Abstract
Background:
Unlike bone, cartilage, or muscle, tendon-specific markers are not well established. The purpose of the study was to investigate expression pattern and level of 6 well-known tendon-specific markers, in various human musculoskeletal tissues, tenocytes, and mesenchymal stem cells (MSCs).
Methods:
Musculoskeletal tissue samples of tendon, bone, cartilage, nerve, muscle, and fat were obtained from patients undergoing orthopedic surgery. Tenocytes, MSCs from bone marrow, adipose tissue, and umbilical cord were isolated from each tissue and cultured. Six tendon-specific markers, scleraxis (Scx), tenomodulin (TNMD), thrombospondin-4 (TSP-4), tenascin-C (TNC), type I collagen (Col I), and type III collagen (Col III) were investigated in tendon tissue, tenocytes, and MSCs.
Results:
mRNA levels of 6 tendon-specific markers were significantly higher in tendon tissue that in other connective tissues levels of Scx, TNMD, TSP-4, and Col III immediately decreased after plating tenocytes in culture dishes whereas those of TNC and Col I did not. In comparison with tendon tissue, mRNA levels pattern of Scx, TNMD, and TSP-4 in tenocytes were significantly higher than that in MSCs, but lower than in tendon tissue whereas expression pattern of TNC, Col I and III showed different pattern with each other.
Conclusion:
This study demonstrated that 6 commonly used tendon-specific markers were mainly expressed in tendon tissue, but that expression level and pattern of the tendon-specific markers with respect to kinds of tissues, culture duration of tenocytes and sources of MSCs.
Keywords: Tendons, Scleraxis, Biomarkers, Thrombospondin-4, Mesenchymal stem cells
Introduction
Along with increase in life expectancy and physical or sports activity, tendon disease and injury has become a very common problem for general population. About 30 million tendon and ligament injuries per year occur worldwide, and around 200,000 tendon and ligament repairs per year are performed in the US [1, 2]. Acute or chronic tendon injuries do not heal spontaneously due to slow tissue turnover and cell proliferation rate, and low vessel and nervy supply, and little contribution of degenerated tendon tissue to healing [3–5], but incompletely heal via scar formation [6]. It is well-known that healed tendon via scar formation had significant week mechanical strength because of smaller crimp pattern of the collagen fibers and diameter of fibril [3, 7]. As mechanism of tendon injury and regeneration is poorly understood, treatment strategy is mainly symptomatic including rest, non-steroidal anti-inflammatory drug, injection, and physical therapy. Therefore, fundamental treatment strategy capable of regeneration of tendon tissue should be necessary and has been sought during last several decades.
Various kinds or combinations of cells including mesenchymal stem cells (MSCs), scaffolds, and signal materials have been researched for their potentials for tendon regeneration, and demonstrated promising results [6, 8]. For the evaluation of potentials of tendon regeneration, its (or their) in vitro tenogenic differentiation and the ability to form neo-tendon-like tissue in vitro or in vivo have been used [9, 10]. However, one of major hurdles is the lack of tendon-specific markers. For osteogenic differentiation, Runt-related transcription factor-2 (Runx-2) or core-binding factor subunit alpha-1 (CBFα1), bone alkaline phosphatase (BAP), and osteocalcin (OC) have been used. For chondrogenic differentiation, Sry-related high mobility group box-9 (Sox-9), type II collagen (Col II), and aggrecan have been widely used, and for myogenic differentiation, paired box protein-7 (Pax-7), myogenic factor 5 (Myf5), myogenic determination factor 1 (MyoD), and myogenin have been used [6, 11]. For tenogenic differentiation, previous studies generally used matrix genes such as Col I and III, TNC, decorin (DCN), fibronectin (FN), and cartilage oligomeric matrix protein (COMP) to evaluate the tenogenic differentiation [10, 12, 13]. However, expressions of these extracellular matrix genes are not restricted to tendon, but also expressed in many other musculoskeletal tissues including bone, fat, cartilage, and muscle. Especially, lots of proteoglycans and glycoproteins are highly expressed in both cartilage and tendon tissues [6]. More recently reported tendon associated genes identified during tendon development include Scx, TNMD, TSP-4, follistatin, ephrin receptors A4 (EphA4), eyes absent homologue 1 and 2 (Eya1 and Eya2), sineoculis homeobox homolog 1 and 2 (Six1 and Six2) [14, 15]. But, still several studies expressed concerns over the validity of such genes as tendon-specific markers [11, 13, 16–18]. Furthermore, most studies used tissue samples from animals which should be interpreted cautiously because findings might be different with tencocytes, rather than tissue, and with samples from human [18].
Another important but ignored issue of tendon-specific gene markers is about the level of their expression that could determine the efficacy of tenogenic differentiation. Majority of studies investigating efficacy of certain treatment on cells determined the success or failure of tenogenic differentiation with statistically significant changes in comparison with untreated controls [19–21]. However, statistical significant changes of mRNA levels of tendon-specific markers do not necessarily mean that the treatment could differentiate sufficiently enough to mature tenocytes. Therefore, in addition to identification of tendon-specific markers, assessment of their expression level in MSCs in comparison with tenocytes as well as native tendon tissue would provide valuable information.
The purpose of the study was to investigate expression pattern and level of 6 well-known tendon-specific markers, Scx, TNMD, TSP-4, TNC, Col I, and Col III, in various human musculoskeletal tissues, tenocytes of different passages, and bone marrow (BM), adipose tissue (AD) and umbilical cord (UC) MSCs. Our hypothesis was that there were differences in expression pattern and level of the tendon-specific markers with respect to kinds of tissues, culture duration of tenocytes and sources of MSCs.
Materials and methods
Tissue samples from various musculoskeletal tissues
The study protocol was approved by the institutional review board at our institution, and was conducted in accordance with the approved guidelines. All patients from whom tissue specimens were harvested provided informed consent. Tissue samples (n = 3 per each kind) of biceps long head tendon, bone, cartilage, muscle, and fat were obtained from patients undergoing open reduction and internal fixation or hemiarthroplasty following proximal humerus fractures (mean age 67.7 ± 10.2; 1 male and 2 females). Biceps long head tendon, deltoid muscle, and subcutaneous fat was identified and harvested during the deltopectoral approach to the proximal humerus. Bone and cartilage were identified and harvested before and during reduction of fracture fragments. Nerves (n = 3) were harvested from patients who undergoing amputation due to arteriosclerosis obliterans complications (mean age 537 ± 13.7; 3 males).
Isolation and culture of tenocyte and mesenchymal stem cells
Tendon tissues were washed twice in calcium-and magnesium-free phosphate-buffered saline (DPBS) and finely minced into 1–2 mm fragments. Cells were isolated by treating with 0.3% collagenase II for 2 h in Dulbecco’s modified Eagle medium (DMEM; Welgene, Daegu, Korea) containing antibiotic solution (100 U/mL penicillin and 100 mg/mL streptomycin) with gentle agitation [22, 23]. After the same volume of DPBS was added, undigested tissue was removed using a 100-mm nylon sieve, and cells were collected by centrifugation, washed twice, resuspended in DMEM supplemented with 10% fetal bovine serum (FBS; Welgene) and antibiotic solution (growth medium), and seeded in 100-mm tissue culture dishes at a density of 2–5 × 104 cells/cm2 at 37 °C in a humidified 5% CO2 atmosphere. The medium was replaced every 2–3 days. When cells reached 60–80% confluence, they were detached by incubation for 5 min with 0.25% trypsin (Welgene), washed, and then replated at a ratio of 1:3. Proliferation rate of tenocytes were measured with the population doubling time (PDT) with the following formulae: PD = Log (Nf/Ni)/Log2, PDT = CT/PD, where PD is the populationdoubling time, CT is cell culture time, Nf is the final number of cells, and Ni is the initial number of cells [24].
BM, AD, and UC MSCs were isolated as previously described [25]. Extracted bone marrow was diluted with 2 × DPBS, layered on top of the Ficoll-Paque™ Premium (GE Healthcare, Uppsala, Sweden) at a ratio of 1:2. This was then centrifuged at 400 g (with brake off) for 30 min at 20 °C. The upper most layer was aspirated and discarded. Then, the mononuclear layer was collected and diluted with 3 × DPBS, This was centrifuged at 400 g for 5 min and washed again with DPBS. The supernatant was discarded and the pellet was resuspended with 10 mL of growth media (low glucose DMEM containing 10% inactivated FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin), then this was centrifuged at 400 g for 5 min. The cellular pellet was resuspended in growth media. Cells were plated onto conventional tissue culture plates at a concentration of 1 × 106 cells/cm2 and incubated in a 5% CO2 incubator with humidified air at 37 °C. Media were replaced every 3 days. When cells reached to 80% confluence, cells were split at a ratio of 1:4.
Adipose tissue were washed with equal volumes of DPBS containing 5% penicillin and streptomycin to remove blood products and were finely minced by using surgical scissors and scalpels. This was intermittently agitated at 37 °C, then treated with 0.1% collagenase type I for 60 min. The collagenase was inactivated with an equal volume of DPBS, then centrifuged at 1200 g for 10 min at 20 °C, separating the stromal vascular fraction from the adipocytes. The cellular pellet was resuspended in DPBS and filtered through a 100 µm mesh filter to remove debris. The filtrate was centrifuged as detailed above, and resuspended with growth media and the number of cells were counted. Cells were plated onto conventional tissue culture plates at a concentration of 1 × 106cells/cm2 and incubated in a 5% CO2 incubator with humidified air at 37 °C. Media were replaced every 3 days. When cells reached to 80% confluence, cells were split at a ratio of 1:4.
Umbilical cord was washed twice in DPBS and was finely minced into 1-mm to 2-mm fragments by using surgical scissors and scalpels. Cells were released by treating the fragments with 0.075% collagenase type I for 2 h at 37 °C with gentle agitation. After the addition of the same volume of DPBS, undigested tissues were removed by using a 100-μm nylon sieve. Cells were collected by centrifugation, washed twice, resuspended in growth media, and plated in 100-mm tissue culture dishes at a density of 1 × 104 cells/cm2 at 37 °C in a humidified 5% CO2 atmosphere for 3–4 days. Medium was replaced every 3 days. At 80% confluence, cells were split at a ratio of 1:4.
RNA isolation and quantitative reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted, and reverse transcription and amplification were performed as previously described [24]. Briefly, total RNA was extracted using RNeasy® mini kit (QIAGEN, Hilden, Germany) and quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop, Wilmington, DE, USA). First-strand complementary DNA (cDNA) was synthesized with 1μg of mRNAs using Superscript III Reverse Transcription kit (Invitrogen, Carlsbad, CA, USA). Synthesized cDNA was diluted to 300 μL with PCR grade water and then stored at − 20 °C till use.
To perform real-time PCR utilizing a LightCycler480 (Roche Applied Science, Mannhein, Germany), TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) were used as a probe/primer set. The PCRs were performed in a final volume of 20 μL containing 10 μL 2xLightCycler® 480 Probes Master (Roche Applied Science), 1 μL TaqMan® Gene Expression Assay (Applied Biosystems), 5 μL cDNA as the template, and 4 μL PCR grade water using the following program: 95 °C for 10 min, 40 cycles of 95 °C for 10 s and 60 °C for 1 min, followed by 72 °C for 4 s, and a final cooling at 40 °C for 30 s. Experiments were performed in triplicate, and averaged values were calculated for normalized expression levels. During PCR amplification, amplified product amounts were monitored by continuous measurement of fluorescence. mRNA levels were normalized versus GAPDH as follows; the cycle number at which the transcript of each gene was detectable (threshold cycle, Ct) was normalized against Ct of GAPDH, which is referred to as ΔCt. mRNA levels relative to GAPDH are expressed as 2−ΔCt, where ΔCt = CTgene of interest − CTGAPDH.
Western blot analysis
Cell protein extracts were prepared from tenocytes seeded at a density of 2 × 104 cells/cm2 in the 6-well plate using PRO-PREP™ protein extraction solution (iNtRON, Sungnam, Korea). Equal amounts of protein extracts for each group were electrophoresed into 10% SDS-PAGE gels. The electrophoresed proteins were blotted onto a PVDF membrane with 0.45μm pore size. The membranes were blocked with TBS-T buffer containing 5% skim milk or 5% BSA for 1 h at room temperature and 1× Tris-buffered saline with 0.1% tween 20 (TBS-T) followed by incubation with primary antibody overnight at 4 °C. Primary antibodies were as follows; Scx antibody (#sc-87425, Santa Cruz Biotechnology, Dallas, TX, USA); TNMD antibody (#sc-49325, Santa Cruz Biotechnology, Dallas, TX, USA); TSP-4 antibody (#sc-28293, Santa Cruz Biotechnology, Dallas, TX, USA); TNC antibody (#sc-25328, Santa Cruz Biotechnology, Dallas, TX, USA); Col I antibody (#ab34710, abcam); Col III antibody (#sc-271249, Santa Cruz Biotechnology, Dallas, TX, USA); and β-actin antibody (#ab170325, abcam). The membranes were washed with TBS-T, and incubated with HRP-conjugated secondary antibody diluted 1:4000 for 45 min at room temperature. After washing with 1× TBS containing 0.1% Tween 20 (TBS-T), the membranes were scanned using ImageQuant LAS4000 mini (GE Healthcare Life Sciences, Little Chalfont, UK). The protein synthesis levels were normalized to those of beta-actin.
Statistical analysis
All data values were expressed as means and standard deviations. The significances of differences were determined using the independent t test and 1-way analysis of variance. For post hoc analysis, Tukey’s and Bonferroni’s test were used to compare the gene and protein expression levels among the groups. The analysis was performed using SPSS software, version 13.0 (SPSS Inc, Chicago, IL, USA) and p < 0.05 was considered to be statistically significant.
Results
Changes of morphology and proliferation rate of tenocytes along with passages
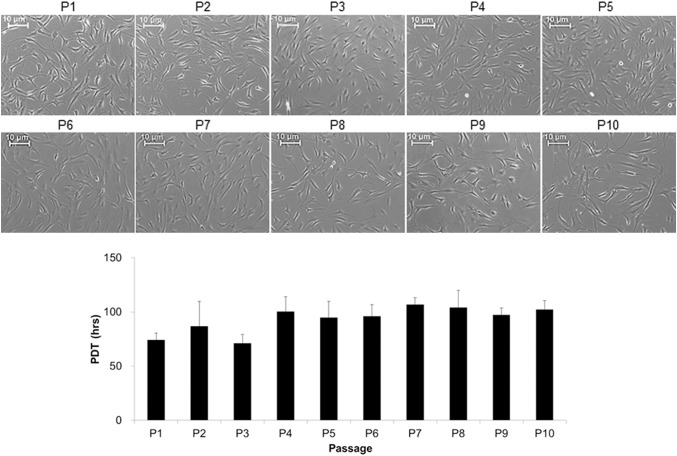
Spindle-shaped fibroblastic appearance of tenocytes were observed and maintained until P10 (Fig. 1). However, there was also a slight tendency of elongating and enlarging over passage, especially after P6. The PDTs from P1 to P3 cells, less than 90 h, was significantly lesser than those from P6 to P10, greater than 90 h.
Fig. 1.
Changes of morphology and proliferation rate of tenocytes along with passages in monolayer culture. Tenocytes were seeded at a density of 5 × 104 cell/cm2 and cultured until they reached 80% of confluence. Scale bars: 10 μm
mRNA levels of tendon-specific markers in various musculoskeletal tissues
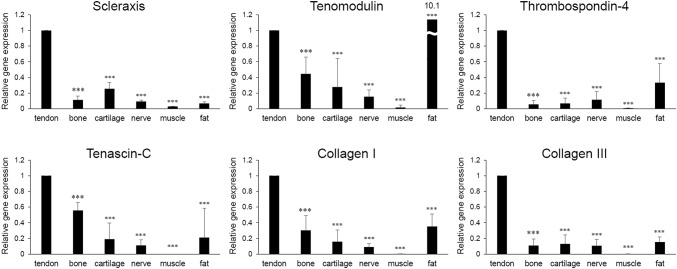
mRNA levels of Scx, TNMD, TSP-4, TNC, Col I, and Col III were significantly lower in bone, cartilage, nerve, muscle and fat tissues compared in tendon except for that of TNMD which was significantly higher in fat tissue than in tendon (10.1-fold increase) (Fig. 2) [26]. However, except in muscle tissues, all of the 6 tendon-specific genes were expressed in bone, cartilage, nerve, and fat tissues, but with varying degrees of expression.
Fig. 2.
mRNA levels of tendon tissue-specific markers in human tendon, bone, cartilage, nerve, muscle, and fat tissues (n = 3 per each tissue). Comparison was performed between mRNA level of each tissue versus that of tendon. *p < 0.05, **p < 0.01, ***p < 0.001
Changes of mRNA levels and protein synthesis of tendon-specific markers in MSCs with respect to passage
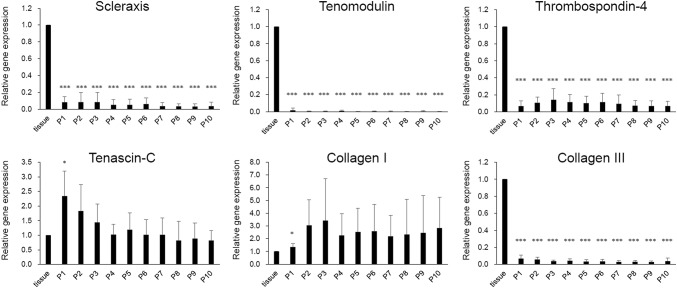
mRNA levels of Scx, TNMD, TSP-4, and Col III immediately decreased after plating tenocytes in culture dishes by less than 0.1-fold, and did not significantly change up to passage 10 (Fig. 3). mRNA levels of TNC and Col I significantly increased at passage 1. But afterward, it demonstrated large variation with respect to sample batches without statistical significance throughout culture while both maintained slightly increased level up to passage 10.
Fig. 3.
Changes of mRNA levels level of tendon tissue-specific markers in human tenocytes during culture. Comparison was performed between mRNA level of each passage versus that of tendon tissue. *p < 0.05, **p < 0.01, ***p < 0.001
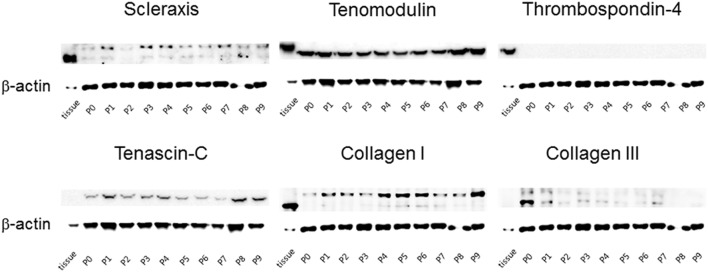
Protein synthesis of Scx, TNMD, TSP-4, TNC, Col I and III showed consistent findings with their mRNA levels (Fig. 4).
Fig. 4.
Changes of protein synthesis level of tendon tissue-specific markers in human tenocytes during culture
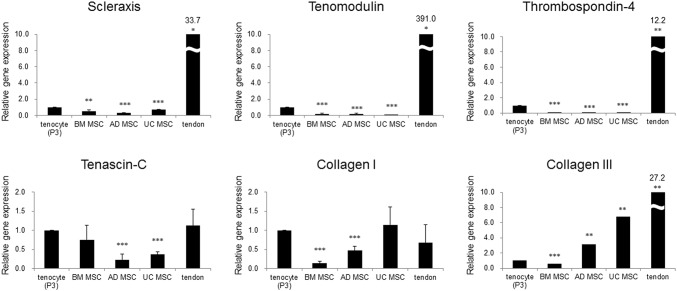
mRNA levels pattern of tendon-specific markers in tenocytes, MSCs, and tendon
mRNA levels of the 6 tendon-specific markers in MSCs in Tenocytes, MSCs, and Tendon showed 3 different patterns. First, mRNA levels of Scx, TNMD, and TSP-4 in tenocytes were significantly higher than those in BM, AD, and UC MSCs whereas they were significantly lower than in tendon tissue (Fig. 5). mRNA levels of Scx in BM, AD, and UC MSCs were lower than that of tenocytes by 0.5-, 0.3-, and 0.7-fold, respectively, and mRNA levels of TNMD were by 0.2-, 0.2, and 0.05-fold, respectively. Especially, mRNA levels of TSP-4 were rarely detected in all 3 kinds of MSCs, all less than by 0.1-fold. Expression levels of Scx, TNMD, and TSP-4 in tendon were higher than those of tenocytes by 33.5-, 391.0-, and 12.2-fold, respectively. Second, mRNA levels of TNC in tenocytes were significantly higher than that in AD and UC MSCs whereas they were not when compared with those in BM MSCs. And, mRNA levels of Col I in BM and AD MSCswere significantly lower than with those in tenocytes by 0.1- and 0.5-fold, respectively. Meanwhile, mRNA levels of TNC and Col I in tendon were not significantly different with those in tenocytes. Third, mRNA levels of Col III in tenocytes were significantly higher than in BM MSCs (0.6-fold) whereas they were significantly lower than that in AD and UC MSCs and in tendon, 3.1-, 6.8, and 27.2-fold, respectively.
Fig. 5.
Comparison of mRNA levels of tendon tissue-specific markers in human tenocytes, MSCs, and tendon tissue. Comparison was performed between mRNA level of tenocytes versus that of each MSC and tendon tissue. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
The most important findings of this study are (1) mRNA levels of the 6 tendon-specific genes, Scx, TNMD, TSP-4, TNC, Col I, and Col III, were significantly higher in tendon tissue than in other connective tissues, bone, cartilage, nerve, and fat tissues; (2) mRNA levels of Scx, TNMD, TSP-4, and Col III immediately decreased after plating tenocytes in culture dishes whereas those of TNC and Col I were not; (3) mRNA levels of Scx, TNMD, TSP-4, and TNC were significantly increased in tenocytes than in BM, AD, and UC MSCs. Especially, TSP-4 was rarely detected in MSCs. (4) In comparison with mRNA levels in tendon, expression of the 6 tendon-specific genes showed 3 patterns. First, mRNA levels of Scx, TNMD and TSP-4 were higher than those in MSCs, but lower than in tendon tissue. Second, mRNA levels of TNC and Col I were higher than in some MSCs, but were not different with that in tendon tissue. Third, mRNA levels of Col III were higher than in BM MSCs, but lower than in AD and UC MSC, and tendon tissue. These results demonstrated that whereas 6 commonly used tendon-specific markers are found mainly in tendon tissue in comparison with other musculoskeletal tissues. However, it should be cautious when they are used as criteria for tenogenic differentiation of MSCs as pattern of mRNA levels of each marker is much different with respect to sources of MSCs and expression levels of Scx, TNMD, TSP-4 and Col III in tenocytes were far less than those in tendon tissues. These results suggest that assessment of tenogenic differentiation of stem cells with commonly used tenogenic markers should not be used solely, but needs to be one of several evaluation methods with different angles including histological and biomechanical evaluation.
In this study, we included 6 most commonly used tendon-specific markers, Scx, TNMD, TSP-4, TNC, Col I and III. While these markers have been widely used to assess phenotype of tenocytes or tenogenic differentiation of stem cells from human sources [27–29], only one study, to our knowledge, investigated the validity of these markers with human samples. Using RNA microarray analysis, Jelinsky et al. [18] reported approximately 1600 and 300 tendon-selective transcripts in rat and human tendon tissues, respectively and found that TNMD and TSP-4 were found to be the highest tendon-selective gene, but Scx, TNC, Col I and III were not. However, their study was performed only with tissues, but not with cells, which may be contaminated by other structures, such as nerves and vessels inside tendon tissue [6]. Furthermore, they did not report protein expression of the biomarkers. Meanwhile, they described that the great reduction in the number of tendon-selective genes in human (300 transcripts) compared to rat (1600 transcripts) may be due to the greater variability of the cadaveric samples as well as collection and sampling processing. We agree with them. And as the tissues in this study have been only processes for mRNA and Western blotting, findings of this study with relatively fresh human tissues and cells would provide useful information with respect to tendon regeneration especially using human resources.
It has been previously reported tenocyte phenotypic drift after prolonged maintenance in monolayer cell culture that may lead to present less tendon-specific proteins at the levels present in tendon tissue [30–32]. Some authors reported that cells within the first 3 passages could be used for in vitro monolayer cell models with less signs of phenotypic drift [33]. However, most studies compared phenotype drift of tenocytes between early and late passages, and no study compared phenotype drift of tenocytes with tendon tissue. The results of this study demonstrated that morphology of tenocytes did not change significantly up to P10. However, proliferation rate became significantly slower after P3. Furthermore, when compared with tendon tissue, phenotype drift or dedifferentiation of tenocytes occurred immediately after plating, and showed different pattern with respect to tendon-specific genes; pattern of Scx, TNMD, TSP-4 and Col III showed immediate decrease and was maintained during subsequent culture whereas that of TNC and Col I showed immediate increase and relatively large variation during subsequent culture. These findings suggest that functional phenotypic drift occurs early after culture despite minimal changes in morphological phenotypic drift up to P10 which should be considered in various studies using tenocytes.
A majority of previous studies assessed tenogenic differentiation of MSCs with statistically significant changes of mRNA levels of tendon-specific marker genes [19, 27, 28, 34]. Meanwhile, results of this study demonstrated that mRNA levels of most tendon-specific markers, Scx, TNMD, TSP-4, and TNC, in BM, AD, and UC MSCs was at least threefold less than those in tenocytes (Fig. 5). Especially, mRNA levels of TSP-4 in MSCs were minimum 240-fold less than that in tenocytes. These results suggest that change of mRNA levels that are statistically significant but not sufficiently large might not be adequate for the criteria of tenogenic differentiation. Furthermore, as mRNA levels of most tendon-specific markers in tenocytes significantly decrease immediately after plating in culture dish, expression level in cultured tenocytes should be far less than that in tendon tissue. Thus, mRNA levels profiles of various tendon-specific markers in tenocytes, MSCs, and tendon tissue would provide valuable information for the evaluation of in vitro and in vivo tenogenic differentiation. All of these findings suggest that we should be more cautious in interpretation of tenogenic differentiation data and reports especially comparing mRNA levels of tendon-specific markers.
There are several limitations in this study. First, we included only small numbers (6) of known tendon-specific markers. Whereas more than a dozen of tenogenic markers were reported by researchers, the 6 markers in this study have been most commonly used in both human and animal studies which suggest that the findings of this study would provide useful information for researchers conducting relevant studies. Second, this study included a monolayer cell culture system only. There have been reported other culture systems [35, 36], and they would show different phenotype drift with the results of this study. Nonetheless, still a monolayer culture of tenocytes and MSCs are most commonly used culture method among various culture conditions. Any further studies would be necessary after a certain culture condition become widely accepted between researchers. Third, this study compared tendon-specific genes in naïve MSCs, but not in tenogenic differentiation conditions. Direct comparison of tendon-specific marker of tenocyte, MSCs induced to tenogenic differentiation, and tendon tissue would be necessary in further studies.
In conclusion, this study demonstrated that 6 commonly used tendon-specific markers were mainly expressed in tendon tissue, but that expression level and pattern of the tendon-specific markers with respect to kinds of tissues, culture duration of tenocytes and sources of MSCs.
Acknowledgements
This research was supported by the Basic Science Research Program and the Bio and Medical Technology Development Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2015M3A9E6028412 and NRF-2017R1A2B2010995).
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
The study protocol was approved by the institutional review board at our institution, and was conducted in accordance with the approved guidelines (Seoul National University Boramae Medical Center Institutional Review Board No. 20120405/06-2012-78/118). All patients from whom tissue specimens were harvested provided informed consent.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maffulli N, Wong J, Almekinders LC. Types and epidemiology of tendinopathy. Clin Sports Med. 2003;22:675–692. doi: 10.1016/S0278-5919(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 2.Pennisi E. Tending tender tendons. Science. 2002;295:1011. doi: 10.1126/science.295.5557.1011. [DOI] [PubMed] [Google Scholar]
- 3.Battaglia TC, Clark RT, Chhabra A, Gaschen V, Hunziker EB, Mikic B. Ultrastructural determinants of murine achilles tendon strength during healing. Connect Tissue Res. 2003;44:218–224. doi: 10.1080/03008200390248452. [DOI] [PubMed] [Google Scholar]
- 4.Uhthoff HK, Trudel G, Himori K. Relevance of pathology and basic research to the surgeon treating rotator cuff disease. J Orthop Sci. 2003;8:449–456. doi: 10.1007/s10776-002-0624-5. [DOI] [PubMed] [Google Scholar]
- 5.Heinemeier KM, Schjerling P, Heinemeier J, Magnusson SP, Kjaer M. Lack of tissue renewal in human adult Achilles tendon is revealed by nuclear bomb (14)C. FASEB J. 2013;27:2074–2079. doi: 10.1096/fj.12-225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lui PP, Rui YF, Ni M, Chan KM. Tenogenic differentiation of stem cells for tendon repair-what is the current evidence? J Tissue Eng Regen Med. 2011;5:e144–e163. doi: 10.1002/term.424. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita H, Ochi M, Ikuta Y. Histological and biomechanical observations of the rabbit patellar tendon after removal of its central one-third. Arch Orthop Trauma Surg. 1997;116:454–462. doi: 10.1007/BF00387577. [DOI] [PubMed] [Google Scholar]
- 8.Docheva D, Müller SA, Majewski M, Evans CH. Biologics for tendon repair. Adv Drug Deliv Rev. 2015;84:222–239. doi: 10.1016/j.addr.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen WH, Lai MT, Wu AT, Wu CC, Gelovani JG, Lin CT, et al. In vitro stage-specific chondrogenesis of mesenchymal stem cells committed to chondrocytes. Arthritis Rheum. 2009;60:450–459. doi: 10.1002/art.24265. [DOI] [PubMed] [Google Scholar]
- 10.Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 11.Kuemmerle JM, Theiss F, Okoniewski MJ, Weber FA, Hemmi S, Mirsaidi A, et al. Identification of novel equine (Equus caballus) tendon markers using RNA sequencing. Genes (Basel) 2016;7:E97. doi: 10.3390/genes7110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JY, Zhou Z, Taub PJ, Ramcharan M, Li Y, Akinbiyi T, et al. BMP-12 treatment of adult mesenchymal stem cells in vitro augments tendon-like tissue formation and defect repair in vivo. PLoS One. 2011;6:e17531. doi: 10.1371/journal.pone.0017531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandau O, Meindl A, Fässler R, Aszódi A. A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev Dyn. 2001;221:72–80. doi: 10.1002/dvdy.1126. [DOI] [PubMed] [Google Scholar]
- 14.Edom-Vovard F, Duprez D. Signals regulating tendon formation during chick embryonic development. Dev Dyn. 2004;229:449–457. doi: 10.1002/dvdy.10481. [DOI] [PubMed] [Google Scholar]
- 15.Wolfman NM, Hattersley G, Cox K, Celeste AJ, Nelson R, Yamaji N, et al. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-beta gene family. J Clin Invest. 1997;100:321–330. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SE, Vaughan-Thomas A, Clements DN, Pinchbeck G, Macrory LC, Smith RK, et al. Gene expression markers of tendon fibroblasts in normal and diseased tissue compared to monolayer and three dimensional culture systems. BMC Musculoskelet Disord. 2009;10:27. doi: 10.1186/1471-2474-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298:234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Jelinsky SA, Archambault J, Li L, Seeherman H. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 2010;28:289–297. doi: 10.1002/jor.20999. [DOI] [PubMed] [Google Scholar]
- 19.Mazzocca AD, McCarthy MB, Chowaniec D, Cote MP, Judson CH, Apostolakos J, et al. Bone marrow-derived mesenchymal stem cells obtained during arthroscopic rotator cuff repair surgery show potential for tendon cell differentiation after treatment with insulin. Arthroscopy. 2011;27:1459–1471. doi: 10.1016/j.arthro.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Xiong Y, Jiang Y, Zhang Z, Zhou G, Zhang W, et al. Conditional tenomodulin overexpression favors tenogenic lineage differentiation of transgenic mouse derived cells. Gene. 2017;598:9–19. doi: 10.1016/j.gene.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Brown JP, Galassi TV, Stoppato M, Schiele NR, Kuo CK. Comparative analysis of mesenchymal stem cell and embryonic tendon progenitor cell response to embryonic tendon biochemical and mechanical factors. Stem Cell Res Ther. 2015;6:89. doi: 10.1186/s13287-015-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo CH, Kim JE, Yoon KS, Shin S. Platelet-rich plasma stimulates cell proliferation and enhances matrix gene expression and synthesis in tenocytes from human rotator cuff tendons with degenerative tears. Am J Sports Med. 2012;40:1035–1045. doi: 10.1177/0363546512437525. [DOI] [PubMed] [Google Scholar]
- 23.Yoon JY, Lee SY, Shin S, Yoon KS, Jo CH. Comparative analysis of platelet-rich plasma effect on tenocytes from normal human rotator cuff tendon and human rotator cuff tendon with degenerative tears. Clin Shoulder Elbow. 2018;21:3–14. doi: 10.5397/cise.2018.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo CH, Kim OS, Park EY, Kim BJ, Lee JH, Kang SB, et al. Fetal mesenchymal stem cells derived from human umbilical cord sustain primitive characteristics during extensive expansion. Cell Tissue Res. 2008;334:423–433. doi: 10.1007/s00441-008-0696-3. [DOI] [PubMed] [Google Scholar]
- 25.Jo CH, Yoon PW, Kim H, Kang KS, Yoon KS. Comparative evaluation of in vivo osteogenic differentiation of fetal and adult mesenchymal stem cell in rat critical-sized femoral defect model. Cell Tissue Res. 2013;353:41–52. doi: 10.1007/s00441-013-1619-5. [DOI] [PubMed] [Google Scholar]
- 26.Saiki A, Olsson M, Jernås M, Gummesson A, McTernan PG, Andersson J, et al. Tenomodulin is highly expressed in adipose tissue, increased in obesity, and down-regulated during diet-induced weight loss. J Clin Endocrinol Metab. 2009;94:3987–3994. doi: 10.1210/jc.2009-0292. [DOI] [PubMed] [Google Scholar]
- 27.Lee CH, Shah B, Moioli EK, Mao JJ. CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. J Clin Investig. 2010;120:3340–3349. doi: 10.1172/JCI43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan SL, Ahmad RE, Ahmad TS, Merican AM, Abbas AA, Ng WM, et al. Effect of growth differentiation factor 5 on the proliferation and tenogenic differentiation potential of human mesenchymal stem cells in vitro. Cells Tissues Organs. 2012;196:325–338. doi: 10.1159/000335693. [DOI] [PubMed] [Google Scholar]
- 29.Haddad-Weber M, Prager P, Kunz M, Seefried L, Jakob F, Murray MM, et al. BMP12 and BMP13 gene transfer induce ligamentogenic differentiation in mesenchymal progenitor and anterior cruciate ligament cells. Cytotherapy. 2010;12:505–513. doi: 10.3109/14653241003709652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Güngörmüş C, Kolankaya D. Gene expression of tendon collagens and tenocyte markers in long-term monolayer and high-density cultures of rat tenocytes. Connect Tissue Res. 2012;53:485–491. doi: 10.3109/03008207.2012.694511. [DOI] [PubMed] [Google Scholar]
- 31.Almarza AJ, Augustine SM, Woo SL. Changes in gene expression of matrix constituents with respect to passage of ligament and tendon fibroblasts. Ann Biomed Eng. 2008;36:1927–1933. doi: 10.1007/s10439-008-9565-1. [DOI] [PubMed] [Google Scholar]
- 32.Yao L, Bestwick CS, Bestwick LA, Maffulli N, Aspden RM. Phenotypic drift in human tenocyte culture. Tissue Eng. 2006;12:1843–1849. doi: 10.1089/ten.2006.12.1843. [DOI] [PubMed] [Google Scholar]
- 33.Mazzocca AD, Chowaniec D, McCarthy MB, Beitzel K, Cote MP, McKinnon W, et al. In vitro changes in human tenocyte cultures obtained from proximal biceps tendon: multiple passages result in changes in routine cell markers. Knee Surg Sports Traumatol Arthrosc. 2012;20:1666–1672. doi: 10.1007/s00167-011-1711-x. [DOI] [PubMed] [Google Scholar]
- 34.Yin Z, Guo J, Wu TY, Chen X, Xu LL, Lin SE, et al. Stepwise differentiation of mesenchymal stem cells augments tendon-like tissue formation and defect repair in vivo. Stem Cells Transl Med. 2016;5:1106–1116. doi: 10.5966/sctm.2015-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, He A, Zhang Z, Zhang W, Zhou G, Cao Y, et al. Induction of transient tenogenic phenotype of high-density cultured human dermal fibroblasts. Connect Tissue Res. 2015;56:288–299. doi: 10.3109/03008207.2015.1023433. [DOI] [PubMed] [Google Scholar]
- 36.Stoll C, John T, Endres M, Rosen C, Kaps C, Kohl B, et al. Extracellular matrix expression of human tenocytes in three-dimensional air–liquid and PLGA cultures compared with tendon tissue: implications for tendon tissue engineering. J Orthop Res. 2010;28:1170–1177. doi: 10.1002/jor.21109. [DOI] [PubMed] [Google Scholar]