs compared to GaTRiC (Table 3).
Fig. 9.
Constructed protein models of (a) GaTRiCα, (b) GaTRiCβ, and (c) GaTRiCδ. The structures are shown in ribbon format with subunits labeled. Ap indicates the apical domain, Int is the intermediate domain and Eq indicates the equatorial domain. The position of amino acids in each domain has been published in Yusof et al. (2017)
Table 3.
Comparison of structural features of psychrophilic and mesophilic TRiC chaperonin
| TRiC subunits | Number of ion pairs | |
|---|---|---|
| G. antarctica | S. cerevisiae | |
| α | 38 | 43 |
| β | 35 | 47 |
| δ | 23 | 27 |
In this study, the critical ion pairs were relatively evaluated by analyzing the conserved ion pairs in mesophilic S. cerevisiae TRiC that did not present in GaTRiC. Locations of the residue substitutions were depicted as shown in Fig. 2 for TRiC α-subunit, Fig. 3 TRiC β-subunit and Fig. 4 for TRiC δ-subunit. In the TRiC α-subunit of S. cerevisiae, Leucine-528 was involved in the inter-ring contact of the equatorial domain via ionic interaction. Leucine-528 interacts with Lysine-524, Lysine-529 and distant residues of Leucine 450, Phenylalanine-451, and Leucine-133. However, in GaTRiCα, the substitution of Leucine-528 to Phenylalanine retains the ionic interaction between the near residues but hinders the interaction to the distant residue of Phenylalanine-451 and Leucine-133. Another intriguing finding was the substitution of Glutamine-272 to Alanine at the GaTRicα apical domain. Glutamine-272 is among the conserved residues in the apical domain protrusion lid that is involved in substrate binding of TRiC. Glutamine-272 is involved with the ionic interaction with Glutamine-269, Proline-267, and Leucine-270 that are conserved in the TRiC apical domain. However, in GaTRiCα the substitution of Glutamine-272 to Alanine hinders the ionic interaction with Glutamine-269, Proline-267, and Leucine-270. On the other hand, GaTRiCβ also contains several substitutions that decrease the ionic interaction on the hinge regions about which loops and domains must move in order to accommodate substrate binding and catalytic cycle. In the apical domain of S. cerevisiae TRiCβ, Glutamine-296 is located at the loop region and interacts with Aspartic acid-300 via ionic interaction. The substitution of Glutamine-296 to Serine hindered the ionic interaction between Glutamine-296 and Glutamic acid-300 due to a distance of more than 5 Å. Besides that, amino acid substitutions on the loop region of the intermediate domain that links the apical and equatorial domains decreased the ionic interaction in the domain. Substitution of Glutamine-222 to Cysteine hindered the ionic interaction between Glutamine-222 and Phenylalanine-298 and Isoleucine-303. As for GaTRiCδ, residue substitution of Histidine-303 to Alanine hindered the ionic interaction with Aspartic acid-299 and Glutamic acid-321 in the apical domain. In the equatorial domain, substitution of Glutamine-125 to Glycine hindered the ionic interaction between Glutamine-125 and Glutamic acid-513 that was conserved in the TRiC ATP binding site. Residue substitutions and interactions were summarized in Table 4.
Table 4.
Ionic interactions in GaTRiC and S. cerevisiae TRiC
| Interaction | TRiC subunits | Domain | Organism | |
|---|---|---|---|---|
| G. antarctica | S. cerevisiae | |||
| Ionic interaction | α | Equatorial |
Phe 525-Lys 521 Phe 525-Lys 526 |
Leu 528-Lys 524 Leu 528-Lys 529 Leu 528-Leu 450 Leu 528-Phe 451 Leu 528-Leu 133 |
| Apical |
Ala 270- Ile 271 Ala 270- Glu 267 |
Gln 272- Gln 269 Gln 272- Pro 267 Gln 272- Leu 270 |
||
| β | Apical |
Ser 296- Tyr 293 Ser 296- Leu 297 |
Gln 296- Tyr 293 Gln 296- Leu 297 Gln 296- Asp 300 |
|
| Intermediate | Cys 222- Gly 219 |
Gln 222-Gly 219 Gln 222-Pro 223 Gln 222-Phe 298 Gln 222-Ile 303 |
||
| δ | Apical | Ala 303- Gly 228 |
His 303-Asp 299 His 303-Glu 321 |
|
| Equatorial | Gly 122-none |
Gln 125-Glu 513 Gln 125-Arg 130 |
||
The structural analysis of GaTRiCα showed substitution of hydrophobic residues to less hydrophobic residues. Interestingly, a few conserved residues in the apical, intermediate, and equatorial domains of GaTRiCα were substituted with slightly less hydrophobic or more hydrophilic residues. Hydrophobic interaction within 5 Å analysis showed that substitution at the equatorial domain; Isoleucine-104 to Valine at the ATPase domain and Valine-180 to Alanine at the intermediate domain of GaTRiCα causes lesser hydrophobic interaction compared to its homolog; S. cerevisiae. The apical domain of GaTRiCα also showed residues substitution of Isoleucine-372 to Valine and Glutamic acid-345 to Aspartic acid where this reflects the flexibility for substrate binding. The substitution of Leucine 218 to Valine at the intermediate domain also reflects a more flexible structure with lower hydrophobic residues. On the other hand, GaTRiCβ showed a high substitution of Isoleucine to Valine at these positions; equatorial position 110 and 115; apical position 285 and intermediate position 367. Parallel to these findings, the GaTRiCδ also showed substitution of Isoleucine to Valine at these positions; intermediate 404 and at the apical domain position-321. High homology nature of GaTRiC with other TRiC from mesophilic organism; S. cerevisiae and B. taurus suggests a possible finding of specific residue substitution that alters the hydrophobic interactions especially those that involved the conserved regions of TRiC. The substitution of Isoleucine to Valine at position 372 in GaTRiCα alters the hydrophobic interactions between Isoleucine-372 side chains that fit into hydrophobic pockets made by Cysteine-226, Glutamine-363, Valine-318, and Leucine-373. Substitution of Valine-372 decreases the hydrophobic interactions by increasing the distance between the residues to more than cut off point of 5 Å. In S. cerevisiae TRiCβ, the Isoleucine-285 interacts with Isoleucine-306, Histidine-308, Leucine-289, Alanine-234, and Leucine-238. However, the substitution of Isoleucine-285 to Valine in GaTRiCβ only allows hydrophobic interactions between Valine at position-285 and Isoleucine at position-306. In GaTRiCδ, substitution of Isoleucine-404 to Valine interfere with hydrophobic interactions made by Isoleucine-404 with conserved distant regions of Leucine-148, Valine-179, and Valine-182 that were located in the regions involved in intra-ring contacts. The substitution Valine-404 only allows hydrophobic interaction between Valine-404 and Valine-182. The substitution of hydrophobic to less hydrophobic residues interferes with the hydrophobic interactions and allows the surface to be more solvent-exposed. Residue substitutions and interactions were summarized in Table 5.
Table 5.
Hydrophobic interactions in GaTRiC and S. cerevisiae TRiC
| Interaction | TRiC subunits | Domain | Organism | |
|---|---|---|---|---|
| G. antarctica | S. cerevisiae | |||
| Hydrophobic | α | Apical | Val 372-Tyr 222 |
Ile 372-Cys 226 Ile 372-Gln 363 Ile 372-Val 318 Ile 372-Leu 373 |
| Asp 345-Glu 346 |
Glu 345-Glu 346 Glu 345-Ser 342 Glu 345-Leu 344 |
|||
| Intermediate | Tyr 218-Leu 220 |
Leu 218-Val 219 Leu 218-Ile 385 |
||
| Ala 180-Leu 406 |
Val 180-Phe 175 Val 180-Ser 405 Val 180-Val 181 |
|||
| Equatorial | Val 104- Ser 100 |
Ile 104-Ile 458 Ile 104-Ser 100 Ile 104-Ala 105 |
||
| β | Apical | Val 285-Ile 306 |
Ile 285-Ile 306 Ile 285-His 308 Ile 285-Leu 289 Ile 285-Ala 234 Ile 285-Leu 238 |
|
| Intermediate |
Val 367-Leu 176 Val 367-Leu 208 |
Ile 367-Val 368 Ile 367-Phe 207 |
||
| Equatorial | Val 110-Ala 106 |
Ile 110-Ala 106 Ile 110-Ile 120 Ile 110-Glu 107 |
||
| Val 115-Gly 113 |
Ile 115-Lys 425 Ile 115-Gln 112 Ile 115-Lys 114 Ile 115-Leu 109 |
|||
| δ | Apical |
Val 321-Glu 326 Val 321-Gln 242 |
Ile 321-Gln 242 Ile 321-Gln 293 Ile 321-Arg 322 Ile 321-Glu 326 |
|
| Intermediate | Val 404-Val 182 |
Ile 404- Leu 148 Ile 404-Val 179 Ile 404-Val 182 |
||
Analysis on aromatic dimers within 6 Å showed the presence of seven aromatic dimers interaction in S. cerevisiae TRiC α-subunit while only four interactions in G. antarctica TRiC α-subunit. The four aromatic dimers in GaTRiCα are located at the equatorial ATP binding site which was also found in S. cerevisiae TRiCα. While another two aromatic dimers interaction in S. cerevisiae TRiCα were located at the hinge region of intermediate and apical domains. Analysis of aromatic-sulfur interactions between 5.3 Å shows that S. cerevisiae TRiCα and GaTRiCα contained an aromatic-sulfur interaction at the equatorial ATP binding domain. At the same time, two aromatic-sulfur interactions were detected at the apical and intermediate domains of S. cerevisiae TRiCα but not in GaTRiCα. Another aromatic interaction is the interaction between the electron-rich aromatic ring and positively charged amino acids which are arginine, lysine, and histidine (Slutsky and Marsh 2004). This interaction is namely known as cation-pi interaction where studies have shown that its bonding energies are significant and have the same order of magnitude as hydrogen bonds and salt bridges (Dougherty and Stauffer 1990; Dougherty 1996; Ma and Dougherty 1997; Zacharias and Dougherty 2002). Analysis of the TRiC α, β, and δ subunits within 6 Å showed that S. cerevisiae has more cation-pi interaction compared to G. antarctica. The GaTRiCα contained two cation-pi interactions where else S. cerevisiae TRiCα contained three interactions where all of the interactions were located at the hinge loop of equatorial and apical domains. For β-subunit analysis, G. antarctica has five cation-pi interaction with one interaction at the equatorial, one at the intermediate, and three at the apical domains whereas S. cerevisiae has eight interactions with three interactions at the apical, two interactions at the intermediate, and three at the equatorial domains. Parallel to this analysis, the δ-subunit of G. antarctica has lesser cation-pi interaction with four interactions compared to S. cerevisiae that has six interactions. The δ-subunit of GaTRiC has three interactions at the apical domain similar to S. cerevisiae δ-subunit. However, G. antarctica only has one interaction at the equatorial domain whereas S. cerevisiae has three interactions. Analysis of the residues that are involved with the aromatic interactions with their positions was described in Table 6.
Table 6.
Aromatic interactions in GaTRiC and S. cerevisiae TRiC
| Interactions | TRiC subunits | Domain | Organism | |
|---|---|---|---|---|
| G. antarctica | S. cerevisiae | |||
| Aromatic dimers | α | Apical | None | Phe 350-Tyr 354 |
| Intermediate | None | Phe 60-Tyr 393 | ||
| Equatorial |
Tyr 129-Phe 449 Tyr 139-Tyr 430 Phe 175-Phe 176 Tyr 478- Phe 496 |
Phe 129-Phe 451 Phe 175-Phe 176 Tyr 432-Phe 436 Tyr 480-Tyr 497 Tyr 497-Tyr 500 |
||
| β | Apical, Intermediate, Equatorial | None | None | |
| δ | Apical, Intermediate, Equatorial | None | None | |
| Aromatic-sulfur | α | Apical | None | Phe 366- Cys 371 |
| Intermediate | None | Phe 60-Met 52 | ||
| Equatorial | Phe 176- Met 179 | Phe 176-Met 179 | ||
| β | Apical, Intermediate, Equatorial | None | None | |
| δ | Apical, Intermediate, Equatorial | None | None | |
| Cation-Pi | α | Apical | Tyr 218-Lys 220 | Phe 366-Lys 211 Phe 366-Arg 388 |
| Intermediate | None | None | ||
| Equatorial | Tyr 497-Arg 495 | Phe 14-Arg 9 | ||
| β | Apical |
Phe 311-Arg 287 Tyr 491- Arg 399 Tyr 491- Arg 495 |
Phe 279-Lys 336 Phe 284-Arg 316 Phe 329-Lys 270 |
|
| Intermediate | Tyr 207-Arg 370 |
Phe 212-Arg 316 Phe 212-Lys 355 |
||
| Equatorial | Phe 23-Arg 19 |
Phe 23-Arg 19 Tyr 464-Arg 460 Tyr 491-Arg 399 |
||
| δ | Apical |
Phe 243-Lys 249 Tyr 308-Arg 272 Phe 327-Arg 199 |
Phe 243- Lys 249 Tyr 308-Arg 272 Phe 327- Arg 199 |
|
| Equatorial | Tyr 431-Lys 134 |
Phe 15-Arg 13 Phe 15-Arg 531 Phe 443-Lys 134 |
||
Discussion
In this study, the expression levels of G. antarctica TRiC (GaTRiC) subunits were analyzed when exposed to different temperatures for six hours. In order to study the response of GaTRiC to low and high temperatures, the mRNA levels of TRiC genes were analyzed where the expression of genes shows an approximate constant expression level with the optimum growth temperature. These findings reflect the importance of TRiC in G. antarctica in all temperatures where the expression of TRiC complex is rather high and expressed constitutively despite cold shock or heat shock. In any event, the constitutive expression of G. antarctica TRiC complex mRNA at 0 °C and − 12 °C suggests that TRiC is important in G. antarctica to accommodate cell regulation in a cold, low-energy environment. In this report, we describe the purification of the native G. antarctica TRiC chaperonin and characterize its functional and structural properties. We found that purified GaTRiC is able to restore denatured luciferase to produce the protein in its functional stage up to 77% when incubated at a low temperature of 4 °C. The hallmark of the function of TRiC chaperonin is the ability to assist folding of denatured proteins to a functional state (Cuellar et al. 2014). TRiC complex is well known to exhibit ATPase activity in the presence of its substrate and the hydrolysis cycle promotes a conformational change of the folding complex (Melki and Cowan 1994). The highest recorded activity for renatured luciferase is when the denatured luciferase is incubated with GaTRiC at 4 °C, followed by 12 °C, 0 °C, and 30 °C. The most plausible explanation of these results is that GaTRiC has the ability of a chaperonin where denatured luciferase is able to be folded to the functional stage with its presence. The luciferase activity also demonstrates that GaTRiC works efficiently at low temperature compared to the high temperature of 30 °C where this shows that GaTRiC has cold-adaptive alterations of structural properties that allow efficient protein folding projected in low temperatures. It is possible that GaTRiC has an optimal folding activity at 4 °C which indicates the maintenance of structural flexibility, whereas there is a loss of flexibility at temperatures higher or lower than 4 °C. This finding is also supported with the study Gobionotothen gibberifrons TRiC/CCT where the psychrophilic TRiC is consistent with maintenance of structural flexibility at 4 °C, whereas that for bovine TRiC/CCT indicates a loss of flexibility at 4 °C (Cueller et al. 2014). According to Somero, 2004, comparative analyses of orthologous psychrophilic, mesophilic, and thermophilic enzymes have shown that the rate-limiting step in enzymatic catalysis is determined by the loops flexibility for substrate binding and product release during the catalytic cycle. Therefore, we hypothesized that amino acid substitutions play a vital role in facilitating the flexibility of the hinge and loop regions for catalytic conformational changes. In order to evaluate the functional finding with the structural analysis, a comparative study was done between G. antarctica TRiC subunits and mesophilic orthologs from S. cerevisiae. In psychrophilic organisms, studies on their enzymes have shown that the rate-limiting step in the enzymatic catalysis is the flexibility of the loops that must move to accommodate substrate binding and product release during catalytic cycle (Somero 2004). The comparative analysis between psychrophilic G. antarctica TRiC chaperonin and mesophilic S. cerevisiae TRiC have shown the presence of residue substitutions at the apical domain that is involved with substrate binding, an intermediate domain that links the apical and equatorial domain and also on an equatorial domain that facilitates ATP hydrolysis for protein conformation change. The residue substitutions which were found in these subunits—α, β, and δ: (i) bulky/polar side chains to alanine or valine, (ii) charged residues to alanine, and (iii) isoleucine to valine are expected to increase intramolecular flexibility within the GaTRiC complex as seen in other cold-adapted variants of several proteins (Alimenti et al. 2003; Fields and Somero 1998; Pucciarelli et al. 2005). Besides the increase in structural flexibility, observation of high residues substitution of alanine on the substrate binding domain showed that non-polar, polar and charged residues of the apical domain may play a role for substrate binding specificity and kinetics binding (Zhuravleva and Radford 2014). Mechanistically, TRiC chaperonin is reported to interact with its substrate via hydrophobic, electrostatic, and polar motifs (Dunn et al. 2001; Kalisman et al. 2012). Pucciarelli et al. 2006 have shown that Antarctic salmon, Notothenia corriceps β and θ-subunits of its TRiC chaperonin contain multiple flexibility-enhancing residue substitutions such as bulky polar residues in the temperate fish replaced by Alanine or Glycine in the psychrophilic fish at the locations that allow conformational changes for binding and release of client proteins at activation energies lower than mesophilic TRiC. Here, we managed a comprehensive study on the intra- and inter-protein interactions in the G. antarctica TRiC subunits in order to refine the understanding of the molecular interactions. For adaptation to cold temperatures, analysis on GaTRiC showed that reduction in electrostatic interactions of ion pairs may play an important role for efficient protein folding. The ionic interactions in G. antarctica TRiC are lower than in S. cerevisiae TRiC due to residue substitutions that hinder the interactions or residues distances that are too far from each other. The higher number of ion pairs in mesophilic S. cerevisiae TRiC compared to G. antarctica TRiC shows that ion pairs contribute to protein stability at elevated temperatures (Elcock 1998). In addition to counting the number of ion pairs with a definite distance cut off, a more accurate comparison of ion pairs between the mesophilic and psychrophilic TRiC was done by evaluating the critical ion pairs. Critical ion pairs were defined as ionic interaction between distance regions of polypeptides with more than five residues and where the corresponding residues in the homologous counterpart do not form an ion pair either because at least one of the two residues was substituted or because the two residues were too far from each other with a distance cutoff of 6 Å (Bae and Phillips Jr, 2004). The analysis on the critical ion pairs shows that several conserved distant ion pairs in the GaTRiC subunits are hindered by residue substitution where it is reasonable as these ionic interactions dominantly contribute to stability in the protein folded state. Thus, in order to relax the rigidity of the protein complex, fewer ion pairs are needed in the structure which leads to high flexibility for efficient catalytic activity in low-energy environments (Perl et al. 2000; Vieille and Zeikus 2001). Another intriguing finding that may contribute to cold adaptation in GaTRiC subunits is the residue substitutions that alter the hydrophobic interactions. A strong correlation between thermal stability and apolar buried surface area is that the more rigid the structure is, the more hydrophobic buried surface area it has (Bae and Phillips Jr, 2004). Hydrophobic interaction is one of the driving forces that allow for spontaneous protein folding into 3D structures (Matthews 2001). In thermophilic organisms, hydrophobic interaction has been thought to be responsible for an increase in thermal stability (Chriswell et al. 2003). Amino acids with large nonpolar or largely nonpolar side chains such as Tryptophan, Phenylalanine, Isoleucine, Leucine, and Cysteine are among the most hydrophobic residues with high solvent transfer free energy (Matthews 2001). The least hydrophobic amino acids are those that are charged and are largely polar such as arginine, lysine, aspartic acid, glutamic acid, histidine, and serine where these residues are grouped as the hydrophilic residues. Therefore, conserved residues in the apical, intermediate and equatorial domains of GaTRiC α, β, and δ were analyzed for hydrophobic interaction which is found to correlate with thermal stability. GaTRiC subunits contain lesser hydrophobic interaction compared to S. cerevisiae TRiC subunits. Substitution of hydrophobic residues to lesser hydrophobic or hydrophilic residues in G. antarctica TRiC decreases the hydrophobic interaction by increasing the residues distance to more than 5 Å. In G. antarctica TRiC subunits, residues that are involved in the hydrophobic interactions with residues in a distant region of polypeptide are replaced with less hydrophobic or more hydrophilic residues. Alteration of hydrophobic interaction by residue substitution at the conserved region may contribute to temperature adaptation. Substitution of Isoleucine to Valine at GaTRiCα position 372, GaTRiCβ position 285, and GaTRiCδ position 404 decreases the residues hydrophobicity and hydrophobic interactions that occur between distance polypeptides. In S. cerevisiae TRiC, the Isoleucine in α, β and δ subunits at position 372, 285 and 404, respectively, is involved in the hydrophobic interaction where the Isoleucine fits into hydrophobic pockets made by residues in a distant region of the polypeptide. The substitution of Isoleucine to Valine in GaTRiCα, GaTRiCβ, and GaTRiCδ hinder the hydrophobic interaction between conserved residues making the hydrophobic surfaces substantially more solvent-exposed compared to S. cerevisiae TRiC subunits. In other mesophilic TRiCδ, there are three hydrophobic interactions that are involved in the intra-ring contact at position Isoleucine-404; however, residue substitutions in GaTRiCδ hinder or maybe lower the hydrophobic interaction by distant regions of more than 5 Å. These substitutions may affect the overall stability and flexibility and probably contribute to the cold adaptation of the psychrophilic G. antarctica TRiC. Moreover, another prevalent interaction that plays a vital role in protein folding and stabilization is the aromatic interactions (Ringer et al. 2007). Aromatic amino acids are usually found in the hydrophobic core of a protein and are involved in the van der Waals contact whether with another aromatic ring forming aromatic dimers (Lanzarotti et al. 2011) or cations which are known as cation-pi interaction (Burley and Petsko 1986) or sulfur atom forming sulfur-aromatic interactions (Ringer et al. 2007). Each interaction of this non-covalent interaction is formed by aromatic rings capable of making an enthalpic contribution to protein structure stability and structure determination (Zauhar et al. 2000). Studies have shown that aromatic residues are one of the main contributors in protein structures for structure stability, folding, protein-protein recognition and ligand binding (Goldstein 2007). Isolated aromatic residues tend to favor the formation of high order clusters such as trimers, tetramers, and pentamers that adopt particular well-defined structures (Lanzarottie et al. 2011). The interactions of pairs of aromatic dimers tend to form larger clusters of trimers and tetramers conformations where these have been associated with a possible role in folding and protein-protein interaction. Our analysis of the aromatic interaction within 6 Å shows that GaTRiC subunits have fewer aromatic interactions compared to S. cerevisiae TRiC subunits. Regulating stability by adjusting the interior interactions of proteins is probably one of the efforts to maintain proper flexibility to perform an efficient catalytic activity in low-energy environment. Most of the aromatic interactions in both G. antarctica TRiC and S. cerevisiae TRiC are located at the equatorial and apical domains. The cation-pi interaction in GaTRiCα consists of only two interactions of adjacent residues with each at the apical and equatorial domain whereas in S. cerevisiae TRiCα, three interactions are found where one of the interactions is formed by distant residues. As the cation-pi interaction in the GaTRiCα and S. cerevisiae TRiCα are located at the hinge loop of respective domains, it is expected that lesser interactions are found in the GaTRiC subunits to facilitate flexibility for efficient folding in cold. Amino acid substitutions observed in the GaTRiCα, β and δ that decrease the ionic and hydrophobic parallel with lesser aromatic interactions compared to S. cerevisiae TRiC subunits are consistent with the evolution of polypeptide flexibility in order to accommodate efficient folding activity in low-energy cold environments (Pucciarelli et al. 2006). For example, the equatorial domain in TRiC is involved in the ATP hydrolysis for conformational change in protein binding and release. The increased flexibility probably allows mobility for the ATP binding site to undergo conformational changes during protein folding by lowering the activation energy of the intra and inter-ring contact between the subunits. It is also reasonable to observe less intra and inter-protein interaction in the apical domain as this allows more flexibility for the helical ‘lid’ to open and close during binding of non-native substrates and release of properly folded substrates. The modest interactions in the intermediate domain that connects the apical and equatorial domains also may play an inevitable role as a flexible hinge to allow the rotation of domains during protein folding. Therefore, we propose that the cold adaptive ability to function efficiently at low temperatures of GaTRiC chaperonin may be based upon the flexibility-enhancing residue substitutions on the apical, intermediate and equatorial domains of the structures. The penalty for having a too flexible structure can lead to cold denaturation due to the entropy contrast between the interior and exterior of the protein (Privalov 1990). Therefore, it is proposed that the residue substitutions observed in the psychrophilic structure undergo positive selection or strategic design selection to prevent the risk of losing native configuration due to cold denaturation. In conclusion, our structural analysis suggests that positive selections of modifications in residue substitutions enhance protein structural flexibility and mobility for efficient folding in cold.
Electronic supplementary material
(DOC 5343 kb)
Acknowledgments
We are grateful to the Ministry of Science, Technology and Innovation, Malaysia, (MOSTI) for funding our project under grant number 02-05-20-SF0007. We thank Prof Jamie Rossjohn for the opportunity to use his lab at Monash University and synchrotron facilities in Australia. Special thanks to Dr. Travis Beddoe from La Trobe University who assisted us with his advice and technical guidance. We are thankful to the Ministry of Higher Education Malaysia for the FRG0463-2017 grant for funding the continuous culturing of Glaciozyma antarctica in Biotechnology Research Institute, Universiti Malaysia Sabah. We also thank those who were involved in the structural project of Glaciozyma antarctica in Malaysia Genome Institute and others who assisted with technical assistance and helpful discussions.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al-Fageeh MB, Smales CM (2006) Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochemical Journal (2):247–259. 10.1042/BJ20060166 [DOI] [PMC free article] [PubMed]
- Alimenti C, Ortenzi C, Carratore V, Luporini P. Structural characterization of En-1, a cold-adapted protein pheromone isolated from the Antarctic ciliate Euplotes nobilii. Biochim Biophys Acta. 2003;1621:17–21. doi: 10.1016/S0304-4165(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Bae E, Phillips GN., Jr Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem. 2004;279(27):28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- Bergholz PW, Bakermans C, Tiedje JM. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol. 2009;191:2340–2352. doi: 10.1128/JB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo SY, Wong CMVL, Rodrigues KF, Najimudin N, Murad AMA, Mahadi NM. Thermal stress responses in Antarctic yeast, Glaciozyma antarctica PI12, characterized by real-time quantitative PCR. Polar Biol. 2013;36:381–389. doi: 10.1007/s00300-012-1268-2. [DOI] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Amino-aromatic interactions in proteins. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-X. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:4–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Campanaro S, Williams TJ, Burg DW, De Francisci D, Treu L, Lauro FM, Cavicchioli R. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Env Microb. 2011;13(8):2018–2038. doi: 10.1111/j.1462-2920.2010.02367.x. [DOI] [PubMed] [Google Scholar]
- Casanueva A, Tuffin M, Cary C, Cowan DA. Molecular adaptations to psychrophily: the impacts of ‘omic’ technologies. Trends Microbiol. 2010;8:129–138. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Criswell AR, Bae E, Stec B, Konisky J, Phillips GN., Jr Structures of thermophilic and mesophilic adenylate kinases from the genus Methanococcus. J Mol Biol. 2003;330:1087–1099. doi: 10.1016/S0022-2836(03)00655-7. [DOI] [PubMed] [Google Scholar]
- Cuellar J, Yébenes H, Parker SK, Carranza G, Serna M, Valpuesta JM, Zabala JC, Detrich HW., III Assisted protein folding at low temperature: evolutionary adaptation of the Antarctic fish chaperonin CCT and its client proteins. Biol Open. 2014;3(4):261–270. doi: 10.1242/bio.20147427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR. The interaction network of the chaperonin CCT. EMBO J. 2008;27:1827–1839. doi: 10.1038/emboj.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty DA (1996) Cation-pi Interactions in Chemistry and Biology: A new view of benzene, phe, tyr, and trp. Science 271(5246):163–168. 10.1126/science.271.5246.163 [DOI] [PubMed]
- Dougherty DA, Stauffer DA (1990) Acetylcholine binding by a synthetic receptor. Implications for biological recognition. Science 250(4987):1558–1560. PMID:2274786 [DOI] [PubMed]
- Dunn AY, Melville MW, Frydman J. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol. 2001;135:176–184. doi: 10.1006/jsbi.2001.4380. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Luthy R, Bowie JU. Verify3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Elcock AH (1998) The stability of salt bridges at high temperatures: implications for hyperthermophilic proteins. J Mol Biol 284:489–502. PMID:9813132 [DOI] [PubMed]
- Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/S1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- Fields PA, Somero GN. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci USA. 1998;95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdaus-Raih M, Hashim NHF, Bharudin I, Abu Bakar MF, Huang KK, Alias H, et al. The Glaciozyma antarctica genome reveals an array of systems that provide sustained responses towards temperature variations in a persistently cold habitat. PLoS ONE. 2018;13(1):e0189947. doi: 10.1371/journal.pone.0189947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Fulda S, Gorman AM, Hori O & Samali A (2010) Cellular stress responses: cell survival and cell death. International Journal of Cell Biology, vol. 2010, 214074, 23. 10.1155/2010/214074 [DOI] [PMC free article] [PubMed]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD and Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: John WM (ed) The proteomics protocols handbook, Humana press, pp 571-607
- Goodchild A, Raftery M, Saunders NFW, Guilhaus M, Cavicchioli R. Biology of the cold adapted archaeon, Methanococcus burtonii determined by proteomics using liquid chromatography-tandem mass spectrometry. J Proteome Res. 2004;3(6):1164–1176. doi: 10.1021/pr0498988. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16(6):574–581. 10.1038/nsmb.1591 [DOI] [PubMed]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hashim N, Bharudin I, Nguong D, Higa S, Bakar F, Nathan S, Rabu A, Kawahara H, Illias R, Najimudin N, Mahadi N, Murad A. Characterization of Afp1, an antifreeze protein from the psychrophilic yeast Glaciozyma antarctica PI12. Extremophiles. 2013;17:63–73. doi: 10.1007/s00792-012-0494-4. [DOI] [PubMed] [Google Scholar]
- Hensen SM, Heldens L, van Enckevort CM, van Genesen ST, Pruijn GJ, Lubsen NH. Activation of the antioxidant response in methionine deprived human cells results in an HSF1-independent increase in HSPA1A mRNA levels. Biochimie. 2013;95:1245–1251. doi: 10.1016/j.biochi.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Jolly C & Morimoto RI (2000) Role of the heat shock response and molecular chaperones in oncogenesis and cell Death. J Natl Cancer Inst. 92(19): 1564–1572. 10.1093/jnci/92.19.1564 [DOI] [PubMed]
- Jones PG, Inouye M. The cold-shock response-a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Kalisman N, Adams CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry and combinatorial homology modeling. Proc Natl Acad Sci U S A. 2012;109:2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisman N, Schröder GF, Levitt M (2013) The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure 21(4):540–549. 10.1016/j.str.2013.01.017 [DOI] [PMC free article] [PubMed]
- Kalman M, Ben-Tal N. Quality assessment of protein model-structures using evolutionary conservation. Bioinformatics. 2010;26(10):1299–1307. doi: 10.1093/bioinformatics/btq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto J, Kurihara T, Kitagawa M, Kato I, Esaki N. Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extremophiles. 2007;11:819–826. doi: 10.1007/s00792-007-0098-6. [DOI] [PubMed] [Google Scholar]
- Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG. Aromatic-aromatic interactions in proteins: beyond the dimer. J Chem Inf Model. 2011;51:1623–1633. doi: 10.1021/ci200062e. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS & Thornton JM (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. 10.1107/S0021889892009944
- Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Struc. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JC, Dougherty DA (1997) The Cation−π interaction. Chem Rev 97(5):1303–1324. 10.1021/cr9603744 [DOI] [PubMed]
- Matthews BW (2001) Hydrophobic interactions in proteins. In: eLS. John Wiley & Sons Ltd, Chichester. 10.1038/npg.els.0002975
- Melki R, Cowan NJ (1994) Facilitated folding of actins and tubulins occurs via a nucleotide- dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol Cell Biol 14:2895-2904. PMCID: PMC358657 [DOI] [PMC free article] [PubMed]
- Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. Proc Int Conf Intell Syst Mol Bio. 1997;5:187–190. doi: 10.1093/nar/gkh440. [DOI] [PubMed] [Google Scholar]
- Melo F, Feytmans E. Assessing protein structures with a non-local atomic interaction energy. J Mol Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM (1992) Stereochemical quality of protein structure coordinates. Proteins Struct Funct Genet 12(4):345–364. 10.1002/prot.340120407 [DOI] [PubMed]
- Phadtare S. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol. 2004;6:125–136. [PubMed] [Google Scholar]
- Phadtare S, Inouye M (2008) The cold shock response. EcoSal Plus 3. 10.1128/ecosalplus.5.4.2 [DOI] [PubMed]
- Phadtare S, Alsina J, Inouye M. Cold shock response and cold shock proteins. Curr Opin Microbiol. 1999;2(2):175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- Perl D, Mueller U, Heinemann U, Schmid FX. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat Struct Bio. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Dgoddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Thomas EF. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piette F, D’Amico S, Mazzucchelli G, Danchin A, Leprince P, Feller G. Life in the cold: a proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl Environ Microbiol. 2011;77(11):3881–3883. doi: 10.1128/AEM.02757-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov PL. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25(4):281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- Pucciarelli S, Marziale F, Di Giuseppe G, Barchetta S, Miceli C. Ribosomal cold- adaptation: characterization of the genes encoding the acidic ribosomal P0 and P2 proteins from the Antarctic ciliate Euplotes focardii. Gene. 2005;360(2):103–110. doi: 10.1016/j.gene.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Pucciarelli S, Parker SK, Detrich HW, III, Melki R. Characterization of the cytoplasmic chaperonin containing TCP-1 from the Antarctic fish Nototheinia coriiceps. Extremophiles. 2006;10(6):537–549. doi: 10.1007/s00792-006-0528-x. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33(2):116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramli A, Mahadi N, Shamsir M, Rabu A, Joyce-Tan K, Murad A, Illias R. Structural prediction of a novel chitinase from the psychrophilic Glaciozyma antarctica PI12 and an analysis of its structural properties and function. J Comput Aided Mol Des. 2012;26(8):947–961. doi: 10.1007/s10822-012-9585-7. [DOI] [PubMed] [Google Scholar]
- Ringer AL, Senenko A, Sherrill CD. Models of S/π interactions in protein structures: comparison of the H2S–benzene complex with PDB data. Prot Soc. 2007;16(10):2216–2223. doi: 10.1110/ps.073002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;3113:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky MM, Marsh ENG (2004) Cation-π interactions studied in a model coiled-coil peptide. Protein Sci 13(8):2244–2251. 10.1110/ps.04702104 [DOI] [PMC free article] [PubMed]
- Somero GN. Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol. 2004;139B:321–333. doi: 10.1016/j.cbpc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28(3):405–420. doi: 10.1093/nar/30.1.276. [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14(11):598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO Jour. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubino P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Tsai C, Aslam K, Drendel HM, Asiago JM, Goode KM, Paul LN, Rochet JC, Hazbun TR. Hsp31 is a stress response chaperone that intervenes in the protein Misfolding process. J Biol Chem. 2015;290(41):24816–24834. doi: 10.1074/jbc.M115.678367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta JM, Martín-Benito J, Gómez-Puertas P, Carrascosa JL, Willison KR. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529:11–16. doi: 10.1016/S0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for Thermostability. Microbiol Mol Biol Rev. 2001;65(1):143–143. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct & Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. Chaperonins: two rings for folding. Trends Biochem Sci. 2011;36:424–432. doi: 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Yusof NA, Abu Bakar FD, Mahadi NM, Murad AMA. Comparative modeling of TCP1 ring complex (TRiC) from a psychrophilic yeast, Glaciozyma antarctica. Transactions on Science and Technology. 2017;4(3–3):324–329. [Google Scholar]
- Yusof NA, Abu Bakar FD, Illias RM, Mahadi NM, Murad AMA. In silico characterisation of the Glaciozyma antarctica genome: mining the molecular chaperones. Malaysian Applied Biology. 2015;44(1):161–165. [Google Scholar]
- Zacharias N, Dougherty DA (2002) Cation–π interactions in ligand recognition and catalysis. Trends Pharmacol Sci 23(6):281–287. 10.1016/s0165-6147(02)02027-8 [DOI] [PubMed]
- Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ. Evidence for a strong sulfur–aromatic interaction derived from crystallographic data. Biopolymers. 2000;53:233–248. doi: 10.1002/(SICI)1097-0282(200003)53:3<233::AID-BIP3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Zheng S, Ponder MA, Shih JY, Tiedje JM, Thomashow MF, Lubman DM. A proteomic analysis of Psychrobacter articus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis. 2007;28:467–488. doi: 10.1002/elps.200600173. [DOI] [PubMed] [Google Scholar]
- Zhuravleva A, Radford SE. How TRiC folds tricky proteins. Cell. 2014;159(6):1251–1252. doi: 10.1016/j.cell.2014.11.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 5343 kb)