Abstract
Purpose
Fluorescence in situ hybridization (FISH) in spermatozoa provides an estimate of the frequency of chromosomal abnormalities, but there is not a clinical consensus on how to statistically analyze sperm FISH results. We therefore propose a statistical approach to establish sperm aneuploidy thresholds in a fertile population.
Methods
We have determined the distribution and variation of the frequency of nullisomy, disomy, and diploidy for a set of 13 chromosomes (1, 2, 9, 13, 15, 16, 17, 18, 19, 21, 22, X, and Y) in sperm nuclei from 14 fertile men by means of automatized FISH. The dispersion of data has been analyzed by the non-parametric Wilcoxon Rank Sum test. We have established the threshold values for each chromosome and aneuploidy type on the basis of the confidence interval values (99.9%).
Results
Nullisomy thresholds ranged from 0.49% for chromosome 19 to 3.09% for chromosome 22; disomy thresholds ranged from 0.30% for chromosome 21 to 1.47% for chromosome 15; diploidy thresholds ranged from 0.24% for the 9/19 chromosome set to 1.21% for the 13/21 chromosome set.
Conclusions
Applying this approach with clinical purposes will enable us to categorize the patient as altered or normal regarding his sperm aneuploidy. Any result surpassing the cited threshold values indicates a 99.9% probability of being significantly different from fertile controls.
Keywords: Human sperm aneuploidy, Fertile donors, Automatized analysis, Fluorescence in situ hybridization (FISH), Aneuploidy thresholds, Nullisomy, Disomy, Diploidy
Introduction
Aneuploidy is the main cause of early pregnancy loss, cognitive impairment, developmental disorders, and infertility in humans. An estimated 10–30% of fertilized human eggs have the “wrong” number of chromosomes, most being either trisomic or monosomic [1]. It has been proposed that embryo aneuploidy can originate at two possible levels: at germ cell (due to meiotic errors) and post-zygotic level (due to mitotic missegregation) [2].
Insofar as germ cells are concerned, it is well established that sperm aneuploidy is strongly correlated with embryo aneuploidy [3, 4]. Over the last two decades, a close correlation between male infertility/subfertility and sperm aneuploidy has been described [5–9]. It has been widely reported that infertile males produce gametes showing a higher rate of chromosomal abnormalities than those of the general population [10]. Specifically, aneuploidy and diploidy frequencies have been directly related to a reduction in the number and progressive motility of sperm [11–13], high levels of FSH [13], and previous IVF failures in normozoospermic patients [14], as reviewed in [7]. An increased sperm aneuploidy rate has also been registered in patients with severe teratozoospermia and is particularly high in macrocephalic spermatozoa [15–18]. Furthermore, it has been reported that some patients with normal semen parameters may also produce higher sperm aneuploidy levels which could be associated with recurrent pregnancy lost (RPL) [19].
Moreover, various studies confirm that the incidence of sperm aneuploidy increases proportionally with male factor severity, as reviewed in [20]. This fact indicates that, in particular cases, the paternal contribution to aneuploidy may be more relevant than that suggested by general data on products of conception and live births [21–24]. This is particularly relevant in cases of assisted reproductive techniques (ART), such as intracytoplasmic sperm injection (ICSI), which have improved the chances of achieving pregnancy, even among patients with severe seminal anomalies [25, 26].
During meiosis, there are two mechanisms that can cause chromosome segregation errors: non-disjunction and anaphase lag. Non-disjunction is considered the main mechanism leading to aneuploidy in sperm [27, 28], and includes chromosome gains and losses, and the production of nullisomic (22 chromosomes) and disomic (24 chromosomes) gametes. On the other hand, anaphase lag produces only nullisomic sperm.
In summary, several male factor conditions could be related with embryo lethality or reproductive inefficacy. However, a clear clinical correlation is pending. Thus, at this moment, we can identify certain patients who may benefit from sperm aneuploidy screening as carriers of chromosome aberrations, severe oligoasthenoteratozoospermia (OAT), non-obstructive azoospermia (NOA), severe teratozoospermia, and unexplained RPL or recurrent implantation failure (RIF).
In this scenario, in order to estimate the risk of embryo aneuploidy in a given sperm sample, the assessment of aneuploidy frequencies by means of fluorescence in situ hybridization (FISH) has been proposed as a prognostic indicator prior to ART.
Nevertheless, the study of sperm aneuploidy with clinical purposes involves certain drawbacks to routine implementation: (i) a large number of single sperm nuclei are required to reach statistical significance. Due to the low incidence of sperm aneuploidy, scoring a large quantity of nuclei for analysis is essential in order to achieve the statistical power needed for accurate quantification. In recent years, automated or semi-automated scoring of sperm multicolor FISH approaches has been compared to manual scoring and has been validated for routine practice in couples consulting for infertility problems [29–35]. In this sense, automated systems can automatically analyze a large number of sperm cells per sample, thus making the sperm-FISH technique more accessible. (ii) In addition to disomy, nullisomy and diploidy rates should also be properly assessed and reported in order to obtain an accurate as possible aneuploidy profile. (iii) The more chromosomes tested, the more accurate the test should be. Routinely, at the clinical level, only a few chromosomes are studied. It is typically stated that FISH studies in sperm should include five chromosomes, usually those implicated in aneuploidies that are compatible with survival, namely, autosomal trisomies (13, Patau syndrome; 18, Edwards’s syndrome; 21, Down syndrome) and sex chromosome aneuploidies, including X chromosome monosomy (the only monosomy viable in humans). Very few laboratories offer to study additional chromosomes (usually chromosomes 15, 16, 17, and 22, which are related to male infertility and implantation failure), thus providing a seven- or nine-chromosome test. (iv) Finally, one of the most critical unresolved points remains which is the threshold at which sperm aneuploidy is clinically significant. Once sperm aneuploidy frequencies from infertile/subfertile male have been assessed, they must be compared with a fertile control group. However, there is not yet a consensus on how a single sample data should be statistically treated versus baseline values in order to determine its normality or abnormality regarding the fertile population. This is the reason why we propose a statistical setup to achieve this comparison allowing each laboratory to generate its own dynamic system of thresholds from baseline values. It is also important to highlight that published aneuploidy baseline values should be used with caution to establish control thresholds, since analytical workflows are not normalized (e.g., different sperm-head decondensation protocols, several chromosomal combinations of probes sets, differences in probe design for a specific chromosome, inter/intra-observer subjectivity in manual systems). In 2004, Tempest and Griffin [8] reviewed the literature focussing on the frequency and distribution of sperm disomy. They already stated that it is difficult, if not impossible, to compare results of the different papers because the analyzed population of males is so heterogeneous. Moreover, there are differences related to scoring criteria and systems, FISH protocol, demographics, and statistical approaches. Therefore, it is always recommendable that each laboratory establishes its own aneuploidy baseline values in fertile controls.
The increases in sperm abnormalities, despite being significant, are usually moderate; however, they are enough to affect the fertile potential of patients. Sperm FISH study constitutes a cytogenetic tool to classify patients according to risk—i.e., those showing a greater incidence of chromosomal abnormalities than those exhibiting baseline values according to the control population—and should be interpreted as an indication of Preimplantation Genetic Test for aneuploidy (PGT-A).
Objective
The aim of our work is to propose a statistical approach based on strict criteria to state statistical differences between a sample from an infertile/subfertile male and the fertile population. To this end, we first describe the distribution of chromosome-specific sperm aneuploidy rates for a set of 13 chromosomes (1, 2, 9, 13, 15, 16, 17, 18, 19, 21, 22, X, and Y) in 14 fertile men. This set includes not only the chromosomes conventionally analyzed by FISH (13, 15, 16, 17, 18, 21, 22, X, and Y) but also the subset of chromosomes 1, 2, 9, and 19. More than 4000 spermatozoa per probe set are analyzed. In addition, an accurate estimation of disomy, nullisomy, and diploidy rates is provided using an automatized scan platform. This allows the description of an accurate aneuploidy baseline for the fertile population.
Secondly, we propose to state an aneuploidy threshold following statistical criteria based on confidence intervals per chromosome and the type of numerical abnormality (nullisomy, disomy, or diploidy) associated, which represents of the main novelties of this work.
Materials and methods
Donor recruitment
Fourteen healthy fertile sperm donors were recruited from CREA Reproductive Medicine Centre’s donor sperm bank program in Valencia (Spain). They were all Caucasian, and all of them met the inclusion criteria established by Spanish’s regulations governing sperm banks. In brief, they had previously fathered at least one child and they had no history of chemotherapy, radiotherapy, or chronic illness. They were all normozoospermic according to World Health Organization (WHO) guidelines [36], with normal karyotype and normal values in serologic tests. They all underwent blood tests to screen for Thalassemia, Von Willerbrand, hypercholesterolemia, and diabetes. Sexually transmitted infections were ruled out (STI’s such as HIV, HBV, HCV, syphilis; measles, toxoplasma, Cytomegalovirus, and herpes were screened in blood samples, and Chlamydia and Neisseria gonorrhoeae were screened in urine). Urine samples also had to prove free of traces of designer drugs. All the subjects declared themselves to be non-smokers and to not consume alcohol regularly. A sperm culture was then obtained in order to rule out infection. Dynamic DNA fragmentation and active caspase 3 assessment by Terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay were performed. Varicocele was also ruled out.
An informed consent was signed by all the donors participating in this study. The study was approved by the Institutional Research Ethics Committee.
Sample collection
Donor semen samples were obtained by masturbation after 2–3 days of sexual abstinence. Once routine sperm parameters were assessed, semen samples were frozen following a slow protocol with liquid nitrogen vapors using TYB with Glycerol (Irvine Scientific, USA) as cryoprotectants. Then, samples were stored in high security CBS canisters (Cryo Bio System, France) in liquid nitrogen (− 196 °C). At the moment of the study, semen samples were thawed by 15–20 min at room temperature, washed in Sperm Washing Medium G- MOPS Plus (Vitrolife, Sweden) (1800 rpm × 10′), and fixed.
Sample fixation
The samples were washed twice (1800 rpm × 10′) in phosphate-buffered saline (PBS; Gibco; Life Technologies, Madrid, Spain) with 0.1% polyvinyl alcohol (PVA; Sigma-Aldrich, Madrid, Spain), fixed in Carnoy’s solution (methanol/acetic acid, 3:1; Merck, Madrid, Spain), spread onto several slides, and air-dried. Afterwards, the slides were washed twice in 0.5 M NaOH (Panreac, Barcelona, Spain) solution for 4 min each, then in 2× SSC (Gibco, Life Technologies) for 10 min, and subsequently dehydrated in an ethanol series (70%, 90%, and 100%) for 1 min each and air-dried for sperm membrane permeabilization and nuclei decondensation.
Fluorescence in situ hybridization
Molecular cytogenetic analysis was performed on 13 chromosomes: 1, 2, 9, 13, 15, 16, 17, 18, 19, 21, 22, X, and Y. Centromeric DNA probes for chromosomes 1, 2, 9, 15, 16, 17, 18, 19, 21, X, and Y and locus-specific probes for chromosomes 13 and 22 were combined in six different sets as follows (fluorophore colors in brackets: S, Spectrum; G, green; O, orange; A, aqua): (i) chromosomes 13[SG]/21[SO], (ii) 18[SA]/X[SG]/Y[SO], (iii) 15[SO]/17[SG], (iv) 16[SA]/22[SG], (v) 1[SG]/2[SO], and (vi) 9[SG]/19[SA]. Probes for chromosomes 13, 21, 18, X, and Y were supplied by Metasystems, Aneuscore-Metasystems kit (IZASA, Barcelona Spain) and probes for chromosomes 2, 9, 15, 16, 17, and 22 by Vysis (Abbott Laboratories, IL, USA). The probe for chromosome 1 was supplied by Cytocell (Cytocell, Cambridge, UK), and the probe for chromosome 19 was supplied by Agilent (Agillent Technologies, Santa Clara, CA, USA). The corresponding probe mix was added to each slide containing a previously decondensed and permeabilized sperm sample. Afterwards, slides were denatured (74 °C for 3 min) and hybridized (37 °C for 4 h) using the Thermobrite Slide Processing System (Abbott Laboratories). Post-hybridization washes were performed to remove any unbound DNA probe, according to the manufacturer’s instructions. Finally, slides were counterstained with 4,6-diamidino-2-phenylindole (DAPI-II; Abbott Laboratories).
Aneuploidy and diploidy assessment
FISH analysis was carried out using an automated system. Scanning, relocation, capture, and fluorescence analysis were performed using an Imager Z2 fluorescence microscope (Zeiss; IZASA, Barcelona Spain) connected to a Metafer-Metacyte® system (Metasystems International; IZASA, Barcelona Spain). The functionality of the software for evaluating sperm nuclei has previously been validated [29]. The MetaCyte imaging software package is based on a flexible cell classifier system that can be calibrated by the user. An automated classifier was programmed to operationalize nuclei selection based on area, contour ratio, sphericity, and eccentricity following strict criteria, as previously described [9, 29, 34]. Cells with abnormal scores were re-analyzed manually by two experienced technicians applying the same strict criteria. In short, the acceptance criteria were (i) hybridization efficiency > 95%, (ii) intact nuclei of similar size that did not overlap and were clearly located, and (iii) nuclei needed to have a similar degree of decondensation, with clear, similarly intense and non-fragmented signals. Only sperm heads showing a regular contour and well-defined limits were evaluated. Disomy and diploidy were confirmed when all signals were of the same intensity and when the separation between signals was at least one diameter size of the signal. Nullisomy was defined when no signal was detected for only one chromosome in a set, in the presence of other chromosome signals. Disomy was defined as two clearly separated signals for the same chromosome and one signal for the other chromosome in a set. Diploidy was defined as two signals from each chromosome in a set. For the particular set of 18/X/Y, diploidy was defined as two signals for gonosomes and two signals for chromosome 18 in the same nucleus.
Statistical analysis
Sperm aneuploidy results were analyzed using the non-parametric Wilcoxon Rank Sum with one tail. The analysis was performed with the “R” statistical package.
Results
A group of 14 healthy fertile donors was studied. Their mean age (± standard deviation, SD) was 25.4 ± 6.0. Their semen parameters are summarized in Table 1, revealing values in a normal range according to WHO guidelines [36]. In brief, mean sperm concentration was 81.6 ± 21.2 million/ml, mean sperm motility was 60 ± 6% a + b, and sperm morphology revealed a mean of 8.2 ± 2.9% of normal forms.
Table 1.
Semen parameters of fertile donors
| Samples | A | B | C | D | E | F | G | H | I | J | K | L | M | N | Mean | SD | WHO reference values |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volume (ml) | 2.0 | 2.5 | 3.3 | 8.2 | 4.3 | 2.0 | 3.9 | 3.0 | 3.2 | 2.6 | 3.2 | 2.7 | 2.8 | 3.9 | 3.4 | 1.5 | 1.5 |
| Total sperm count (million) | 192.0 | 192.5 | 247.5 | 393.6 | 339.7 | 164.0 | 354.9 | 306.0 | 166.4 | 180.6 | 195.2 | 270.0 | 358.4 | 273.0 | 259.6 | 79.9 | 39 |
| Concentration (million/ml) | 96 | 77 | 75 | 48 | 79 | 82 | 91 | 102 | 52 | 81 | 61 | 100 | 128 | 70 | 81.6 | 21.2 | 15 |
| Motility (% a + b) | 59 | 62 | 58 | 60 | 53 | 51 | 65 | 61 | 66 | 65 | 70 | 52 | 65 | 53 | 60.0 | 6.0 | 40 |
| Morphology (% normal forms) | 6 | 7 | 5 | 9 | 10 | 8 | 6 | 7 | 10 | 16 | 8 | 6 | 11 | 6 | 8.2 | 2.9 | 4.0 |
| Age (years) | 30 | 30 | 32 | 19 | 20 | 31 | 26 | 20 | 32 | 18 | 19 | 20 | 29 | 34 | 25.4 | 6.0 | – |
| Body mass index | 21.9 | 27.3 | 24.8 | 25.4 | 25.0 | 22.8 | 28.9 | 23.2 | 20.0 | 20.8 | 24.2 | 24.9 | 24.2 | 26.3 | 24.3 | 2.4 | – |
Sperm aneuploidy and diploidy rates
A total of 281,490 sperm nuclei were analyzed in order to establish aneuploidy rates. An average of 20,106 ± 1725.1 sperm nuclei were analyzed per donor and 3351.1 ± 216.7 sperm nuclei per probe set. Aneuploidy (nullisomy and disomy) rates per chromosome and per donor are shown in Table 2. The mean rate of aneuploidy varied depending on chromosome and aneuploidy type (nullisomy or disomy). The minimum value was 0.16 ± 0.13 (chromosome 21), and the maximum value was 1.42 ± 0.80 (chromosome 16). These lower and upper mean values correspond to nullisomy rate scores. In the same way, the mean rate of disomy ranged from 0.06 (± 0.04) for chromosome 9, to 0.43 (± 0.27) for chromosome 19. Interestingly, the normalized standard distribution range was lower in disomy (from 0.04 to 0.42%) than in nullisomy (from 0.13 to 0.80%). Table 3 shows diploidy rates per chromosome set and per donor analyzed. In this case, the mean rate of diploidy ranged from 0.14 (± 0.08) (for the 9/19 chromosome set) to 0.44 (± 0.30) (for the 13/21 chromosome set). Moreover, the proportion of X-bearing (50.2%) and Y-bearing (49.8%) sperm calculated was not significantly different from the 1:1 ratio (P value = 0.4631; Wilcoxon signed rank test).
Table 2.
Nullisomy and disomy rates per chromosome in 14 fertile donors
| Nullisomy* | Disomy** | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | 1 | 2 | 9 | 13 | 15 | 16 | 17 | 18 | 19 | 21 | 22 | X/Y | 1 | 2 | 9 | 13 | 15 | 16 | 17 | 18 | 19 | 21 | 22 | X/Y | |
| Donor | Nuclei analyzed | ||||||||||||||||||||||||
| A | 18,992 | 0.76 | 0.96 | 0.52 | 0.86 | 0.19 | 2.59 | 0.27 | 1.04 | 0.12 | 0.08 | 0.75 | 1.73 | 0 | 0 | 0.06 | 0.08 | 0.22 | 0.31 | 0.05 | 0.31 | 0.37 | 0.11 | 0.12 | 0.38 |
| B | 19,021 | 1.20 | 0.81 | 0.94 | 0.03 | 0.79 | 1.35 | 0.21 | 0.17 | 0.24 | 0.00 | 0.99 | 0.30 | 0.13 | 0.10 | 0.14 | 0.33 | 1.15 | 0.57 | 0.26 | 0.17 | 0.83 | 0.30 | 0.30 | 0.67 |
| C | 19,588 | 0.31 | 0.41 | 0.71 | 0.86 | 0.43 | 0.40 | 0.12 | 0.38 | 0.25 | 0.17 | 1.67 | 0.93 | 0.03 | 0.03 | 0.03 | 0.09 | 1.47 | 0.46 | 0.25 | 0.03 | 0.17 | 0.11 | 0.25 | 0.19 |
| D | 20,856 | 0.23 | 0.52 | 0.55 | 0.05 | 0.24 | 1.59 | 0.35 | 0.20 | 0.03 | 0.11 | 3.09 | 0.94 | 0.00 | 0.07 | 0.06 | 0.53 | 0.13 | 0.27 | 0.08 | 0.20 | 0.95 | 0.19 | 0.24 | 0.40 |
| E | 20,415 | 0.24 | 0.20 | 0.98 | 1.06 | 0.22 | 0.73 | 0.30 | 0.09 | 0.06 | 0.05 | 1.27 | 0.88 | 0.17 | 0.00 | 0.11 | 0.05 | 0.14 | 0.03 | 0.16 | 0.06 | 0.52 | 0.27 | 0.06 | 0.26 |
| F | 18,961 | 0.53 | 0.34 | 0.94 | 1.56 | 0.61 | 2.08 | 0.76 | 0.37 | 0.32 | 0.26 | 1.11 | 0.81 | 0.00 | 0.19 | 0.06 | 0.11 | 0.55 | 0.58 | 0.09 | 0.34 | 0.83 | 0.09 | 0.83 | 0.28 |
| G | 20,063 | 0.20 | 0.51 | 0.44 | 0.22 | 0.21 | 0.86 | 0.27 | 0.26 | 0.06 | 0.03 | 2.17 | 0.46 | 0.00 | 0.00 | 0.00 | 0.08 | 0.06 | 0.21 | 0.42 | 0.06 | 0.41 | 0.20 | 0.24 | 0.32 |
| H | 19,289 | 0.24 | 0.38 | 0.47 | 0.65 | 0.59 | 0.31 | 0.35 | 0.42 | 0.12 | 0.25 | 1.18 | 0.52 | 0.03 | 0.00 | 0.09 | 0.20 | 0.09 | 0.22 | 0.29 | 0.49 | 0.44 | 0.23 | 0.31 | 0.17 |
| I | 18,796 | 1.38 | 1.38 | 1.07 | 0.51 | 0.82 | 0.80 | 0.88 | 0.57 | 0.21 | 0.11 | 1.30 | 1.24 | 0.32 | 0.45 | 0.03 | 0.06 | 0.25 | 0.23 | 0.16 | 0.12 | 0.46 | 0.20 | 0.47 | 0.39 |
| J | 17,157 | 1.88 | 1.44 | 0.84 | 1.17 | 1.04 | 0.87 | 1.22 | 0.76 | 0.27 | 0.50 | 1.10 | 1.44 | 0.13 | 0.22 | 0.03 | 0.17 | 0.37 | 1.10 | 0.11 | 0.25 | 0.20 | 0.27 | 0.28 | 0.51 |
| K | 19,779 | 0.30 | 0.19 | 0.58 | 0.19 | 0.22 | 1.34 | 0.11 | 0.16 | 0.09 | 0.11 | 1.43 | 0.28 | 0.04 | 0.00 | 0.09 | 0.03 | 0.22 | 0.52 | 0.11 | 0.28 | 0.41 | 0.08 | 0.18 | 0.35 |
| L | 22,681 | 0.23 | 0.44 | 0.40 | 0.55 | 1.79 | 1.99 | 0.46 | 0.00 | 0.49 | 0.10 | 0.97 | 0.19 | 0.05 | 0.14 | 0.11 | 0.02 | 0.17 | 0.27 | 0.21 | 0.12 | 0.22 | 0.12 | 0.15 | 0.16 |
| M | 22,742 | 0.45 | 0.21 | 0.76 | 0.18 | 1.82 | 2.24 | 1.89 | 0.18 | 0.23 | 0.27 | 0.99 | 0.36 | 0.05 | 0.00 | 0.00 | 0.02 | 0.68 | 0.74 | 0.07 | 0.03 | 0.12 | 0.00 | 0.14 | 0.18 |
| N | 23,150 | 0.32 | 0.58 | 0.65 | 0.97 | 2.54 | 2.71 | 0.86 | 0.70 | 0.22 | 0.24 | 0.50 | 1.20 | 0.02 | 0.07 | 0.09 | 0.12 | 0.54 | 0.25 | 0.13 | 0.03 | 0.16 | 0.17 | 0.28 | 0.14 |
| Total | 281,490 | ||||||||||||||||||||||||
| Mean | 20.106 | 0.59 | 0.60 | 0.70 | 0.63 | 0.82 | 1.42 | 0.58 | 0.38 | 0.19 | 0.16 | 1.32 | 0.81 | 0.07 | 0.09 | 0.06 | 0.14 | 0.43 | 0.41 | 0.17 | 0.18 | 0.43 | 0.17 | 0.27 | 0.31 |
| SD | 1725.1 | 0.53 | 0.41 | 0.22 | 0.47 | 0.74 | 0.80 | 0.50 | 0.29 | 0.13 | 0.13 | 0.65 | 0.48 | 0.09 | 0.13 | 0.04 | 0.14 | 0.42 | 0.27 | 0.10 | 0.14 | 0.27 | 0.08 | 0.19 | 0.15 |
*No. of nullisomic nuclei/analyzed nuclei × 100
**No. of disomic nuclei/analyzed nuclei × 100
Table 3.
Diploidy rates per chromosome set in 14 fertile donors
| Diploidy* | |||||||
|---|---|---|---|---|---|---|---|
| Chromosome set | 1/2 | 9/19 | 13/21 | 15/17 | 16/22 | 18/X/Y | |
| Donor | Nuclei analyzed | ||||||
| A | 18,992 | 0.03 | 0.09 | 0.39 | 0.27 | 0.20 | 0.24 |
| B | 19,021 | 0.36 | 0.24 | 1.21 | 0.76 | 0.96 | 1.17 |
| C | 19,588 | 0.10 | 0.03 | 0.29 | 0.12 | 0.12 | 0.35 |
| D | 20,856 | 0.36 | 0.23 | 0.90 | 0.43 | 0.33 | 0.40 |
| E | 20,415 | 0.14 | 0.17 | 0.32 | 0.38 | 0.16 | 0.53 |
| F | 18,961 | 0.42 | 0.06 | 0.34 | 0.23 | 0.11 | 0.25 |
| G | 20,063 | 0.00 | 0.03 | 0.20 | 0.27 | 0.12 | 0.26 |
| H | 19,289 | 0.35 | 0.09 | 0.59 | 0.59 | 0.34 | 0.63 |
| I | 18,796 | 0.24 | 0.06 | 0.36 | 0.28 | 0.56 | 0.45 |
| J | 17,157 | 0.34 | 0.20 | 0.50 | 0.48 | 0.47 | 0.54 |
| K | 19,779 | 0.34 | 0.20 | 0.56 | 0.63 | 0.33 | 0.60 |
| L | 22,681 | 0.28 | 0.24 | 0.26 | 0.25 | 0.10 | 0.25 |
| M | 22,742 | 0.10 | 0.12 | 0.09 | 0.00 | 0.17 | 0.10 |
| N | 23,150 | 0.24 | 0.22 | 0.21 | 0.06 | 0.22 | 0.11 |
| Total | 281,490 | ||||||
| Mean | 20,106.43 | 0.24 | 0.14 | 0.44 | 0.34 | 0.30 | 0.42 |
| SD | 1725.14 | 0.14 | 0.08 | 0.30 | 0.22 | 0.24 | 0.27 |
*No. of diploid nuclei/analyzed nuclei × 100
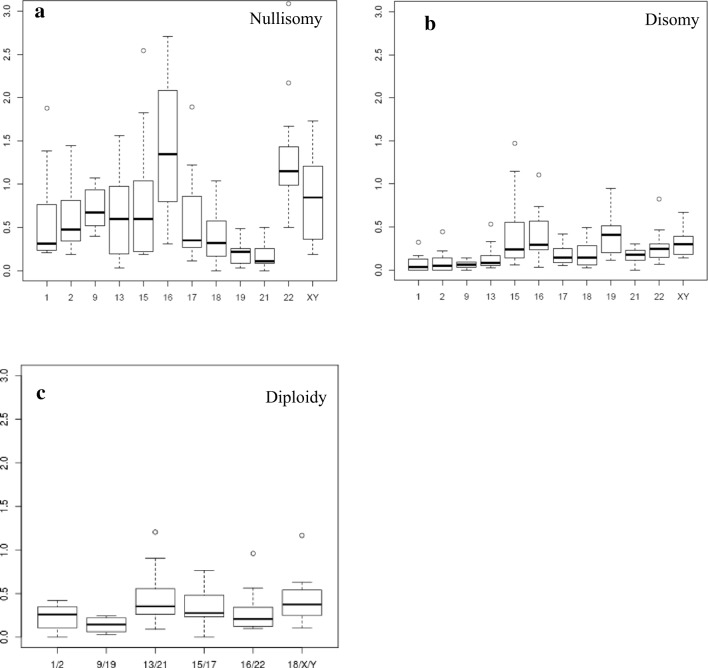
Box plot diagrams were employed to assess the distributional characteristics and dispersion of the percentage of abnormalities per chromosome. Figure 1 shows the distribution of nullisomy, disomy, and diploidy scores per chromosome. In relation to nullisomy, it is important to note the high dispersion of data in an average range per chromosome and the intra-chromosomal variability. The shape of the box plot corresponding to chromosome 16 shows the greatest dispersion, evidencing high variability in this chromosome. Attention should be drawn to the fact that nullisomy data for non-commonly studied chromosomes (i.e., chromosomes 1, 2, 9, 16, and 22) were highly consistent in terms of dispersion. Chromosomes 21 and 19, on the other hand, showed lower variability. In relation to disomy, the average varied among chromosomes, while the data showed a more stable distribution than for nullisomy. Chromosomes 15, 16, and 19 displayed a greater level of dispersion. On the other hand, the distribution of diploidy was stable among the different chromosome sets. Figure 2 represents mean rates of nullisomy, disomy, and diploidy per chromosome and chromosome set, respectively.
Fig. 1.
Box plot of aneuploidy frequencies per chromosome in fertile donors. a Nullisomy and b disomy frequencies per chromosome. c Diploidy frequencies per chromosome set
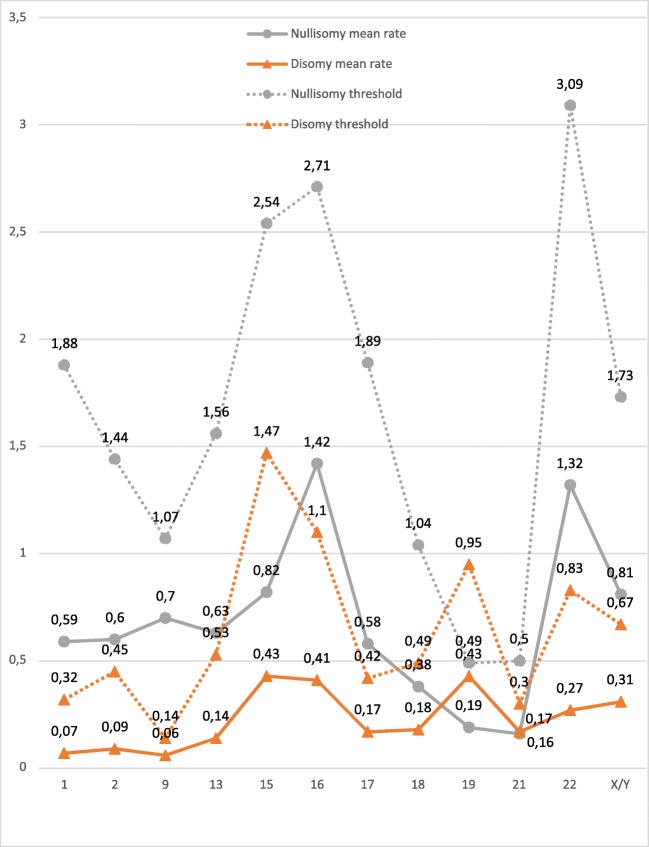
Fig. 2.
Nullisomy and disomy mean rates and thresholds per chromosome
Sperm aneuploidy and diploidy thresholds
The analysis of nullisomy, disomy, and diploidy data shows the data dispersion and allows thresholds to be established for each chromosome and abnormality type in order to determine their clinical relevance. The non-parametric Wilcoxon Rank Sum test was applied in order to determine the lower and upper threshold of the average distribution considering three confidence intervals (CI), 90%, 95%, and 99.9% (α = 0.10, α = 0.05, α = 0.01). Table 4 shows the upper limits of the analysis per chromosome and abnormality type (nullisomy, disomy, and diploidy) with different CIs. Selecting a confidence interval of 99.9% implies that any aneuploidy result obtained that surpasses the threshold values stated will have a 99.9% probability of being classified as significantly different to the fertile control group. In short, nullisomy thresholds range from 0.49% for chromosome 19 to 3.09% for chromosome 22; disomy thresholds range from 0.30% for chromosome 21 to 1.47% for chromosome 15; and diploidy thresholds range from 0.24% for the 9/19 chromosome set to 1.21% for the 13/21 chromosome set (Fig. 3).
Table 4.
Upper values of the confidence interval computed per chromosome and per abnormality type in fertile donors. Results for different α levels 0.1, 0.05, and 0.01 are displayed
| Chromosome | 1 | 2 | 9 | 13 | 15 | 16 | 17 | 18 | 19 | 21 | 22 | XY | |
| Nullisomy (%) | α = 0.01 | 1.88 | 1.44 | 1.07 | 1.56 | 2.54 | 2.71 | 1.89 | 1.04 | 0.49 | 0.50 | 3.09 | 1.73 |
| α = 0.05 | 0.87 | 0.88 | 0.84 | 0.91 | 1.30 | 1.91 | 0.84 | 0.56 | 0.26 | 0.25 | 1.67 | 1.08 | |
| α = 0.10 | 0.82 | 0.82 | 0.81 | 0.86 | 1.20 | 1.79 | 0.76 | 0.52 | 0.25 | 0.21 | 1.57 | 1.05 | |
| Disomy (%) | α = 0.01 | 0.32 | 0.45 | 0.14 | 0.53 | 1.47 | 1.10 | 0.42 | 0.49 | 0.95 | 0.30 | 0.83 | 0.67 |
| α = 0.05 | 0.11 | 0.14 | 0.09 | 0.21 | 0.68 | 0.57 | 0.24 | 0.26 | 0.62 | 0.22 | 0.35 | 0.40 | |
| α = 0.10 | 0.10 | 0.13 | 0.09 | 0.18 | 0.64 | 0.52 | 0.21 | 0.26 | 0.58 | 0.21 | 0.31 | 0.39 | |
| Chromosome set | 1/2 | 9/19 | 13/21 | 15/17 | 16/22 | 18/X/Y | |||||||
| Diploidy (%) | α = 0.01 | 0.42 | 0.24 | 1.21 | 0.76 | 0.96 | 1.17 | ||||||
| α = 0.05 | 0.34 | 0.20 | 0.61 | 0.46 | 0.40 | 0.54 | |||||||
| α = 0.10 | 0.32 | 0.19 | 0.58 | 0.44 | 0.37 | 0.53 | |||||||
Fig. 3.
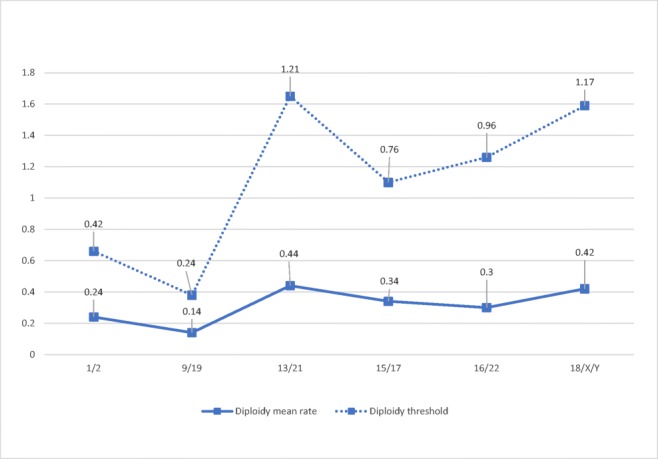
Diploidy mean rates and thresholds per chromosome set
Discussion
According to several reports [6, 37–40], our FISH results indicate different incidences of aneuploidy for each chromosome and also for aneuploidy and ploidy type (nullisomy or disomy and diploidy). While it is recognized that all chromosomes are predisposed to non-disjunction, certain chromosomes are more prone to display an increased frequency of non-disjunction. The mechanisms proposed for the prevalence of certain aneuploidies include the presence and/or variants of nucleolar organizer regions (NORs), presence and/or variants of heterochromatin, altered frequencies of recombination, differences in size, and non-homologous pairing of sex chromosomes in males [41]. Therefore, it seems logical to establish a particular threshold for each chromosome and aneuploidy type (nullisomy and disomy). In the same way, diploidy thresholds must be specifically assessed for each chromosome set. In this sense, a total aneuploidy threshold for all the chromosomes studied should be established with caution, since it could mask the abnormality observed for single chromosomes.
Disomy and nullisomy are a result of mal-segregation during meiosis. However, nullisomy is not always assessed by laboratories when applying aneuploidy FISH study, claiming that it can be confused with hybridization errors. In this sense, simultaneous scoring of two fluorochromes in the same nucleus provides an internal control to differentiate nullisomy from failure hybridization. The assessment of nullisomy involves ensuring that each nucleus is checked one by one, at least three times, once for each fluorochrome. The use of automated systems allows to superimpose images captured with different fluorescence filters avoiding manual counting errors. This assures that nullisomy is scored as the lack of a signal in the presence of another signal in the same nucleus. This also ensures that there is no failure in decondensation and permeabilization process nor in hybridization. Regarding nullisomy values, our results reveal higher rates of nullisomy (ranging from 0.16 to 1.42%) than those of disomy (ranging from 0.07 to 0.43%) for most of the chromosomes tested. This is true for all the chromosomes studied, except for chromosomes 19 and 21, which showed higher rates of disomy than nullisomy (0.19 and 0.16 versus 0.43 and 0.17% respectively). Meiosis is a highly regulated process, and errors in any of the steps involved activate meiotic checkpoints. A direct consequence of the activation of checkpoints is a decrease in the number of cells between the beginning and the end of spermatogenesis (reviewed in [42]). Thus, a higher prevalence of nullisomy over disomy in sperm is attributable to more effective checkpoint mechanisms against disomy than against nullisomy during spermatogenesis. Moreover, it has previously been suggested that differences in the incidence of aneuploidy between spermatozoa and oocytes are not due to differences in chromosome segregation errors, but rather to more effective checkpoint mechanisms during spermatogenesis than during oogenesis [1, 28]. In accordance with our results, Pellestor reported a significant excess of hypohaploidy over hyperhaploidy in spermatic karyotypes [40]. Pang and co-workers also reported an excess of nullisomy over disomy and proposed that equal frequencies would only be expected if all forms of aneuploidy arose through non-disjunction [39]. However, the excess of nullisomy over disomy might be explained by the fact that nullisomy also arises from the loss of chromosomes after a delay in anaphase. Considering that the frequency of nullisomy can be equal to or greater than that of disomy, it is important to highlight the relevance of assessing nullisomy and not only disomy when testing for aneuploidies in sperm at the clinical level. It must be taken into account that sperm nullisomy leads to embryo monosomy and that monosomies are more deleterious for embryo development than trisomies. This harmful effect manifests itself as spontaneous abortions, infertility, or sterility. In 2004, Munné and co-workers [43] observed a significant excess of monosomies over trisomies when assessing cleavage stage aneuploidy, disproving the previously assumed 1:1 ratio. Monosomies mostly affected acrocentric chromosomes suggesting that they are more prone to non-disjunction mechanisms. In 2012, Rabinowitz and co-workers [44] corroborated these results. Moreover, they stated that monosomies were of paternal well as maternal origin, with paternal monosomies for chromosomes 1, 2, 4, 8, 16, 19, 21, and 22 being more frequent.
These results, together with the differences observed between the chromosomes we have tested for incidence and dispersion of aneuploidy, stress the need to define which chromosomes need to be studied. Obviously, increasing the number of chromosomes studied will make testing more accurate. According to our results, some chromosomes that are not routinely included in sperm aneuploidy tests at the clinical level show a nullisomy or disomy frequency similar to that of chromosomes routinely included in the said tests. This is particularly true for nullisomy frequency in chromosomes 19 and 21, and for disomy frequency in chromosomes 2 and 13, for example (Fig. 1). Furthermore, aneuploidy data dispersion helps us to determine which chromosomes are more stable regarding aneuploidy and which chromosomes present higher variability. Our results (Fig. 1) show that disomy and diploidy are more stable variants (since they show less dispersion) than nullisomy. On the other hand, disomy for chromosomes 15, 16, 19, 22, X, and Y and nullisomy for chromosomes 16, 22, X, and Y show higher values and greater dispersion at the fertile population baseline level.
As far as we know, our aneuploidy study includes the largest chromosome series analyzed by means of automatized FISH. Templado and co-workers surveyed prior studies from 1995 to 2010 covering percentages of disomy per chromosome in sperm from normal men [45]. Only multicolor FISH studies with a minimum of five donors and strict scoring criteria were included, and none of the studies reviewed included chromosome 17 or 19. Despite this, the weighted mean values of disomy reported [45] are in line with our results. In terms of diploidy, our results show a quite balanced distribution among chromosomes exhibiting a more stable behavior, as expected. Thus, if diploidy is detected in a given pair of chromosomes, we can assume there is diploidy in the rest of the chromosome complement. The differences observed in the whole could be due to the study of different spermatozoa for each pair of chromosomes. Taking all this into account, an accurate sperm aneuploidy test for clinical purposes should include more chromosomes (other than the five that are routinely tested) and nullisomy assessment. Moreover, variability in the frequencies of meiotic chromosome abnormalities among fertile men has also been described [46–49]. The existence of normal fertile men with increased non-disjunction in the general population makes it advisable to study an increased number of individuals in order to minimize the effect of potential outlier rates of sperm chromosome numerical abnormalities [48]. In this sense, the statistical approach we propose to determine threshold values can and should be updated regularly by adding new fertile donor aneuploidy data in order to increase the fitness and accuracy of the test.
The statistical approaches routinely used to detect differences between two groups when the population is normally distributed are variance analysis tests, a t test or ANOVA; this would be the case if our two groups had a similar n. However, when we want to determine sperm aneuploidy in the sperm sample of an individual patient, we cannot assume our population is normally distributed. Indeed, what we are comparing is one individual (patient) to one control group (fertile men). For this reason, as an alternative to a t test or ANOVA, we propose a Wilcoxon non-parametric test and a confidence interval in order to determine aneuploidy threshold values. In this way, we obtain an accurate probability, which allows a prognostic result. At this point, by selecting a CI of 99.9%, any aneuploidy data surpassing any threshold value stated for said CI will have a 99.9% probability of being significantly different from the fertile control group, thus classifying it as altered. These aneuploidy threshold values constitute an aneuploidy distribution model in a fertile population, making it possible to compare patients with the control group. Furthermore, we can then assume that any patient surpassing any sperm aneuploidy threshold value is at risk of generating more aneuploidy gametes than fertile counterparts.
One of the limitations of this study is that the number of controls was low. In fact, an accurate power analysis based on Gaussian distribution establishes that the minimum sample size required to ascertain the maximum reliability to set a threshold will depend on chromosome and aneuploidy type, because each aneuploidy type has its own deviation for each chromosome. However, this kind of design is out of the scope of this initial work. In this sense, it is important to highlight the importance of adding new control data in order to increase the fitness and accuracy of the test when categorizing infertile men into normal or altered regarding sperm aneuploidy. However, as a preliminary study, its relevance lies in the fact that it proposes a novel statistical approach—based on strict criteria—to statistically stress differences between an infertile/subfertile male and the fertile population.
The application of this tool in infertile/subfertile men could help us to deepen the etiological knowledge of reproductive male impairment. In this sense, the reproductive male failure is an extremely complex trait that includes overlapping phenotypes. This circumstance complicates the study of its biological basis. For this reason, it is important to note the relevance of collecting semen parameters and accurate clinical data along with sperm aneuploidy levels. Thus, the establishment of accurate correlations between aneuploidy level and clinical traits could help to improve reproductive genetic counselling in male factor.
Conclusion
We have determined the distribution and variation in frequency of aneuploidy (nullisomy, disomy) and diploidy for a set of 13 chromosomes (1, 2, 9, 13, 15, 16, 17, 18, 19, 21, 22, X, and Y) in sperm nuclei from 14 fertile men. We also suggest a new statistical approach to address aneuploidy results obtained by FISH based on the establishment of chromosome and aneuploidy type thresholds, in order to categorize the patient as altered or normal regarding his sperm aneuploidy. As far as we know, this is the first study to propose upper CI values (99.9%) of nullisomy, disomy, and diploidy frequencies obtained from fertile men as thresholds. This statistical approach also allows the determination of a unique criterion to statistically stress differences between a sample from one infertile/subfertile male and those of the fertile population that goes beyond a cutoff point. Applying this approach with clinical purposes will enable us to categorize the patient as altered or normal regarding his sperm aneuploidy. Any result surpassing the cited threshold values indicates a 99.9% probability of being significantly different from fertile controls. We are aware that the number of controls studied in this case is limited; however, this is offset by one of advantages of our approach, which is that these threshold values can be updated regularly by adding new data for fertile men, with the goal of increasing the fitness and accuracy of the test and the subsequent categorization of infertile men as normal or altered, regarding sperm aneuploidy. Also, since analytical workflows between laboratories are not normalized, we want to emphasize the necessity for individual laboratories to determine their own sperm aneuploidy thresholds according to their own aneuploidy baseline values from control population and their own FISH protocol. In this sense, our proposal constitutes a tool for each laboratory to setup aneuploidy thresholds from baseline values according to their conditions. Finally, to gain a better understanding of the clinical significance of increased sperm aneuploidy, prospective sperm aneuploidy studies must be performed. These further studies will require more case and control subjects, in order to ascertain specific sperm dysfunctions, which are the prevalent cause of aneuploidy.
Compliance with ethical standards
An informed consent was signed by all the donors participating in this study. The study was approved by the Institutional Research Ethics Committee.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 2.Egozcue S, Blanco J, Vendrell JM, Garcia F, Veiga A, Aran B, et al. Human male infertility: chromosome anomalies, meiotic disorders, abnormal spermatozoa and recurrent abortion. Hum Reprod Update. 2000;6(1):93–105. doi: 10.1093/humupd/6.1.93. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigo L, Peinado V, Mateu E, Remohi J, Pellicer A, Simon C, et al. Impact of different patterns of sperm chromosomal abnormalities on the chromosomal constitution of preimplantation embryos. Fertil Steril. 2010;94(4):1380–1386. doi: 10.1016/j.fertnstert.2009.05.061. [DOI] [PubMed] [Google Scholar]
- 4.Sills ES, Li X, Frederick JL, Khoury CD, Potter DA. Determining parental origin of embryo aneuploidy: analysis of genetic error observed in 305 embryos derived from anonymous donor oocyte IVF cycles. Mol Cytogenet. 2014;7(1):68. doi: 10.1186/s13039-014-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrello N, Arcidiacono G, Vicari E, Asero P, Di Benedetto D, De Palma A, et al. Morphologically normal spermatozoa of patients with secretory oligo-astheno-teratozoospermia have an increased aneuploidy rate. Hum Reprod. 2004;19(10):2298–2302. doi: 10.1093/humrep/deh438. [DOI] [PubMed] [Google Scholar]
- 6.Durakbasi-Dursun HG, Zamani AG, Kutlu R, Görkemli H, Bahce M, Acar A. A new approach to chromosomal abnormalities in sperm from patients with oligoasthenoteratozoospermia: detection of double aneuploidy in addition to single aneuploidy and diploidy by five-color fluorescence in situ hybridization using one probe set. Fertil Steril. 2008;89(6):1709–1717. doi: 10.1016/j.fertnstert.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 7.Sarrate Z, Blanco J, Anton E, Egozcue S, Egozcue J, Vidal F. FISH studies of chromosome abnormalities in germ cells and its relevance in reproductive counseling. Asian J Androl. 2005;7(3):227–236. doi: 10.1111/j.1745-7262.2005.00061.x. [DOI] [PubMed] [Google Scholar]
- 8.Tempest HG, Griffin DK. The relationship between male infertility and increased levels of sperm disomy. Cytogenet Genome Res. 2004;107(1–2):83–94. doi: 10.1159/000079575. [DOI] [PubMed] [Google Scholar]
- 9.Vendrell X, Ferrer M, García-Mengual E, Muñoz P, Triviño JC, Calatayud C, Rawe VY, Ruiz-Jorro M. Correlation between aneuploidy, apoptotic markers and DNA fragmentation in spermatozoa from normozoospermic patients. Reprod BioMed Online. 2014;28(4):492–502. doi: 10.1016/j.rbmo.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Egozcue J, Blanco J, Anton E, Egozcue S, Sarrate Z, Vidal F. Genetic analysis of sperm and implications of severe male infertility—a review. Placenta. 2003;24(Suppl B):S62–S65. doi: 10.1016/s0143-4004(03)00186-3. [DOI] [PubMed] [Google Scholar]
- 11.Collodel G, Capitani S, Baccetti B, Pammolli A, Moretti E. Sperm aneuploidies and low progressive motility. Hum Reprod. 2007;22(7):1893–1898. doi: 10.1093/humrep/dem099. [DOI] [PubMed] [Google Scholar]
- 12.Vegetti W, Van Assche E, Frias A, Verheyen G, Bianchi MM, Bonduelle M, et al. Correlation between semen parameters and sperm aneuploidy rates investigated by fluorescence in-situ hybridization in infertile men. Hum Reprod. 2000;15(2):351–365. doi: 10.1093/humrep/15.2.351. [DOI] [PubMed] [Google Scholar]
- 13.Vendrell JM, Garcia F, Veiga A, Calderon G, Egozcue S, Egozcue J, Barri PN. Meiotic abnormalities and spermatogenic parameters in severe oligoasthenozoospermia. Hum Reprod. 1999;14(2):375–378. doi: 10.1093/humrep/14.2.375. [DOI] [PubMed] [Google Scholar]
- 14.Egozcue S, García F, López-Teijón M, Olivares R, Serra O, Aura M, et al. Estudio de meiosis en biopsia testicular y su correlación con el patrón seminológico. Rev Iberoam Fertil Reprod Hum. 2004;(Supl 1):252.
- 15.Brahem S, Mehdi M, Elghezal H, Saad A. Analysis of sperm aneuploidies and DNA fragmentation in patients with globozoospermia or with abnormal acrosomes. Urology. 2011;77(6):1343–1348. doi: 10.1016/j.urology.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 16.Brahem S, Mehdi M, Elghezal H, Saad A. Study of aneuploidy rate and sperm DNA fragmentation in large-headed, multiple-tailed spermatozoa. Andrologia. 2012;44(2):130–135. doi: 10.1111/j.1439-0272.2010.01115.x. [DOI] [PubMed] [Google Scholar]
- 17.Collodel G, Moretti E. Sperm morphology and aneuploidies: defects of supposed genetic origin. Andrologia. 2006;38(6):208–215. doi: 10.1111/j.1439-0272.2006.00742.x. [DOI] [PubMed] [Google Scholar]
- 18.Mehdi M, Gmidene A, Brahem S, Guerin JF, Elghezal H, Saad A. Aneuploidy rate in spermatozoa of selected men with severe teratozoospermia. Andrologia. 2012;44(Suppl 1):139–143. doi: 10.1111/j.1439-0272.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- 19.Ramasamy R, Scovell JM, Kovac JR, Cook PJ, Lamb DJ, Lipshultz LI. Fluorescence in situ hybridization detects increased sperm aneuploidy in men with recurrent pregnancy loss. Fertil Steril. 2015;103(4):906–909. doi: 10.1016/j.fertnstert.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piomboni P, Stendardi A, Gambera L. Chromosomal aberrations and aneuploidies of spermatozoa. In: Baldi E, Muratori M, editors. Genetic damage in human spermatozoa. Advances in experimental medicine and biology 791. New York: LLC Springer; 2014. pp. 27–52. [DOI] [PubMed] [Google Scholar]
- 21.Gianaroli L, Magli MC, Cavallini G, Crippa A, Nadalini M, Bernardini L, Menchini Fabris GF, Voliani S, Ferraretti AP. Frequency of aneuploidy in sperm from patients with extremely severe male factor infertility. Hum Reprod. 2005;20(8):2140–2152. doi: 10.1093/humrep/dei033. [DOI] [PubMed] [Google Scholar]
- 22.Harton GL, Tempest HG. Chromosomal disorders and male infertility. Asian J Androl. 2012;14(1):32–39. doi: 10.1038/aja.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahraman S, Findikli N, Biricik A, Oncu N, Ogur C, Sertyel S, Karlikaya G, Karagozoglu H, Saglam Y. Preliminary FISH studies on spermatozoa and embryos in patients with variable degrees of teratozoospermia and a history of poor prognosis. Reprod BioMed Online. 2006;12(6):752–761. doi: 10.1016/s1472-6483(10)61087-5. [DOI] [PubMed] [Google Scholar]
- 24.Magli MC, Gianaroli L, Ferraretti AP, Gordts S, Fredericks V, Crippa A. Paternal contribution to aneuploidy in preimplantation embryos. Reprod BioMed Online. 2009;18(4):536–542. doi: 10.1016/s1472-6483(10)60131-9. [DOI] [PubMed] [Google Scholar]
- 25.Tesarik J, Mendoza C. Treatment of severe male infertility by micromanipulation-assisted fertilization: an update. Front Biosci. 2007;12:105–114. doi: 10.2741/2051. [DOI] [PubMed] [Google Scholar]
- 26.Verpoest W, Tournaye H. ICSI: hype or hazard? Hum Fertil. 2006;9(2):81–92. doi: 10.1080/14647270500422158. [DOI] [PubMed] [Google Scholar]
- 27.Márquez C, Egozcue J, Martorell MR, Moreno V, Templado C. Colcemid increases the frequency of chromosome abnormalities in human sperm. Cytogenet Cell Genet. 1996;72(2–3):164–170. doi: 10.1159/000134177. [DOI] [PubMed] [Google Scholar]
- 28.Templado C, Uroz L, Estop A. New insights on the origin and relevance of aneuploidy in human spermatozoa. Mol Hum Reprod. 2013;19(10):634–643. doi: 10.1093/molehr/gat039. [DOI] [PubMed] [Google Scholar]
- 29.Carrell DT, Emery BR. Use of automated imaging and analysis technology for the detection of aneuploidy in human sperm. Fertil Steril. 2008;90(2):434–437. doi: 10.1016/j.fertnstert.2007.06.095. [DOI] [PubMed] [Google Scholar]
- 30.Lammers J, Splingart C, Barrière P, Jean M, Fréour T. Double-blind prospective study comparing two automated sperm analyzers versus manual semen assessment. J Assist Reprod Genet. 2014;31(1):35–43. doi: 10.1007/s10815-013-0139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez G, Gillois P, Le MM, Borye R, Esquerré-Lamare C, Satre V, et al. FISH and tips: a large scale analysis of automated versus manual scoring for sperm aneuploidy detection. Basic Clin Androl. 2013;23:13. doi: 10.1186/2051-4190-23-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina O, Sarrate Z, Vidal F, Blanco J. FISH on sperm: spot-counting to stop counting? Not yet. Fertil Steril. 2009;92(4):1474–1480. doi: 10.1016/j.fertnstert.2008.07.1779. [DOI] [PubMed] [Google Scholar]
- 33.Netten H, Young IT, van Vliet LJ, Tanke HJ, Vroljik H, Sloos WC. FISH and chips: automation of fluorescent dot counting in interphase cell nuclei. Cytometry. 1997;28(1):1–10. [PubMed] [Google Scholar]
- 34.Perry MJ, Chen X, Lu X. Automated scoring of multiprobe FISH in human spermatozoa. Cytometry A. 2007;71(11):968–972. doi: 10.1002/cyto.a.20468. [DOI] [PubMed] [Google Scholar]
- 35.Tempest HG, Cheng SY, Gillott DJ, Handyside AH, Thornhill AR, Griffin DK. Scoring of sperm chromosomal abnormalities by manual and automated approaches: qualitative and quantitative comparisons. Asian J Androl. 2010;12(2):257–262. doi: 10.1038/aja.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. Examination and processing of human semen. 2010, 5 edition. Retrieved from https://www.who.int/reproductivehealth/publications/infertility/9789241547789/en/.
- 37.Martin RH, Ko E, Rademaker A. Distribution of aneuploidy in human gametes: comparison between human sperm and oocytes. Am J Med Genet. 1991;39(3):321–331. doi: 10.1002/ajmg.1320390315. [DOI] [PubMed] [Google Scholar]
- 38.Neusser M, Rogenhofer N, Dürl S, Ochsenkühn R, Trottmann M, Jurinovic V, Steinlein O, von Schönfeldt V, Müller S, Thaler CJ. Increased chromosome 16 disomy rates in human spermatozoa and recurrent spontaneous abortions. Fertil Steril. 2015;104(5):1130–1137. doi: 10.1016/j.fertnstert.2015.07.1160. [DOI] [PubMed] [Google Scholar]
- 39.Pang MG, Hoegerman SF, Cuticchia AJ, Moon SY, Doncel GF, Acosta AA, Kearns WG. Detection of aneuploidy for chromosomes 4, 6, 7, 8, 9, 10, 11, 12, 13, 17, 18, 21, X and Y by fluorescence in-situ hybridization in spermatozoa from nine patients with oligoasthenoteratozoospermia undergoing intracytoplasmic sperm injection. Hum Reprod. 1999;14(5):1266–1273. doi: 10.1093/humrep/14.5.1266. [DOI] [PubMed] [Google Scholar]
- 40.Pellestor F. Differential distribution of aneuploidy in human gametes according to their sex. Hum Reprod. 1991;6(9):1252–1258. doi: 10.1093/oxfordjournals.humrep.a137522. [DOI] [PubMed] [Google Scholar]
- 41.Spriggs EL, Rademaker AW, Martin RH. Aneuploidy in human sperm: the use of multicolor FISH to test various theories of nondisjunction. Am J Hum Genet. 1996;58(2):356–362. [PMC free article] [PubMed] [Google Scholar]
- 42.Sarrate Z, Vidal F, Blanco J. Meiotic abnormalities in metaphase I human spermatocytes from infertile males: frequencies, chromosomes involved, and the relationships with polymorphic karyotype and seminal parameters. Asian J Androl. 2014;16(6):838–844. doi: 10.4103/1008-682X.135126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munné S, Bahçe M, Sandalinas M, Escudero T, Márquez C, Velilla E, Colls P, Oter M, Alikani M, Cohen J. Differences in chromosome susceptibility to aneuploidy and survival to first trimester. Reprod BioMed Online. 2004;8(1):81–90. doi: 10.1016/s1472-6483(10)60501-9. [DOI] [PubMed] [Google Scholar]
- 44.Rabinowitz M, Ryan A, Gemelos G, Hill M, Baner J, Cinnioglu C, Banjevic M, Potter D, Petrov DA, Demko Z. Origins and rates of aneuploidy in human blastomeres. Fertil Steril. 2012;97(2):395–401. doi: 10.1016/j.fertnstert.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 45.Templado C, Vidal F, Estop A. Aneuploidy in human spermatozoa. Cytogenet Genome Res. 2011;133(2–4):91–99. doi: 10.1159/000323795. [DOI] [PubMed] [Google Scholar]
- 46.Rubes J, Vozdova M, Robbins WA, Rezacova O, Perreault SD, Wyrobek AJ. Stable variants of sperm aneuploidy among healthy men show associations between germinal and somatic aneuploidy. Am J Hum Genet. 2002;70(6):1507–1519. doi: 10.1086/340791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tempest HG, Ko E, Rademaker A, Chan P, Robaire B, Martin RH. Intra-individual and inter-individual variations in sperm aneuploidy frequencies in normal men. Fertil Steril. 2009;91(1):185–192. doi: 10.1016/j.fertnstert.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Templado C, Bosch M, Benet J. Frequency and distribution of chromosome abnormalities in human spermatozoa. Cytogenet Genome Res. 2005;111(3–4):199–205. doi: 10.1159/000086890. [DOI] [PubMed] [Google Scholar]
- 49.Uroz L, Rajmil O, Templado C. Meiotic chromosome abnormalities in fertile men: are they increasing? Fertil Steril. 2011;95(1):141–146. doi: 10.1016/j.fertnstert.2010.06.042. [DOI] [PubMed] [Google Scholar]