Abstract
Despite high anti-HBV efficacies, while the nucleoside analogs (e.g., lamivudine) lead to the emergence of drug-resistance, interferons (e.g., IFN-α causes adverse side-effects. Comparatively, various natural or plant products have shown similar or even better efficacy. Hence, new antiviral strategies must focus not only on synthetic molecules but also on potential natural compounds. In this report, we have combined the in vitro cell culture and in silico molecular docking methods to assess the novel anti-HBV activity and delineate the inhibitory mechanism of selected plant-derived pure compounds of different classes. Of the tested (2.5-50 μg/ml) twelve non-cytotoxic compounds, ten (10 μg/ml) were found to maximally inhibit HBsAg production at day 5. Compared to quercetin (73%), baccatin III (71%), psoralen (67%), embelin (65%), menisdaurin (64%) and azadirachtin (62%) that showed high inhibition of HBeAg synthesis, lupeol (52%), rutin (47%), β-sitosterol (43%) and hesperidin (41%) had moderate efficacies against HBV replication. Further assessment of quercetin in combination with the highly active compounds, enhanced its anti-HBV activity up to 10%. Being the most important drug target, a 3-D structure of HBV polymerase (Pol/RT) was modeled and docked with the active compounds, including lamivudine as standard. Docking of lamivudine indicated strong interaction with the modeled HBV Pol active-site residues that formed stable complex (∆G = −5.2 kcal/mol). Similarly, all the docked antiviral compounds formed very stable complexes with HBV Pol (∆G = −6.1 to −9.3 kcal/mol). Taken together, our data suggest the anti-HBV potential of the tested natural compounds as novel viral Pol/RT inhibitors.
Keywords: Keywords, Hepatitis B virus, Antiviral, HBV polymerase, Natural compounds, Molecular docking, Pol/RT inhibitors
1. Introduction
Hepatitis B virus (HBV) causes acute and chronic liver diseases, including fulminant liver failure, cirrhosis and hepatocellular carcinoma, affecting about two billion of world population (Teo and Locarnini, 2010). Of these, an estimated 240 million individuals with chronic hepatitis B (CHB) live in high to moderate endemic regions in Asia, sub-Sharan Africa, Eastern Europe and Latin America (WHO, 2017). HBV is the smallest known DNA virus with a circular, double-stranded genome (∼3.2 kb) having four overlapped open reading frames encoding surface or envelope (S), core (C), pre-core (pre-C/e), polymerase/reverse-transcriptase (Pol/RT) and X proteins (Beck and Nassal, 2007). Very uniquely, HBV replicates its DNA via an RNA intermediate molecule through reverse-transcription, and hence nicknamed ‘pararetrovirus’.
Owing to similarity with retroviral replication mechanism, almost all the approved drugs against human immunodeficiency virus (HIV) and herpes simplex virus (HSV) have been proved effective against HBV. Unfortunately, despite their high efficacies, majority of approved anti-HBV drugs have some therapeutic limitations. While long-term treatment with HBV Pol inhibitors, the nucleot(s)ide analogs (e.g., lamivudine, adepovir, entecavir etc.) leads to the emergence of drug-resistance in the Pol ‘Tyr-Met-Asp-Asp (YMDD)’ motif, the interferon (pegIFN-α) chemotherapy causes adverse side-effects in some patients (Lok et al., 2007; Locarnini et al., 2008). In addition, despite the global success of vaccinations, reports of vaccine-escape HBV mutants present another hurdle (Teo and Locarnini, 2010, Torresi, 2008). Moreover, in low-income countries, various political and socio-economic factors further hamper the prevention and control of CHB.
Traditional herbal products and drugs of indigenous origin have an ancient history of curing several chronic and infective diseases. A range of various plant metabolites are shown to impede virus replication without affecting the host physiology or with limited side-effects (Martin and Ernst, 2003, Hussain et al., 2017). In addition to their ability to interfere directly with viral replication, these natural products may also modulate the host immune response against infection (Kurokawa et al., 2010). In line with this, several natural or plant-derived compounds belonging to different chemical classes have been reported to have potential anti-HBV activities (Chen et al., 1995, Chang et al., 2005, Wang, 2000; Huang et al., 2008; Chou et al., 2012, Qiu and Chen, 2013, Wu, 2016, Parvez et al., 2016). Compared to interferons or lamivudine treatment, some plant products are reported with similar or even better efficacy (Chen and Zhu, 2013, Arbab et al., 2017, Zhang et al., 2017). Interestingly therefore, approximately 80% of the CHB patients in China rely on traditional herbal medicines.
The HBV-reporter cell line, HepG2.2.15 is one of the established systems for preliminary screening of anti-HBV drugs in vitro. The in silico molecular docking tool enables to assess the binding ability of a ligand to protein active-site as well as to compare the binding modes of different ligands to the active site-pocket (Leach, 2001). Therefore, in this study, we evaluated the novel anti-HBV activity as well as delineated mechanism of action of selected plant-derived compounds of different classes, using cell culture and molecular docking methods.
2. Materials and methods
2.1. Natural compounds and drugs
Twelve natural antiviral compounds of plant origin were selected on the basis of their published in vitro or vivo antiviral activities. All cell culture grade natural compounds viz. rutin, quercetin, menisdaurin, β-sitosterol, hesperidin, psoralen, bergenin, azadirachtin, baccatin III, lupeol, embelin and naringenin were purchased (Sigma-Aldrich, Germany). The approved anti-HBV nucleoside analog, lamivudine (3TC; Sigma-Aldrich, Germany) served as standard (positive control).
2.2. Cell culture
HBV-reporter cells, HepG2.2.15 (human hepatoblastoma line, HepG2-derived) were kindly obtained from Dr. S. Jameel (International Center for Genetic Engineering & Biotechnology, New Delhi, India). HepG2.2.15 cell were maintained in RPMI-1640 medium (Invitrogen, USA), supplemented with 10% heat-inactivated bovine calf serum (Gibco, USA), 1x penicillin-streptomycin (Invitrogen, USA), and 1x sodium pyruvate (HyClone Laboratories, USA) at 37 0C in a humidified chamber with 5% CO2 supply.
2.3. Cytotoxicity assay
All compounds were first tested for in vitro cytotoxic effects, if any, on cultured HepG2.2.15 cells. Cells were seeded (0.5 × 105 well, in triplicate) in a 96-well flat-bottom culture plate (Becton-Dickinson Labware) and grown over night. Compounds were first dissolved in 50 μl of dimethyl sulfoxide (DMSO) following final preparation in culture media (1 mg/ml). Five doses (2.5, 5, 10, 20 and 50 μg/ml) of each compound were further prepared by diluting in culture media. The final concentration of DMSO used never exceeded >0.1%, and therefore, had no cytotoxicity. RPMI with 0.1% DMSO served as untreated or negative control. The cultures were replenished with media containing a dose of the drug, and incubated for 48 h followed by MTT assay (TACS MTT Cell Proliferation Assay Kit, Trevigen) as per the manufacturer’s manual. The 50% cytotoxic concentrations (CC50) for all compounds were determined using regression curve in Excel software. The experiment was repeated to confirm the reproducibility.
2.4. Microscopy
At 24 and 48 h post-treatment, cells were visually monitored for morphological changes like, lesions of cell membrane and the compactness of cytoplasmic components under an inverted microscope (Optica, Italy) with 200× magnification.
2.5. HBsAg inhibitory activities of natural compounds
The HepG2.2.15 cells were seeded in flat-bottom 96 well plates (0.5 × 105/well in triplicate) and incubated overnight. Based on the cytotoxicity results, and previously reported assays, the treatment dose of 10 μg/ml was chosen for all pure compounds in anti-HBV experiments. Lamivudine (2 μM) prepared in DMSO and RMPI, was included as positive control (Parvez et al., 2006) while RPMI with 0.1% DMSO served as untreated control. Next day, the old media were replaced with fresh media containing compounds and controls, and the culture was incubated for 5 days as described elsewhere (Arbab et al., 2017). Culture supernatants were replaced with fresh dose every alternate day and samples collected on day 1, 3, and 5 were saved at −20 °C. The viral HBsAg production in the culture supernatant was analyzed using Monolisa HBsAg ULTRA Elisa Kit (BioRad, USA) as per the manufacturer’s instructions. All data were presented as % inhibition of HBsAg expression in relation to the untreated control. The experiment was repeated to confirm the reproducibility.
2.6. Inhibition of viral HBeAg production
Further inhibitory activities of the selected compounds on HBV replication were tested as above on HepG2.2.15 cells. Culture supernatants of each sample were collected on day 5, and analyzed for the viral HBeAg synthesis, using HBeAg/Anti-HBe Elisa Kit (DIASource, Belgium) following the manufacturer’s instructions. All data were presented as % inhibition of HBeAg expression in relation to the untreated control. The experiment was repeated to confirm the reproducibility.
2.7. Assessment of combination treatment
Quercetin (10 μg/ml) was further assessed for its enhanced anti-HBV effect, if any, in combination with baccatin III, psoralen, embelin, menisdaurin and azadirachtin alone or all together (1:1/w/w). Culture supernatants collected on day 5 were analyzed for HBeAg synthesis, and data were presented as above.
2.8. Transient transfection and reporter-protein assay
To rule out the inhibitory effect of the tested antiviral compounds on host-synthesized proteins, luciferase assay was performed as described previously (Al-Dosari and Parvez, 2018). Briefly, HepG2.2.15 cells were transfected with Renilla-luciferase plasmid (pRL-TK; Promega, USA) using FuGENE6 (Promega, USA) in a 48-well culture plate, following treatment (10 μg/ml; in triplicate) with all compounds, including DMSO (0.1%) as untreated control. After 2 days of incubation (i.e., day 3 post-transfection), cell lysates were prepared and luciferase expression was measured (Luciferase Reporter Assay System; Promega, USA), and data were presented in relation to the negative control.
2.9. Homology modeling of HBV Pol
The sequence of HBV Pol/RT (GenBank: AGA95798.1) was retrieved from NCBI database (https://www.ncbi.nlm.nih.gov/genbank/). The HBV amino acid residues 347–698 responsible for the Pol activity (Daga et al., 2010) was used as query to search protein database for the identification of a suitable template with known X-ray crystal structure. The X-ray crystal structure of genetically-close HIV-RT (PDB Id: 1RTD) was selected as template to model the HBV Pol structure as reported previously (Huang et al., 1998). Swiss model server was used for the prediction of three-dimensional structure of HBV Pol (Arnold et al., 2006). The quality of the predicted structure was validated by Ramachandran plot using RAMPAGE server (Lovell et al., 2003).
2.10. Molecular docking
The modeled HBV Pol was used as receptor for screening the binding modes of antiviral natural compounds, including lamivudine (Fig. 1) in Autodock Vina and PyRx virtual screening tools (Trott and Olson, 2010, Dallakyan and Olson, 2015). All compounds were energy minimized using Universal Force Field (UFF). The compounds showing ΔG ≥ −6.0 kcal/mol were re-docked to HBV Pol to evaluate their detailed binding mechanisms using Autodock 4.2 as described elsewhere (Morris et al., 2009, Al-Yousef et al., 2017). For molecular docking, target protein and ligands were prepared as described elsewhere (Rehman et al., 2014, Rabbani et al., 2017). Briefly, the target model protein HBV Pol was prepared by removing any heterogeneous compounds and water molecules. Polar hydrogen atoms and Kollman charges were added to the protein using Autodock tool (ADT). Affinity grid maps were generated in such a way as to cover the complete active sites of the protein. Molecular docking was performed using Lamarkian Genetic Algorithm (LGA) to calculate the possible conformations of the ligands that bind to the target protein. Here, the ligand was set free to search and bind at the active site of the protein in the most favorable or minimum energy conformation. Initial positions and orientations of the ligands were set randomly while the torsions were set to a maximum of 6. Each run of the docking was performed to calculate 2,500,000 energy evaluations. The population size, translational step, quaternion and torsion steps were set to 150, 0.2 and 5 respectively. On the basis of binding energy (ΔG), best docked structures were saved and analyzed for receptor-ligand interactions using Discovery Studio 4.0 (Accelrys Software Inc., 2012). The binding constant (Kb) for protein-ligand interaction was calculated using the following relation (Rehman et al., 2016): (R = gas constant, 1.987 cal/mol/K; T = temperature, 298 K).
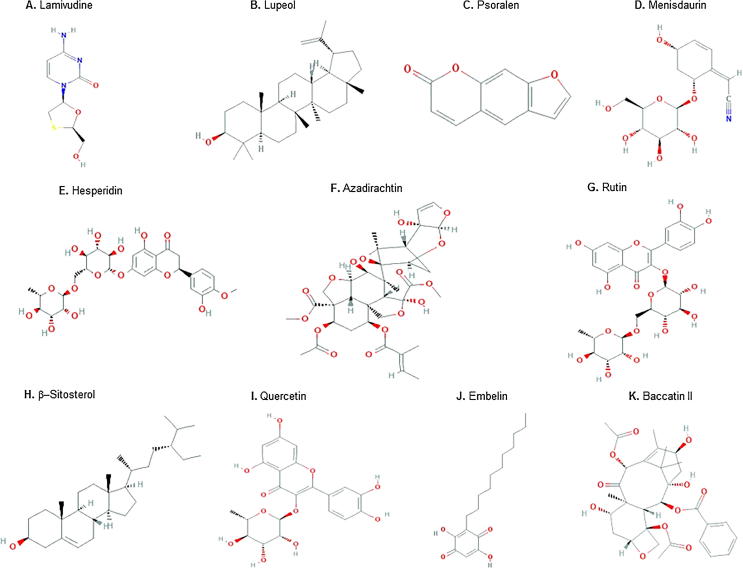
Fig. 1.
Structures of the studied antiviral compounds (lamivudine, quercetin, rutin, hesperidin, lupeol, azadirachtin, β-sitosterol, psoralen, embelin, menisdaurin, and baccatin III).
Source: https://pubchem.ncbi.nlm.nih.gov/.
2.11. Statistical analysis
All data were presented as the mean ± standard deviation (S.D.) of triplicate experiments normalized to untreated control, and analyzed by one-way analysis of variance. Dunnett's t-test was used to calculate statistical significance using Excel software. The significance was defined as p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Results
3.1. Cytotoxicity assay
Rutin, quercetin, menisdaurin, β-sitosterol, hesperidin, psoralen, bergenin, azadirachtin, baccatin III, lupeol, embelin and naringenin showed no signs of cytotoxicity even at the maximal dose (50 μg/ml), confirmed by microscopy and MTT assay.
3.2. Time-course inhibition of HBsAg expression by selected compounds
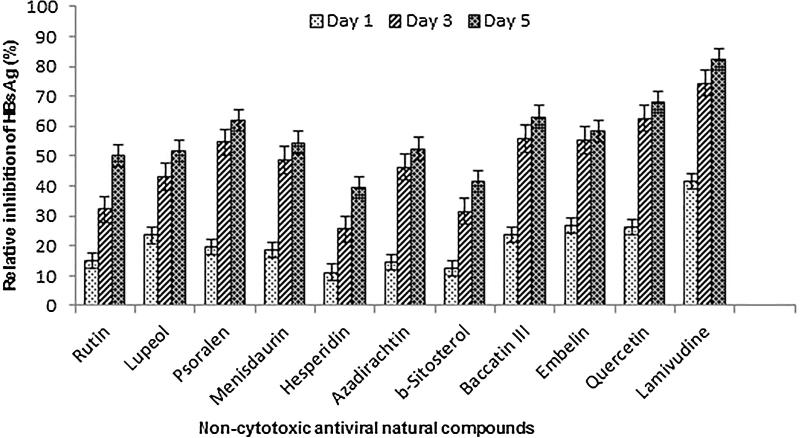
To investigate the anti-HBV potential, the selected compounds were subjected to a time-course (day 1, 3 and 5) study, using HepG2.2.15 cells (Fig. 2). Notably, prolonged treatment beyond day 5 did not show any significant effect, and further continuation of the culture resulted in cell overgrowth and death (Arbab et al., 2017). The optimal anti-HBV activities on day 5 post-treatment were: quercetin (68%) > baccatin III (63%) > psoralen (62%) > embelin (58.5%) > menisdaurin (54.5%) > azadirachtin (52.5%) > lupeol (51.5%) > rutin (50%) > β-sitosterol (51.5%) > hesperidin (39.5%) as compared to untreated control. Notably, further treatment with the maximal 50 μg/ml dose did not suppress HBsAg significantly (data not shown). The two compounds, bergenin and naringenin did not show any effect on inhibition of HBsAg, and therefore excluded from further antiviral analysis.
Fig. 2.
Time–course anti–HBV activity of selected natural compounds (10 μg/ml), showing inhibition of HBsAg expressions relative to untreated control in HepG2.2.15 culture supernatants. Lamivudine (2 μM) was used as a reference anti–HBV drug. Values (Y–axis): means of three determinations.
3.3. Down regulation of HBV replication by the active compounds
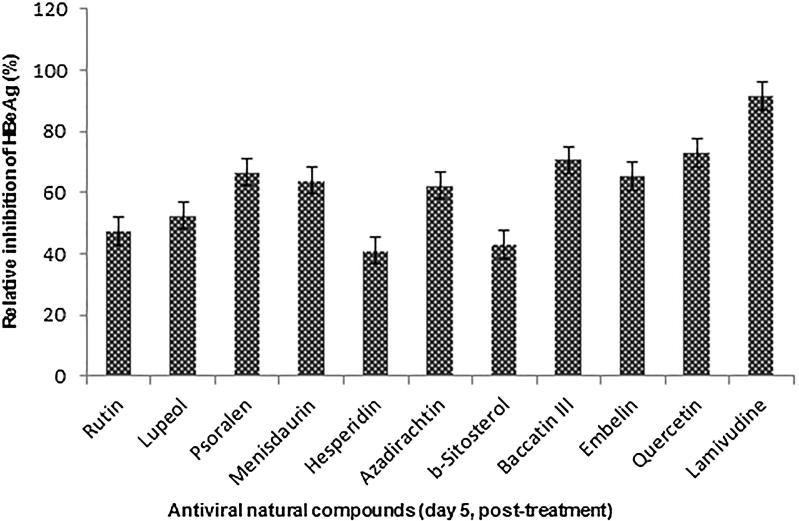
The HBV secretory protein ‘e’ antigen (HBeAg) is a processed product of the ‘pre-Core’ gene that is co-translated with ‘Core’ by a bicistronic subgenomic-RNA. Therefore, in natural infection, seropositivity of HBeAg is a hallmark of active viral DNA replication (Shafritz and Lieberman, 1984, Beck and Nassal, 2007). Notably, this is analogous to HIV ‘p24′ antigen where ELISA is a valid tool to monitor retroviral RNA replication (Cunningham et al., 1993). We therefore, further evaluated the antiviral compounds on their inhibitory effect on HBeAg production for five days (Fig. 3). Notably, prolonged treatment beyond day 5 did not show any significant effect and further continuation resulted in cell overgrowth and death (data not shown). The estimated antiviral activities of the compounds on day 5 post-treatment were: quercetin (73%) > baccatin III (71%) > psoralen (67%) > embelin (65.5%) > menisdaurin (64%) > azadirachtin (62%) > lupeol (52.5%) > rutin (47.5%) > β-sitosterol (43%) > hesperidin (41%). Of these, while six compounds (quercitin, psoralen, baccatin III, menisdaurin, embelin and azadirachtin) showed promising efficacies, four (lupeol, rutin, β-sitosterol and hesperidin) had moderate anti-HBV activities.
Fig. 3.
Inhibition of HBeAg expressions by selected natural compounds relative to untreated control in HepG2.2.15 culture supernatants at day 5. Lamivudine was used as a reference anti–HBV drug. Values (Y–axis): means of three determinations.
3.4. Enhanced anti-HBV effect of combination treatment
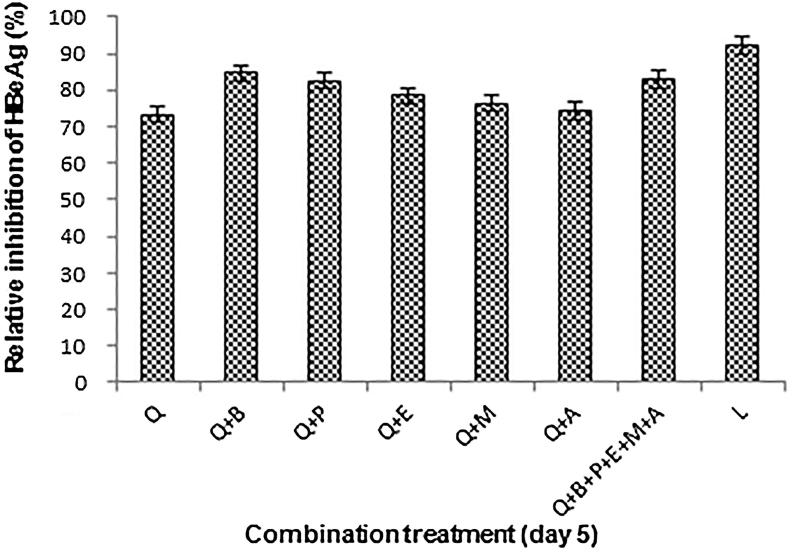
Compared to quercetin treatment alone, its combinations (1:1, w/w) with individual compounds (baccatin III, psoralen, embelin, menisdaurin and azadirachtin) further enhanced the antiviral activity by 8–10% (Fig. 4). There was however, no further enhancement in the combinatorial effect when quercetin was used with a mixture of all compounds.
Fig. 4.
Antiviral effects of combination treatment on HBV replication, showing inhibition of HBeAg expressions relative to untreated control at day 5. Values (Y–axis): means of three determinations. Q: Quercetin, B: Baccatin III, P: Psoralen, E: Embelin, M: Menisdaurin, A: Azadirachtin, and L: Lamivudine.
3.5. Transient transfection and reporter-gene assay
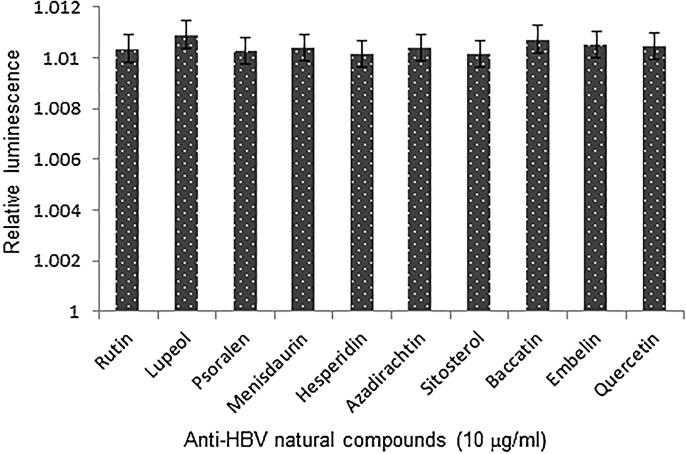
Our reporter-protein assay showed non-significant change in the levels of Renilla-luciferase protein synthesis in HepG2.2.15 cells upon treatment with the optimal dose (10 μg/ml) of all anti-HBV natural compounds (Fig. 5). This further confirmed the specific inhibition of viral proteins (HBsAg and HBeAg), without affecting the non-viral/host protein expression by the tested compounds.
Fig. 5.
Reporter-gene assay, showing relative expression of Renilla-luciferase in HepG2.2.15 cells treated with anti-HBV compounds for 2 days (day 3 post-transfection). Values (Y–axis): means of three determinations.
3.6. Homology modeling and validation of HBV Pol
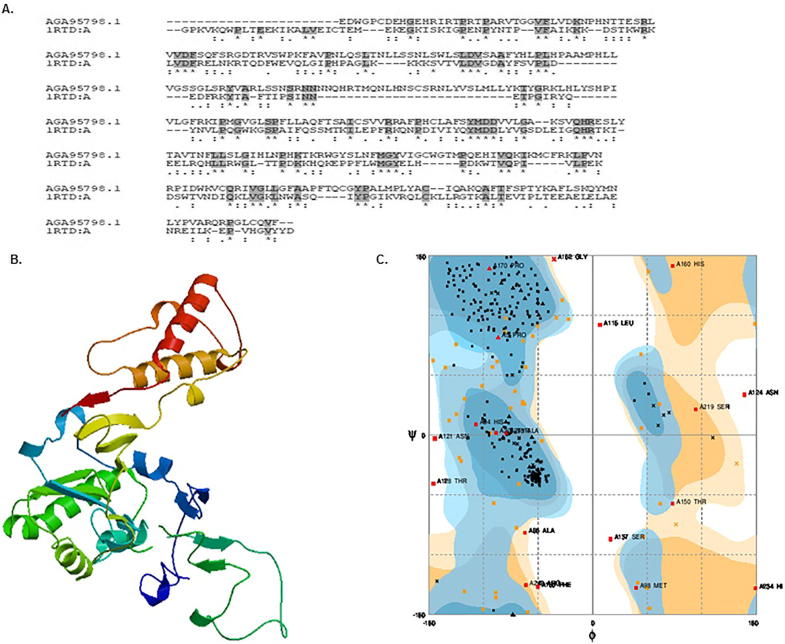
HBV Pol has been the most desired target protein to develop potential anti-HBV nucleot(s)ide drugs despite emergence of drug-resistant mutations in the Pol ‘YMDD’ motif (Lok et al., 2007; Locarnini et al., 2008). Since the crystal structure of HBV Pol was not available, we used homology modeling to predict its three-dimensional structure. It has been reported that the amino acid residues from 347 to 698 determine the reverse transcriptase activity of HBV Pol. We therefore, used the above sequence as query to search protein database for the identification of the most suitable template. Based on the primary sequence alignment (Fig. 6A), we used HIV-1 RT (PDB Id: 1RTD) as a template for HBV Pol homology modeling because the two viral proteins demonstrated highly conserved residues important for enzymatic activities. The homology model of HBV Pol was generated using the Swiss model server (Fig. 6B), and validated by Ramachandran plot. The Ramachandran plot showed 75.2% of the residues occupying the favored region, while 19.0% and 3.9% residues occupied the allowed and generously allowed region, respectively. Only, 2.0% of the residues were located in the disallowed outlier region (Fig. 5C). Overall the Ramachandran plot suggested that the generated model of HBV Pol was of reasonably good quality and therefore, used for molecular docking.
Fig. 6.
Molecular modeling of HBV Pol/RT protein. (A) Amino acid sequence homology of HBV Pol/RT (AGA95798.1) and HIV-1 RT (IRDA:A). (B) Deduced 3-D structure of HBV pol. (C) Ramachandran plot-validation of HBV Pol structure.
3.7. Molecular docking analysis of HBV Pol
To gain an insight into the binding mechanism of selected natural compounds to HBV Pol, we performed molecular docking using Autodock Vina in PyRx virtual screening tool. The amino acid residues and the type of interactions responsible for forming a stable HBV Pol-inhibitor complex are presented in Table 1. Compounds showing ΔG ≥ −6.0 kcal/mol were re-docked in Autodock 4.2 for detailed binding mechanism.
Table 1.
Binding parameters for the interaction of HBV Pol with plant-derived antiviral compounds.
| Interacting Pol residues | Types of Interaction | Bond length (Å) |
|---|---|---|
| Lamivudine* | ||
| SER202:OG - UNK1:O | Hydrogen Bond | 3.00 |
| UNK1:C - ALA181:O | Carbon Hydrogen Bond | 3.51 |
| UNK1:C - ALA181:O | Carbon Hydrogen Bond | 3.66 |
| PRO59 - UNK1 | Hydrophobic (Alkyl) | 5.20 |
| ALA181 - UNK1 | Hydrophobic (Alkyl) | 5.12 |
| ALA181 - UNK1 | Hydrophobic (Alkyl) | 3.84 |
| UNK1 - PRO59 | Hydrophobic (Alkyl) | 4.42 |
| Quercetin | ||
| LYS32:HZ1 - UNK1:O | Hydrogen Bond | 2.87 |
| ASN36:HD21 - UNK1:O | Hydrogen Bond | 2.42 |
| ALA87:HN - UNK1:O | Hydrogen Bond | 2.24 |
| PHE88:HN - UNK1:O | Hydrogen Bond | 2.84 |
| UNK1:H - ASP83:OD1 | Hydrogen Bond | 2.57 |
| UNK1:H - ASP83:OD2 | Hydrogen Bond | 3.08 |
| UNK1:H - VAL84:O | Hydrogen Bond | 2.03 |
| LYS32:NZ - UNK1 | Electrostatic (Pi-Cation) | 4.30 |
| ARG41:NH2 - UNK1 | Electrostatic (Pi-Cation) | 3.54 |
| ASP83:OD2 - UNK1 | Electrostatic (Pi-Anion) | 3.74 |
| ARG41:HE - UNK1 | Hydrogen Bond | 3.04 |
| ALA86:HN - UNK1 | Hydrogen Bond | 3.12 |
| UNK1 - ARG41 | Hydrophobic (Pi-Alkyl) | 4.74 |
| UNK1 - MET171 | Hydrophobic (Pi-Alkyl) | 5.35 |
| Rutin | ||
| ASN36:ND2 - UNK1:O | Hydrogen Bond | 2.96 |
| ARG41:NH2 - UNK1:O | Hydrogen Bond | 3.13 |
| ASP83:N - UNK1:O | Hydrogen Bond | 3.17 |
| MET250:N - UNK1:O | Hydrogen Bond | 3.24 |
| GLY251:N - UNK1:O | Hydrogen Bond | 3.10 |
| UNK1:H - ASN248:O | Hydrogen Bond | 1.99 |
| UNK1:H - SER81:O | Hydrogen Bond | 1.79 |
| UNK1:H - ASN33:O | Hydrogen Bond | 2.53 |
| ASP83:OD2 - UNK1 | Electrostatic (Pi-Anion) | 4.46 |
| ASP83:OD2 - UNK1 | Electrostatic (Pi-Anion) | 4.49 |
| UNK1 - LYS241 | Hydrophobic (Pi-Alkyl) | 4.56 |
| Hesperidin | ||
| SER40:N - UNK1:O | Hydrogen Bond | 2.79 |
| SER40:OG - UNK1:O | Hydrogen Bond | 2.89 |
| SER85:OG - UNK1:O | Hydrogen Bond | 2.86 |
| SER117:OG - UNK1:O | Hydrogen Bond | 2.70 |
| UNK1:H - SER117:OG | Hydrogen Bond | 2.14 |
| UNK1:H - SER40:OG | Hydrogen Bond | 2.22 |
| UNK1:H - GLU39:OE2 | Hydrogen Bond | 1.98 |
| HIS156:CE1 - UNK1:O | Carbon Hydrogen Bond | 3.39 |
| TRP3 - UNK1 | Hydrophobic (Pi-Pi Stacked) | 5.28 |
| TRP3 - UNK1 | Hydrophobic (Pi-Pi Stacked) | 5.18 |
| UNK1:C - LEU147 | Hydrophobic (Alkyl) | 4.17 |
| UNK1 - ALA86 | Hydrophobic (Pi-Alkyl) | 5.23 |
| Lupeol | ||
| ARG120 - UNK1 | Hydrophobic (Alkyl) | 4.59 |
| UNK1:C - LEU42 | Hydrophobic (Alkyl) | 5.39 |
| UNK1:C - ARG120 | Hydrophobic (Alkyl) | 3.87 |
| UNK1:C - PRO5 | Hydrophobic (Alkyl) | 4.29 |
| TRP3 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.99 |
| HIS156 - UNK1 | Hydrophobic (Pi-Alkyl) | 5.22 |
| HIS156 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.59 |
| HIS156 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.19 |
| TYR158 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.20 |
| TYR158 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.83 |
| Azadirachtin | ||
| SER117:OG - UNK1:O | Hydrogen Bond | 2.97 |
| ARG120:NE - UNK1:O | Hydrogen Bond | 3.10 |
| ARG153:NH1 - UNK1:O | Hydrogen Bond | 2.82 |
| ARG153:NH1 - UNK1:O | Hydrogen Bond | 3.25 |
| UNK1:C - GLU1:OE2 | Carbon Hydrogen Bond | 3.52 |
| UNK1:C - PRO5 | Hydrophobic (Alkyl) | 4.37 |
| HIS156 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.77 |
| HIS156 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.08 |
| β-Sitosterol | ||
| UNK1:C - TRP3 | Hydrophobic (Pi-Sigma) | 3.98 |
| UNK1:C - ARG41 | Hydrophobic (Alkyl) | 3.95 |
| UNK1:C - MET171 | Hydrophobic (Alkyl) | 4.16 |
| UNK1:C - LEU42 | Hydrophobic (Alkyl) | 4.98 |
| UNK1:C - MET171 | Hydrophobic (Alkyl) | 5.19 |
| UNK1:C - PRO4 | Hydrophobic (Alkyl) | 4.05 |
| TRP3 - UNK1 | Hydrophobic (Pi-Alkyl) | 5.13 |
| TRP3 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.00 |
| TRP3 - UNK1 | Hydrophobic (Pi-Alkyl) | 4.05 |
| TRP3 - UNK1 | Hydrophobic (Pi-Alkyl) | 4.10 |
| TRP3 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.25 |
| PHE88 - UNK1:C | Hydrophobic (Pi-Alkyl) | 5.17 |
| TYR89 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.85 |
| TYR89 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.83 |
| HIS160 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.39 |
| HIS160 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.42 |
| Psoralen | ||
| LYS32:HZ3 - UNK1: O | Hydrogen Bond | 2.30 |
| ARG41: HE - UNK1:O | Hydrogen Bond | 2.48 |
| ARG41:HH22 - UNK1: O | Hydrogen Bond | 2.34 |
| HIS160:NE2 - UNK1 | Electrostatic (Pi-Cation) | 4.94 |
| HIS160 - UNK1 | Hydrophobic (Pi-Pi T-shaped) | 5.39 |
| UNK1 - ALA86 | Hydrophobic (Pi-Alkyl) | 5.17 |
| UNK1 - MET171 | Hydrophobic (Pi-Alkyl) | 5.22 |
| UNK1 - PRO5 | Hydrophobic (Pi-Alkyl) | 5.39 |
| UNK1 - ALA86 | Hydrophobic (Pi-Alkyl) | 4.76 |
| UNK1 - ALA86 | Hydrophobic (Pi-Alkyl) | 4.44 |
| Embelin | ||
| LYS32:HZ1 - UNK1: O | Hydrogen Bond | 2.70 |
| HIS160:HE2 - UNK1: O | Hydrogen Bond | 2.77 |
| HIS160:CE1 - UNK1: O | Carbon Hydrogen Bond | 3.34 |
| UNK1:C - PRO5 | Hydrophobic (Alkyl) | 4.25 |
| TRP3 - UNK1:C | Hydrophobic (Pi-Alkyl) | 4.73 |
| UNK1 - ARG41 | Hydrophobic (Pi-Alkyl) | 4.63 |
| UNK1 - ALA86 | Hydrophobic (Pi-Alkyl) | 5.18 |
| Menisdaurin | ||
| MET250:HN - UNK1:O | Hydrogen Bond | 2.85 |
| GLN267:HE21 - UNK1:O | Hydrogen Bond | 2.10 |
| UNK1:C - ASN248:O | Carbon Hydrogen Bond | 3.55 |
| LYS241 - UNK1 | Hydrophobic (Alkyl) | 4.52 |
| Baccatin III | ||
| ASN33:HD21 - UNK1:O | Hydrogen Bond | 2.33 |
| ASN36:HD21 - UNK1:O | Hydrogen Bond | 2.40 |
| UNK1:H - ASN248:O | Hydrogen Bond | 2.75 |
| LYS241:CE - UNK1:O | Carbon Hydrogen Bond | 3.62 |
| NK1:C - ASP83:OD2 | Carbon Hydrogen Bond | 3.38 |
| UNK1:C - MET250 | Hydrophobic (Alkyl) | 4.81 |
Standard/positive control.
3.7.1. Lamivudine-Pol interaction
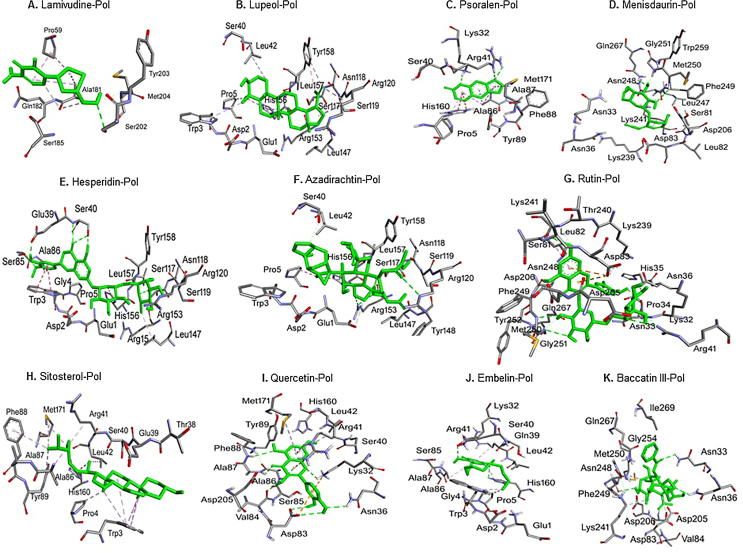
Lamivudine (Fig. 7A) is the first generation nucleot(s)ide-based anti-retroviral drug generally used in the treatment of HIV/AIDS and CHB. Docking of lamivudine (as control) to HBV Pol indicated that it was bound at the active site mainly through hydrophobic (alkyl) interactions. It interacted strongly with HBV Pol by forming four hydrophobic interactions with Pro59 and Ala181 (Fig. 7A). It also formed one conventional hydrogen bond with Ser202 and two carbon-hydrogen bonds with Ala181. Other residues that surrounded lamivudine were Gln182 and Ser185 as well as Tyr203 and Met204 of the ‘YMDD’ motif. Molecular docking study revealed that lamivudine-HBV Pol complex was stabilized by an estimated free energy of −5.2 kcal/mol, corresponding to the binding affinity of 6.5 × 103/mol (Table 2).
Fig. 7.
In silico molecular docking analysis, showing strong interactions of HBV Pol active site residues with lamivudine, lupeol, psoralen, menisdaurin, hesperidin, azadirachtin, rutin, β-sitosterol, quercetin, embelin, and baccatin III.
Table 2.
Molecular docking parameters of the interacting plant-derived compounds with HBV Pol.
| Compounds | Chemical class | ΔG^ (kcal/mol) | Kd^^ (/mol) |
|---|---|---|---|
| Lamivudine* | Nucleoside analog | −5.2 | 6.5 × 103 |
| Quercetin | Flavanoid | −7.4 | 2.7 × 105 |
| Rutin | Flavanoid | −9.2 | 5.6 × 106 |
| Hesperidin | Flavanoid | −9.3 | 6.6 × 106 |
| Lupeol | Triterpenoid | −8.1 | 8.7 × 105 |
| Azadirachtin | Terpenoid | −8.1 | 8.7 × 105 |
| β-Sitosterol | Phytosterol | −8.3 | 1.2 × 106 |
| Psoralen | Furocaumarin | −6.4 | 4.9 × 104 |
| Embelin | Benzoquinone | −6.0 | 2.5 × 104 |
| Menisdaurin | Cyanoglycoside | −6.5 | 8.9 × 104 |
| Baccatin III | Taxol | −7.0 | 1.4 × 105 |
Standard/positive control.
Binding free energy.
Binding affinity.
3.7.2. Lupeol-Pol interaction
Lupeol (Fig. 1B) interacted with HBV Pol by forming four alkyl hydrophobic interactions with Pro5, Leu42 and Arg120, and six Pi-alkyl hydrophobic interactions with Trp3, His156 and Tyr158 (Fig. 7B). Other residues that surrounded lupeol were Glu1, Asp2, Ser40, Ser117, Asn118, Ser119 and Leu157. The Gibb's free energy of lupeol-Pol interaction was predicted to be −8.1 kcal/mol which corresponded to a binding constant of 8.7 × 105/mol (Table 2).
3.7.3. Psoralen-Pol interaction
Psoralen (Fig. 1C) interacted with HBV Pol by forming one hydrogen bond with Lys32 and two hydrogen bonds with Arg41. It also formed one Pi-cation electrostatic interaction with His160. Notably, His160 was also involved in forming one Pi-Pi T-shaped hydrophobic interaction with Psoralen. Moreover, the complex of psoralen and HBV Pol was stabilized by one Pi-alkyl hydrophobic interactions with Pro5 and Met171 while Ala86 formed three Pi-alkyl hydrophobic interactions (Fig. 7C). Other residues that surrounded psoralen were Ser40, Tyr89, Phe88 and Ala87. The Gibb's free energy of psoralen-Pol interaction was predicted to be −6.4 kcal/mol which corresponded to a binding constant of 4.9 × 104/mol (Table 2).
3.7.4. Menisdaurin-Pol interaction
Menisdaurin (Fig. 1D) interacted with HBV Pol by forming three hydrogen bonds with Asn248, Met250 and Gln267. It also formed one alkyl hydrophobic interaction with Lys241 (Fig. 7D). Other residues that surrounded menisdaurin were Asn33, Asn36, Ser81, Leu82, Asp83, Asp206 of YMDD motif, Lys239, Lys241, Leu247, Phe249, Trp259, Gly251. The Gibb's free energy of menisdaurin-Pol interaction was predicted to be −6.5 kcal/mol which corresponded to a binding constant of 8.9 × 104/mol (Table 2).
3.7.5. Hesperidin-Pol interaction
Hesperidin (Fig. 1E) interacted with HBV Pol by forming seven hydrogen bonds with Glu39, Ser40, Ser85 and Ser117, and one carbon hydrogen bond with His156 (Fig. 7E). It also formed two Pi-Pi stacked hydrophobic interactions with Trp3, one alkyl hydrophobic interaction with Leu147 and one Pi-alkyl hydrophobic interaction with Ala86. Other residues that surrounded hesperidin were Glu1, Asp2, Gly4, Pro5, Arg15, Ala86, Asn118, Ser119, Arg120, Leu147, Arg153, His156, Leu157 and Tyr158. The Gibb's free energy of hesperidin-HBV Pol interaction was predicted to be −9.3 kcal/mol, corresponding to a binding constant of 6.6 × 106/mol (Table 2).
3.7.6. Azadirachtin-Pol interaction
Azadirachtin (Fig. 1F) was bound at the active site of HBV Pol through four hydrogen bonds with Ser117, Arg120 andArg153 (Fig. 7F). It also forms one carbon hydrogen bond with Glu1. Moreover, the complex between azadiractin and HBV Pol was further stabilized by three hydrophobic interactions with Pro5 and His156. Other residues that surrounded azadiractin were Asp2, Trp3, Ser40, Leu42, Asn118, Ser119, Leu147, Leu157 and Tyr158. The Gibb's free energy of azadirachtin-Pol interaction was found to be −8.1 kcal/mol, corresponding to a binding constant of 8.7 × 105/mol (Table 2).
3.7.7. Rutin-Pol interaction
Rutin (Fig. 1G) also binds at the active site of HBV Pol by forming seven hydrogen bonds with Asn33, Asn36, Arg41, Ser81, Asp83 and Asn248 (Fig. 7G). It also forms two electrostatic interactions (Pi-anion) with Asp83 and one hydrophobic interaction (Pi-alkyl) with Lys241. Other residues that surrounded rutin were Lys32, Pro34, His35, Leu82, Lys239, Thr240, Asn248, Phe249, Met250, Gly251, Tyr252 and Gln267. Notably, rutin interacted with Asp205 and Asp206 of YMDD motif of HBV Pol. The Gibb's free energy of rutin-Pol interaction was predicted to be −9.2 kcal/mol that corresponded to a binding constant of 5.6 × 106/mol (Table 2).
3.7.8. β-Sitosterol-Pol interaction
β-Sitosterol (Fig. 1H) interacted with HBV Pol by forming one Pi-sigma hydrophobic interaction with Trp3, five alkyl hydrophobic interactions with Pro4, Arg41, Leu42 and Met171, and ten Pi-alkyl hydrophobic interactions with Trp3, Phe88, Tyr89 and His160 (Fig. 7H). Other residues that surrounded β-sitosterol were Thr38, Glu39, Ser40, Ala86 and Ala87. The Gibb's free energy of β-sitosterol-Pol interaction was found to be −8.3 kcal/mol that corresponded to a binding constant of 1.2 × 106/mol (Table 2).
3.7.9. Quercetin-Pol interaction
Quercetin (Fig. 1I) binds at the active site of HBV Pol by forming nine hydrogen bonds with Lys32, Asn36, Arg41, Ala86, Ala87, Phe88, Asp83 and Val84 (Fig. 7I). It also forms two Pi-cation (with Lys32 and Arg41) and one Pi-anion electrostatic interactions (with Asp83). Further, it forms two hydrophobic interactions (Pi-alkyl) with Arg41 and Met171. Other residues that surrounded quercetin were Ser40, Leu42, Val84, Ser85, Tyr89, His160 and Asp205 of YMDD motif. The Gibb's free energy of quercetin-Pol interaction was predicted to be −7.4 kcal/molwhich corresponded to a binding constant of 2.7 × 106/mol (Table 2).
3.7.10. Embelin-Pol interaction
Embelin (Fig. 1J) interacted with HBV Pol by forming three hydrogen bonds with Lys32 and His160. It also formed one alkyl hydrophobic interaction with Pro5 and three Pi-alkyl hydrophobic interactions with Trp3, Arg41 and Ala86 (Fig. 7J). Other residues that surrounded embelin were Glu1, Asp2, Gly4, Gln39, Ser40, Leu42, Ser85 and Ala87. The Gibb's free energy of embelin-Pol interaction was predicted to be −6.1 kcal/mol which corresponded to a binding constant of 2.4 × 104/mol (Table 2).
3.7.11. Baccatin III-Pol interaction
Baccatin III (Fig. 1K) interacted with HBV Pol by forming three hydrogen bonds with Asn33, Asn36 and Asn248. It also formed two carbon hydrogen bonds with Asp83 and Lys241 in addition to one alkyl hydrophobic interaction with Met250 (Fig. 7K). Other residues that surrounded baccatin III were Val84, Phe249, Gly254, Gln267 and Ile269. Interestingly, Baccatin III also interacted with Asp205 and Asp206 of YMDD motif of HBV Pol. The Gibb's free energy of baccatin III-Pol interaction was predicted to be −7.0 kcal/mol which corresponded to a binding constant of 1.4 × 105/mol (Table 2).
4. Discussion
During the past decades, antiviral drug research technology has helped establish many efficacious anti-HBV drugs that eventually lead to emergence of drug-resistant strains. Hence, new antiviral strategies must focus not only on synthetic molecules but also on potential natural or plant products. Antiviral drug discovery and development passes through several stages, of which the in vitro cell culture systems and in silico molecular modeling represent first and fastest drug screening steps. Absence of a robust cell culture system for the propagation of HBV impeded the in vitro antiviral studies until the advent of HBV DNA cloning and establishment of stable cell lines. The HBV-reporter cell line, HepG2.2.15 is one of the established systems for preliminary screening of anti-HBV drugs. In the present study, the anti-HBV potential of specifically selected natural compounds was assessed by their inhibitory effects on HBsAg and HBeAg expressions in the HepG2.2.15 culture supernatants. Of these, quercetin, psoralen, baccatin III, menisdaurin, embelin and azadirachtin showed the high activities whereas lupeol, rutin, β-sitosterol and hesperidin had moderate efficacies against HBV replication. HBV Pol is the most important viral protein as potential drug-target despite emergence of drug-resistant mutations in and around the Pol/RT ‘YMDD’ motif (Locarnini et al., 2008). We used homology modeling to generate a 3-D structure of HBV Pol and validated it by Ramachandran plot wherein the ‘YMDD’ corresponded to the Tyr203, Met204, Asp205 and Asp206 residues. To further delineate the binding mechanism of the anti-HBV active compounds and lamivudine (ligands) to the modeled HBV Pol (receptor), we performed molecular docking. Docking of lamivudine indicated that it strongly interacted with the active site of HBV Pol through bonding with Pro59, Ala181, Ser202 and Ala181, forming a stable lamivudine-Pol complex with an estimated Gibb’s free energy of −5.2 kcal/mol. Interestingly therein, the two Pol ‘YMDD’ motif residues, Tyr203 and Met204 were among the lamivudine surrounding amino acids.
Quercetin, a widely distributed plant flavonol is reported to have in vitro antiviral activities against meningovirus, HSV, HIV, parainfluenza type 3, pseudorabies virus, respiratory syncytial virus (RSV) and Sindbis virus (Kaul et al., 1985, Mucsi, 1984, Vrijsen et al., 1998, Choi et al., 2009, Kelly, 2011, Chiow et al., 2016). Though a single study has shown its anti-HBV potential in vitro (Cheng et al., 2015), the mechanism of action is not reported. In line with this, our in vitro data confirmed the high anti-HBV potential of quercetin. The docking of quercetin with HBV Pol revealed formation of bonds with Lys32, Asn36, Arg41, Ala86, Ala87, Phe88, Asp83 and Val84 that stabilized the quercetin-pol complex with an estimated free energy of −7.4 kcal/mol. Notably therein, Asp205 belonging to the Pol ‘YMDD’ motif was among the quercetin surrounding residues.
Psoralen is a natural caumarin that has been recently reported to inhibit Epstein–Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) replication (Cho et al., 2013). To add further, our data presented the novel antiviral potential of psoralen against HBV replication. Molecular docking showed the interaction of psoralen with HBV Pol through Lys32, Arg41 and His160 where the stable psoralen-Pol complex had a free energy of −6.4 kcal/mol.
Baccatin III is isolated from the needles of the Pacific yew tree, and synthetically available as 10-deacetylbaccatin III. Baccatin III is widely assumed to be an inactive derivative of taxol, isolated from Taxus brevifolia bark. Though in a single in vitro study, Baccarin III has shown anti-HSV activity at non-cytotoxic dose (Krawczyk et al., 2005); its mechanism of action is not known. In conformity, our results, for the first time, showed a high anti-HBV activity of baccatin III. Docking of baccatin III with HBV pol showed the interaction through Asn33, Asn36, Asp83, Lys241, Asn248 and Met250 residues that formed the baccatin-Pol complex with a free energy of −7.0 kcal/mol. Notably, Asp205 and Asp206 of the ‘YMDD’ motif was among the baccatin III surrounding amino acids.
Menisdaurin, a plant cyanoglycoside has very limited information on its antiviral activities. The two reports on anti-HBV activity of menisdaurin isolated from Saniculiphyllum guangxiense (Geng et al., 2012) and mangrove Bruguiera gymnorrhiza (Yi et al., 2015) are recently published. In line with this, our data endorsed its in vitro anti-HBV potential. Contrarily however, in our study menisdaurin’s activity was lesser as compared to its reported higher activity to lamivudine (Yi et al., 2015). In the docking analysis, menisdaurin-HBV Pol interaction was through Lys241, Asn248, Met250 and Gln267 that predicted the free energy of −6.5 kcal/mol. Notably therein, one of the Pol ‘YMDD’ residues, Asp206 was among the menisdaurin surrounding amino acids.
The active principle of Embelia schimperi, embelin is shown to inhibit hepatitis C virus (HCV) serine protease activity (Hussein et al., 2000). In this report, our in vitro assessment has revealed the novel antiviral property of embelin against HBV. In molecular docking, embelin formed complex with HBV Pol through Trp3, Pro5, Lys32 and His160 with an estimated free energy −6.1 kcal/mol.
Azadirachtin is the secondary metabolite of Indian neem (Azadirachta indica). In a single report, while the neem leaves extract showed inhibition of dengue virus (DNV) replication, the pure azadirachtin did not reveal any inhibitory effect in vitro and in vivo (Parida et al., 2002). In this report we demonstrated the novel anti-HBV activity of azadirachtin. Our docking study showed interaction of azadiractin with HBV Pol through Glu1, Pro5, His156, Ser117, Arg120 and Arg153, resulting in a stable azadiractin-Pol complex with a free energy of −8.1 kcal/mol.
Though the triterpenoid lupeol has shown weak antiviral activities in several studies, lupeol isolated from Strobilanthes cusia root has demonstrated a robust anti-HSV activity in vitro (Flekhter et al., 2004). It has therefore, served as a lead drug for the generation of more effective compounds against HSV and Influenza A (Zheng et al., 2009). In line with its antiviral activities, we showed the moderate anti-HBV potential of lupeol in vitro. In our in silico analysis, lupeol interacted with HBV Pol through Trp3, Pro5, Leu42 and Arg120, His156 and Tyr158, and stabilized the lupeol-Pol complex with a free energy −8.1 kcal/mol.
Rutin, a well-known therapeutic flavonol has been reported for in vitro antiviral activities against avian influenza virus (H5N1) and murine norovirus (MNV) (Carvalho et al., 2013, Ibrahim et al., 2013, Chéron et al., 2015). Very recently, we have demonstrated the anti-HBV activity of G. senegalensis leaves extract with high content of rutin (Arbab et al., 2017, Alam et al., 2017). Though moderate, the anti-HBV activity of rutin in the present study strongly supported our previous finding. When subjected to docking, rutin interacted with HBV Pol by forming bonds with Asn33, Asn36, Arg41, Ser81, Asp83 and Asn248 residues where the rutin-pol complex showed a free energy of −9.2 kcal/mol. Notably therein, Asp 205 and Asp206, the ‘YMDD’ residues were among the rutin surrounding amino acids.
Plant β-sitosterol has been shown to exhibit in vitro and in vivo anti-HIV activity (Bouic, 1997). Recently, antiviral effect of β-sitosterol isolated from cottonseed oil against tobacco mosaic virus (TMV) is also reported (Zhao et al., 2015). This is the first report showing the in vitro anti-HBV activity of β-sitosterol. This was further supported by our in silico analysis where β-sitosterol interacted with HBV Pol through Trp3, Pro4, Arg41, Leu42, Phe88, Tyr89, His160 and Met171 where the free energy of β-sitosterol-Pol complex was −8.3 kcal/mol.
Hespiridin is a flavonoid with very limited knowledge of antiviral activities. A water-soluble derivative of hesperidin, glucosyl hesperidin has been reported to have in vitro inhibitory activities against human and avian influenza A viruses (Saha et al., 2009). Our present data demonstrated the novel anti-HBV potential of hesperidin. In molecular docking, hesperidin formed bonds with Glu39, Ser40, Ser85, Ser117 and His156 where the hesperidin-Pol complex had a free energy of −9.3 kcal/mol.
5. Conclusion
In the present study, our in vitro anti-HBV assessment of plant-derived non-cytotoxic compounds showed promising inhibitory effects on HBsAg and HBeAg expressions. Of these, quercetin, baccatin III, psoralen, embelin, menisdaurin and azadirachtin showed high anti-HBV activity whereas lupeol, rutin, β-sitosterol and hesperidin had moderate effects. Moreover, a combination of quercetin with highly active compounds enhanced the therapeutic efficacy. This was further supported by in silico molecular docking analysis where all tested anti-HBV molecules strongly interacted with the modeled HBV Pol active site residues. Taken together, our data suggested the novel anti-HBV potential of these natural compounds as viral pol/RT-inhibitors. Nonetheless, further experimental and pre-clinical studies on the most active compounds are warranted.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
The authors thankfully acknowledge the financial support by the Deanship of Scientific Research, King Saud University, Riyadh (Project No. RG-1435-053).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mohammad K. Parvez, Email: mohkhalid@ksu.edu.sa.
Perwez Alam, Email: aperwez@ksu.edu.sa.
References
- Alam P., Parvez M.K., Arbab A.H., Al-Dosari M.S. Quantitative analysis of rutin, quercetin, naringenin and gallic acid by validated RP- and NP-HPTLC methods for the quality control of anti-HBV active extract of Guiera senegalensis. Phrm. Biol. 2017;55:1317–1323. doi: 10.1080/13880209.2017.1300175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dosari M.S., Parvez M.K. Novel plant inducers of PXR-dependent cytochrome P450 3A4 expression in HepG2 cells. Saudi Pharm. J. 2018;26:1069–1072. doi: 10.1016/j.jsps.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Yousef H.M., Ahmed A.F., Al-Shabib N.A., Laeeq S., Khan R.A., Rehman M.T. Onion peel ethylacetate fraction and its derived constituent quercetin 4'-o-beta-d glucopyranoside attenuates quorum sensing regulated virulence and biofilm formation. Front. Microbiol. 2017;8:1675–1678. doi: 10.3389/fmicb.2017.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbab A.H., Parvez M.K., Al-Dosari M.S., Al-Rehaily A.J. In vitro evaluation of novel antiviral activities of 60 medicinal plants extract against hepatitis B virus. Exp. Ther. Med. 2017;14:626–634. doi: 10.3892/etm.2017.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. The Swiss-Model workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Beck J., Nassal M. Hepatitis B virus replication. World J. Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho O.V., Botelh C.V., Ferreir C.G., Ferreir H.C., Santo M.R., Dia M.A. In vitro inhibition of canine distemper virus by flavonoids and phenolic acids: implications of structural differences for antiviral design. Res. Vet. Sci. 2013;95:717–724. doi: 10.1016/j.rvsc.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Chang J.S., Liu H.-W., Wang K.-C., Chen M.-C., Chiang L.-C. Ethanol extract of polygonum cuspidatum inhibits hepatitis B virus in a stable HBV-producing cell line. Antiviral Res. 2005;66:29–34. doi: 10.1016/j.antiviral.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhu J. Anti-HBV effect of individual traditional chinese herbal medicine in vitro and in vivo: An analytic review. J. Viral Hep. 2013;20:445–452. doi: 10.1111/jvh.12112. [DOI] [PubMed] [Google Scholar]
- Chen H.C., Chou C.K., Lee S.D., Wang J.C., Yeh S.F. Active compounds from saussurea lappa clarks that suppress hepatitis B virus surface antigen gene expression in human hepatoma cells. Antiviral Res. 1995;27:99–109. doi: 10.1016/0166-3542(94)00083-k. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Sun G., Guo W., Huang Y., Sun W., Zhao F., Hu K. Inhibition of hepatitis B virus replication by quercetin in human hepatoma cell lines. Virol. Sin. 2015;30:261–268. doi: 10.1007/s12250-015-3584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chéron N., Yu C., Kolawole A.O., Shakhnovich E.I., Wobus C.E. Repurposing of rutin for the inhibition of norovirus replication. Arch. Virol. 2015;160:2353–2358. doi: 10.1007/s00705-015-2495-y. [DOI] [PubMed] [Google Scholar]
- Choi H.J., Kim J.H., Lee C.H., Ahn Y.J. Antiviral activity of quercetin 7-rhamnoside against porcine epidemic diarrhea virus. Antiviral Res. 2009;81:77–81. doi: 10.1016/j.antiviral.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiow K.H., Phoon M.C., Putti T., Tan B.K., Chow V.T. Evaluation of antiviral activities of Houttuynia cordata Thunb. extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pac. J. Trop. Med. 2016;9:1–7. doi: 10.1016/j.apjtm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-J., Jeong S.-G., Park J.-E., Han J.-A., Kang H.-R., Lee D., Song M.J. Antiviral activity of angelicin against gammaherpesviruses. Antiviral Res. 2013;100:75–83. doi: 10.1016/j.antiviral.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Chou S.C., Huang T.J., Lin E.H., Huang C.H., Chou C.H. Anti-hepatitis B virus constituents from solanum erianthum. Planta Med. 2012;78 1190–1190. [PubMed] [Google Scholar]
- Cunningham A.L., Dwyer D.E., Dowton D.N. Viral markers in HIV infection and AIDS. J. Acquir. Immune Defic. Syndr. 1993;6:S32–S35. [PubMed] [Google Scholar]
- Daga P.R., Duan J.S., Doerksen R.J. Computational model of hepatitis B virus DNA polymerase: Molecular dynamics and docking to understand resistant mutations. Protein Sci. 2010;19:796–807. doi: 10.1002/pro.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Meth. Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Flekhter O.B., Boreko E.I., Nigmatullina L.P., Pavlova N.I., Medvedeva N.I. Synthesis and antiviral activity of lupane triterpenoids and their derivatives. Pharm. Chem. J. 2004;38:355–358. [Google Scholar]
- Geng C.A., Huang X.Y., Lei L.G., Zhang X.M., Chen J.J. Chemical constituents of Saniculiphyllum guangxiense. Chem. Biodivers. 2012;9:1508–1516. doi: 10.1002/cbdv.201100270. [DOI] [PubMed] [Google Scholar]
- Hussein G., Miyashiro H., Nakamura N., Hattori M., Kakiuchi N., Shimotohno K. Inhibitory effects of sudanese medicinal plant extracts on hepatitis C virus (HCV) protease. Phytother. Res. 2000;14:510–516. doi: 10.1002/1099-1573(200011)14:7<510::aid-ptr646>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Hussain W., Haleem K.S., Khan I., Tauseef I., Qayyum S., Ahmed B., Riaz M.N. Medicinal plants: a repository of antiviral metabolites. Fut. Virol. 2017 [Google Scholar]
- Huang H., Chopra R., Verdine G.L., Harrison S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Ibrahim A.K., Youssef A.I., Arafa A.S., Ahmed S.A. Anti-H5N1 virus flavonoids from Capparis sinaica Veill. Nat. Prod. Res. 2013;27:2149–2153. doi: 10.1080/14786419.2013.790027. [DOI] [PubMed] [Google Scholar]
- Krawczyk E., Luczak M., Kniotek M., Nowaczyk M. Cytotoxic, antiviral (in-vitro and in-vivo), immunomodulatory activity and influence on mitotic divisions of three taxol derivatives: 10-deacetyl-baccatin III, methyl (N-benzoyl-(2'R,3'S)-3'-phenylisoserinate) and N-benzoyl-(2'R,3'S)-3'-phenylisoserine. J. Pharm. Pharmacol. 2005;57:791–797. doi: 10.1211/0022357056235. [DOI] [PubMed] [Google Scholar]
- Kelly G.S. Quercetin: monograph. Altern. Med. Rev. 2011;16:172–194. [PubMed] [Google Scholar]
- Kaul T.N., Middleton E., Ogra P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- Kurokawa M., Shimizu T., Watanabe W., Shiraki K. Development of new antiviral agents from natural products. Open Antimicrob. Agents J. 2010;2:49–57. [Google Scholar]
- Leach A. second Ed. Prentice Hall; NJ, USA: 2001. Molecular Modelling: Principles and Applications. [Google Scholar]
- Lok A.S., Zoulim F., Locarnini S., Bartholomeusz A., Ghany M.G., Pawlotsky J.M., Liaw Y.F., Mizokami M., Kuiken C. Hepatitis B Virus Drug Resistance Working Group. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I., Word J.M. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Martin K.W., Ernst E. Antiviral agents from plants and herbs: a systematic review. Antivir. Ther. 2003;8:77–90. [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comp. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucsi I. Combined antiviral effects of flavonoids and 5-ethyl-2′-deoxyuridine on the multiplication of herpesviruses. Acta Virol. 1984;28:395–400. [PubMed] [Google Scholar]
- Parida M.M., Upadhyay C., Pandya G., Jana A.M. Inhibitory potential of neem (Azadirachta indica Juss) leaves on dengue virus type-2 replication. J. Ethnopharmacol. 2002;79:273–278. doi: 10.1016/s0378-8741(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Parvez M.K., Sehgal D., Sarin S.K., Basir S.F., Jameel S. Inhibition of hepatitis B virus DNA replicative intermediate forms by recombinant interferon-gamma. World J. Gastroenterol. 2006;12:3006–3014. doi: 10.3748/wjg.v12.i19.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez M.K., Arab A.H., Al-Dosari M.S., Al-Rehaily A.J. Antiviral natural products against chronic hepatitis B: recent developments. Curr. Pharm. Des. 2016;3:286–293. doi: 10.2174/1381612822666151112152733. [DOI] [PubMed] [Google Scholar]
- Qiu L.P., Chen K.-P. Anti-HBV agents derived from botanical origin. Fitoterapia. 2013;84:140–157. doi: 10.1016/j.fitote.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Rabbani N., Tabrez S., Islam B.U., Rehman M.T., Alsenaidy A.M., Alajmi M.F., Khan R.A., Alsenaidy M.A., Khan M.S. Characterization of colchicine binding with normal and glycated albumin: in-vitro and molecular docking analysis. J. Biomol. Struct. Dyn. 2017 doi: 10.1080/07391102.2017.1389661. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., Ahmed S., Khan A.U. Interaction of meropenem with 'N' and 'B' isoforms of human serum albumin: a spectroscopic and molecular docking study. J. Biomol. Struc. Dyn. 2016;34:1849–1864. doi: 10.1080/07391102.2015.1094411. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., Shamsi H., Khan A.U. Insight into the binding of Imipenem to Human Serum Albumin by spectroscopic and computational approaches. Mol. Pharm. 2014;11:1785–1797. doi: 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- Saha R.K., Takahashi T., Suzuki T. Glucosyl hesperidin prevents Influenza A virus replication in vitro by Inhibition of viral sialidase. Biol. Pharm. Bull. 2009;32:1188–1192. doi: 10.1248/bpb.32.1188. [DOI] [PubMed] [Google Scholar]
- Shafritz D.A., Lieberman H.M. The molecular biology of hepatitis B virus. Annu. Rev. Med. 1984;35:219–232. doi: 10.1146/annurev.me.35.020184.001251. [DOI] [PubMed] [Google Scholar]
- Teo C.G., Locarnini S.A. Potential threat of drug-resistant and vaccine-escape HBV mutants to public health. Antivir. Ther. 2010;15:445–449. doi: 10.3851/IMP1556. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijsen R., Everaert L., Boeye A. Antiviral activity of flavones and potentiation by ascorbate. J. Gen. Virol. 1998;69:1749–1751. doi: 10.1099/0022-1317-69-7-1749. [DOI] [PubMed] [Google Scholar]
- Wang B.E. Treatment of chronic liver diseases with traditional chinese medicine. J. Gastroenterol. Hepatol. 2000;15:E67–E70. doi: 10.1046/j.1440-1746.2000.02100.x. [DOI] [PubMed] [Google Scholar]
- WHO, 2017. Hepatitis: Factsheets. http://www.who.int/mediacentre/factsheets/fs204/en/.
- Wu Y.H. Naturally derived anti-hepatitis B virus agents and their mechanism of action. World J. Gastroenterol. 2016;22:188–204. doi: 10.3748/wjg.v22.i1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X.X., Deng J.G., Gao C.H., Hou X.T., Li F., Wang Z.P. Four new cyclohexylideneacetonitrile derivatives from the hypocotyl of mangrove (Bruguiera gymnorrhiza) Molecules. 2015;20:14565–71455. doi: 10.3390/molecules200814565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.B., Zhang X.L., Chen N.H., Wu Z.N., Ye W.C., Li Y.L., Wang G.C. Four matrine-based alkaloids with antiviral activities against HBV from the seeds of Sophora alopecuroides. Org. Lett. 2017;19:424–427. doi: 10.1021/acs.orglett.6b03685. [DOI] [PubMed] [Google Scholar]
- Zhao L., Feng C., Hou C., Hu L., Wang Q., Wu Y. First discovery of acetone extract from cottonseed oil sludge as a novel antiviral agent against plant viruses. PLoS One. 2015;10:e0117496. doi: 10.1371/journal.pone.0117496. [DOI] [PMC free article] [PubMed] [Google Scholar]