Abstract
Diversity comparison and phylogenetic analyses of carbohydrate-active enzymes (CAZymes), auxiliary activities (AAs) and cytochromes P450 among 40 fungi, which are based on different nutritional pathways, help clarify and explain their divergence and improvement of various life-styles. Molecular clock analyses allow us to understand the evolutionary and developmental rules in decomposition gene families. Our results suggested that fungi in different ecological types acquired an obvious preference on specific decomposing gene families during evolutionary selection. White rot and litter saprotrophic fungi possessed more complete types of varied degradation gene families and were superior in quantities. With evolution and development of lignocellulose decomposition mechanism, certain families (like CBM1, GH6, GH7, GH10, and CYP53) disappeared in brown rot fungi and symbiotic fungi. In addition, the earlier time of phylogenetic divergence determined the more integrated and larger decomposition families. And various gains and losses in gene quantity of varied decomposition families led in particularly phylogenetic clades or nodes, then accelerated in forming varied ecotypes of species.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1689-5) contains supplementary material, which is available to authorized users.
Keywords: Degrading gene families, Edible and medical mushrooms, Gene evolution, Phylogeny, Eco-lifestyle
Introduction
Cultivation media have significant influence in the mushroom yield, mycelial growth rate, biological efficiency and nutritional parameters of edible and medicinal mushrooms (Philippoussis et al. 2001; Isikhuemhen and Mikiashvilli 2009). Most of the popular edible and medicinal mushrooms are saprophytic fungi. A majority of saprophytic fungi are white-rot fungi, while a few saprophytic fungi are brown-rot fungi and litter decomposers. As wood-destroying fungi, white-rot mushrooms, such as Flammulina velutipes (Curtis) Singer and Pleurotus ostreatus (Jacq.) Quél., can degrade all plant cell wall components (Floudas et al. 2012); while lignin is modified but not appreciably degraded by brown-rot mushrooms (Floudas et al. 2012), such as Wolfiporia cocos (F.A. Wolf) Ryvarden & Gilb and Coniophora olivacea (Fr.) P. Karst. (Castanera et al. 2017). Litter decomposers are saprotrophic fungi in soil, such as Volvariella volvacea (Bull.) Singer and Agaricus bisporus (J.E. Lange) Imbach, are considered to have strong activities in straw degradation but are weak for degrading the lignin component of lignocelluloses (Cai et al. 1994). Another part of edible and medicinal mushrooms is symbiotic with other species, such as Laccaria bicolor (Maire) P.D. Orton and Boletus edulis Bull. All the edible and medicinal mushrooms can decompose and utilize lignocellulose as nutrition resources to some extent. However, studies of wood-decay organisms have focused on model fungal systems. Examples are the white-rot fungi Phanerochaete chrysosporium Burds. and Ceriporiopsis subvermispora (Pilát) Gilb. & Ryvarden and the brown-rot fungus Postia placenta (Fr.) M.J. Larsen & Lombard (Martinez et al. 2004, 2009; Fernandez-Fueyo et al. 2012), while only a few studies focus on how edible and medicinal mushrooms decompose and utilize lignocellulose.
Lignocellulose is a composite of the polysaccharide cellulose and hemicellulose, and lignin, which is poly-phenolic (Broda et al. 1996). The three types of polymers strongly intermeshed and chemically bonded by non-covalent forces and by covalent cross-linkages (Pérez et al. 2002). These complex constructions make enzymatic attack more difficult than that on other glucose-based polymers such as starch (Broda et al. 1996). The conversion of lignocellulose by fungi is quite complicated, with a set of hydrolytic enzymes including laccase, cellulose, xylanase, and mannanase (Wu et al. 2015). The cellulose, hemicellulose, and lignin in the plant cell wall are degraded through synergistic action of multiple enzymes (Pérez et al. 2002). Genomics of many model edible and medicinal mushrooms have been sequenced, such as L. bicolor (Martin et al. 2008), Coprinopsis cinerea (Schaeff.) Redhead (Stajich et al. 2010), Schizophyllum commune Fr. (Ohm et al. 2010), A. bisporus (Morin et al. 2012), V. volvacea (Bao et al. 2013; Chen et al. 2013), and F. velutipes (Park et al. 2014), which permit comparative and functional genomic analyses of nutritional niche adaptation and decomposition of lignocellulose by edible and medicinal mushrooms (Eastwood et al. 2011).
Lignocellulolytic enzymes comprise families that fall into two categories: CAZymes and oxidoreductases (Broda et al. 1996; Floudas et al. 2012). CAZymes are responsible for the breakdown, biosynthesis or modification of glycoconjugates, oligo- and polysaccharides (Zhao et al. 2013) and play a key role in the synthesis and breakdown of plant cell wall as well as in host–pathogen interactions (Cantarel et al. 2009; Zhao et al. 2013). Based on the structurally related catalytic modules or functional domains, CAZymes cover approximately 300 protein families in five classes of enzyme activities: glycoside hydrolases (GHs), glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and carbohydrate-binding modules (CBMs) (Cantarel et al. 2009). The CE, GH, and PL superfamilies are also known as cell wall degrading enzymes (CWDE) due to their role in the disintegration of the plant cell wall by bacterial and fungal pathogens (Ospina-Giraldo et al. 2010). In addition to the catalytic modules, CBMs are the most common non-catalytic modules associated with enzymes active in cell-wall hydrolysis (Cantarel et al. 2009), for example, enhancing the localization of the enzymes that bear them on the surface of crystalline cellulose (Guillén et al. 2010) or disrupting the crystallinity of cellulose (Wang et al. 2008). Oxidoreductases are comprised by auxiliary activities (AAs) and cytochromes P450 (Floudas et al. 2012). AAs accommodate a range of enzyme mechanisms and substrates related to lignocellulose conversion, including lignocellulolytic enzymes, such as laccase, manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidase (VP) (Levasseur et al. 2013). Cytochromes P450 monooxygenases are mixed functional oxidoreductases ubiquitously distributed throughout living organisms (Floudas et al. 2012), playing important roles in the biosynthesis of numerous secondary metabolites and in downstream events of the lignin degradation process (Ospina-Giraldo et al. 2010; Morin et al. 2012).
Fungi can produce all kinds of CAZymes and oxidoreductases to achieve various biological functions (Wu et al. 2015). Saprotrophic fungi gain nutrition for fungal growth and reproduction mainly through releasing carbohydrates from lignocellulose in the environment; fungal pathogens secrete a variety of CAZymes and oxidoreductases to penetrate and successfully infect their hosts (Zhao et al. 2013). When cultured on different substrates, various plant biomass degrading enzymes were produced by different fungi, i.e. Fusarium graminearum Schwabe, Rhizopus oryzae Went & Prins. Geerl. (Battaglia et al. 2011), and the model filamentous fungus Neurospora crassa Shear & B.O. Dodge (Tian et al. 2009). In recent years, the fast expansion of basidiomycete genomic and functional genomics data (e.g. transcriptomics, proteomics), which facilitated exploration of key genes and regulatory mechanisms of biomass degradation was reported (Cai et al. 2017; Peng et al. 2018). As a result, the activities of hydrolytic enzymes from different fungi showed preferences for different types of plant biomass and adaption to their lifestyles (Zhao et al. 2013).
To reconstruct the evolution of lignocellulose decay mechanisms, 40 diverse edible and medicinal mushroom genomes were selected, including species representing each of a range of functional niches: white rot and brown rot wood decay, litter saprotrophic, and mutualistic ectomycorrhizal symbiosis. In this study, the CAZymes and oxidoreductases related with lignocellulolytic decomposition from different nutrition modes of representative edible and medicinal fungi were annotated and compared to estimate phylogenetic relationships.
Materials and methods
Data collection and annotation
A data set including 40 fungi involved in edible or/and medicinal mushrooms from both Basidiomycota and Ascomycota was used to estimate the timing of phylogenetic divergence events (Table 1) (Boa 2004; Dai and Yang 2008; Dai et al. 2010). The genomic data of V. volvacea and F. velutipes were kindly provided by Prof. Baogui Xie from Mycological Research Center, College of Life Sciences, Fujian Agriculture and Forestry University. Also, the genomic data of others were downloaded from JGI database (DOE Joint Genome Institute, https://genome.jgi.doe.gov/).
Table 1.
The classification, ecology, functions and accession numbers of 40 fungi in this study
| Phylum | Class | Order | Ecology | Species | Functions | JGI project ID |
|---|---|---|---|---|---|---|
| Ascomycota | Pezizomycetes | Pezizales | Symbiotic | Choiromyces venosus | Edible | 1016738 |
| Morchella conica | Edible | 1023999 | ||||
| Tuber melanosporum | Edible | 1047810 | ||||
| Basidiomycota | Agaricomycetes | Agaricales | White rot | Pleurotus ostreatus | Edible and medicinal | 1184896 |
| Flammulina velutipes | Edible and medicinal | – | ||||
| Armillaria mellea | Edible and medicinal | NCBI Project Id: 196228 | ||||
| Schizophyllum commune | Edible and medicinal | 1076475 | ||||
| Panellus stipticus | Medicinal | 1018598 | ||||
| Brown rot | Fistulina hepatica | Edible and medicinal | 1079823 | |||
| Symbiotic | Tricholoma matsutake | Edible and Medicinal | 1079273 | |||
| Laccaria amethystina | Edible | 1006252 | ||||
| Laccaria bicolor | Edible | 1091345 | ||||
| Amanita muscaria | Medicinal | 1078184 | ||||
| Straw rot | Agaricus bisporus | Edible and Medicinal | 1076821 | |||
| Coprinopsis cinerea | Medicinal | 1004745 | ||||
| Volvariella volvacea | Edible and medicinal | ANCH00000000 | ||||
| Auriculariales | White rot | Auricularia subglabra | Edible | 402897 | ||
| Boletales | Brown rot | Serpula lacrymans | Medicinal | 1076883 | ||
| Symbiotic | Boletus edulis | Edible and medicinal | 1006377 | |||
| Gyrodon lividus | Edible | 1016730 | ||||
| Paxillus involutus | Edible and medicinal | 1076818 | ||||
| Pisolithus tinctorius | Medicinal | 402035 | ||||
| Scleroderma citrinum | Medicinal | 1079271 | ||||
| Suillus brevipes | Edible | 1011225 | ||||
| Suillus luteus | Edible and medicinal | 1006870 | ||||
| Gloeophyllales | White rot | Neolentinus lepideus | Edible | 1006918 | ||
| Brown rot | Gloeophyllum trabeum | Medicinal | 1077715 | |||
| Polyporales | White rot | Pycnoporus cinnabarinus | Edible and medicinal | 1111350 | ||
| Pycnoporus sanguineus | Edible and medicinal | 1024951 | ||||
| Polyporus arcularius | Edible and medicinal | 1006898 | ||||
| Lentinus tigrinus | Edible | 1020065 | ||||
| Trametes versicolor | Edible and medicinal | 1077971 | ||||
| Brown rot | Fomitopsis pinicola | Medicinal | 1078004 | |||
| Laetiporus sulphureus | Edible and medicinal | 1006914 | ||||
| Wolfiporia cocos | Edible and medicinal | 1078005 | ||||
| Russulales | White rot | Stereum hirsutum | Medicinal | 402896 | ||
| Heterobasidion annosum | Medicinal | 1076437 | ||||
| Dacrymycetes | Dacrymycetales | White rot | Calocera viscosa | Edible | 1017988 | |
| Brown rot | Calocera cornea | Edible | 1006894 | |||
| Tremellomycetes | Tremellales | Symbiotic | Tremella mesenterica | Edible and medicinal | 1084394 |
– Indicated no JGI project ID or other public accession number
The annotation of carbohydrate active enzymes (CAZymes) was based on HMM Model (Yin et al. 2012) of dbCAN database (database for automated carbohydrate active enzyme annotation). The potential function structure regions of protein sequences were analyzed when the genomic data of 40 fungi involved in edible or/and medicinal mushrooms were compared using the Hidden Markov model (HMM), then the CAZymes were annotated and classified.
The protein sequences of AAs and P450 were downloaded from Carbohydrate-Active enZYmes Database (https://www.cazy.org/) and Cytochrome P450 Server (https://drnelson.uthsc.edu/CytochromeP450.html), respectively. The AAs and P450 superfamilies were annotated and classified by blasting with the downloaded sequences.
Phylogenetic analyses
A total of 1438 single-copy genes from 125 species based on OrthoDB 7 database (BUSCO Benchmarking sets of Universal Single-Copy Orthologs) (Waterhouse et al. 2013) were aligned using MAFFT 6.717b (Katoh and Toh 2008), and the Hidden Markov Model (HMM) was constructed using HMMER 3.0 (Eddy 2009). The orthologous gene families were selected by blasting the genomes of 40 edible and medicinal mushrooms in this study using the constructed HMM. The orthologous gene families were defined as the highest HMM value when e-values no more than 1E-50. All the selected orthologous gene families were analyzed using MAFFT 6.717b (Katoh and Toh 2008), and the conserved regions were selected using Gblocks (Castresana et al. 2000). A total of 413 orthologous gene families with gene length of conserved regions no less than 20% of whole gene were selected and linked to a single file with custom Perl program (Floudas et al. 2012). The best evolutionary model of nucleotide substitution in evolution of genes was selected using ProtTest 3.2 (Darriba et al. 2011) before phylogenetic study. Maximum likelihood estimate of modal parameters can be analyzed by ProtTest 3.2 using Akaike information criterion (AIC), Bayesian information criterion (BIC), and decision theory criterion (DT). The best evolutionary model selected is “LG + I + F”. The rate matrices are “LG”, rate variation of site is “ + I” (invariable sites), and the observed amino acid frequencies are “ + F” (observed amino acid frequencies). Then the phylogenetic tree was constructed using RAxML 7.2.8 with PROTGAMMAILGF model and bootstrap of 100 (Stamatakis et al. 2006).
Molecular clock analysis
To study the molecular clock of edible and medicinal mushrooms, three well-characterized fossil calibrations were used to calibrate the minimum ages of Boletales, Agaricales, and Ascomycota clades in the Dikarya, respectively (Eastwood et al. 2011; Floudas et al. 2012). The 50-mya permineralized suilloid ectomycorrhiza fossil associated with pine roots from the middle Eocene Princeton chert of British Columbia, Canada (LePage et al. 1997) was used to calibrate the Boletales (Eastwood et al. 2011; Floudas et al. 2012). The Archaeomarasmius legetti fossil from mid-Cretaceous amber discovered in a well-characterized layer of clay in New Jersey, USA of 94–90 mya (Hibbett et al. 1997; Eastwood et al. 2011) was used to calibrate the Agaricales for the resembled morphology of modern agarics (small mushrooms with a stipe and a cap) (Floudas et al. 2012). A 400-mya-old ascomycete fossil Paleopyrenomycites devonicus from the early Devonian associated with the extinct vascular plant Asteroxylon mackiei (Taylor et al. 2005) was used to calibrate Ascomycota (Floudas et al. 2012).
The divergence time between species was estimated with the Langley-Fitch method with r8s (Sanderson et al. 2006). The three fossil calibrations followed a lognormal distribution, where the minimum age for the calibrations afore-described provided the offset values.
The CAFÉ program (De Bie et al. 2006) and BadiRate program (Librado et al. 2012) were used to analyze the evolution of protein family size variation (expansion or contraction) in CAZymes and oxidoreductases in 40 edible and medicinal mushrooms. CAFÉ used a stochastic model of gene birth and death to infer statistically significant gain and loss in gene families, given gene copy numbers in each organism and ancestor. A family-wide significance threshold of 0.05 was used. BadiRate provides the appropriate statistical framework for testing biologically relevant hypothesis on the input data, consisting of the species’ phylogenetic tree and the size of each gene family in each extant species (Carretero-Paulet et al. 2015), with Birth–Death–and–Innovation (BDI, a.k.a. Birth-Death-and-Immigration) stochastic models as implemented.
Accession numbers for the downloaded datasets
The accession numbers for the downloaded genomic data from JGI database used in the present study have been shown in Table 1 (Ohm et al. 2010; Eastwood et al. 2011; Olson et al. 2012; Collins et al. 2013; Nordberg et al. 2014; Branco et al. 2015; Floudas et al. 2012, 2015; Kohler et al. 2015; Alfaro et al. 2016; Nagy et al. 2016). Among them, the genomic data of Armillaria mellea could be downloaded from JGI without the JGI project ID, but the NCBI Project Id (196228) had been listed. The genomic data of V. volvacea and F. velutipes were kindly provided by Prof. Baogui Xie from Mycological Research Center, College of Life Sciences, Fujian Agriculture and Forestry University. The genomic data of V. volvacea were deposited at DDBJ/EMBL/GenBank under the accession no. ANCH00000000 (Chen et al. 2013), but no accession numbers of genomic data of F. velutipes could be listed here.
Results and discussion
Phylogeny and evolution of edible and medicinal mushrooms
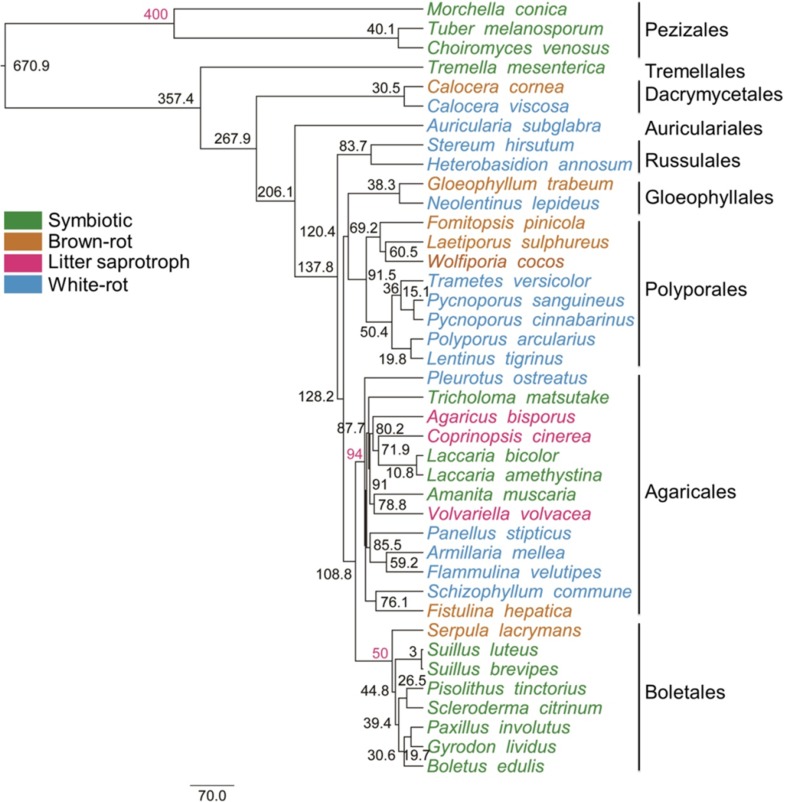
To estimate phylogenetic relationships among 40 edible and medicinal mushrooms, data sets using 413 single-copy genes were constructed, with 149,592 amino acids. The phylogenetic tree of edible and medicinal mushrooms was strongly supported, with all but six nodes receiving maximal support values (Figure S1); and the tree topology was consistent with prior published phylogenetic inferences using multigene analysis (Fig. 1) (Matheny et al. 2007). All the symbiotic mushrooms clustered in Ascomycota and Boletales, Agaricales and Tremellales of Agaricomycetes, Basidiomycota. The tree also resolved four independent brown rot lineages as a previous study (Floudas et al. 2012). All brown-rot mushrooms were in Basidiomycota, clustered in Boletales, Agaricales, Polyporales, and Gloeophyllales of Agaricomycetes, and Dacrymycetales of Dacrymycetes. The white-rot mushrooms distributed in Agaricales, Polyporales, and Russulales of Agaricomycetes; some others were in Gloeophyllales and Auriculariales of Agaricomycetes, and Dacrymycetales of Dacrymycetes. Only a few edible and medicinal mushrooms are litter composers, including A. bisporus, C. cinerea, and V. volvacea, which all fell into Agaricales of Agaricomycetes.
Fig. 1.
Phylogenetic tree of edible and medicinal mushrooms based on genomes. Time divergence estimates (in millions of years) are presented as mean node ages besides the nodes. The mean node ages of fossil calibrations are in red
The relaxed molecular clock analyses with normal fossil calibration priors produced broadly spaced estimates for the posterior probability of the non-calibrated nodes (Eastwood et al. 2011). The basal split between the major fungi documented here occurred 670.9 mya (Fig. 1), consistent with previous studies: 662 mya (Floudas et al. 2012) or 727 mya (Douzery et al. 2004). Agaricomycotina have occurred around 357.4 mya, Agaricomycetes originated 137.8 mya, and Agaricomycetidae branched off 108.8 mya. The values are much lower than some of the previously published studies (Eastwood et al. 2011; Floudas et al. 2012), but are similar to those obtained by previous analyses using Beast (Garnica et al. 2016).
Diversities of CAZymes and oxidoreductase in edible and medicinal mushrooms
A total of 389 predicted protein families related with lignocellulose decomposition were found in 40 edible and medicinal mushroom genomes, including 78 glycoside hydrolases (GHs), 49 glycosyltransferases (GTs), 15 polysaccharide lyases (PLs), 13 carbohydrate esterases (CEs), 45 carbohydrate-binding modules (CBMs), 12 auxiliary activities (AAs), and 177 cytochromes P450s.
Diversities of CAZymes
The lowest number of CAZY gene families selected for study in basidiomycetes was found in Tremella mesenterica Retz. (221 gene families), while ascomycetes Tuber melanosporum Vittad. (216 gene families) had the least CAZY genes. The highest number of selected CAZY gene families in basidiomycetes and ascomycetes was identified in Auricularia subglabra Looney, Birkebak & Matheny (787 gene families) and Morchella conica Pers. (421 gene families), respectively (Fig. 2).
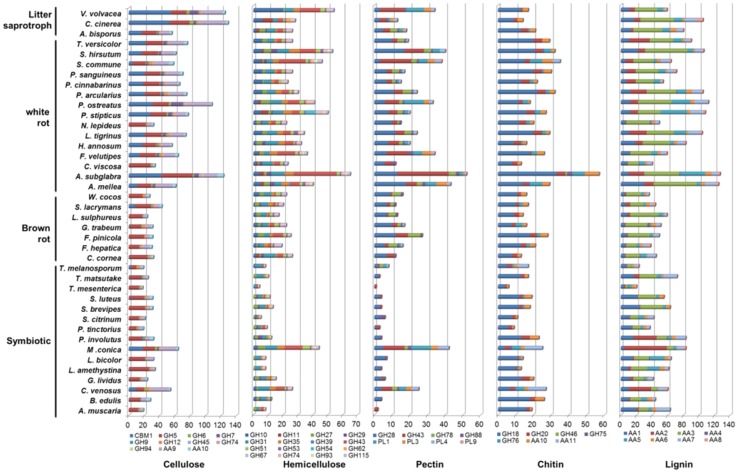
Fig. 2.
Gene contents in cellulose, hemicellulose, pectin, chitin, and lignin decomposition families in the genomes of 40 edible and medicinal mushrooms. The length of bars indicates number of each gene families in the predicted genome
The distribution of gene copies was not homogeneous among the genomes and among the gene families (Eastwood et al. 2011). Some CAZY families, such as GH5, GH18, GH20, GH28, GH31, GH43, PL1 and PL4 were detected in all the fungal species examined (Fig. 2), while some others, such as CBM1, GH10, GH11, GH76 and GH46 occurred only in several fungi. Interestingly, the distribution of some CAZyme families appeared to be phylum-specific. For example, 13 CAZY families were only found in the Basidiomycota, including GH9, GH29, GH89, GH30, GH79, GT65, GT68, PL14, PL8, CBM19 and CE9. In contrast, eight CAZY families appeared to be Ascomycetes-specific, including GH25, GH26, GH81, GT62, CBM16 and PL3.
Diversities of AAs
The highest number of copies for AAs gene families was present in Auricularia subglabra (156 copies) followed by A. mellea (148 copies), while the lowest number of copies were identified in T. mesenterica (21 copies) and T. melanosporum (34 copies). The differences among the genomes in total gene copies were less prominent in the dataset compared to the CAZY dataset, with the exception of AA9, AA2, AA0, AA11, and AA5. AA0 family was detected only in strains of Ascomycetes, and AA11 occurred in Basidiomycota solely. AA2, AA5, and AA9 were enriched in white-rot, litter saprotrophic and some symbiotic fungi, while only absent in brown rot fungi.
Diversities of P450s
The highest number of copies for P450s gene families was present in P. involutus (223 copies) followed by A. subglabra (217 copies), while the lowest number of copies was identified in T. mesenterica (9 copies) and T. melanosporum (26 copies). Similarly, the distribution of some P450 families appeared to be phylum-specific. For instance, 18 families were only found in the Basidiomycota, including CYP63A1, CYP63A4, CYP512A1, CYP67A2, CYP530A1, CYP66B1 and CYP53C2. On the contrary, one family (CYP503B4) appeared to be Ascomycetes-specific.
Gene evolution of edible and medicinal mushrooms
A clear separation can be found among four ecotype-fungi, independently from their phylogenetic positions, demonstrating a strong relationship between lifestyle and expansion/contraction of functional domains in the genomes of these fungi (Fig. 1).
Evolution of CAZymes
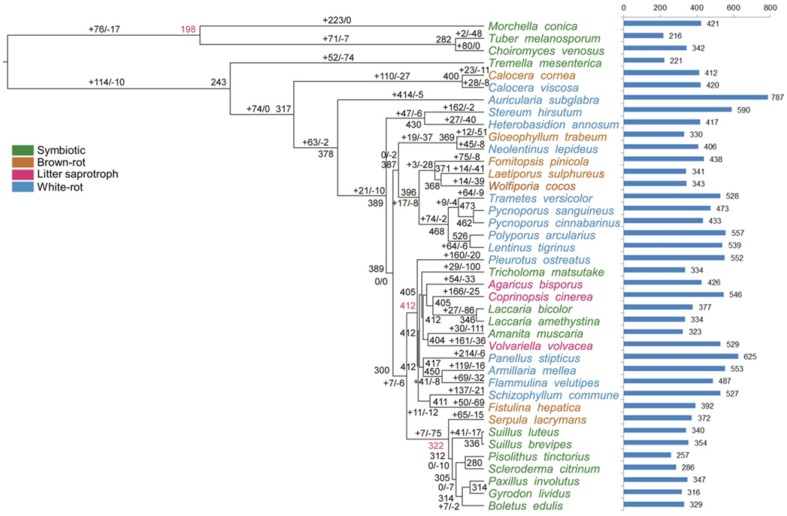
CAFÉ analysis identified gene families that had undergone significant expansion and contraction (Fig. 3). A more detailed analysis of genes involved in lignocellulose decomposition revealed a link between brown rot evolution and gene repertories. A total of 783 protein models were retrieved and used in the phylogenetic and reconciliation analyses. A large number of gene losses can be grasped in the expansion and contraction analysis among four ecotype-fungi. In general, loss of gene function may represent a common evolutionary response (Olson et al. 1999). For symbiotic fungi, T. mesenterica (expansion 2 copies and contraction 48 copies) and C. venosus (expansion 80 copies and contraction 0 copies), which had a significant difference in gene evolution, were diverse into two different nodes (around 40.1 mya), and variant gene families were GH5, CE1, CBM1, GH43, GH35, GH76 and GH31. Other two white rot fungi, S. hirsutum (expansion 162 copies and contraction 2 copies) and H. annosum (expansion 27 copies and contraction 40 copies), also were divided into two nodes (83.7 mya) with a number of gene families (like GH5, CE1, GH10, GH43, GH35, GH76, GH18, and GH28) whose number differed strongly from each other. Both of A. muscaria (symbiotic fungus, expansion 30 copies and contraction 11 copies) and V. volvacea (litter saprotrophic fungus, expansion 161 copies and contraction 36 copies) belonged to Agaricales, while they had different eco-lifestyles and were divergent into two nodes (78.8 mya), with the significantly different gene families GH7, PL3, GH5, CE1, GH10, CBM1, GH43, GH6, GH31, PL1, and GH18. Brown rot fungus G. trabeum (expansion 12 copies and contraction 51 copies) and white rot fungus N. lepideus (expansion 45 copies and contraction 8 copies) belonging to Gloeophyllales were divided into two nodes (38.8 mya) as well, and gene family GH18 were different in number. Another combination, S. commune (white rot fungus, expansion 137 copies and contraction 21 copies) and F. hepatica (brown rot fungus, expansion 50 copies and contraction 69 copies), which were assigned to Agaricales, included eight gene families (PL3, GH5, CE1, CBM1, GH43, GH76 and GH62) greatly different in number and were attributing into two nodes (76.1 mya). Among these gene families whose number are greatly different with gene evolution, CBM1 (binding to cellulase), GH5 (endo-b-1,4-glucanase), GH6 (cellobiohydrolase) and GH7 (reducing end-acting cellobiohydrolase) families were correlative with cellulose decomposition. Gene families, like GH10 (endo-1,3-b-xylanase), GH31 (a-xylosidase), GH35 (exo-b-glucosaminidase), GH43 (a-l-arabinofuranosidase), GH62 (a-l-arabinofuranosidase) and CE1 (acetyl xylan esterase), were functional in hemicellulose degradation. Pectin-relative gene families were GH28 (exo-polygalacturonase), PL1 (exo-pectate lyase) and PL3 (pectate lyase). Chitin-relative gene families were GH18 (chitinase) and GH76 (a-1,6-mannanase).
Fig. 3.
Patterns of gene duplication and loss in CAZymes gene families. Numbers at internal nodes indicate the estimated copy numbers of ancestral genes as retrieved under the best-fit BadiRate gene family turnover model (BDI-GR-ML). Numbers on the branches denote the minimum number of gains and losses estimated from the data. Bars indicate copy numbers in sampled genomes
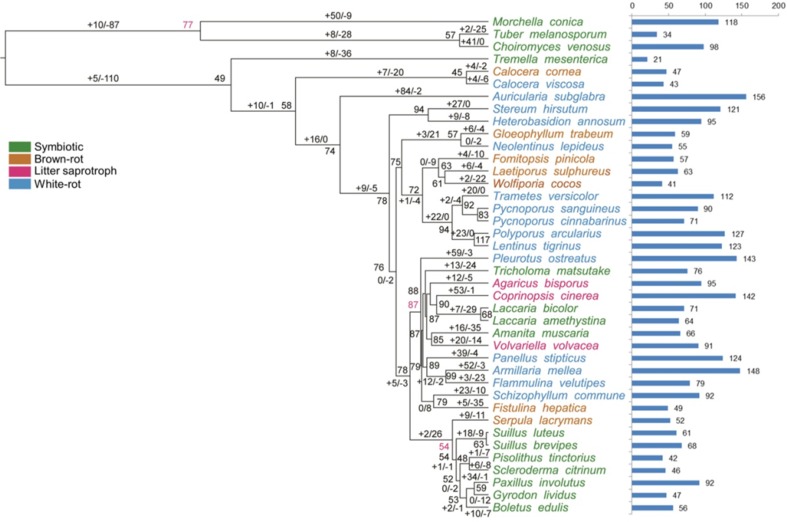
Fourteen gene families were found to have significant departures from a random birth–death process of gene family evolution (p < 0.05; Table 2), including three CAZY families (CMB1, CMB13 and CE10), suggesting that there have been lineage-specific shifts in rates of gene duplication and loss (Floudas et al. 2012). The patterns of gene duplication and loss in CBM1 (carbohydrate-binding modules) gene families were shown in Fig. 4. Obviously, CBM1 family contracted strongly in brown rot fungi while expanded in other species, which revealed and demonstrated a family evolution.
Table 2.
Gene families that have significant departures from a random birth–death process of gene family evolution in edible and medicinal mushroom genomes
| Enzyme families | p value | Substrate preference |
|---|---|---|
| AA1 | 0.001 | Lignin |
| AA2 | 0.001 | Lignin |
| AA4 | 0 | Lignin |
| AA5 | 0.001 | Lignin |
| AA6 | 0.001 | Lignin |
| AA7 | 0.001 | Lignin |
| AA8 | 0.001 | Lignin |
| AA9 | 0.001 | Cellulose |
| AA10 | 0.001 | Cellulose |
| AA11 | 0.003 | Chitin |
| AA12 | 0 | – |
| CE10 | 0.046 | – |
| CMB1 | 0.046 | Cellulose |
| CMB13 | 0.031 | – |
– Indicate unknown substrate
Fig. 4.
Patterns of gene duplication and loss in CBM1 (carbohydrate-binding modules) gene families. Numbers at internal nodes indicate the estimated copy numbers of ancestral genes as retrieved under the best-fit BadiRate gene family turnover model (BDI-GR-ML). Numbers on the branches denote the minimum number of gains and losses estimated from the data. Bars indicate copy numbers in sampled genomes
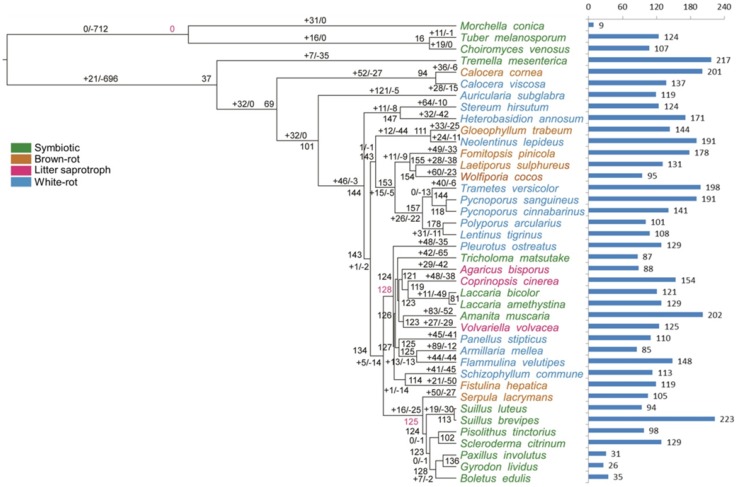
Evolution of AAs
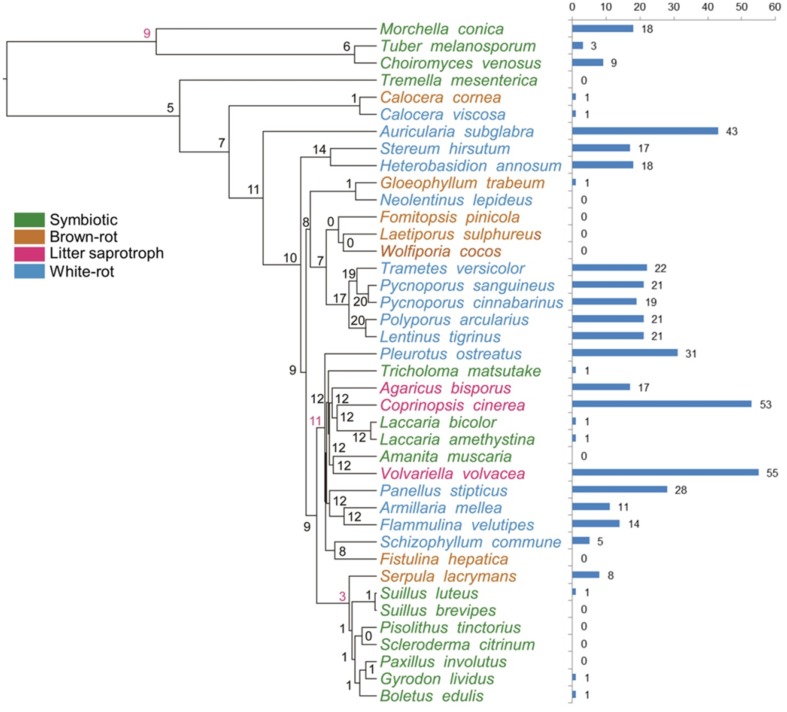
To gain access to cellulose, wood-decaying fungi must overcome or circumvent lignin (Floudas et al. 2012). Consistent with a central role of AAs in lignin degradation, genes of the AAs family were present in all the edible and medicinal mushroom genomes, and the common ancestor on node 7 was estimated to have 154 copies (Fig. 5), suggesting that the diversification of the family predated the separation of Dikarya (Floudas et al. 2012).
Fig. 5.
Patterns of gene duplication and loss in AAs (auxiliary activities) gene families. Numbers at internal nodes indicate the estimated copy numbers of ancestral genes as retrieved under the best-fit BadiRate gene family turnover model (BDI-GR-ML). Numbers on the branches denote the minimum number of gains and losses estimated from the data. Bars indicate copy numbers in sampled genomes
For symbiotic fungi, T. mesenterica (expansion 5 copies and contraction 110 copies) and C. venosus (expansion 10 copies and contraction 87 copies) had the same eco-lifestyle while they were divided into two significant different nodes. Every number of these gene families differed to a large degree, such as AA9, AA1, AA2, AA7, AA11 and AA3. Similarly, A. muscaria (symbiotic fungus, expansion 16 copies and contraction 35 copies), V. volvacea (litter saprotrophic fungus, expansion 20 copies and contraction 14 copies), S. commune (white rot fungus, expansion 23 copies and contraction 10 copies), F. hepatica (brown rot fungus, 5 copies and 35 copies), C. cinerea (litter saprotrophic fungus, expansion 53 copies and contraction 1 copies) and L. bicolor (symbiotic fungus, expansion 7 copies and contraction 29 copies) evolved into different nodes, respectively, and gene families AA9, AA2, AA3, AA7 and AA5 are involved in the evolution.
In terms of AA9 (GH61) family, lytic polysaccharide monooxygenase, showed significant expansions in white rot lineages, with a family-wide p value of < 0.001 in CAFÉ analyses (Table 2, Fig. 5). Originally described as cellulases, GH61 was recognized as a family with weak endoglucanase activity; however, GH61 enzymes have recently been reported to be copper-dependent lytic polysaccharide monooxygenases (LPMO) that enhance cellulose degradation in concert with classical cellulases and was classified as “Auxiliary Activities” (AA) (Levasseur et al. 2013). Other Auxiliary Activity families, like AA1 (multicopper oxidase, managing laccase, ferroxidase and laccase-like multicopper oxidase), AA2 (Class II peroxidase, concluding manganese peroxidase, lignin peroxidase and versatile peroxidase), AA3 (GMC oxidoreductase, containing cellobiose dehydrogenase, alcohol oxidase, alcohol oxidase and glucose oxidase), AA5 (radical-copper oxidase, taking on glyoxal oxidase and galactose oxidase) and AA7 (1,4-benzoquinone reductase), revealed diverse distribution in various eco-type fungi. For instance, Class II peroxidase AA2 family, composed of LiP, MnP and VP families, was largely present in white rot and litter saprotrophic fungi and absent in brown rot fungi. Genes of the AA9, AA1, AA3, AA5 and AA7 families were present in all the edible and medicinal mushroom genomes, but AA5 and AA9 families were absent from T. mesenterica and C. cornea.
Evolution of P450s
CAFÉ analysis revealed that families (from minimum 9 copies in M. conica to maximum 223 copies in S. brevipes) had expanded in the P450 gene evolution (Fig. 6) and indicated a considerable protein family expansion. The number of expanded gene family was significantly higher in white rot and symbiotic fungi than those in brown rot and litter saprotrophic fungi. A. muscaria (symbiotic fungus, expansion 83 copies and contraction 52 copies) and V. volvacea (litter saprotrophic fungus, expansion 27 copies and contraction 29 copies) were separated into two nodes and P450 families expand to some degree (i.e. CYP512A1, CYP512B1, CYP502A1, CYP53C2, CYP510A3, CYP512G2 and CYP534D2). G. trabeum (brown rot fungus, expansion 33 copies and contraction 25 copies) and N. lepideus (white rot fungus, expansion 24 copies and contraction 11 copies) also divided into two nodes and some gene families expanded greatly, such as CYP530A11 and CYP502A1. Two white rot fungi, S. hirsutum (expansion 64 copies and contraction 10 copies) and H. annosum (expansion 32 copies and contraction 42 copies) had different lignin-degrading capacity and gene families (i.e. CYP67A2, CYP502A1, CYP66B1, CYP509E1, CYP509F3 and CYP512G2) significantly expanded in number.
Fig. 6.
Patterns of gene duplication and loss in P450s gene families. Numbers at internal nodes indicate the estimated copy numbers of ancestral genes as retrieved under the best-fit BadiRate gene family turnover model (BDI-GR-ML). Numbers on the branches denote the minimum number of gains and losses estimated from the data. Bars indicate copy numbers in sampled genomes
A large expansion corresponding to cytochrome P450 superfamily could reveal the complexity of lignin-relative derivatives and correlative aromatic compounds. The expanded gene families of P450 genome seemed to be mainly involved in processes like secondary metabolism, pheromone response, detoxification, and virulence. When it came to CYP51 clan, which was conserved and primarily acts on cell wall ergosterol biosynthesis reaction, it did not expand and contract significantly. For CYP502A1 superfamily belonging to CYP64 clan, it partially expand in white rot and litter saprotrophic fungi, which was mainly influential on various catalyzing reactions involved in biosynthesis aflatoxins and other such secondary metabolites. CYP53C2 that was attributed to CYP53 and possibly encoded benzoate para-hydroxylase was absent in brown rot and symbiotic fungi, like G. trabeum and A. muscaria.
Gene evolution of lignocellulose decomposition families in 40 edible and medicinal mushrooms
To clarify the gene evolution of lignocellulose decomposition families, we analyzed 40 genomes of edible and medicinal mushrooms and created a dataset, which spans many orders and represents cellulose, hemicellulose, pectin, chitin and lignin degrading strains, combined with previously sequenced genomes. Our result showed that edible and medicinal mushrooms contained 11 CAZy families related to the degradation of cellulose, and 16 CAZy families related to the degradation of hemicellulose, 8 pectin, 7 chitin, and 8 lignin (Figs. 1, 2).
Cellulose decomposition families
Genes of GH5, AA9, and AA10 families existed commonly in all the40 mushrooms which belong to four ecotypes. The presence of GH5 family, which encoded endoglucanases, possibly increased the abilities of these mushrooms to deconstruct cellulose and plant biomass. During the evolution process, GH5 family expanded in white rot and litter saprotrophic fungi (earliest dating back to 206.1 mya) and contracted in brown rot fungi. And genes in AA9 and AA10 families, which were interpreted into lytic polysaccharide monooxygenases, were important in deconstruction of recalcitrant polysaccharides, and significant to form a complete suite of decomposition enzymes in order to strengthen the degrading ability of strains effectively. These two families were absent in symbiotic fungi except for strains in Pezizales (ancestor can be tracked back to 400 mya). Therefore, GH5, AA9 and AA10 declined with eruption of different ecotypes of fungi but might be the versatile and essential cellulose decomposition families during the whole evolution history. However, genes of CBM1, GH6 and GH7 families mainly appeared in white rot and litter saprotrophic fungi and were absent in brown rot and symbiotic lineages. Two white rot fungi, N. lepideus (38.3 mya) and C. ciscosa (30.5 mya), that lack genes of CBM1 family were classified into Gloeophyllales and Dacrymcetales, which mostly consisted of brown rot fungi. Therefore, with the complex lifestyle and degrading pattern selection, the early decomposing cellulose process (enhancing and accumulating enzymes on the surface of crystalline cellulose with CBM1 family and then attack crystalline cellulose with GH6 and GH7 families) might be abandoned during gene evolution and development from 40 mya. Similar to situation of strains in GH5, AA9, and AA10 families’ distribution, M. conica (400 mya), C. venosus (40.1 mya) and T. melanosporum (40.1 mya), which had a common divergency point and were part of Pezizales, have genes of CBM1. The reason was probably that early symbiotic fungi may decompose lignocellulose by themselves in need for cooperating with a smaller number of plants. Therefore, in general, the earlier the divergence occurred, the more complete the cellulose families were. And litter saprotrophic and white rot fungi got a longer history in cellulose decomposition.
Hemicellulose decomposition families
Predominantly, genes related to hemicellulose were involved in 20 glycoside hydrolases families among 40 edible and medicinal mushrooms. GH10 and GH62 families, which primarily coded xylanases and deconstruct xylan, only existed in some white rot and litter saprotrophic fungi and Pezizales. Other families, like GH43 and GH35, were of extensive existence among four ecotypes. A similar case in cellulose decomposition evolution, white rot and litter saprotrophic fungi possessed a larger number of genes and a more complete gene category. Differently, the hemicellulose degradation process among different eco-lifestyle fungi mainly differed in gene quantity increasing and decreasing while the decomposing pattern still focused on secreting xylanase, xylosidase, arabinofuranosidase and xylan esterase in the evolution history.
Pectin and chitin decomposition families
Pectin lyase, pectate lyase, pectin esterase, and polygalacturonase can break down pectin (Alghisi and Favaron 1995; Lagaert et al. 2009). Our results showed white rot and litter saprotrophic fungi were enriched in the total number of pectin families and had higher retention rates than brown rot and symbiotic fungi. In addition, GH28 family was a reserved family for pectin digestion among almost all the edible and medicine mushrooms while PL1 and PL3 families were discarded in symbiotic fungi (except strains in Pezizales). That demonstrates late divergency (after 40.1 mya) symbiotic strains did not digest pectin through pectin lyase and pectate lyase, which may result from the symbiotic variety of plants. On the other hand, for chitin digestion, there was no significant difference in chitin families’ quantity and category. GH18 and GH76 families only showed some contraction in brown rot fungi, which may avoid chitin and mannan to some extent.
Lignin decomposition families
Commonly, peroxidases were known to be key participators in breaking down lignin among various fungal species. These enzymes typically were heme-containing proteins and belonged to the class II peroxidase superfamily (Hammel and Cullen 2008; Martınez 2002). Class II peroxidase AA2 family composed of LiP, MnP and VP families, and AA2 families were largely present in white rot and litter saprotrophic fungi and absent in brown rot fungi. In agreement with previous results the LiP, MnP and VP families involved in lignin degradation were distributed in the white rot species and absent from the brown rot species (Floudas et al. 2012; Martínez et al. 2004, 2009). And AA3 family (encoding oxidoreductase) was in the same case with AA2. For symbiotic fungi, they owned larger gene expansion in AA1, AA5 and AA7 families, which either depend on copper ions or act on benzoquinone redox process. That meant the late evolution species may have a preference in copper-enzymes and copper-depend decomposition while the enzymes encoded by AA2 family may still be principle in lignin-degradation pattern.
Apart from AA families, lignin degradation also hinged on P450 families. Also, CYP502A1 (belonging to CYP64 clan) and CYP53 got a large distribution in white rot, symbiotic and litter saprotrophic fungi (no matter when the species diverges, and the earliest time point was 400 mya), which mainly did with the secondary metabolism, like benzoate and lignin derivative decomposition. Interestingly, these two families only existed in strains of Basidiomycota.
Conclusion
Fungi in different ecotypes acquired an obvious preference on specific decomposing gene families during evolutionary selection. Overall, white rot and litter saprotrophic fungi possessed more complete types of varied degradation gene families and were superior in quantity. With the evolution and development of lignocellulose decomposition mechanism, certain families (like CBM1, GH6, GH7, GH10 and CYP53) disappeared in brown rot and symbiotic fungi. In addition, the earlier time of divergence point determined the more integrated and larger decomposition families, like M. conica (400 mya, symbiotic fungus). On the other hand, various degradation families were able to expand and contract in evolution history. Those gains and losses in gene quantities of varied decomposition families resulted in particularly phylogenetic clades or nodes, then led in varied eco-lifestyles of species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the National Basic Research Program of China (973 Program, no. 2014CB138301). We thank Prof. Baogui Xie (Mycological Research Center, College of Life Sciences, Fujian Agriculture and Forestry University) for providing genomic data of V. volvacea and F. velutipes.
Compliance with ethical standards
Conflict of interest
The authors declare that no conflicts of interest exist.
Contributor Information
Qi An, Email: fungiqian@yahoo.com.
Xue-Jun Wu, Email: samantha0128@163.com.
Yu-Cheng Dai, Email: yuchengd@yahoo.com.
References
- Alfaro M, Castanera R, Lavin JL, Grigoriev IV, Oguiza JA, Ramirez L, Pisabarro AG. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environ Microbiol. 2016;18:4710–4726. doi: 10.1111/1462-2920.13360. [DOI] [PubMed] [Google Scholar]
- Alghisi P, Favaron F. Pectin-degrading enzymes and plant–parasite interactions. Eur J Plant Pathol. 1995;101:365–375. [Google Scholar]
- Bao DP, Gong M, Zheng HJ, Chen MJ, Zhang L, Wang H, Jiang JP, Wu L, Zhu YQ, Zhu G, Zhou Y, Li CH, Wang SY, Zhao Y, Zhao GP, Tan Q. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS One. 2013;8:e58294. doi: 10.1371/journal.pone.0058294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genom. 2011;12:38. doi: 10.1186/1471-2164-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boa E. Wild edible fungi: a global overview of their use and importance to people. Non-wood forest products 17. Rome: Food and Agriculture Organization of the United Nation; 2004. [Google Scholar]
- Branco S, Gladieux P, Ellison CE, Kuo A, LaButti K, Lipzen A, Grigoriev IV, Liao HL, Vilgalys R, Peay KG, Taylor JW, Bruns TD. Genetic isolation between two recently diverged populations of a symbiotic fungus. Mol Ecol. 2015;24:2747–2758. doi: 10.1111/mec.13132. [DOI] [PubMed] [Google Scholar]
- Broda P, Birch PR, Brooks PR, Sims PF. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol Microbiol. 1996;19:923–932. doi: 10.1046/j.1365-2958.1996.474966.x. [DOI] [PubMed] [Google Scholar]
- Cai YJ, Buswell JA, Chang ST. Production of cellulases and hemicellulases by the straw mushroom, Volvariella volvacea. Mycol Res. 1994;98:1019–1024. [Google Scholar]
- Cai Y, Gong Y, Liu W, Hu Y, Chen L, Yan L, Zhou Y, Bian Y. Comparative secretomic analysis of lignocellulose degradation by Lentinula edodes grown on microcrystalline cellulose, lignosulfonate and glucose. J Proteom. 2017;163:92–101. doi: 10.1016/j.jprot.2017.04.023. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acdis Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Librado P, Chang TH, Ibarra-Laclette E, Herrera-Estrella L, Rozas J, Albert VA. High gene family turnover rates and gene space adaptation in the compact genome of the carnivorous plant Utricularia gibba. Mol Biol Evol. 2015;32:1284–1295. doi: 10.1093/molbev/msv020. [DOI] [PubMed] [Google Scholar]
- Castanera R, Pérez G, López-Varas L, Amselem J, LaButti K, Singan V, Lipzen A, Haridas S, Barry K, Grigoriev IV, Pisabarro AG, Ramírez L. Comparative genomics of Coniophora olivacea reveals different patterns of genome expansion in Boletales. BMC Genom. 2017;18:883. doi: 10.1186/s12864-017-4243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chen BZ, Gui F, Xie BG, Deng YJ, Sun XY, Lin MY, Tao YX, Li SJ. Composition and expression of genes encoding carbohydrate-active enzymes in the straw-degrading mushroom Volvariella volvacea. PLoS One. 2013;8:e58780. doi: 10.1371/journal.pone.0058780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Keane TM, Turner DJ, O'Keeffe G, Fitzpatrick DA, Doyle S. Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: toward a new infection model system. J Proteome Res. 2013;12:2552–2570. doi: 10.1021/pr301131t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YC, Zhou LW, Yang ZL, Wen HA, Bau T, Li TH. A revised checklist of edible fungi in China. Mycosystema. 2010;29:1–21. [Google Scholar]
- Dai YC, Yang ZL. A revised checklist of medicinal fungi in China. Mycosystema. 2008;27:801–824. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee YH, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie XF, Kües U, Hibbett DS, Hoffmeister D, Högberg N, Martin F, Grigoriev IV, Watkinson SC. The plant cell wall—decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Fernandez-Fueyo E, Ruiz-Dueñas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Ryu JS, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, John FJST, Wymelenbergn AV, Sabat G, BonDurant SS, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavín JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kües U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, Salamov AA, Hoffmeister D, Schwenk D, Hadar Y, Yarden O, de Vries RP, Wiebenga A, Stenlid J, Eastwood D, Grigoriev IV, Berka RM, Blanchette RA, Kersten P, Martinez AT, Vicuna R, Cullen D (2012) Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci USA 109:5458–5463 [DOI] [PMC free article] [PubMed]
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Arun Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, John FST, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- Floudas D, Held BW, Riley R, Nagy LG, Koehler G, Ransdell AS, Younus H, Chow J, Chiniquy J, Lipzen A, Tritt A, Sun H, Haridas S, LaButti K, Ohm RA, Kues U, Blanchette RA, Grigoriev IV, Minto RE, Hibbett DS. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol. 2015;76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica S, Riess K, Schön ME, Oberwinkler F, Setaro SD. Divergence times and phylogenetic patterns of Sebacinales, a highly diverse and widespread fungal lineage. PLoS One. 2016;11:e0149531. doi: 10.1371/journal.pone.0149531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén D, Sánchez S, Rodríguez-Sanoja R. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biot. 2010;85:1241–1249. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- Hammel KE, Cullen D. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol. 2008;11:349–355. doi: 10.1016/j.pbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Hibbett D, Grimaldi D, Donoghue M. Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. Am J Bot. 1997;84:981. [PubMed] [Google Scholar]
- Isikhuemhen OS, Mikiashvilli NA. Lignocellulolytic enzyme activity, substrate utilization, and mushroom yield by Pleurotus ostreatus cultivated on substrate containing anaerobic digester solids. J Ind Microbiol Biotechnol. 2009;36:1353–1362. doi: 10.1007/s10295-009-0620-1. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canback B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Dore J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Hogberg N, Johansson T, Khouja HR, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Tunlid A, Grigoriev IV, Hibbett DS, Martin F. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- Lagaert S, Beliën T, Volckaert G. Plant cell walls: Protecting the barrier from degradation by microbial enzymes. Semin Cell Dev Biol. 2009;20:1064–1073. doi: 10.1016/j.semcdb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- LePage B, Currah R, Stockey R, Rothwell G. Fossil ectomycorrhizae from the Middle Eocene. Am J Bot. 1997;84:410. [PubMed] [Google Scholar]
- Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Vieira FG, Rozas J. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics. 2012;28:279–281. doi: 10.1093/bioinformatics/btr623. [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahrén D, Brun A, Danchin EGJ, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Tacon FLE, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kües U, Lucas S, de Peer YVan, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouzé P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452:88–92 [DOI] [PubMed]
- Martınez AT. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb Technol. 2002;30:425–444. [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Wymelenberg AV, Gaskell J, Lindquist E, Sabat G, BonDurant SS, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavín JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harriso P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kües U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, Rokhsar D, Berka R, Cullen D. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Frøslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi) Mol Biol Evol. 2007;43:430–451. doi: 10.1016/j.ympev.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hildén K, Kües U, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HAB, Xu JP, Eastwood DC, Foster GD, Sonnenberg ASM, Cullen D, de Vriesh RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. P Natl Acad Sci USA. 2012;109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy LG, Riley R, Tritt A, Adam C, Daum C, Floudas D, Sun H, Yadav JS, Pangilinan J, Larsson KH, Matsuura K, Barry K, Labutti K, Kuo R, Ohm RA, Bhattacharya SS, Shirouzu T, Yoshinaga Y, Martin FM, Grigoriev IV, Hibbett DS. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol Biol Evol. 2016;33:959–970. doi: 10.1093/molbev/msv337. [DOI] [PubMed] [Google Scholar]
- Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, De Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker S, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FWMR, vanKuyk PA, Horton JS, Grigoriev IV, Wösten HAB. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canback B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen LT, Dunand C, Duplessis S, Durling M, Gonthier P, Grimwood J, Fossdal CG, Hansson D, Henrissat B, Hietala A, Himmelstrand K, Hoffmeister D, Hogberg N, James TY, Karlsson M, Kohler A, Kues U, Lee YH, Lin YC, Lind M, Lindquist E, Lombard V, Lucas S, Lunden K, Morin E, Murat C, Park J, Raffaello T, Rouze P, Salamov A, Schmutz J, Solheim H, Stahlberg J, Velez H, de Vries RP, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, Martin F, Grigoriev IV, Stenlid J. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 2012;194:1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C. The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genom. 2010;11:525. doi: 10.1186/1471-2164-11-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Baek JH, Lee S, Kim C, Rhee H, Kim H, Seo JS, Park HR, Yoon DE, Nam JY, Kim HI, Kim JG, Yoon H, Kang HW, Cho JY, Song ES, Sung GH, Yoo YB, Lee CS, Lee BM, Kong WS. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS One. 2014;9:e93560. doi: 10.1371/journal.pone.0093560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Victoria Aguilar-Pontes M, Hainaut M, Henrissat B, Hildén K, Mäkelä MR, de Vries RP. Comparative analysis of basidiomycete transcriptomes reveals a core set of expressed genes encoding plant biomass degrading enzymes. Fungal Genet Biol. 2018;112:40–46. doi: 10.1016/j.fgb.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Pérez J, Munoz-Dorado J, de la Rubia TDLR, Martinez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol. 2002;5:53–63. doi: 10.1007/s10123-002-0062-3. [DOI] [PubMed] [Google Scholar]
- Philippoussis A, Zervakis G, Diamantopoulou P. Bioconversion of agricultural lignocellulosic wastes through the cultivation of the edible mushrooms Agrocybe aegerita, Volvariella volvacea and Pleurotus spp. World J Microb Biot. 2001;17:191–200. [Google Scholar]
- Sanderson MJ. r8s version 1.71. Analysis of rates (“r8s”) of evolution. Davis: Section of Evolution and Ecology, University of California; 2006. [Google Scholar]
- Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, Deng JX, Dietrich FS, Fargo DC, Farman ML, Gathman AC, Goldberg J, Guigó R, Hoegger PJ, Hooker JB, Huggins A, James TY, Kamada T, Kilaru S, Kodira C, Kües U, Kupfer D, Kwan HS, Lomsadze A, Li WX, Lilly WW, Ma LJ, Mackey AJ, Manning G, Martin F, Muraguchi H, Natvig DO, Palmerini H, Ramesh MA, Rehmeyer CJ, Roe BA, Shenoy N, Stanke M, Ter-Hovhannisyan V, Tunlid A, Velagapudi R, Vision TJ, Zeng QD, Zolan ME, Pukkila PJ. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci USA. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Taylor TN, Hass H, Kerp H, Krings M, Hanlin RT. Perithecial ascomycetes from the 400 million year old Rhynie chert: an example of ancestral polymorphism. Mycologia. 2005;97:269–285. [PubMed] [Google Scholar]
- Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LS, Zhang YZ, Gao PJ. A novel function for the cellulose binding module of cellobiohydrolase I. Sci China Ser C. 2008;51:620–629. doi: 10.1007/s11427-008-0088-3. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Zhang XL, Cui BK, Dai YC. Comparative genomic analysis of edible (medicinal) fungi reveals different ecological habitats. Mycosystema. 2015;34:742–760. [Google Scholar]
- Yin YB, Mao XZ, Yang JC, Chen X, Mao FL, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZT, Liu HQ, Wang CF, Xu JR. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013;14:274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.