Abstract
Purpose
Clinical pregnancy rate after IVF with eSET stagnates between 30 and 40%. In order to increase pregnancy and live birth rates, multiple embryo transfer is still common practice. Providing additional non-invasive tools to choose the competent embryo for transfer could avoid multiple pregnancy and improve time to pregnancy. Cumulus mRNA analysis with quantitative PCR (QPCR) is a non-invasive approach. However, so far, no gene sets have been validated in prospective interventional studies.
Methods
A prospective interventional single-center pilot study with two matched controls (day-3 and day-5 eSET) was performed in 96 patients consenting to the analysis of the cumulus-corona of their oocytes. All patients were super-ovulated for ICSI and eSET at day 3. All oocytes were denuded individually and cumulus was analyzed by quantitative PCR using three predictive genes (EFNB2, SASH1, CAMK1D) and two housekeeping genes (UBC and β2M). Patients (n = 62) with 2 or more day-3 embryos (good or excellent morphology) had their embryo chosen following the normalized expression of the genes.
Results
Corona testing significantly increased the clinical pregnancy and live births rates (63% and 55%) compared to single embryo transfer (eSET) on day 3 (27% and 23%: p < 0.001) and day 5 (43% and 39%: p = 0.022 and p = 0.050) fresh transfer cycle controls with morphology-only selection. Time-to-pregnancy was significantly reduced, regardless of the number of good-quality embryos available on day 3.
Conclusion
Combining standard morphology scoring and cumulus/corona gene expression analysis increases day-3 eSET results and significantly reduces the time to pregnancy.
Trial registration number
This is not an RCT study and was only registered by the ethical committee of the University Hospital UZBRUSSEL of the Vrije Universiteit Brussel VUB (BUN: 143201318000).
Electronic supplementary material
The online version of this article (10.1007/s10815-018-01398-2) contains supplementary material, which is available to authorized users.
Keywords: Cumulus cells, Gene expression, Single embryo transfer, Clinical pregnancy, Non-invasive, Oocyte quality
Introduction
Embryo selection in ART is still largely based on morphological criteria, although large prospective studies indicate that selection of embryos based on morphology remains a suboptimal approach [1, 2]. The recent shift from cleavage-stage embryo to blastocyst transfer improved significantly live birth rates [3, 4], as has the transfer of multiple cleavage-stage embryos on day 3 or blastocysts on day 5. The latter strategy enhances the incidence of multiple pregnancy and its associated risk of neonatal and maternal complications [5, 6]. Optimizing the embryo selection procedure should allow ART practitioners to choose the embryo with the highest chance of live birth and to reduce multiple embryo transfer and costs associated with multiple births for both care providers and parents.
In order to increase the efficiency of IVF/ICSI, several minimally invasive embryo selection approaches were developed, including assays to measure specific proteins [7] or cell-free DNA [8] in follicular fluids, gene expression analysis in the cumulus cells [9], microRNA analysis in spent blastocyst culture media [10], or time-lapse video monitoring of embryo development during incubation [11, 12]. Although some studies initially reported promising results, there is still no evidence from prospective interventional trials of their added value when compared to routine morphology assessment (e.g., for time lapse [13]).
It is well known that with increasing age of the patient, a significant fraction of the morphologically normal embryos chosen for transfer are aneuploid and have lower developmental potential [14]. Such aneuploid embryos most often lead to miscarriages. For this reason, aneuploidy screening has been introduced in the ART clinic to avoid transfer and gestation of an abnormal embryo. In order to identify chromosomal aberrations, invasive pre-implantation genetic screening methods (PGS or PGT-A) based on blastomere biopsy are used including fluorescence in situ hybridization (FISH) [15], array CGH [16], genome-wide single-nucleotide polymorphism (SNP) analysis [17], and NGS [18]. Blastomere biopsy is gradually being replaced by trophectoderm biopsy, and blastocoelic fluid analysis was proposed [19]. A beneficial effect of PGS after blastomere or trophectoderm biopsy on live birth rates has been demonstrated in three smaller randomized clinical studies [14, 16, 20] and two retrospective studies [21, 22]. A large multicenter prospective randomized European study (ESTEEM) with 400 patients showed that with PGS on polar bodies there was no difference in the live birth rates in the experimental arm with PGT-A analysis versus the control arm without PGT-A [23]. This contradicted results of some retrospective studies.
In addition, PGS is invasive [24], expensive, and can only be performed in specialized laboratories. Embryos need to be cultured until day 5 or 6 and embryo vitrification is an essential part of the process. The effect of mosaicisms in the trophectoderm of embryos adds another complexity to PGS [18, 25]. In addition, in some countries, PGS is not allowed by law. Hence, the search for a non-invasive and cost-effective method to test embryonic developmental competence is still ongoing.
Analysis of the cumulus cells (CC) which are tightly interconnected with the oocyte until after ovulation [26, 27] constitutes a suitable non-invasive alternative. CC are dispensable shortly after the cumulus-oocyte complex is retrieved and the analysis of CC is therefore non-invasive. The expression of multiple oocyte competence markers in CC can be analyzed with micro arrays or QPCR, two well-established, sensitive molecular techniques, applicable in the routine clinical diagnostic laboratory.
Multiple gene sets were proposed as predictive biomarkers for oocyte competence and successful embryo development or implantation (for review [28]). However, the proposed “oocyte quality” genes in CC differed between studies and so far, no prospective clinical study demonstrated that CC analysis can result in improved selection of the embryo to transfer compared to routine morphological analysis. In two retrospective studies, we reported that expression analysis of ephrin-B2 (EFNB2), SAM and SH3 domain–containing protein 1(SASH1) and calcium/calmodulin-dependent protein kinase ID (CAMK1D) in cumulus can be used to select a single embryo out of the morphologically chosen suitable embryos for transfer [29, 30]. EFNB2, SASH1, and CAMK1D have been related to cell expansion [31], the Toll-like receptor 4 pathway [32], and the calcium pathway, respectively.
Here, we report the results of a prospective clinical study using the expression data of EFNB2, SASH1, and CAMK1D and two control (normalization) genes in CC. The gene expression test of these five genes was termed the Corona Test because proximal cumulus cells were used (and not the more distal dissected CC or free-floating granulosa cells). The primary endpoints of the study were clinical pregnancy and live birth rates in the fresh single embryo transfer or in the consecutive vitrified/thawed transfer cycles. Results in the experimental study arm were compared to routine clinical practice: eSET on day 3 and eSET on day 5 (two control arms). The efficiency of the Corona Test was analyzed in relation to patient age and number of available good-quality embryos (GQE), two major determinants of live birth after ART treatment.
Materials and methods
Study design
This was an assessor blinded prospective interventional study with matched controls, that comprised one experimental and two control arms. For the patients included in the experimental arm, the Corona Test result complemented the decision (on which embryo to transfer) resulting from routine morphological embryo grading (described in detail below). On day 3, after ICSI, a single cleavage-stage embryo with the highest Corona Test rank out of the group of morphologically good-quality embryos was transferred. The Corona Test assessment was performed evaluator-blinded. Supernumerary embryos of good quality were vitrified and transferred in accordance with the Corona Test rank in consecutive embryo transfer cycles.
The effectiveness of using the Corona Test on top of morphology evaluation was compared to routine practice. Each Corona Test patient with > 2 GQE on day 3 was matched with 2 control patients undergoing in vitro fertilization with ICSI with elective embryo selection only based on morphological embryo grading. In one control arm, the embryo transfer occurred on day 3 and in the other control arm on day 5. Routine patients with only 2 GQE always have embryo transfer on day 3; therefore, Corona Test patients with only 2 GQE were only matched with day-3 controls.
The patients for matching were provided by the data manager who was blinded to the treatment outcome. The matched control patients had to comply with the following criteria: (1) fitting the inclusion and exclusion criteria of the tested patients (see below), (2) being in same age category: ≤ 30 or 31–38 years, (3) having the same or similar number of GQE available on day 3 (2, 3–4, 5–6 or > 6), and (4) among patients meeting the first 3 criteria being closest in time of oocyte pick-up (OPU) to the Corona Test patient. The decision to match each patient with sufficient GQEs with dual controls was justified by the expected higher live birth rates after blastocyst transfer compared to cleavage-stage transfers.
Clinical pregnancy rate was defined as the ultrasonographic visualization of a fetal sac at week 7 or later [33] with normal fetal heartbeat. All patients were followed up during gestation and contacted by a study nurse after the planned delivery date.
Patient population
This study was approved by the Ethics Committee of the University Hospital of the Vrije Universiteit Brussel (BUN: 143201318000). All patients signed an informed consent. The patient enrolment flow is described in Fig. 1. This study included ICSI (intracytoplasmic sperm injection) patients which were between 22 and 38 years old, with a first or second IVF/ICSI treatment cycle who underwent single embryo transfer (SET) on day 3 after oocyte pick-up (OPU) and stimulated with GnRH antagonist and HP-hMG (Menopur, Ferring Pharmaceuticals, Copenhagen, Denmark) or a combination of HP-hMG and rFSH (Elonva, MSD, Oss, The Netherlands) with at least 2 final injections of at least 150 units/day of HP-hMG. Exclusion criteria were as follows: irregular menstrual cycle (< 24 or > 35 days), a BMI < 17 or > 33, smoking > 10 cigarettes per day. Patients with severe endometriosis ≥ III (AFS classification), PCOS (polycystic ovary syndrome; Rotterdam 2003 criteria), known low ovarian response based on Bologna criteria or an oocyte maturation defect, included in any other study, scheduled for PGD (pre-implantation genetic diagnosis), or couples in need of TESE (testicular sperm extraction), or with severe oligoasthenoteratozoospermia (OAT) with sperm count below 100.000/ml were excluded from the study.
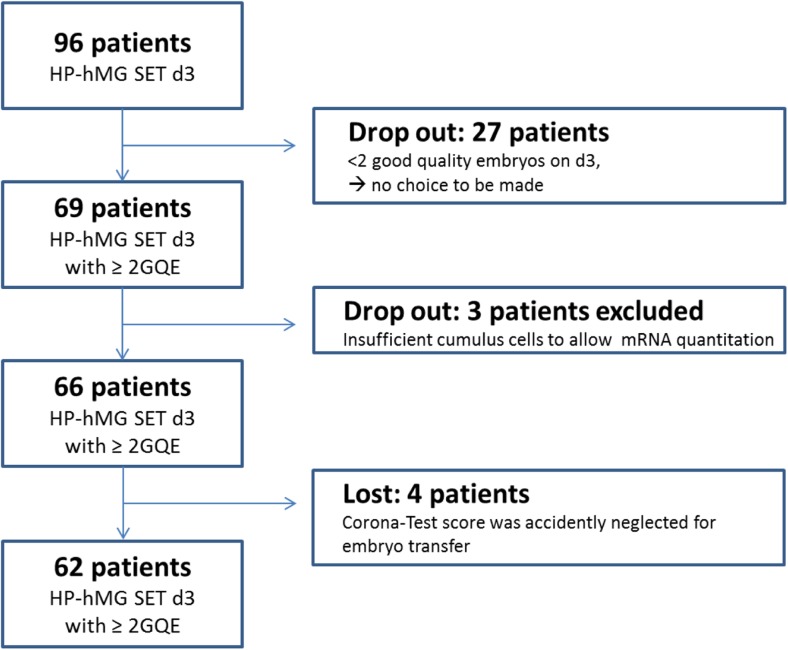
Fig. 1.
Patient enrolment flow diagram for the Corona Test group. Ninety patients were contacted for inclusion in the study group, and 62 patients had the additional Corona Test on their cumulus cells and were informative for the study. Patients with only one transferable embryos on day 3 were considered dropouts in view of the absence of a need for embryo selection. HP-hMG SET d3 refers to the stimulation and transfer protocol applied, namely a short GnRH antagonist protocol with highly purified human menopausal gonadotropin stimulation and single embryo transfer on day 3 after pick-up. GQE, good-quality embryos, are embryos eligible for transfer, based on the morphologic evaluation as described in the material and methods section
Altogether, 96 patients were contacted for inclusion in the study group. Patients with less than two transferable embryos on day 3 were considered dropouts in view of the absence of a need for embryo selection. Finally, 62 patients were informative for the study. They had the additional Corona Test on their cumulus cells performed and the Corona Test was used to rank their GQE. The indications for ART were comparable for patients in the experimental arm receiving the Corona Test and controls patients; these were listed in Supplementary Table 1.
The stimulation scheme and patient ultrasound and blood monitoring were done according to the standard procedure at UZ Brussel [34]. As soon as ≥ 3 follicles ≥ 17 mm were present, 5000–10,000 IU of hCG (human chorionic gonadotropin, Pregnyl, MSD, Oss, The Netherlands) was administered and oocytes were retrieved 36 h later.
Patient characteristics are described in Table 1. Patients that were not pregnant from the transfer in the fresh cycle had supernumerary embryos frozen on the day of transfer. At their request, frozen embryos were warmed and transferred singly according to the Corona Test rank in a consecutive artificial cycle [35].
Table 1.
Patient characteristics for the Corona Test population and matched control groups
| Matched controls | Corona Test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day-13 transfer | Day-5 transfer | Day-3 transfer | |||||||||
| Unit | Mean | SD | n | Mean | SD | n | Mean | SD | n | One-way ANOVA | |
| Age | Years | 32.8 | 3.5 | 62 | 33.7 | 3.4 | 44 | 33.4 | 2.9 | 62 | ns |
| BMI | kg/m2 | 24.4 | 5 | 62 | 23.3 | 5.1 | 43 | 23.3 | 4.5 | 62 | ns |
| AMH | μg/l | 2.0 | 1.4 | 52 | 2.6 | 1.8 | 36 | 2.22 | 1.55 | 50 | ns |
| Days stimulation | n | 10.2 | 1.6 | 62 | 10.2 | 1.5 | 44 | 10.4 | 1.44 | 62 | ns |
| Total dose stimulation | IU | 2021.2 | 632.8 | 62 | 1865 | 637.6 | 44 | 1840.7 | 578.1 | 62 | ns |
| Serum FSH | IU/l | 17.3 | 6.5 | 51 | 17.2 | 6.1 | 38 | 16 | 6.0 | 51 | ns |
| serum LH | IU/l | 2.4 | 2.9 | 51 | 2.6 | 2.6 | 38 | 2.8 | 2.4 | 51 | ns |
| serum E2 | ng/l | 1771 | 764 | 51 | 2103 | 895.5 | 38 | 1977 | 801 | 51 | ns |
| Serum Progesterone | μg/l | 0.8 | 0.4 | 51 | 1 | 0.3 | 38 | 0.89 | 0.25 | 51 | p < 0.05 |
| Number COC at pick-up | n | 8.2 | 4.7 | 62 | 9.3 | 5.3 | 44 | 8.4 | 4.3 | 62 | ns |
| Maturation rate | % | 84 | 17 | 62 | 85 | 13 | 44 | 88 | 12.7 | 62 | ns |
| Fertilization rate | % | 83 | 16 | 62 | 89 | 15 | 44 | 85 | 18.3 | 62 | ns |
| Number GQE day 3 | n | 4.7 | 3.4 | 62 | 6.1 | 3.9 | 44 | 4.5 | 2.9 | 62 | p < 0.05 |
Serum values refer to the concentration measured 2 days prior to oocyte pick-up. AMH anti-Müllerian hormone. COC cumulus-oocyte complexes, E2 estradiol, GQE, good-quality embryos = embryos eligible for transfer, n number of observations, ns not significant
Collection of cumulus cells, ICSI, and culture and morphological classification of embryos
Cumulus-oocyte complexes (COC) were collected and rinsed in HTF HEPES buffered medium (Gynotec, Malden, The Netherlands) and incubated in Sage fertilization medium (CooperSurgical, Leisegang Medical, Berlin, Germany) or Sequential Fertilization medium with phenol red (Origio, Malov, Denmark) until denudation. Shortly before the ICSI procedure, oocytes were denuded individually by pipetting in a 80 IU/ml Cumulase (Origio, Malov, Denmark) droplet and two sequential wash droplets. Metaphase II oocytes were used for ICSI [36], and CC were snap frozen in liquid nitrogen immediately after denudation. The most proximal and clean CC of the COC were collected. These comprise the entire corona cell population which might differ from the more distally mechanically biopsied CC or GC. For this reason, the current test is further referred to as Corona Test. From denudation onwards, the oocytes and embryos were tracked individually. Embryo culture for Corona Test and its two matched controls was performed in the same embryo culture media, either Quinns Advantage Cleavage and Blastocyst medium (Cooper Surgical) or Sequential Cleav and Blast medium (Origio). Embryos underwent routine morphological evaluation. This evaluation comprised the scoring of pronuclei, early cleavage, and full embryo grading on day 3 and if applicable day 5 as described earlier [37]. In brief, the embryo evaluation performed on day 3 resulted in the selection of the embryos suitable for transfer (=GQE). The parameters included the blastomere number and size according to the division pattern, multinucleation, the presence of vacuoles, granularity, or fragmentation. Top-quality embryos were considered normal for all criteria and had at least 7 blastomeres with ≤ 10% fragmentation while good-quality embryos had at least 7 blastomeres, < 20% fragmentation, or 6 blastomeres with ≤ 10% fragmentation. Matched controls in the day-5 arm had all embryos grown up to the blastocyst stage and were additionally scored for morphology on the morning of day 5 [38]; selection of the best embryo for transfer was based on all observations during development.
Design of the predictive model and gene selection
The current predictive gene combination is a result of multiple micro array and QPCR experiments. Potentially interesting genes were selected based on micro array experiments and were validated in independent PCR experiments. In each PCR experiment, the best predictive genes of the previous study were validated together with potential novel predictive genes. This iterative validation strategy and using intra-patient comparisons (CC of patients not pregnant after the fresh transfer but getting pregnant after the frozen SET) led to a model including two genes EFNB2 and CAMK1D exon 1 [30]. Using the same model building strategy and cumulus samples of 21 retrospective patients (33 cumulus samples), SASH1 was identified as a highly predictive gene and added to a model with EFNB2 and CAMK1D exon 1. The new model contains three genes: EFNB2 and SASH1 were positive correlated and CAMK1D exon 1 was negatively correlated with clinical pregnancy. The predictive power of the model was assessed by means of a receiver operating characteristic curve (ROC) analysis. The ROC obtained in the dataset had an AUC (area under the curve) of 0.8081.
RT-QPCR of cumulus cells
Total RNA extraction and reversed transcription were performed as described before [29] and cDNA was frozen at − 80 °C until QPCR analysis. Two stably expressed endogenous control genes, β2M and UBC [29], were analyzed with the 3 specific genes EFNB2, SASH1, and CAMK1D. The mean of β2M and UBC expression was used as the normalization factor. All quantitative PCR reactions, standard curves, and negative controls were performed as described before [30].
Combining cumulus cell gene expression with morphological evaluation
One day after collection, the expression of specific marker genes (Supplementary Figure 1) was quantified in the analytical laboratory (which was blinded for embryo quality) in CC of normally fertilized oocytes. The Corona Test results for all fertilized oocytes were communicated to the embryology lab within 2 days after oocyte retrieval. If on day 3 the patient had 2 or more top or good-quality embryos available according to the morphologic criteria (see higher), the embryo with the highest Corona Test rank was transferred in the fresh treatment cycle. Supernumerary GQE were vitrified on the day of transfer [35]. Patients with less than 2 good-quality embryos were not informative for the study as no choice could be made. These patients were considered as dropout (Fig. 1) and underwent transfer based on morphology alone.
Statistics
Patient characteristics were compared using an ANOVA with subsequent comparisons between groups that were corrected for simultaneous hypothesis testing according to Tukey. Kaplan-Meier statistical analysis was performed to calculate the effect on time-to-pregnancy between the Corona Test patients and the 2 control groups. A Chi square test was used to compare the fresh transfer and cumulative clinical pregnancy rates. The study was powered to demonstrate a significant difference of 20% of the clinical pregnancy rate—the main outcome variable of the study—between the experimental arm (Corona Test group) and the control arms (day-3 and day-5 groups) with a power of 80%. For assessing secondary endpoints (the effect of patient age, and number of GQE), Chi square analysis for pregnancy rates was performed. For all analyses significance was considered when p < 0.05.
Results
The patient population
Patients for this prospective interventional study were recruited from October 2013 until May 2016. In total, 96 HP-hMG-stimulated patients (88 patients received HP-hMG and 8 patients received HP-hMG and rFSH via a mixed protocol) were included for the Corona Test group (experimental arm) (Fig. 1). For 27 patients (28%), only one or no GQE was available on day 3 which meant there was no choice to be made. These patients were non-informative and considered dropouts. From these 27 non-informative patients, 16 patients had transfer of a good- or top-quality embryo, 4 had only low-quality embryos available, while 7 had no embryo to transfer. In this “dropout” group (N = 27), only 6 clinical pregnancies (22%) were recorded.
In addition, for 3 patients (3%), the Corona Test was not informative as there were too few cumulus cells to detect one of the endogenous controls (β2M and UBC). For 4 patients (4%), the Corona Test results were unintentionally not taken into account at embryo transfer and these patients were thus considered non-informative.
Finally, 62 patients in the experimental arm which had more than 2 GQE and Corona Test results were available for final analysis (Fig. 1). All 62 informative Corona Test patients of the experimental arm were matched with routine patients with day-3 SET transfer (day-3 control arm, n = 62) and one with routine patients with day-5 SET transfer (day-5 control arm, n = 44). In both control groups, embryo selection was based on morphology only. The three groups (Corona Test group and the two control groups) were compared for clinical features, stimulation characteristics, and general performance after ICSI (Table 1). As per routine strategy in the ART clinic, patients with only 2 GQE on day 3 will always have transfer on day 3. The inherent absence of this subgroup of patients in the day-5 control group can explain the slightly higher serum estradiol (p > 0.05), progesterone (p < 0.05), mean number of oocytes at pick-up (p > 0.05), and the higher mean number of GQE available on day 3 (p < 0.05) in the control day-5 transfer patients compared to the 2 day-3 transfer groups.
Corona Test effect in the fresh single embryo transfer
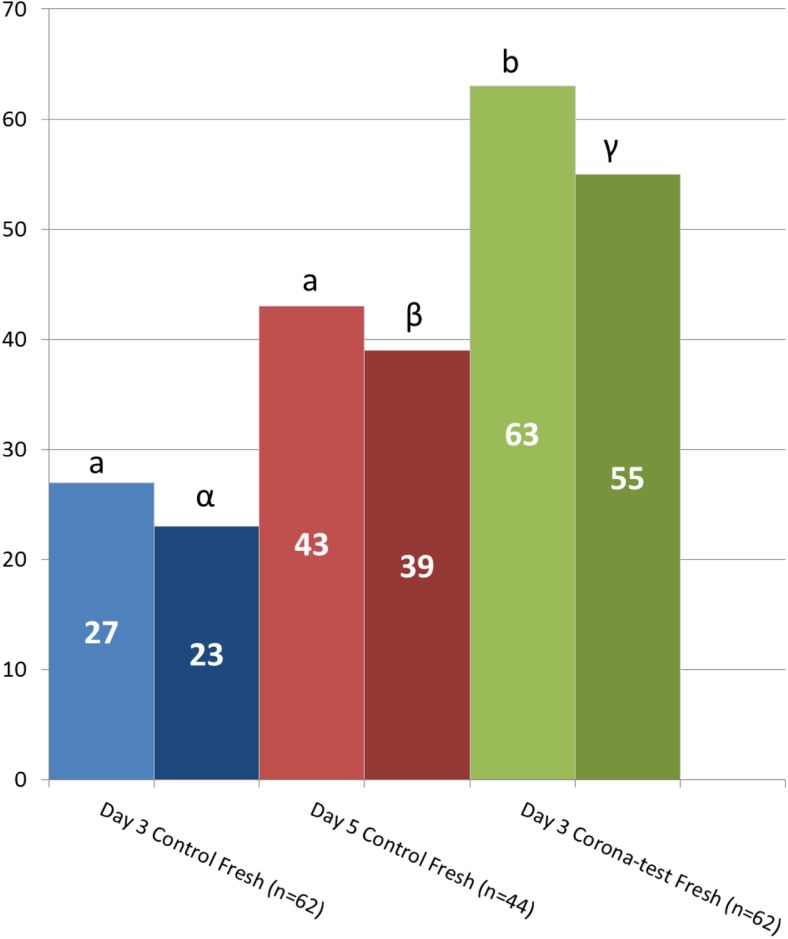
The clinical pregnancy rate after fresh transfer was significantly higher in the Corona Test group (63%, n = 62) when compared to the matched controls with day-3 transfer (27%, n = 62, p < 0.001) or day-5 transfer (43%, n = 44, p = 0.022), respectively (Fig. 2).
Fig. 2.
Clinical pregnancy and live birth after fresh transfer. Graph depicting the clinical pregnancy (light color), and live birth rates (dark color) obtained in the 3 groups after fresh transfer: day 3 transfer based on embryo morphology and the Corona Test (green bars), and control patients with only morphology-based embryo selection for transfer on day 3 (blue bars) and day 5 after pick-up (red bars). Numbers in the bars are the % clinical pregnancies or live birth respectively. Comparisons were performed using the Chi square analysis between the experimental group and the 2 specific control groups and different letters indicate a statistical difference with p < 0.05
In the experimental arm (Corona Test group), a total of 39 clinical pregnancies were observed in the fresh transfer cycle. For all of these patients, the transfer date was at least 9 months ago: 34 healthy singletons were born, two patients had an extra-uterine pregnancy, and three patients miscarried. This results in a 55% live birth rate for the patients in the experimental arm. For the day-3 and day-5 control arms, live birth rates were 23% (p < 0.001) and 39% (p = 0.050), respectively. Miscarriage rates after week 7 were not statistically different in the 3 groups.
Effect of Corona Test on cumulative pregnancy rate
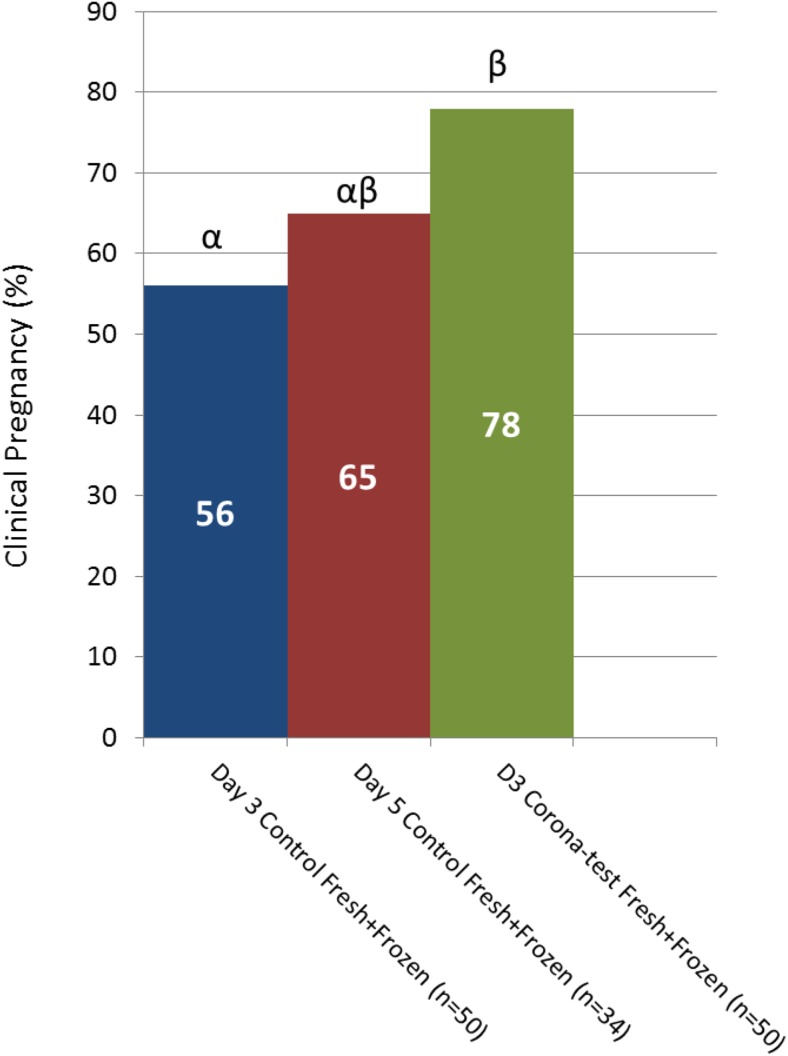
Since the CC of all fertilized oocytes were analyzed, outcome data for the embryos which were transferred in the subsequent cryo-thaw cycles was also available. Cumulative clinical pregnancy rates were calculated for all patients who either became pregnant (from a fresh or a frozen embryo) or had all their embryos transferred one by one as of today. For the experimental arm (Corona Test group), this applied to the first 50 patients with a fresh transfer for whom information from 26 frozen embryo transfers could be added. For the day-3 and day-5 matched control groups, 66 and 37 frozen transfers could be added to the 50 and 34 fresh transfers for the cumulative pregnancy analysis, respectively. In the matched control groups, 93% of non-pregnant patients had used all their available embryos; only two day-3 and two day-5 patients have still embryos cryopreserved.
For the 50 patients in the experimental arm (Corona Test group), the cumulative clinical pregnancy rate reached 78% which was significantly higher than for the matched day-3 control group (56%, p = 0. 010) (Fig. 3). Comparison with the day-5 control, 65% revealed a similar trend (p = 0.089). Even if the two day-3 matched control patients with remaining vitrified embryos would become pregnant, the cumulative pregnancy rate would remain inferior to the Corona Test patients (60%, p = 0.026).
Fig. 3.
Cumulative clinical pregnancy rates. Graph depicting the clinical pregnancy from the fresh transfer and eventual clinical pregnancies obtained after the sequential frozen embryo transfers in patients which were not pregnant from the fresh transfer. The 3 groups are day 3 transfer based on embryo morphology and the Corona Test (green bars), and control patients with only morphology based embryo selection for transfer on day 3 (blue bars) and day 5 after pick-up (red bars), the numbers in the bars are the % clinical pregnancies. Comparisons were performed using the Chi square analysis between the experimental group and the 2 specific control groups and different letters indicate a statistical difference with p < 0.05
Effect of Corona Test on time to pregnancy
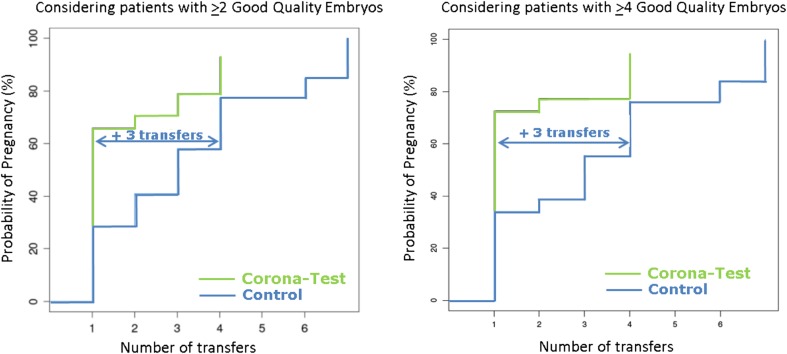
A Kaplan-Meier statistical analysis (Fig. 4) was performed on the Corona Test patients (n = 50, those patients who became pregnant either from the fresh or frozen embryo transfer and those that had all their transferrable embryos replaced) compared to the matched controls (n = 50 for day-3 and n = 34 for day-5 transfer). A significantly shorter time-to-pregnancy was observed in the experimental arm (Corona Test group), compared to the control groups with day-3 and day-5 transfers, (Table 2). This analysis took into consideration the number of GQE available on day 3. The Kaplan-Meier analysis was significant for patients with up to 5 GQE and > 7 GQE. In patients with > 6 GQE, a positive trend was observed but was not statistically significant (p = 0.0656). In every category (> 2GQE to > 8GQE), three more embryo transfer cycles were needed in both control groups to reach a similar pregnancy rate compared to the fresh transfer cycle of the patients who underwent the Corona Test.
Fig. 4.
Effect of the implementation of the Corona Test on the time to pregnancy. Graphs depict the time to pregnancy, expressed by the number of single embryo transfers required, for the Corona Test patients (n = 50) compared to control patients without Corona Test (n = 50 + 34 respectively day 3 and day 5 transfer controls) based on the Kaplan-Meier statistical analysis. This analysis was performed taking into consideration the number of good-quality embryos (GQE) that was available on day 3. Detailed results are available in Table 2. As an example, the graphs depicting the results obtained for the analysis of patients with > 2 GQE (A, n = 134) and the analysis of patients with > 4 GQE (B, n = 72) are represented here. Three more embryo transfer cycles were needed in the control group to reach a similar pregnancy rate as in the fresh transfer cycle of the Corona-tested patients
Table 2.
Effect of the Corona Test on time to pregnancy. Based on the Kaplan-Meier statistical analysis, a significantly shorter time-to-pregnancy is observed when a Corona Test is utilized, compared to the control groups with day 3 and day 5 transfers, regardless the number of good-quality embryos (GQE) that were available on day 3. In every category (> 2GQE up to > 8GQE), three more embryo transfer cycles were needed in the control group to reach a similar pregnancy rate as in Corona Test patients
| Minimal number of GQE on day 3 | Embryos (n) | p value | Number of extra transfers in the day 3 + day 5 control patients to obtain the same pregnancy rate as in the fresh transfer Corona Test group |
|---|---|---|---|
| ≥ 2 GQEmbryos | 134 | < 0.0001 | 3 |
| ≥ 3 GQEmbryos | 97 | 0.0002 | 3 |
| ≥ 4 GQEmbryos | 72 | 0.0009 | 3 |
| ≥ 5 GQEmbryos | 49 | 0.0041 | 3 |
| ≥ 6 GQEmbryos | 31 | 0.0656 | 3 |
| ≥ 7 GQEmbryos | 25 | 0.0021 | 3 |
| ≥ 8 GQEmbryos | 14 | 0.0984 | 3 |
Performance of the Corona Test as a function of the number of good-quality embryos available on day 3
In Table 3, results of fresh transfers were compared in relation to the number of available transferable embryos. In the Corona Test group, the clinical pregnancy rate was at least 55%, independent of the number of GQE available. In both controls, the clinical pregnancy rate was for each category lower than the rate observed in the Corona Test group. This resulted in significantly higher clinical pregnancy rates when patients had 2, 3–4, or > 6 GQE on day 3. The subgroup with 5 or 6 GQE had the lowest number of patients and showed only a trend at this point p = 0.097.
Table 3.
Effect of the number of good-quality embryos available on day 3. Clinical pregnancies in fresh transfer cycles in the Corona Test group and control groups were compared in relation to the number of good-quality embryos (GQE), to choose from on day 3
| Clinical pregnancies in the fresh transfer cycle | Chi square comparing Corona Test to day 3 + day 5 controls | |||
|---|---|---|---|---|
| # GQE | Day 3 control | Day 5 control | Day 3 Corona Test | |
| (n) | (n) | (n) | ||
| 2 | 22% (4/18) | 61% (11/18) | p = 0.009 | |
| 3–4 | 23% (5/22) | 36% (8/22) | 55% (12/22) | p = 0.024 |
| 5–6 | 40% (4/10) | 50% (5/10) | 70% (7/10) | p = 0.097 |
| >6 | 33% (4/12) | 50% (6/12) | 75% (9/12) | p = 0.030 |
| Overall | 27% (17/62) | 43% (19/44) | 63% (39/62) | |
Performance of the Corona Test by patient age in the fresh transfer cycle
Clinical pregnancy rates of Corona Test patients were 65% and 60% for age groups < 35 years and 35–38 years, respectively (Table 4). Within each age category, pregnancy outcome was superior in Corona-tested group. The group < 35 years with Corona Test selection on day-3 embryos had a clinical pregnancy rate of 65%, while the day-3 control had only 29% (p < 0.01). In the age group 35–38 years, the day-3 control had a 24% clinical pregnancy rate and Corona Test increased it to 60% (p < 0.05). Compared to day-5 controls, the Corona Test led to almost a 50% increase in clinical pregnancy rates (NS).
Table 4.
Effect of the patient age on the Corona Test in fresh cycle outcome. The Corona Test patients (n = 62) and the day 3 (n = 62) and day 5 (n = 44) transfer control group were subdivided based on the patients’ age. Chi Square test compared Corona Test to Day 3 or Day 5 control group, different letters indicate statistical difference, p < 0.05
| Clinical pregnancies in the fresh transfer cycle | |||
|---|---|---|---|
| Patient age | Day 3 control | Day 5 control | Day 3 Corona Test |
| (years) | (n) | (n) | (n) |
| < 35 | 29%a (12/41) | 44%ab (11/25) | 65%b (24/37) |
| 35–38 | 24%a (5/21) | 42%ab (8/19) | 60%b (15/25) |
Does Corona Test scoring deviate from standard morphology–based selection?
In 27% of the patients, the Corona Test and morphology grading were concordant, leading to a clinical pregnancy rate of 65%. In 26% of the patients, the Corona Test and morphological scoring were discordant. The Corona Test’s first choice was not the best embryo as graded by morphology. For the remaining 47% of the patients, there was at least one other embryo available with an equal morphology grade. The Corona Test helped to distinguish between the morphologically similar embryos of this category.
In summary, in 73% (26% + 47%) of the cases, the Corona Test triggered the choice for transfer at day 3, leading to clinical pregnancy rate of 56% and 66%, respectively. This outcome is significantly higher than when no Corona Test was implicated in the decision of day-3 eSET (27% clinical pregnancy rate).
Discussion
This is the first prospective clinical study evaluating the potential clinical value of cumulus-gene expression testing in addition to morphology scoring to increase outcome of eSET. A cumulus-corona testing strategy is non-invasive, performed early in the ICSI procedure, and would therefore influence options in the workflow of an embryology laboratory and influence decisions for the choice of the embryo to transfer.
For the 62 informative study patients, the Corona Test was performed prospectively on all cumulus samples from the normally fertilized oocytes. The embryos were scored by routine morphology and for the GQE the choice of the single embryo to transfer was based on the Corona Test result. This decision order doubled the clinical pregnancy rate from 27 to 63% compared to day-3 morphology-only selection (p < 0.05). The clinical pregnancy rate increased by nearly 50% from 43 to 63% compared to day-5 morphology-only selection (p < 0.05, Fig. 2).
The live birth rate for day-3 eSET after Corona Test selection was significantly higher (55%) than morphology based on day 3 (23%) or day 5 (39%), respectively. Hence, use of the Corona Test results in a significantly shorter time-to-pregnancy (Fig. 4), independent of the number of GQE available (Table 3). The application of the Corona Test led to an avoidance of three cryotransfer cycles for the patients (Fig. 4). The cumulative clinical pregnancy rate of Corona Test patients was 40% higher (p = 0.01) compared to the day-3 transfer control group. A similar trend was observed in comparison to the day-5 transfer control group with the Corona Test having a 20% higher cumulative pregnancy rate but this did not reach significance (p = 0.089) (Fig. 3).
The increased efficiency of IVF/ICSI when applying the Corona Test for day-3 single embryo selection in fresh and frozen cycles is most likely multifactorial. Typically, about 75% of the fertilized oocytes form a good-quality embryo by day 3, but only half of them develop to a good-quality blastocyst and are available for transfer on day 5 [39–41]. Additionally, not all embryos will survive the cryopreservation. For cleavage and blastocyst-stage embryos, warming survival rates of 94% [35] and 85% [42] were reported. Subsequently, cleavage and re-expansion after thawing can be also affected [35, 43]. In cleavage-stage embryos, cleavage after thawing depends on the cell stage of the embryo and the cell loss after thawing. The fraction of embryos affected after vitrification and thawing range from as little as 6% to over 50% [35]. Overall, the latter reports recapitulate that extended culture might inflict invisible damage and vitrification as well. Cognizant of this, it is currently not possible to exclude that some embryos competent of giving a baby with a fresh day-3 transfer might survive but have reduced competence inflicted by vitrification and/or 5-day culture although judged morphologically normal.
Effect of patient age and number of transferable embryos on the outcome of Corona testing
Female patient’s age and follicular reserve are strong predictors of outcome [44]. Having fewer embryos available for selection also influences the chance for positive outcome. For this reason, controls were matched for age and for number of embryos available. Pregnancy rates between 60 and 65% were achieved irrespective of age when Corona testing was done. Similarly, high pregnancy rates between 55 and 75% were observed irrespective the different numbers of available GQE.
There are a number of potential limitations of the study. The current study only reports results in a highly selected population of patients. Patients and controls were in first or second IVF treatment cycle and were predicted good responders to HP-hMG treatment. We and others demonstrated that HP-hMG and rFSH stimulation resulted in different CC gene expression levels and patterns in human [45–47]. For this reason, the patients consented to an antagonist HP-hMG protocol. Also, only ICSI patients were included to assure a maximal amount of cumulus cells for molecular analysis. It remains to be seen if Corona testing also works for conventional IVF patients, where the total mass of cumulus cannot be removed, but can only be biopsied. This study only included patients between 22 and 38 years. It is well known that in patients over 35 years there is a significant increase in chromosomal abnormalities and aneuploidy [48]. The Corona Test might be of added value for patients of 35 years and older, as it was observed that transferring euploid embryos in this group did not increase implantation potential to a similar extent as in younger age group [16].
Conclusion
This prospective interventional study showed a significant improvement (doubling) of the clinical pregnancy and live birth rate in a day-3 eSET policy in the experimental arm when embryo selection is based on a combination of morphological scoring and gene expression testing using the Corona Test of three predictive genes. Control arms were matched for age and number of available embryos on day 3 and routine morphology evaluation. The Corona Test strategy proved also superior to a day-5 transfer policy with morphology-only embryo selection. Reduction of the time-to-pregnancy allows to be more cost-efficient as three cryotransfer cycles can be avoided. The Corona Test improved the selection capacity for the best embryo independent of patients’ age and number of GQE available. The Corona Test is non-invasive, can be easily implemented in routine practice, and is more objective compared to the current practice of morphological embryo selection. The Corona Test avoids transfer of multiple embryos and completely eliminates the risk of multiple pregnancy with its higher prevalence of neonatal and postnatal clinical complications. Application of the Corona Test in routine practice will contribute to a reduction of the emotional stress of infertile couples in need of IVF and cost savings to patients and health care providers.
Electronic supplementary material
(DOCX 14 kb)
(DOCX 210 kb)
Acknowledgments
The authors would like to thank their colleagues of the Centre for Reproductive Medicine, UZ Brussel, for their cooperation in this clinical study, the clinical data manager Walter Meul, and Prof. Dr. André Rosenthal for critical reading and suggestions.
Funding
This study was funded by IWT/VLAIO Innovation Mandate 130327 and 140568 and by the Vrije Universiteit Brussel IOFPOC26.
Footnotes
T. Adriaenssens and I. Van Vaerenbergh are joint first authors
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guerif F, Le Gouge A, Giraudeau B, Poindron J, Bidault R, Gasnier O, et al. Limited value of morphological assessment at days 1 and 2 to predict blastocyst development potential: a prospective study based on 4042 embryos. Hum Reprod. 2007;22:1973–1981. doi: 10.1093/humrep/dem100. [DOI] [PubMed] [Google Scholar]
- 2.Guerif F, Lemseffer M, Leger J, Bidault R, Cadoret V, Chavez C, Gasnier O, Saussereau MH, Royere D. Does early morphology provide additional selection power to blastocyst selection for transfer? Reprod BioMed Online. 2010;21:510–519. doi: 10.1016/j.rbmo.2010.06.043. [DOI] [PubMed] [Google Scholar]
- 3.Papanikolaou EG, Kolibianakis EM, Tournaye H, Venetis CA, Fatemi H, Tarlatzis B, Devroey P. Live birth rates after transfer of equal number of blastocysts or cleavage-stage embryos in IVF. A systematic review and meta-analysis. Hum Reprod. 2008;23:91–99. doi: 10.1093/humrep/dem339. [DOI] [PubMed] [Google Scholar]
- 4.Glujovsky D, Blake D, Farquhar C, Bardach A. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Glujovsky D, editor. Cochrane database Syst Rev. Chichester, UK: John Wiley & Sons, Ltd; 2012;CD002118. [DOI] [PubMed]
- 5.Blondel B, Kogan MD, Alexander GR, Dattani N, Kramer MS, Macfarlane A, Wen SW. The impact of the increasing number of multiple births on the rates of preterm birth and low birthweight: an international study. Am J Public Health. 2002;92:1323–1330. doi: 10.2105/AJPH.92.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heino A, Gissler M, Hindori-Mohangoo AD, Blondel B, Klungsøyr K, Verdenik I, et al. Variations in multiple birth rates and impact on perinatal outcomes in Europe. Baud O, editor. PLoS One. 2016;11:e0149252. [DOI] [PMC free article] [PubMed]
- 7.Lédée N, Gridelet V, Ravet S, Jouan C, Gaspard O, Wenders F, et al. Impact of follicular G-CSF quantification on subsequent embryo transfer decisions: a proof of concept study. Hum Reprod. 2013;28:406–413. doi: 10.1093/humrep/des354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scalici E, Traver S, Molinari N, Mullet T, Monforte M, Vintejoux E, Hamamah S. Cell-free DNA in human follicular fluid as a biomarker of embryo quality. Hum Reprod. 2014;29:2661–2669. doi: 10.1093/humrep/deu238. [DOI] [PubMed] [Google Scholar]
- 9.McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–2874. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- 10.Capalbo A, Ubaldi FM, Cimadomo D, Noli L, Khalaf Y, Farcomeni A, et al. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril. 2016;105:225–35.e1–3. [DOI] [PubMed]
- 11.Kirkegaard K, Hindkjaer JJ, Grøndahl ML, Kesmodel US, Ingerslev HJ. A randomized clinical trial comparing embryo culture in a conventional incubator with a time-lapse incubator. J Assist Reprod Genet. 2012;29:565–572. doi: 10.1007/s10815-012-9750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman LR, Goldberg J, Falcone T, Austin C, Desai N. Does the addition of time-lapse morphokinetics in the selection of embryos for transfer improve pregnancy rates? A randomized controlled trial. Fertil Steril. 2016;105:275–85.e10. doi: 10.1016/j.fertnstert.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong S, Arroll N, Cree LM, Jordan V, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane database Syst Rev. 2015;CD011320. [DOI] [PubMed]
- 14.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100:100–7.e1. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 15.Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8:2185–2191. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- 16.Scott RT, Ferry K, Su J, Tao X, Scott K, Treff NR. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril. 2012;97:870–875. doi: 10.1016/j.fertnstert.2012.01.104. [DOI] [PubMed] [Google Scholar]
- 17.Harper JC, Harton G. The use of arrays in preimplantation genetic diagnosis and screening. Fertil Steril. 2010;94:1173–1177. doi: 10.1016/j.fertnstert.2010.04.064. [DOI] [PubMed] [Google Scholar]
- 18.Munné S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108:62–71.e8. doi: 10.1016/j.fertnstert.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Magli MC, Pomante A, Cafueri G, Valerio M, Crippa A, Ferraretti AP, et al. Preimplantation genetic testing: polar bodies, blastomeres, trophectoderm cells, or blastocoelic fluid? Fertil Steril. 2016;105:676–683.e5. doi: 10.1016/j.fertnstert.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills E, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H-J, Melnick AP, Stewart JD, Xu K, Rosenwaks Z. Preimplantation genetic screening: who benefits? Fertil Steril. 2016;106:597–602. doi: 10.1016/j.fertnstert.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Simon AL, Kiehl M, Fischer E, Proctor JG, Bush MR, Givens C, Rabinowitz M, Demko ZP. Pregnancy outcomes from more than 1,800 in vitro fertilization cycles with the use of 24-chromosome single-nucleotide polymorphism–based preimplantation genetic testing for aneuploidy. Fertil Steril. 2018;110:113–121. doi: 10.1016/j.fertnstert.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Verpoest W, Staessen C, Bossuyt PM, Goossens V, Altarescu G, Bonduelle M, et al. Preimplantation genetic testing for aneuploidy by microarray analysis of polar bodies in advanced maternal age: a randomized clinical trial. Hum Reprod. 2018;33:1767–1776. doi: 10.1093/humrep/dey262. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S, Luo K, Cheng D, Tan Y, Lu C, He H, et al. Number of biopsied trophectoderm cells is likely to affect the implantation potential of blastocysts with poor trophectoderm quality. Fertil Steril. 2016;105:1222–1227.e4. doi: 10.1016/j.fertnstert.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Gleicher N, Orvieto R. Is the hypothesis of preimplantation genetic screening (PGS) still supportable? A review. J Ovarian Res. 2017;10:21. doi: 10.1186/s13048-017-0318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev. 1996;8:485–489. doi: 10.1071/RD9960485. [DOI] [PubMed] [Google Scholar]
- 27.Cakmak H, Franciosi F, Zamah AM, Cedars MI, Conti M. Dynamic secretion during meiotic reentry integrates the function of the oocyte and cumulus cells. Proc Natl Acad Sci U S A. 2016;113:2424–2429. doi: 10.1073/pnas.1519990113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 2014;20:1–11. doi: 10.1093/humupd/dmt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26:1035–1051. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 30.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van Landuyt L, Coucke W, et al. Pregnancy prediction in single embryo transfer cycles after ICSI using QPCR: validation in oocytes from the same cohort. Lambalk CB, editor. PLoS One. 2013;8:e54226. [DOI] [PMC free article] [PubMed]
- 31.Buensuceso AV, Deroo BJ. The ephrin signaling pathway regulates morphology and adhesion of mouse granulosa cells in vitro. Biol Reprod. 2013;88:25. doi: 10.1095/biolreprod.112.100123. [DOI] [PubMed] [Google Scholar]
- 32.Dauphinee SM, Clayton A, Hussainkhel A, Yang C, Park Y-J, Fuller ME, Blonder J, Veenstra TD, Karsan A. SASH1 is a scaffold molecule in endothelial TLR4 signaling. J Immunol. 2013;191:892–901. doi: 10.4049/jimmunol.1200583. [DOI] [PubMed] [Google Scholar]
- 33.Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Vanderpoel S, International Committee for Monitoring Assisted Reproductive Technology. World Health Organization International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril. 2009;92:1520–1524. doi: 10.1016/j.fertnstert.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Devroey P, Pellicer A, Nyboe Andersen A, Arce J-C, Menopur in GnRH Antagonist Cycles with Single Embryo Transfer Trial Group A randomized assessor-blind trial comparing highly purified hMG and recombinant FSH in a GnRH antagonist cycle with compulsory single-blastocyst transfer. Fertil Steril. 2012;97:561–571. doi: 10.1016/j.fertnstert.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Van Landuyt L, Van de Velde H, De Vos A, Haentjens P, Blockeel C, Tournaye H, et al. Influence of cell loss after vitrification or slow-freezing on further in vitro development and implantation of human day 3 embryos. Hum Reprod. 2013;28:2943–2949. doi: 10.1093/humrep/det356. [DOI] [PubMed] [Google Scholar]
- 36.Van Landuyt L, De Vos A, Joris H, Verheyen G, Devroey P, Van Steirteghem A. Blastocyst formation in in vitro fertilization versus intracytoplasmic sperm injection cycles: influence of the fertilization procedure. Fertil Steril. 2005;83:1397–1403. doi: 10.1016/j.fertnstert.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 37.Segers I, Mateizel I, Van Moer E, Smitz J, Tournaye H, Verheyen G, et al. In vitro maturation (IVM) of oocytes recovered from ovariectomy specimens in the laboratory: a promising "ex vivo" method of oocyte cryopreservation resulting in the first report of an ongoing pregnancy in Europe. J Assist Reprod Genet. 2015;32:1221–1231. doi: 10.1007/s10815-015-0528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner D, Schoolcraft W. In-vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Towar reprod certain fertil genet beyond 1999. Carnforth: Parthenon Press; 1999. pp. 378–388. [Google Scholar]
- 39.Utsunomiya T, Ito H, Nagaki M, Sato J. A prospective, randomized study: day 3 versus hatching blastocyst stage. Hum Reprod. 2004;19:1598–1603. doi: 10.1093/humrep/deh288. [DOI] [PubMed] [Google Scholar]
- 40.Ziebe S, Lundin K, Janssens R, Helmgaard L, Arce J-C. MERIT (Menotrophin vs Recombinant FSH in vitro Fertilisation Trial) Group. Influence of ovarian stimulation with HP-hMG or recombinant FSH on embryo quality parameters in patients undergoing IVF. Hum Reprod. 2007;22:2404–2413. doi: 10.1093/humrep/dem221. [DOI] [PubMed] [Google Scholar]
- 41.Sfontouris IA, Kolibianakis EM, Lainas GT, Petsas GK, Tarlatzis BC, Lainas TG. Blastocyst development in a single medium compared to sequential media: a prospective study with sibling oocytes. Reprod Sci. 2017;24:1312–1318. doi: 10.1177/1933719116687653. [DOI] [PubMed] [Google Scholar]
- 42.Rienzi L, Gracia C, Maggiulli R, LaBarbera AR, Kaser DJ, Ubaldi FM, Vanderpoel S, Racowsky C. Oocyte, embryo and blastocyst cryopreservation in ART: systematic review and meta-analysis comparing slow-freezing versus vitrification to produce evidence for the development of global guidance. Hum Reprod Update. 2017;23:139–155. doi: 10.1093/humupd/dmw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veeck LL, Bodine R, Clarke RN, Berrios R, Libraro J, Moschini RM, Zaninovic N, Rosenwaks Z. High pregnancy rates can be achieved after freezing and thawing human blastocysts. Fertil Steril. 2004;82:1418–1427. doi: 10.1016/j.fertnstert.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 44.Stoop D, Ermini B, Polyzos NP, Haentjens P, De Vos M, Verheyen G, et al. Reproductive potential of a metaphase II oocyte retrieved after ovarian stimulation: an analysis of 23 354 ICSI cycles. Hum Reprod. 2012;27:2030–2035. doi: 10.1093/humrep/des131. [DOI] [PubMed] [Google Scholar]
- 45.Adriaenssens T, Mazoyer C, Segers I, Wathlet S, Smitz J. Differences in collagen expression in cumulus cells after exposure to highly purified menotropin or recombinant follicle-stimulating hormone in a mouse follicle culture model. Biol Reprod. 2009;80:1015–1025. doi: 10.1095/biolreprod.107.067462. [DOI] [PubMed] [Google Scholar]
- 46.Grøndahl ML, Borup R, Lee YB, Myrhøj V, Meinertz H, Sørensen S. Differences in gene expression of granulosa cells from women undergoing controlled ovarian hyperstimulation with either recombinant follicle-stimulating hormone or highly purified human menopausal gonadotropin. Fertil Steril. 2009;91:1820–1830. doi: 10.1016/j.fertnstert.2008.02.137. [DOI] [PubMed] [Google Scholar]
- 47.Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25:1259–1270. doi: 10.1093/humrep/deq049. [DOI] [PubMed] [Google Scholar]
- 48.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101:656–663.e1. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 14 kb)
(DOCX 210 kb)