Abstract
Stress-activated and mitogen-activated protein kinases (MAPKs) regulate gene expression by post-translational modifications of transcription factors. Elk-1, a transcription factor that regulates the expression of immediate early genes, is amenable to regulation by all the three mammalian MAPKs. In the present report, using inhibitors specific for different MAPK pathways, we show that during exposure of HeLa cells to heat stress, Elk-1 is SUMOylated with SUMO1 by p38 MAPK pathway-dependent mechanisms. Elk-1-phosphorylation levels were significantly reduced under similar conditions. We also show that transcriptional activity of Elk-1 as assessed by luciferase reporter expression and qPCR estimation of the expression of genes regulated by Elk-1 was downregulated upon exposure to heat stress; this downregulation was reversed when heat exposure was performed in the presence of either SB203580 (p38 MAPK inhibitor) or ginkgolic acid (inhibitor of SUMOylation). Elk-1 induced transcription is also regulated by PIAS2 which acts as a coactivator upon the activation of extracellular signal-regulated kinases (ERKs) and as a corepressor upon its phosphorylation by p38 MAPK. Since heat stress activates the p38 MAPK pathway, we determined if PIAS2 was phosphorylated in heat-stressed HeLa cells. Our studies indicate that in HeLa cells exposed to heat stress, PIAS2 is phosphorylated by p38 MAPK pathway-dependent mechanisms. Collectively, the results presented demonstrate that in heat-stressed HeLa cells, p38 MAPK pathway-dependent SUMOylation of Elk-1 and phosphorylation of PIAS2 correlate with the downregulation of transactivation by Elk-1.
Electronic supplementary material
The online version of this article (10.1007/s12192-019-00974-4) contains supplementary material, which is available to authorized users.
Keywords: Heat stress, p38 MAPK, Elk-1, SUMOylation, PIAS2, Phosphorylation
Introduction
Exposure of cells to heat stress induces numerous deleterious effects which include denaturation and aggregation of proteins and enzymes, impairment of cytoskeletal structures, cell cycle arrest, and downregulation of macromolecular biosynthesis (Cuenda and Rousseau 2007). Prolonged exposure to heat stress may also lead to cell death (Gu et al. 2014). Cells exposed to heat stress elicit a wide variety of responses aimed at maintaining normal cellular homeostasis; these include the induction of heat shock proteins (Hsps) (Lindquist 1986) and upregulation of the ubiquitin-proteasomal system (Glickman and Ciechanover 2002) which work together in maintaining proteostasis. The response to various stressors is mediated by the activation of different mitogen-activated protein kinase (MAPK) pathways which profoundly affect numerous cellular functions (Pearson et al. 2001). In mammals, there are three MAPK pathways, (i) The extracellular signal-regulated kinases (ERKs): mainly activated by growth factors and mitogens (McKay and Morrison 2007; Shaul and Seger 2007), (ii) JUN amino-terminal kinases (JNKs): activated by ionizing radiation, osmotic stress, DNA damage, inflammatory cytokines as well as growth factors (Morrison 2012), and (iii) p38 MAPKs: strongly activated by environmental stress and inflammatory cytokines (Rouse et al. 1994). Central to the activation of all the MAPK pathways is a three-tiered signaling cascade. The cascade consists of MAPK kinase kinases (MKKKs), MAPK kinases (MKKs) and MAPKs. The first components activated in this module are the MKKKs, which in turn phosphorylate specific MKKs. Activated MKKs next phosphorylate specific MAPKs by dual phosphorylation of tyrosine and threonine residues in a conserved Thr-Xaa-Tyr (where Xaa is any amino acid) motif in the activation loop of kinase subdomain VIII (Cuenda and Rousseau 2007; Morrison 2012). The p38 MAPK is activated by MKK3 and MKK6 and the JNK MAPK is activated by MKK4 and MKK7 (Cuenda et al. 1996; Cuenda et al. 1997). Stress-activated MAPK pathways further activate downstream targets that include transcription factors, cytoskeletal proteins, protein kinases, translational machinery components, and glycogen synthase (Duncan and Hershey 1984; Knebel et al. 2001; Knebel et al. 2002; Kuma et al. 2004; Roux and Blenis 2004).
It is now well established that covalent attachment of small ubiquitin-like modifier (SUMO) moieties to proteins also play an important role in the maintenance of cellular homeostasis during exposure to numerous extracellular stressors that include heat stress, DNA damage, viral infection, endoplasmic reticulum stress, nutrient limitation, and oxidative stress (Zhou et al. 2004; Guo and Henley 2014; Enserink 2015). Stress-induced SUMOylation is thought to target spatially related substrates thereby influencing entire complexes/pathways. The effect of SUMOylation has been particularly well studied on the transcription machinery and has been shown to either activate or repress numerous transcription factors (Chymkowitch et al. 2015). SUMO conjugation of proteins is a multistep process; in the first step, an adenylated SUMO moiety is attached to the E1 enzyme (a dimer consisting of Sae1 and Sae2) which is subsequently transferred to Ubc9 (E2 enzyme). Substrate SUMOylation takes place in the presence of an E3 enzyme which acts as a facilitator in the transfer of the SUMO moiety from Ubc9 to the substrate. In mammalian cells, there are numerous SUMO ligases, the best characterized of them are the PIAS (protein inhibitor of activated STAT) proteins (PIAS1, PIAS2 (PIASxα and PIASxβ), PIAS3 (PIAS3 and PIAS3L) and PIAS4 (PIASy and PIASyE6)). In addition to their SUMO-ligase activity, the PIAS proteins interact with and regulate the activity of a variety of other proteins particularly transcription factors (Rytinki et al. 2009).
Transcription factor Elk-1 (E twenty-six-like-1) is a downstream target of all the three MAPK pathways (Shaw and Saxton 2003). It is a member of the ternary complex factor (TCF) subfamily of proteins under the ETS (E twenty-six) domain transcription factor family and has been studied extensively since it regulates the transcription of immediate-early (IE) genes (Townsend et al. 1999; Vickers et al. 2004; Kasza et al. 2005) and is important in the context of various pathological conditions including cancer and neurological disorders. Like other TCFs, Elk-1 acts through a ternary DNA-protein complex with a dimer of SRF (Treisman 1994; Sharrocks 2002). Elk-1 contains a C-terminal transcription activation domain (TAD/C domain) which is phosphorylated by ERK at multiple phosphorylation sites of which S383 plays an important role in the activation of Elk-1 (Gille et al. 1995; Janknecht et al. 1993; Marais et al. 1993). Recent reports indicate that phosphorylation of multiple sites on Elk-1 proceeds with different rates classified as fast, intermediate, and slow; while phosphorylation of the fast and intermediate sites result in transactivation by Elk-1 that of the slow sites inhibits Elk-1 transcriptional activity (Mylona et al. 2016). Elk-1 also contains a transcriptional repression (R) domain which is located N-terminal to the TAD domain; modification of the R motif by SUMOylation at K249 leads to repression of Elk-1 activity (Yang et al. 2003). In addition, SUMOylation of Elk-1 is also modulated by the acetylation of Ubc9 where increased Ubc9-acetylation reduces its activity leading to a reduction in the SUMOylation levels of Elk-1 (Hsieh et al. 2013). In the uninduced state, Elk-1 is SUMOylated and remains as a complex with PIAS2 and HDAC2 and is localized to Elk-1 responsive promoters. The recruitment of HDAC2 to Elk-1 promoters is facilitated by the SUMOylation of Elk-1 which leads to the repression of Elk-1 mediated transcription. Upon activation of the ERK pathway, Elk-1 is phosphorylated by ERK and is deSUMOylated in a PIAS2 and SENP1 (SUMO-specific protease)-dependent manner which leads to the loss of HDAC2 from the complex and consequent activation of Elk-1-dependent transcription (Yang and Sharrocks 2005; Witty et al. 2010). PIAS2 thus acts as a coactivator of Elk-1-dependent transactivation. In contrast, anisomycin (activator of both JNK and p38 MAPKs) induces phosphorylation of PIAS2 at S113 and S116, which in turn inhibits the loss of SUMO and HDAC2 from Elk-1 thus inhibiting its transcriptional activity (Yang and Sharrocks 2006). Elk-1 has also been shown to be ubiquitinated; however, the role of Elk-1-ubiquitination vis à vis its phosphorylation and SUMOylation remains unknown (Teixeira et al. 2013). In addition to the mechanism of regulation of Elk-1 activity as described above, it is also regulated by alternative mechanisms in prostate cancer cells wherein the androgen receptor binds to the ERK-binding site (D domain, located between the R and TAD domains) of Elk-1 and induces the expression of genes that support cancer cell growth (Rosati et al. 2016).
To identify mechanisms that regulate gene expression under stress conditions, we isolated Saccharomyces cerevisiae mutants that could maintain reporter gene expression in cells exposed to heat stress. Subsequent subtractive hybridization cloning of genes that were overexpressed in a mutant led to the cloning of a number of genes including SIZ1 (E3 SUMO ligase; unpublished data from this laboratory). Following up on the above observation, we have investigated if and how SUMOylation influences gene expression in mammalian cells exposed to heat stress. Since MAPK pathways are among the first responders to heat stress in mammalian cells, we decided to investigate if stress signaling and consequent stress-induced gene expression are influenced by SUMOylation of MAPK pathway components and its downstream effectors. Our studies show that Elk-1-SUMOylation is increased and its phosphorylation is decreased in Hela cells exposed to heat stress. The increase in SUMOylation of Elk-1 is dependent on the p38 MAPK pathway and correlates with the loss of Elk-1-mediated transactivation. We further show that under conditions as indicated above, the p38 MAPK pathway induces phosphorylation of PIAS2 which has been reported to repress Elk-1 activity. The present study thus provides a framework for understanding as to how the p38 MAPK pathway regulates Elk-1 activity during exposure to heat stress.
Methods
Cell culture, plasmids, transfection, and experimental treatments
HeLa cells (obtained from the National Centre for Cell Sciences; Pune, India) were grown in Minimum Essential Medium (MEM; Sigma) supplemented with 10% fetal bovine serum (FBS; GIBCO), 2.2 gl−1 sodium bicarbonate, antibiotics, and antimycotic agents (100 Uml−1 penicillin, 100 μgml−1 streptomycin, and 0.25 μgml−1 Amphotericin B) (HiMedia). Cells were maintained at 37 °C with 5% CO2. For transfection with pEZ-M06 (expressing HA-SUMO1 or HA-SUMO2 from the CMV promoter; neomycin selection; Genecopoeia Cat. No. EX-I0435-M06 and EX-I0567-M06 respectively), cells were plated in 6 wells plate and grown to 50–60% confluence. Transfection was done with Xfect Transfection reagent (Clonetech, TAKARA) according to the manufacturer’s instruction. After transfection, the medium was replaced with medium containing 500 μgml−1 neomycin (Sigma) for the selection of transfected cells. Stably transfected cells were further grown in complete MEM media supplemented with neomycin (500 μgml−1). For each experiment, HeLa cells transfected with pEZ-M06 were grown to 70–80% confluence and then exposed to treatments as indicated below. Hereafter, HeLa cells transfected with SUMO1 and SUMO2 expressing plasmids are referred to as HeLaS1 and HeLaS2 cells respectively. For PIAS2 phosphorylation assays, HeLa cells were transfected with pEZ-M14 vector (expressing PIAS2-3xFLAG from the CMV promoter; neomycin selection; Genecopoeia Cat. No. EX-I0268-M14) as per protocol described above.
Before exposure to heat, cells were serum starved for 18 h and subsequently treated with the following inhibitors, 10 μM SB203580 (p38 MAPK inhibitor), 10 μM U0126 (ERK kinase inhibitor) and 10 μM SP600125 (JNK inhibitor) for the following time periods: 60 min for immunoprecipitation (IP) and western blotting, 6 h for reporter gene assay, and 120 min for qPCR. For phorbol myristate acetate (PMA) and anisomycin treatment, serum-starved cells were treated with 10 nM PMA or 250 ngml−1 anisomycin for the following time periods: 60 min for IP and western blotting and 6 h for reporter gene assay. Alterations to the above are indicated in the “results” section.
For heat exposure, 2.1 × 106 cells were plated in T75 flasks for IP and western blotting, 1 × 104 cells were plated in 96 well plates for reporter gene assay and 0.3 × 106 cells were plated in 6 well plates for extraction of RNA for qPCR; all cultures were grown to 70–80% confluence before exposure to heat as indicated above. Heat exposure was carried out by incubating cultures in the incubator set at the required temperature. Cultures reached 42 °C and 45 °C within 3 and 7 min of incubation respectively at the desired temperature.
Immunoprecipitation, phosphatase treatment, and western blotting
Harvested cells were washed with ice-cold phosphate buffered saline (PBS) containing 10 mM N-ethylmaleimide (NEM) and lysed in lysis buffer (50 mM Tris-HCl (7.8), 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 0.5% NP-40, 0.3% Triton X-100, 1 mM DTT, 1:200 phosphatase inhibitor cocktail (Thermo), 10 mM NEM). The lysate was centrifuged at 15000 rpm for 20 min at 4 °C and the supernatant was collected. 1 mg of supernatant protein was used for immunoprecipitation (IP). IP was carried out by standard protocol using 1 μg of the indicated antibody and Protein A-Sepharose beads (Cavigelli et al. 1995). The beads were washed three times with the lysis buffer, SDS PAGE sample loading buffer was then added and boiled for 5 min, and proteins were resolved by SDS PAGE. For phosphatase treatment, the cell extract or immunoprecipitated was treated with 200 unit of λ phosphatase (Sigma Cat. No. P9614) for 30 min at 30 °C in buffer supplied by the manufacturer prior to the addition of SDS PAGE sample loading buffer.
Western blotting was performed by standard procedure. Antibodies used were anti-HA antibody (Ab) (Life Technologies 715,500, 1:1000), anti-SUMO1 Ab (Abcam ab49767, 1 μgml−1), anti-SUMO2/3 Ab (Abcam ab3742, 1 μgml−1), anti-Elk-1 Ab (Abcam ab131465, 1:1000), anti-phospho-Elk-1 (Ser383) Ab (Cell signaling 9181, 1:1000), anti-p38 Ab (Thermo PAI-30391, 1:200), anti-p38 (phospho Y182) Ab (Abcam ab47363, 1:1000), anti-JNK1,2 Ab (Abcam ab112501, 1 μgml−1), anti JNK1,2 (phospho Thr183, phospho Tyr 185) Ab (Thermo MA5-14943, 1:1000), anti-ERK 1,2 Ab (Abcam ab17942, 1:1000), anti-ERK1, 2 (phospho T202, phospho Y204) Ab (BD transduction laboratories 612,358, 1:1000), anti-β actin Ab (Thermo PAI-183, 1:2000), and anti-PIAS2 Ab (Thermo PA5-20952, 1 μgml−1). The intensity of the signals was quantified with Image Lab 5.2.1 (BIORAD) or ImageJ software.
Reporter gene assay
pEGR-1-Luc expressing the firefly luciferase gene under the control of the EGR-1 promoter was used for reporter gene assay. pSV β-Galactosidase (Promega) Control Vector was used as a control for transfection. For luciferase and β-galactosidase assay, HeLa cells were co-transfected with 0.5 μg of pEGR-1-Luc and 0.25 μg of pSV-β-galactosidase. After treatment as mentioned above, cells were harvested and processed for luciferase and β-galactosidase assay. Luciferase assays were performed with firefly luciferase assay kit (Thermo) according to the instructions of the manufacture, and β-galactosidase assay was carried out as described in Uchil et al. 2017. Light emission was measured for 10 s with SYNERGY/H1 microplate reader, Biotek.
mRNA estimation by reverse transcription followed by qPCR
RNA was extracted from HeLaS1 cells with TRIZOL reagent (Ambicon 15596-026) according to the instruction of the manufacturer. The purified RNA was treated with DNAse (Nucleo-pore) at 37 °C for 45 min and its concentration was quantified using μDrop reader (Thermo scientific MULTISKAN GO). RNA integrity was checked by running 2 μl of purified RNA on a 1% agarose gel and 28S:18S rRNA band intensity ratio was found to be 2:1. A PCR was done with the purified RNA using 18S rDNA gene-specific primers to check for genomic DNA contamination following which cDNA was prepared from 5 ng of RNA using Verso cDNA synthesis kit (Thermo scientific AB-1453/A) according to the instruction of the manufacturer. qPCR was performed on a CFX connect real time system (Bio-Rad), using the DyNAmocolor flash SYBR green qPCR kit (F416 L Thermo scientific). Each reaction was performed in duplicate, in a total reaction volume of 20 μl. The cycling program (35 cycles) was set as follows: initial denaturation and DNA polymerase activation (95 °C for 7 min), denaturation (95 °C for 10s), annealing (55 °C for 30s), and extension (72 °C for 30s). Relative quantification and normalization were performed with Bio Rad CFX Manager 3.1. Primer sequences were as described in Table 1. The specificity of the primers was checked by using Primer-BLAST software (NCBI) and by performing PCR with individual primers and inspecting the amplicon length by gel electrophoresis. All data were normalized to the levels of 18S rRNA (measured by reverse transcription followed by qPCR) before comparison and statistical analysis.
Table 1.
Primer sequence
| Gene | Sequence | |
|---|---|---|
| Luc+ | Forward | 5′ CGGAAAGACGATGACGGAAA 3′ |
| Reverse | 5′ CGGTACTTCGTCCACAAACA 3′ | |
| mcl-1 | Forward | 5′ CCAGCTCCTACTCCAGCAAC 3′ |
| Reverse | 5′ TCGTAAGGACAAAACGGGAC 3′ | |
| c-fos | Forward | 5′ AGAATCCGAAGGGAAAGGAA 3′ |
| Reverse | 5′ CTTCTCCTTCAGCAGGTTGG 3′ | |
| srf | Forward | 5′ ACGACCTTCAGCAAGAGGAA 3′ |
| Reverse | 5′ AAGCCAGTGGCACTCATTCT 3′ | |
| 18S rDNA | Forward | 5′ GGACATCTAAGGGCATCACAG 3′ |
| Reverse | 5′ TCAAGAACGAAAGTCGGAGGTT 3′ | |
| A20 | Forward | 5′ ATGCACCGATACACACTGGA 3′ |
| Reverse | 5′ GCGTGTGTCTGTTTCCTTGA 3′ | |
| β-Actin | Forward | 5′ AGGCACCAGGGCGTGAT 3′ |
| Reverse | 5′ GCCCACATAGGAATCCTTCTGAC 3′ | |
| LAS1L | Forward | 5′ ACGAAGTCGTTTCTCGGATTT3′ |
| Reverse | 5′ AGTGCTTGACATGTGTGTATCT3′ | |
| ELF2 | Forward | 5′GGTTGGGATGGGTAATCTCATT3′ |
| Reverse | 5′GAGTCTTCACATAGCACCAGTT3′ | |
| SNRPB | Forward | 5′GATGGGAGTTGGGCATTCAA3′ |
| Reverse | 5′GGGAAGGACAGTGGGAAATAAC3′ | |
| TAF1 | Forward | 5′AAGATGGCAGTGGGTTTGAC3′ |
| Reverse | 5′GCTGAGGTTTTGTCCCTTTG3′ | |
| TAF7 | Forward | 5′CTCCTCACGAACTGGAGAGC3′ |
| Reverse | 5′CCATAACACAGGGCAGGTCT3′ | |
| TAF2 | Forward | 5′ATCAGCCCAAAGGAGGTCTT3′ |
| Reverse | 5′AGCAACCATTGCAGCATCTA3′ | |
| GTF2A1 | Forward | 5′TCACCATCATCAGCAAGCTC3′ |
| Reverse | 5′AGCTGTAGCAGCAGCACTCA3′ | |
| GTF2B | Forward | 5′CACATGTCCAAACCATCCAG3′ |
| Reverse | 5′CAAAACTTGCAGCTCCTGTG3′ | |
| TBP | Forward | 5′CTCAGGGTGCCATGACTCCCG3′ |
| Reverse | 5′TTGTTGTTGCTGCTGCTGCCTTTG3′ |
Statistical analysis
Computations were done with the PRISM (ver. 5.0) software (GraphPad Software, LaJolla, CA, USA). For all data, one-way ANOVA followed by the Tukey’s HSD post hoc test was used to determine significant differences between groups.
Results
Elk-1 SUMOylation is increased in heat stressed HeLa cells
To elucidate if and how SUMOylation regulates stress-activated MAPK pathways and their downstream effectors, we first determined the SUMOylation states of MAPKKKs, MAPKKs, and MAPKs in extracts prepared from HeLa cells either exposed to heat stress at 42 and 45 °C for 1 h or left untreated. None of the signaling molecules examined namely, TAOK1, ASK1 and MEKK1 (MAPKKKs), MKK3 and MKK6 (MAPKKs) and p38, and JNK and ERK (MAPKs) were found to be SUMOylated prior to or following exposure to heat stress (data not shown). We next examined SUMOylation states of transcription factors downstream of MAPK pathways. Numerous transcription factors downstream of MAPK pathways are known to be SUMOylated however, only Elk-1 has been shown to be SUMOylated by p38 MAPK dependent mechanisms. We hence determined if Elk-1 was SUMOylated during exposure to heat stress.
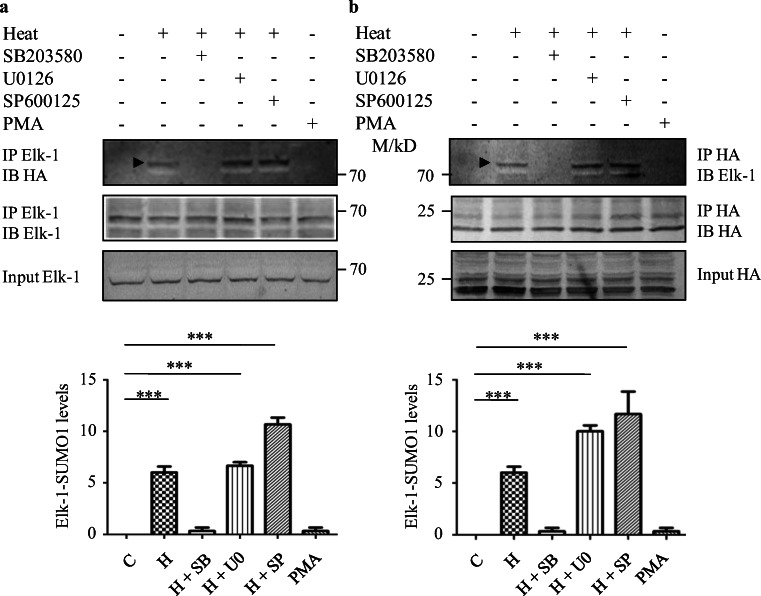
Endogenous Elk-1 was immunoprecipitated with anti-Elk-1 Ab from extract prepared from HeLaS1 and HeLaS2 cells that were exposed to heat stress at 42 °C or 45 °C for 1 h or left untreated (untreated control). Immunoblotting with anti-HA Ab showed that in HeLa cells exposed to 45 °C Elk-1 was SUMOylated with SUMO1 (Fig. 1a) and not with SUMO2 (Fig. S1a). Elk-1-SUMO1 species was not observed in cells exposed to 42 °C. To further confirm that Elk-1 is SUMOylated in heat stressed HeLa cells, we immunoprecipitated HA-SUMO1 with anti-HA Ab from extracts prepared from cells treated as indicated above; subsequent immunoblotting with anti-Elk-1 Ab showed that Elk-1 was SUMOylated in HeLaS1 cells heat-treated at 45 °C for 1 h (Fig. 1b; uncropped image to show additional SUMOylated proteins precipitated by the anti-HA Ab in Fig. S1). The absence of any heat-induced Elk-1-SUMO1 bands in extracts prepared from cells treated with 50 μgml−1 ginkgolic acid (an inhibitor of SUMOylation (Fukuda et al. 2009)) prior to heat exposure further confirmed that Elk-1 is SUMOylated in heat stressed HeLaS1 cells (Fig. 1 a, b).
Fig. 1.
SUMOylation and phosphorylation of Elk-1 in heat stressed HeLa cells. Elk-1 SUMOylation in heat stressed HeLa cells. a Elk-1 was immunoprecipitated from 1 mg extract prepared from HeLaS1 cells that were either exposed to 42 °C or 45 °C for 1 h or left untreated (untreated control) with 1 μg anti-Elk-1 Ab followed by blotting and immunodetection using Abs as indicated in the fig. Arrowheads indicate heat induced SUMOylated species of Elk-1. C unexposed control and GA ginkgolic acid. Input, 10% of the total protein used for immunoprecipitation. M/kD, migration of pre-stained molecular weight markers (Thermo Scientific Page Ruler prestained protein ladder; Cat. No. 26616) is indicated. b Same as a except that immunoprecipitation was carried out with anti-HA Ab and immunodetection with anti-Elk-1 and anti-HA Abs. c Elk-1 phosphorylation levels. Elk-1 phosphorylation levels in HeLaS1 cells exposed to conditions as indicated in the figure was determined in 30 μg whole cell extract by western blotting using anti-phospho-Elk-1 and anti-Elk-1 Abs. All experiments were performed in triplicates and band intensities were estimated using BioRad Image Lab 5.2.1 and Image J software. Data are represented as mean ± SEM of triplicate experiments. ***p < 0.001 relative to control and heat exposed groups was determined by one-way ANOVA followed by adjustment for multiple comparisons using Tukey’s test
In our assays, basal SUMOylation of Elk-1 was not observed consistently; in some assays, low levels of basal Elk-1-SUMO1 were observed (Fig. S2a). SUMOylation of Elk-1 in HeLa cells exposed to 45 °C for 1 h is noteworthy since under similar conditions global SUMOylation of proteins by both SUMO1 and SUMO2/3 was lower as compared to that observed in the unexposed control and in cells exposed at 42 °C for 1 h (Fig. S2b). In agreement with earlier observations (Saito and Hinchey 2000), our results also show that more proteins were SUMOylated by SUMO2/3 as compared with SUMO1 during exposure to moderate heat stress (42 °C lane in Fig. S2b).
Since phosphorylation of Elk-1 by the ERK pathway leads to activation of its transcriptional activity, we also determined the phosphorylation state of Elk-1 in heat-stressed HeLaS1 cells by western blotting using anti-phospho-Elk-1 Ab. Elk-1-phosphorylation was significantly reduced in HeLaS1 cells exposed to conditions as indicated in Fig. 1c. Thus, in heat-stressed HeLaS1 cells, Elk-1 was SUMOylated and dephosphorylated.
All subsequent heat exposure studies were carried out with cells expressing HA-SUMO1 (or as indicated) at 45 °C since (i) SUMOylation levels of Elk-1 in HeLa cells exposed at 45 °C was consistently and significantly higher as compared with the unexposed control (p < 0.001), (ii) Elk-1 was SUMOylated by SUMO1, and (iii) Elk-1 phosphorylation was lower in HeLa cells exposed to similar conditions.
Heat stress-induced SUMOylation of Elk-1 is dependent on the p38 MAPK pathway
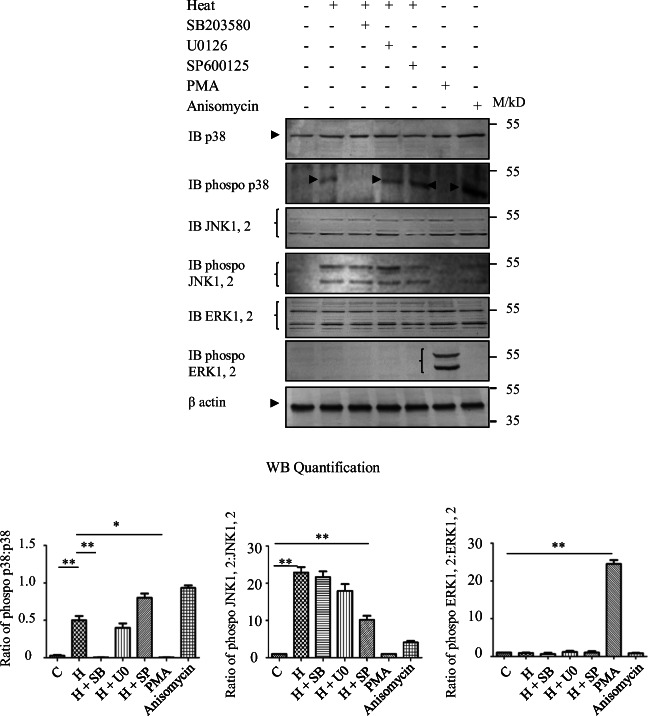
Since Elk-1 is a target of ERK and stress-activated MAPKs, we examined if the increase in SUMOylation of Elk-1 in response to heat stress is dependent on the activation of MAPK pathways and specifically on which MAPK pathway. To elucidate this, we first determined if MAPK pathways were activated in response to heat stress in our experimental system. HeLaS1 cells were either treated with inhibitors of MAPK pathways, 10 μM SB203580 (p38 inhibitor), 10 μM U0126 (ERK inhibitor), and 10 μM SP600125 (JNK inhibitor) for 60 min prior to heat treatment and were then exposed to heat stress at 45 °C for 1 h or left untreated (control). Cells were also treated with PMA (ERK activator) and anisomycin (p38 and JNK activator) as positive controls. Western blotting analysis of extracts prepared from heat-stressed cells treated as indicated above showed an increase in phospho-p38 level in heat-treated cells; phospho-p38 was undetectable in cells treated with SB203580 prior to exposure to heat stress. This confirmed activation of the p38 MAPK pathway on exposure to heat stress. Treatment with anisomycin (positive control) showed an increase in both phospho-p38 and phospho-JNK levels. Heat exposure of HeLa cells treated with either ERK or JNK inhibitors (U0126 and SP600125 respectively) or with the ERK activator PMA did not show any alterations in phospho-p38 levels. ERKs and JNKs thus did not influence phosphorylation of p38 MAPK in heat stressed HeLaS1 cells. Western blotting with anti-p38 Ab showed relatively similar level of p38 protein in all conditions. Phospho-JNK and phospho-ERK levels were also assessed in extracts prepared from cells treated to similar conditions. Phospho-JNK levels increased on exposure to heat stress which decreased in presence of the JNK inhibitor SP600125. We did not observe any increase in phospho-ERK level on exposure to heat stress; it was however increased on PMA treatment (positive control). Results presented in Fig. 2 thus demonstrate activation of p38 and JNK MAPK pathways in HeLa S1 cells exposed to heat stress.
Fig. 2.
Heat stress activates p38 MAPK pathway. 30 μg of extract prepared from HelaS1 cell exposed to 45 °C for 1 h was used to detect proteins as indicated in the figure by western blotting with appropriate Abs. 60 min prior to heat treatment HeLaS1 cells were treated with the following inhibitors at a concentration of 10 μM: SB, p38 inhibitor SB203580; U0, ERK inhibitor U0126; and SP, JNK inhibitor SP600125. PMA (ERK pathway activator) and anisomycin (activator of p38 and JNK pathways) were used as positive controls at a concentration of 10 nM and 250 ngml−1 respectively for 60 min. M/kD migration of pre-stained molecular weight markers is indicated. Data was quantified by first normalizing the band intensities of both phosphoprotein and protein with that of β-actin followed by calculating the ratio of phosphoprotein:protein band intensities. C unexposed control and H Heat stress at 45 °C for 1 h. Results are shown as average ± SEM of triplicate experiments. One-way ANOVA followed by adjustment for multiple comparisons was performed using Tukey’s test. *p < 0.05 and **p < 0.01; p values are relative to the control
We next determined which MAPK pathway was involved in the SUMOylation of Elk-1 (Fig. 1). Elk-1 was immunoprecipitated with anti-Elk-1 Ab from extracts prepared from heat-stressed HeLaS1 cells; subsequent immunoblotting with anti-HA Ab showed that Elk-1 was SUMOylated in heat stressed HeLaS1 cells. However, in cells treated with SB203580 prior to exposure to heat stress, SUMOylation of Elk-1 was undetectable (Fig. 3a). Similar treatment with U0126 and SP600125 did not inhibit heat induced SUMOylation of Elk-1 (Fig. 3a). Precipitation of HA-SUMO1 from extracts prepared from cells treated as indicated above with Anti-HA Ab followed by western blotting with anti-Elk-1 Ab showed essentially similar results (Fig. 3b, an uncropped image in Fig. S3). This further substantiated our conclusion (Fig. 3a) that Elk-1 is SUMOylated by p38 MAPK pathway-dependent mechanisms during exposure to heat stress. Cells exposed to heat stress in the presence of either U0126 or SP600125 did not show any decrease in heat-induced SUMOylation of Elk-1 thus ruling out the possible involvement of the ERK and JNK pathways in SUMOylating Elk-1. PMA was used as a negative control in both the experiments since the activation of the ERK pathway by PMA has been shown to deSUMOylate Elk-1 (Yang et al. 2003). Collectively, the results presented above demonstrate that heat stress-induced SUMOylation of Elk-1 was dependent on the p38 MAPK pathway.
Fig. 3.
Elk-1 is SUMOylated by p38 MAPK-dependent mechanisms. a Elk-1 was immunoprecipitated from 1 mg extract prepared from HeLaS1 cells exposed to conditions as indicated below with 1 μg anti-Elk-1 Ab followed by blotting and immunodetection using Abs as indicated in the fig. Prior to exposure to heat stress cells were treated with MAPK pathway inhibitors (SB, U0 and SP) as described in legend to Fig. 2, PMA was used as negative control. Input: 10% of the total protein incubated for IP. b Same as a except that IP was carried out with anti-HA Ab and immuno-blotting with anti-Elk-1 and anti-HA Abs. C unexposed control and H Heat stress at 45 °C for 1 h. Band intensities were estimated using BioRad Image Lab 5.2.1 and ImageJ software. Data are represented as mean ± SEM of triplicate experiments. ***p < 0.001, relative to control was determined by one-way ANOVA followed by adjustment for multiple comparisons using Tukey’s test. M/kD, migration of pre-stained molecular weight markers is indicated
Elk-1-induced reporter gene expression was downregulated under heat stress by p38 MAPK pathway-dependent mechanisms and by SUMOylation
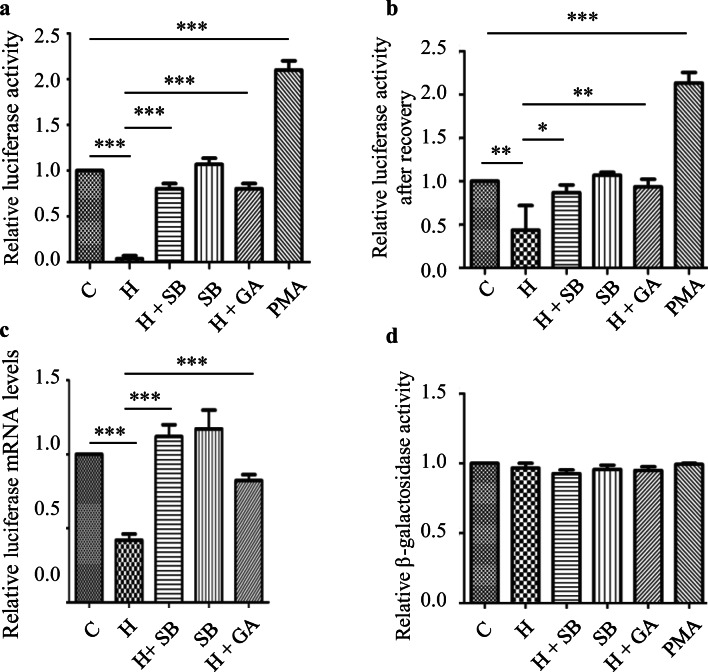
SUMOylation causes both activation and repression of transcription. Activation of the p38 MAPK pathway by anisomycin has been shown to dampen the induction of immediate early gene expression by Elk-1 (Yang and Sharrocks 2006). Since heat stress also activated p38 MAPK pathway and induced Elk-1-SUMOylation, we investigated the transcriptional activity of Elk-1 in response to heat stress. HelaS1 cells were co-transfected with pEGR-1-Luc plasmid (expressing luciferase under the control of the Elk-1 responsive egr1 promoter) and pSV-β-galactosidase plasmid (transfection control). Heat treatment of the transfected cells at 45 °C for 1 h showed reduced luciferase activity relative to the unexposed control. In cells treated with either SB203580 or ginkgolic acid prior to heat stress, luciferase activity was similar to that observed in the unexposed control (Fig. 4a). Exposure to SB203580 alone, however, did not have any effect on the basal activity of luciferase thus ruling out the possibility that inhibition of p38MAPK can induce the expression of Elk-1 responsive genes. Since luciferase is a thermolabile enzyme we also carried out luciferase assays with cells that were allowed to recover for 6 h at 37 °C in the presence of cycloheximide (20 μgml−1) following exposure at 45 °C for 1 h to allow for possible renaturation of heat-inactivated luciferase during the recovery period. Results (Fig. 4b) were essentially similar to those observed when luciferase was assayed immediately following exposure to heat stress (Fig. 4a); we did, however, observe a slight increase in luciferase activity in cells allowed to recover from heat stress (Fig. 4b). We further assessed luciferase mRNA levels in total RNA isolated from cells exposed to condition used for luciferase activity assay by reverse transcription followed by qPCR. Luciferase mRNA levels were reduced in heat-stressed cells which were restored to control levels in cells treated with either SB203580 or ginkgolic acid before exposure to heat stress (Fig. 4c). β-galactosidase activity was estimated in the same cell extracts used for luciferase assay to determine transfection efficiency which was found to be equal in all the groups (Fig. 4d). Expression of luciferase from pEGR-1-Luc plasmid (as estimated by luciferase activity and mRNA levels) was thus reduced in heat-stressed cells. Both luciferase activity and mRNA levels were restored to control levels in heat-stressed cells when heat exposure was carried out in cells pretreated with either SB203580 or ginkgolic acid. Since Elk-1 is SUMOylated in response to heat stress by p38MAPK dependent mechanisms (Fig. 1), it is reasonable to conclude that SUMOylation of Elk-1 in heat-stressed cells is associated with the downregulation of luciferase expression in heat stressed HeLaS1 cells.
Fig. 4.
Elk-1 induced expression of luciferase reporter under heat stress. a Luciferase reporter gene expression in HelaS1 cells cotransfected with pEgr1-luc and pSV-β-Galactosidase. Elk-1 induced expression of luciferase was estimated by measuring luciferase activity in lysates prepared from HeLaS1 cells cotransfected with pEgr1-luc and pSV-β-Galactosidase and exposed to conditions as indicated in the figure. Luciferase activity was estimated with the firefly luciferase assay kit (Thermo) according to the instruction of the manufacture. b Elk-1 induced expression of luciferase reporter under heat stress. Assays were carried exactly as indicated in legend to Fig. 4a except that the cells were allowed to recover for 6 h at 37 °C in the presence of 20 μgml−1 cycloheximide following exposure to heat stress at 45 °C for 1 h. c Reverse transcription followed by qPCR analysis of luciferase mRNA levels in extracts prepared from HeLa cells treated or untreated with heat in the presence or absence of inhibitors as indicated in the figure. Primers used in the experiment with their sequences are listed in Table 1. d β-galactosidase assay (transfection control): β-galactosidase activity was measured by the procedure of Uchil et al. 2017 in the same extract used for measuring luciferase activity. For all the above experiments, three independent experiments were carried out in duplicates and data are shown relative to control which was taken to be 1. C unexposed control, H Heat stress at 45 °C for 1 h, SB indicates cells treated with SB203580 alone, the other abbreviations are as indicated earlier. *p < 0.05, **p < 0.01, and ***p < 0.001 relative to heat exposed cells was determined by one-way ANOVA followed by adjustment for multiple comparisons using Tukey’s test
PMA-activated expression of Elk-1 responsive IE genes was downregulated under heat stress by p38 MAPK pathway-dependent mechanisms and by SUMOylation
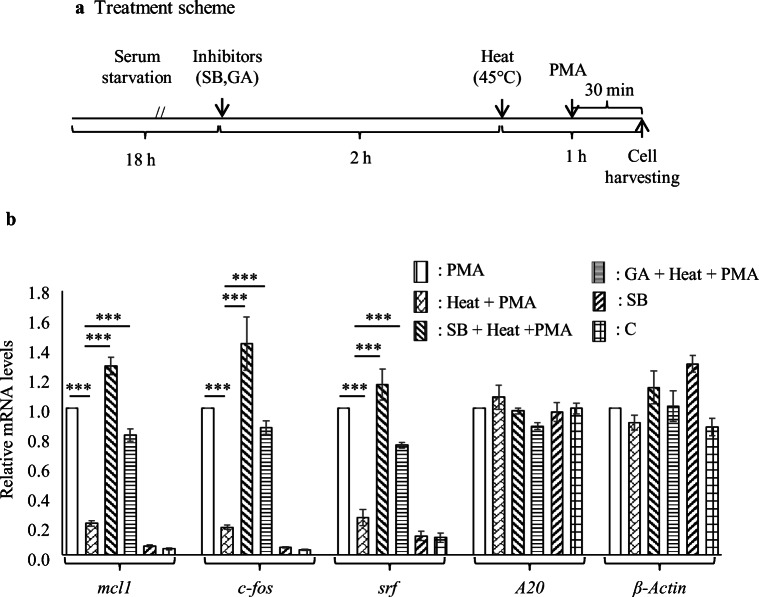
To further study the relationship between heat stress-induced SUMOylation of Elk-1 and its transcriptional activity within cells in real time, we measured mRNA levels of Elk-1 responsive IE genes (srf, mcl1, and cfos) in HeLaS1 cells exposed to heat stress. Reverse transcription followed by qPCR indicated that mRNA levels of the IE genes were too low to study further regulation of their expression under heat stress (Fig. S4). Given the above, the expression of IE genes was induced for the last 30 min of heat stress at 45 °C by the addition of PMA to a final concentration of 10 nM; for the unexposed control, PMA was added to the culture 30 min prior to cell harvesting (Fig. 5a). mRNA levels of all the three IE genes were increased in PMA treated cells as compared to the untreated control (Fig. 5b). PMA-induced increase in the mRNA levels of IE genes was significantly reduced when it was added during exposure to heat stress (30 min into the heat stress protocol of 45 °C for 1 h). Treatment of cells with either SB203580 or ginkgolic acid prior to exposure to heat stress restored the expression levels of the IE gene examined to that observed in cells treated with PMA alone (Fig. 5b). Inhibition of the p38MAPK pathway in unexposed control cells by treatment with SB203580 did not induce the expression of the IE genes thus ruling out the possibility that SB203580 exposure induced the expression of the IE genes (Fig. 5b). Results presented thus indicate that treatment of HeLaS1 cells with either an inhibitor of the p38MAPK pathway or with an inhibitor of SUMOylation before exposure to heat stress restores PMA-induced IE gene expression to near control levels. It is well established that the expression of IE genes is regulated by Elk-1, and that Elk-1 is SUMOylated in response to heat stress (Fig. 1). We thus concluded that SUMOylation of Elk-1 is concomitant with the downregulation of PMA-induced expression of IE genes in heat stressed HeLaS1 cells. Since mRNA levels of IE gene were estimated in cells where both the ERK and p38 MAPK pathways were operative, we further determined the SUMOylation state of Elk-1 in extracts prepared from cells exposed to conditions as in Fig. 5b. Elk-1-SUMO1 species was only detected in cells exposed to heat and PMA (Fig. S5) thus further demonstrating that down-regulation of Elk-1 activity is concomitant with its SUMOylation. We also determined the mRNA level of both A20 and β-Actin genes, which are not under the transcriptional regulation of Elk-1. No change in mRNA level of A20 and β-Actin was found in cells exposed to conditions as indicated in Fig. 5b. The down-regulation of mRNA levels of Elk-1 responsive IE genes in heat-stressed cells was thus not a general effect of exposure to heat stress.
Fig. 5.
PMA induced expression of Elk-1 responsive IE genes in heat stressed HeLaS1 cells. a Treatment scheme b reverse transcription followed by qPCR analysis of mcl1, cfos, srf, A20, and β-Actin mRNA levels in total RNA isolated from HeLaS1cells treated or untreated with heat in the presence or absence of inhibitors as indicated in the fig. Data are average of three experiments performed in duplicates. Primers used in the experiment with their sequences are listed in Table 1. H Heat stress at 45 °C for 1 h and C untreated control, the other abbreviations are as indicated earlier. Data (normalized to 18S rRNA levels) are represented as mean ± SEM of triplicate experiments. ***p < 0.001 relative to control was determined by one-way ANOVA followed by adjustment for multiple comparisons using Tukey’s test
Expression of Elk-1 responsive genes in heat-stressed HeLa cells in the absence of activation by PMA
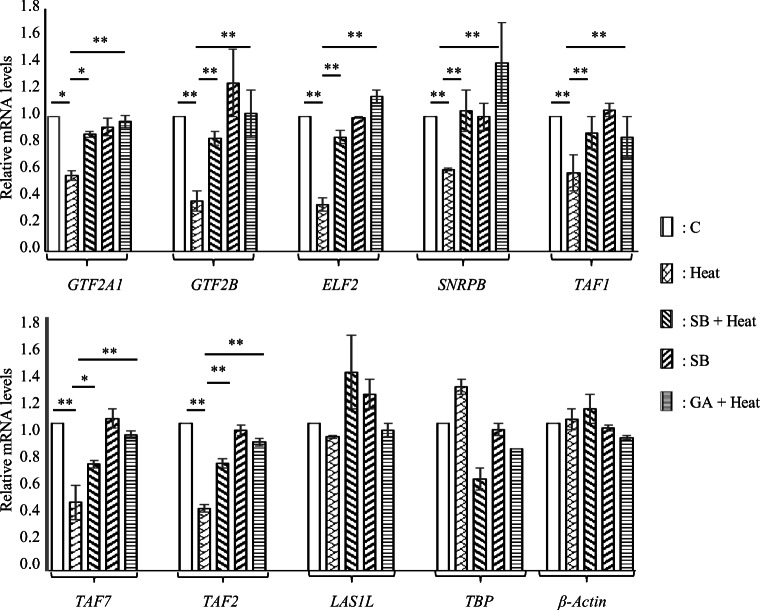
Since the above experiment was carried out under conditions were the expression of IE genes was artificially activated by PMA, we further examined the expression of genes encoding general transcription factors namely, TATA-box binding protein (TBP), TBP associated factor 1 (TAF1), TAF2, TAF7, general transcription factor IIA subunit 1 (GTF2A1), GTF2B, pre-mRNA splicing factor viz. small nuclear ribonucleoprotein polypeptide B (SNRPB), LAS1-like ribosome biogenesis factor (LAS1L), and E74 like ETS transcription factor 2 (ELF2) under conditions as indicated in Fig. 6. Elk-1 binds to the promoters of all the genes indicated above and in addition has been shown to activate the expression of TBP, TAF1, TAF2, TAF7, GTF2A1, and GTF2B (Boros et al. 2009). The expression of all the genes indicated above except for TBP and LAS1L was downregulated upon exposure to heat stress which was restored to control levels when cells were treated with either SB203580 or ginkgolic acid prior to heat exposure at 45 °C for 1 h. Data presented thus far (Figs. 1, 2, 3, 4, 5 and 6) demonstrate that heat stress-induced downregulation in the mRNA levels of Elk-1 responsive genes is concomitant with increased SUMOylation of Elk-1.
Fig. 6.
Expression of additional genes regulated by Elk-1 in heat stressed HeLaS1 cells. Reverse transcription followed by qPCR analysis of TBP, TAF1, TAF2, TAF7, GTF2A1, GTF2B, ELF2, SNRPB, and LAS1L mRNA levels in total RNA isolated from HeLaS1 cells treated or untreated with heat in the presence or absence of inhibitors was performed exactly as described under Fig. 5b except that PMA was not used to induce the expression of the genes examined. Abbreviations are as indicated earlier. Data (normalized to 18S rRNA levels) are average of three experiments performed in duplicates. Statistical considerations are as described in the legend to Fig. 5b
Heat stress-activated p38 MAPK pathway induces PIAS2 phosphorylation
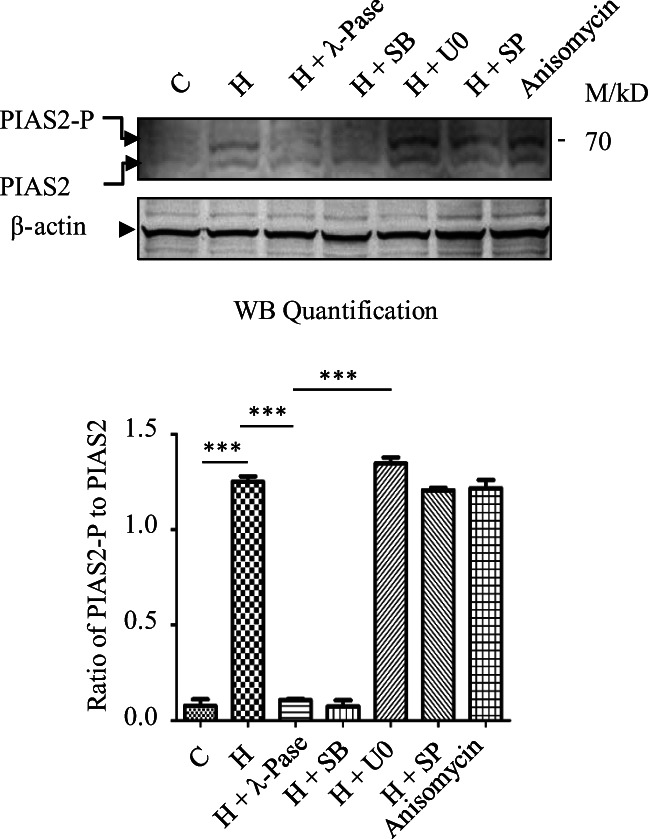
PIAS2 is a member of PIAS family of proteins. It contains a RING finger motif, a SAP domain, and a SUMO binding motif. PIAS2 both activates and represses transcriptional activity of Elk-1 depending on the MAPK pathway that has been activated. We were thus interested to examine if heat stress-mediated activation of p38 MAPK causes phosphorylation of PIAS2, which may be a cause for the inhibition of the transcriptional activity of Elk-1. Exposure of HeLa cells expressing PIAS2-3x-FLAG to heat stress resulted in the appearance of a band of lower mobility that was recognized by anti-PIAS2 Ab (Fig. 7). To examine if the lower mobility band is the phosphorylated form of PIAS2, we treated extracts prepared from heat-exposed cell with λ-phosphatase prior to western analysis with anti-PIAS2 Ab. λ-phosphatase treatment caused disappearance of the slower mobility PIAS2 band, thus confirming that the upward-shifted PIAS2 band was its phosphorylated form. The phosphorylated-PIAS2 band was also not observed in extracts prepared from cells treated with SB203580 prior to exposure to heat stress which indicated that PIAS2 was phosphorylated by p38 MAPK dependent mechanisms. Treatment of cells with U0126 or SP600125 prior to heat exposure showed the presence of phosphorylated form of PIAS2, phosphorylation of PIAS2 was thus not influenced by either the JNK or ERK pathway. We further confirmed the data described above by immunoprecipitation of PIAS2 with anti-PIAS2 Ab from extracts prepared from cells exposed to conditions as indicated above followed by blotting and immunodetection using anti-PIAS2 Ab; where indicated the immunoprecipitate was treated with λ-phosphatase or cells were treated with SB203580 prior to exposure to heat stress. PIAS2 was found to be phosphorylated in heat stress HeLa cells by p38 MAPK dependent mechanisms (Fig. S6). Results presented above (Figs. 7 and S6) indicated that heat activated p38 MAPK pathway induces phosphorylation of PIAS2 which along with SUMOylation of Elk-1 may account for the downregulation of transactivation by Elk-1 in heat-stressed HeLa cells.
Fig. 7.
PIAS2 is phosphorylated in heat stressed HeLa cells by p38 MAPK dependent mechanisms. 30–50 μg of extract prepared from HeLa cells expressing PIAS2-3x-FLAG treated with heat (H) or with heat and 10 μM inhibitors or 250 ngml−1 anisomycin as indicated in legend to Fig. 2 were used to determine phosphorylation of PIAS2 by western blotting using anti PIAS2 antibody. Where indicated the extract from heat treated cells was treated with λ phosphatase (λ-Pase) before adding SDS PAGE loading buffer. Quantification for increase in phosphorylation of PIAS2 was done by determining the ratio of the intensity of the phosphorylated PIAS2 to unphosphorylated PIAS2. Data are represented as mean ± SEM of four independent experiments. Abbreviations are as indicated earlier. ***p < 0.01 relative to heat exposed group was determined by one-way ANOVA followed by adjustment for multiple comparisons using Tukey’s test
PIAS2 interacts directly with Elk-1. Yang et al. demonstrated that the residues in the TAD (375-399) of Elk-1 play an important role in complex formation with PIAS2 (Yang and Sharrocks 2005). The interaction between PIAS2 and Elk-1 is important for regulating transactivation by Elk-1 since PIAS2 can act as both a coactivator and a corepressor for Elk-1 depending on the MAPK pathway that is active in the cell. We further investigated if there was any alteration in the interaction between PIAS2 and Elk-1 during heat stress. PIAS2-Elk-1 interaction was studied by coimmunoprecipitation of PIAS2 and Elk-1 with anti-Elk-1 Ab. IP of Elk-1 with anti-Elk-1 antibody followed by western blotting with anti-PIAS2 Ab did not show any difference in the interaction of PIAS2 and Elk-1 in control and heat-treated cells (Fig. S7). Although p38 MAPK pathway induces phosphorylation of PIAS2 and SUMOylation of Elk-1 during exposure to heat stress with concomitant inhibition of Elk-1 activity, it does not affect the interaction between Elk-1 and PIAS2.
Discussion
All cells respond to heat stress by a general downregulation of cellular transcriptional activity, only a small set of genes including those coding for Hsps are induced during exposure to heat stress. A recent study in Mouse Embryonic Fibroblasts indicates that under heat shock conditions at 42 °C ~ 1500 genes are induced and ~ 8000 genes are downregulated (Mahat et al. 2016). The regulation of gene expression by SUMOylation is particularly significant in the above context. It is well established that global protein SUMOylation is induced upon exposure to a wide variety of stressors and that it profoundly affects cellular transcriptional activity. While the activities of Heat Shock Factor 1 and 2 are positively regulated by SUMOylation that of numerous other transcription factors (p53, STAT1, MEF2A and 2C, c- and N-Myc, Elk-1, ETS1 etc.) downstream to stress-activated MAPK pathways are downregulated by SUMOylation (Chymkowitch et al. 2015). Of the above, the SUMOylation of Elk-1 has been shown to be dependent on the p38 MAPK pathway. Here we report on the effects of heat stress on transcription factor Elk-1. Specifically, we have addressed covalent modifications of Elk-1 during exposure to heat stress and have also determined the effect of heat stress on transactivation by Elk-1.
Transcription factor Elk-1 has been extensively studied since it activates the expression of IE genes in response to growth factors and other stimuli. In addition, it is expressed that various cancer tissues (www.proteinatlas.org/ENSG00000126767-ELK1/pathology) are linked to long-term memory potentiation (Besnard et al. 2011), drug addiction (Sillivan et al. 2013), and depression (Lindecke et al. 2006). Elk-1 has been shown to bind to the promoters of nearly 1000 genes that include IE genes and genes encoding components of basal transcription machinery, spliceosome complex components, and ribosomal proteins. Regulation of Elk-1 activity under heat stress may thus partly account for the effects of heat stress on gene expression.
Our studies demonstrate that in HeLa cells exposed to 45 °C for 1 h Elk-1 is SUMOylated by SUMO1 (Fig. 1a, b). Elk-1-phosphorylation was reduced under similar conditions (Fig. 1c). Elk-1-SUMOylation by SUMO1 at 45 °C is significant, since under similar conditions, global SUMOylation is decreased as compared to the unexposed control and to cells exposed to moderate heat stress at 42 °C (supplemental Fig. 2b). MAPK pathways regulate Elk-1 activity by inducing two posttranslational modifications of Elk-1 namely phosphorylation and SUMOylation which are critically dependent on the MAPK pathway that has been activated. We used inhibitors specific to all the three MAPK pathways to elucidate the possible involvement of a specific MAPK pathway in the heat-induced SUMOylation of Elk-1. Results presented demonstrate that Elk-1 is SUMOylated in heat-stressed HeLaS1 cells by p38 MAPK pathway-dependent mechanisms since Elk-1-SUMOylation was inhibited by SB203580 (p38 MAPK inhibitor) and not by inhibitors of ERKs and JNKs.
Studies on the functional aspects of Elk-1 under heat stress indicated downregulation of Elk-1-dependent luciferase reporter expression in HeLaS1 cells exposed to 45 °C for 1 h. The downregulation of luciferase activity in heat-stressed HeLaS1 cells was dependent on the activation of p38 MAPK pathway and on the SUMOylation pathway since treatment with either p38 inhibitor (SB203580) or with ginkgolic acid (inhibitor of SUMOylation) prior to heat stress restored Elk-1 activity as measured by reporter (luciferase) activity and mRNA levels (Fig. 4a, c). We also estimated mRNA levels of Elk-1-responsive IE genes and found that PMA-activated expression of IE genes was downregulated upon exposure to heat which was restored to near control levels when heat stress was carried in cells treated with either SB203580 or ginkgolic acid. Since the expression of the IE genes was examined in cells treated with PMA, we further examined the expression levels of genes encoding components of the basal transcription machinery, a splicing factor, and a ribosomal biogenesis factor. Of the nine genes examined, the expression of seven genes was downregulated under heat stress; this downregulation was reversed when cells were heat-stressed in the presence of either SB203580 or ginkgolic acid. The expression of TBP and LAS1L mRNA levels that was not downregulated during heat stress may be regulated by additional mechanisms; it is well established that TFIID is necessary for the expression of HSF1 induced genes. It is pertinent to indicate that the modulation of Elk-1 activity during heat stress is different from that observed upon activation of the p38 MAPK pathway by anisomycin (Yang and Sharrocks 2006; Boros et al. 2009). Our observations are consistent with stress specific modulation of p38 MAPK activity (Ferreiro et al. 2010; Faust et al. 2012). Elk-1-induced transcription is also regulated by PIAS2 which acts as a coactivator upon activation of the ERK pathway and as a corepressor upon activation of the p38 MAPK pathway. Our studies indicate that in HeLa cells exposed to heat stress PIAS2 is phosphorylated by p38 MAPK pathway-dependent mechanisms.
Given the following:
-
(i)
In heat stressed HeLa cells, Elk-1 was SUMOylated and dephosphorylated with concomitant downregulation of its activity. The above (Elk-1 SUMOylation and downregulation of its activity) was reversed when cells were treated with either SB203580 or ginkgolic acid prior to heat exposure (Figs. 4, 5, and 6) and
-
(ii)
PIAS2 is phosphorylated in heat stressed HeLa cells by p38 MAPK pathway-dependent mechanisms (Fig. 7),
we conclude that SUMOylation of Elk-1 and phosphorylation of PIAS2 by p38MAPK pathway-dependent mechanisms in heat-stressed HeLa cells is associated with the downregulation of transactivation by Elk-1. Since Elk-1 binds to the promoters of nearly 1000 genes, it is conceivable that the downregulation of Elk-1 activity under conditions of heat stress may partly account for the general downregulation of transcription during exposure to the indicated conditions.
In addition to the above, the strong inhibitory effect of heat stress on Elk-1 activity suggests a module for the regulation of Elk-1 activity in cancer tissue. The hijacking of Elk-1 by the androgen receptor as well as the high Elk-1 expression levels in a number of cancer tissues has led to active research in the area of regulation of Elk-1 in several cancers (Nagalingam et al. 2014; Kawahara et al. 2015; Rosati et al. 2016; Liu et al. 2017; Ahmad et al. 2018). Relevant to the above is the usage of hyperthermia in combination with conventional therapy for cancer treatment which has shown promising results in the treatment of certain cancers (Baronzio et al. 2014).
Electronic supplementary material
(PDF 396 kb)
Acknowledgements
We thank Dr. Andrew D. Sharrocks for kindly providing us with the pEGR-1-luc plasmid and Dr. Ian Stratford, who constructed the plasmid. We are grateful to the director of this institute for his continued support during the course of this work. Thanks are also due to Dr. Lilly Ganju of the Immunomodulation Department for letting us use their qPCR facility and to Ms. Harshita Gupta for technical assistance.
Funding Information
This work was funded by Defense Research & Development Organization (DRDO, Ministry of Defense) Grant No. DIP-265 to the Director of the Defense Institute of Physiology and Allied Sciences. DC and AG are supported by fellowships from DRDO and AS is supported by a fellowship from the Council of Scientific and Industrial Research (CSIR).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ramesh Chand Meena, Email: rcmeena@dipas.drdo.in.
Amitabha Chakrabarti, Email: achakrabarti.dipas.molbiol117@gmail.com.
References
- Ahmad A, Zhang W, Wu M, Tan S, Zhu T (2018) Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes & Genomics 40(3):243–251 [DOI] [PubMed]
- Baronzio G, Parmar G, Ballerini M, Szasz A, Baronzio M, Cassutti V (2014) A brief overview of hyperthermia in cancer treatment. J Integr Oncol 3(1). 10.4172/2329-6771.1000115
- Besnard A, Galan-Rodriguez B, Nanhoutte P, Caboche J. Elk-1 a transcription factor with multiple facets in the brain. Front Neurosci. 2011;5:35. doi: 10.3389/fnins.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros J, Donaldson IJ, Donnell AO, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;11:1963–1973. doi: 10.1101/gr.093047.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli M, Dolfi F, Claret F-X, Karin M. Induction of c-fos expression through JNK-mediatedTCF/Elk-1 phosphorylation. EMBO J. 1995;14(23):5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P, Nguéa PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. Bioessays. 2015;37:1095–1105. doi: 10.1002/bies.201500065. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta (BBA)—Mol Cell Res. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Alonso G, Morrice N, Jones M, Meier R, Cohen P, Nebreda AR. Purification and cDNA cloning of SAPKK3, the major activator of RK/p38 in stress- and cytokine-stimulated monocytes and epithelial cells. EMBO J. 1996;15(16):4156–4164. doi: 10.1002/j.1460-2075.1996.tb00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. Activation of stress activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16(2):295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Hershey JWB (1984) Heat shock-induced translational alterations in HeLa cells initiation factor modifications and the inhibition of translation. J Biol Chem 259(19):11882-11889 [PubMed]
- Enserink JM. Sumo and the cellular stress response. Cell Div. 2015;10:4–16. doi: 10.1186/s13008-015-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust D, Schmitt C, Oesch F, Oesch-Bartlomowicz B, Schreck I, Weiss C, Dietrich C. Differential p38-dependent signalling in response to cellular stress and mitogenic stimulation in fibroblasts. Cell Commun Signal. 2012;10:6–18. doi: 10.1186/1478-811X-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I, Joaquin M, Islam A, Gomez-Lopez G, Barragan M, Lombardia L, Dominguez O, Pisano DG, Lopez-Bigas N, Nebreda AR, Posas F. Whole genome analysis of p38 SAPK-mediated gene expression upon stress. BMC Genomics. 2010;11:144–160. doi: 10.1186/1471-2164-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura KI, Sodeoka M, Yoshida M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16(2):133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–482. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gu ZT, Wang H, Li L, Liu YS, Deng XB, Huo SF, Yuan FF, Liu ZF, Tong HS, Su L. Heat stress induces apoptosis through transcription-independent p53-mediated mitochondrial pathways in human umbilical vein endothelial cell. Sci Rep. 2014;4:4469–4478. doi: 10.1038/srep04469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Henley JM. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014;66(2):71–77. doi: 10.1002/iub.1244. [DOI] [PubMed] [Google Scholar]
- Hsieh Y-L, Kuo H-Y, Chang C-C, Naik MT, Liao P-H, Ho C-C, Huang T-C, Jeng J-C, Hsu P-H, Tsai M-D, Huang T-H, Shih H-M. Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J. 2013;32:791–804. doi: 10.1038/emboj.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza A, O’Donnell A, Gascoigne K, Zeef LAH, Hayes A, Sharrocks AD. The ETS-domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J Biol Chem. 2005;280:1149–1155. doi: 10.1074/jbc.M411161200. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Shareef HK, Aljarah AK, Ide H, Li Y, Kashiwagi E, Netto GJ, Zheng Y, Miyamoto H (2015) ELK1 is up-regulated by androgen in bladder cancer cells and promotes tumor progression. Oncotarget 6(30):29860–29876 [DOI] [PMC free article] [PubMed]
- Knebel A, Morrice N, Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 2001;20(16):4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel A, Haydon CE, Morrice N, Cohen P. Stress-induced regulation of eukaryotic elongation factor 2 kinase by SB 203580-sensitive and insensitive pathways. Biochem J. 2002;367:525–532. doi: 10.1042/bj20020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma Y, Campbell DG, Cuenda A. Identification of glycogen synthase as a new substrate for stress activated protein kinase 2b/p38beta. Biochem J. 2004;379:133–139. doi: 10.1042/bj20031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindecke A, Korte M, Zagrelbesky M, Horejeschi V, Elvers M, Widera D, Prullage M, Pfeiffer J, Kaltschmidt B, Kaltschmidt C. Long-term depression activates transcription of immediate early transcription factor genes: involvement of serum response factor/Elk-1. Eur J Neurosci. 2006;24(2):555–563. doi: 10.1111/j.1460-9568.2006.04909.x. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Liu C-Y, Hsieh F-S, Chu P-Y, Tsai W-C, Huang C-T, Yu Y-B, Huang T-T, Ko P-S, Hung M-H, Wang W-L, Shiau C-W, Chen K-F (2017) Carfilzomib induces leukaemia cell apoptosis via inhibiting ELK1/KIAA1524 (Elk-1/CIP2A) and activating PP2A not related to proteasome inhibition. Br J Haematol 177(5):726-740 [DOI] [PubMed]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-K. [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26(22):3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- Morrison DK. MAPKinase pathways. Cold Spring Harb Perspect Biol. 2012;4:a011254. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona A, Theillet F-X, Foster C, Cheng TM, Miralles F, Bates PA, Selenko P, Treisman R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science. 2016;354:233–237. doi: 10.1126/science.aad1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam A, Kuppusamy P, Singh SV, Sharma D, Saxena NK, (2014) Mechanistic elucidation of the antitumor properties of withaferin a in breast cancer. Cancer Res 74(9):2617-2629 [DOI] [PMC free article] [PubMed]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways:regulation and physiological functions. Endocr Rev. 2001;22(2):153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Rosati R, Patki M, Chari V, Dakshnamurthy S, McFall T, Saxton J, Kidder BL, Shaw PE, Ratnam M. The amino-terminal domain of the androgen receptor co-opts extracellular signal-regulated kinase (ERK) docking sites in Elk-1 protein to induce sustained gene activation that supports prostate cancer cell growth. J Biol Chem. 2016;291(50):25982–25998. doi: 10.1074/jbc.M116.745596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda AR. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAPkinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytinki MM, Kaikkonen S, Pehkonen P, Jääskeläinen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO1 versus SUMO2/3. J Biol Chem. 2000;275(9):6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. Complexities in ETS-domain transcription factor function and regulation; lessons from the TCF subfamily. Biochem Soc Trans. 2002;30:1–9. doi: 10.1042/bst0300001. [DOI] [PubMed] [Google Scholar]
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773(8):1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Shaw PE, Saxton J. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int J Biochem Cell Biol. 2003;35:1210–1226. doi: 10.1016/S1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- Sillivan SE, Whittard JD, Jacobs MM, Ren Y, Mazloom AR, Caputi FF, Horvath M, Keller E, Ma’ayan A, Pan Y-X, Chiang LW, Hurd YL. Elk-1 transcription factor linked to dysregulated striatal mu opoid receptor signalling network and OPRM1 polymorphism in human heroin abusers. Biol Psychiatry. 2013;74(7):511–519. doi: 10.1016/j.biopsych.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FR, Manfiolli AO, Soares CS, Baqui MMA, Koide T, Gomes MD. The F-box protein FBXO25 promotes the proteasome dependent degradation of Elk-1 protein. J Biol Chem. 2013;288(39):28152–28162. doi: 10.1074/jbc.M113.504308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend KJ, Zhou P, Qian L, Bieszczad CK, Lowrey CH, Yen A, Craig RW. Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J Biol Chem. 1999;274(3):1801–1813. doi: 10.1074/jbc.274.3.1801. [DOI] [PubMed] [Google Scholar]
- Treisman R. Ternary complex factors: growth regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437X(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Nagarajan A, Kumar P. Assay for β-galactosidase in extracts of mammalian cells. Cold Spring Harb Protoc. 2017;2017:pdb.top096198. doi: 10.1101/pdb.top096198. [DOI] [PubMed] [Google Scholar]
- Vickers ER, Kasza A, Aksan-Kurnaz I, Seifert A, Zeef LA, O’Donnell A, Hayes A, Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol Cell Biol. 2004;24:10340–10351. doi: 10.1128/MCB.24.23.10340-10351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witty J, Aguilar-Martinez E, Sharrocks AD. SENP1 participates in the dynamic regulation of Elk-1 SUMOylation. Biochem J. 2010;428:247–254. doi: 10.1042/BJ20091948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H, Sharrocks AD. PIASxa differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Yang S-H, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell. 2003;12:63–74. doi: 10.1016/S1097-2765(03)00265-X. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. Global analyses of Sumoylated proteins in Saccharomyces cerevisiae: induction of protein Sumoylation by cellular stresses. J Biol Chem. 2004;279(31):32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 396 kb)