Abstract
Background
Grey matter (GM) atrophy in Alzheimer's disease (AD) is most commonly modeled as a function of time. However, this approach does not take into account inter-individual differences in initial disease severity or changes due to aging. Here, we modeled GM atrophy within individuals across the AD clinical spectrum as a function of time, aging and MMSE, as a proxy for disease severity, and investigated how these models influence estimates of GM atrophy.
Methods
We selected 523 individuals from ADNI (100 preclinical AD, 288 prodromal AD, 135 AD dementia) with abnormal baseline amyloid PET/CSF and ≥1 year of MRI follow-up. We calculated total and 90 regional GM volumes for 2281 MRI scans (median [IQR]; 4 [3–5] scans per individual over 2 [1.6–4] years) and used linear mixed models to investigate atrophy as a function of time, aging and decline on MMSE. Analyses included clinical stage as interaction with the predictor and were corrected for baseline age, sex, education, field strength and total intracranial volume. We repeated analyses for a sample of participants with normal amyloid (n = 387) to assess whether associations were specific for amyloid pathology.
Results
Using time or aging as predictors, amyloid abnormal participants annually declined −1.29 ± 0.08 points and − 0.28 ± 0.03 points respectively on the MMSE and −12.23 ± 0.47 cm3 and −8.87 ± 0.34 respectively in total GM volume (p < .001). For the time and age models atrophy was widespread and preclinical and prodromal AD showed similar atrophy patterns. Comparing prodromal AD to AD dementia, AD dementia showed faster atrophy mostly in temporal lobes as modeled with time, while prodromal AD showed faster atrophy in mostly frontoparietal areas as modeled with age (pFDR < 0.05). Modeling change in GM volume as a function of decline on MMSE, slopes were less steep compared to those based on time and aging (−4.1 ± 0.25 cm3 per MMSE point decline; p < .001) and showed steeper atrophy for prodromal AD compared to preclinical AD in the right inferior temporal gyrus (p < .05) and compared to AD dementia mostly in temporal areas (pFDR < 0.05). Associations with time, aging and MMSE remained when accounting for these effects in the other models, suggesting that all measures explain part of the variance in GM atrophy. Repeating analyses in amyloid normal individuals, effects for time and aging showed similar widespread anatomical patterns, while associations with MMSE were largely reduced.
Conclusion
Effects of time, aging and MMSE all explained variance in GM atrophy slopes within individuals. Associations with MMSE were weaker than those for time or age, but specific for amyloid pathology. This suggests that at least some of the atrophy observed in time or age models may not be specific to AD.
Keywords: Alzheimer's disease, Longitudinal, Atrophy, Aging, Cognition, Amyloid
Abbreviations: AD, Alzheimer's disease; CN, cognitively normal; GM, grey matter; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination
Highlights
-
•
Modeling atrophy as a function of time or aging show similar anatomical patterns.
-
•
GM atrophy as a function of time or aging seems nonspecific for amyloid pathology.
-
•
GM atrophy as a function of MMSE shows involvement of different anatomical patterns.
-
•
Atrophy modeled based on time or age was steeper than modeled based on MMSE.
-
•
Atrophy patterns based on MMSE were specific for amyloid pathology.
1. Introduction
Alzheimer's disease (AD) is a neurodegenerative disorder and the most common cause of dementia (Lobo et al., 2000; Plassman et al., 2007). The disease presumably starts with the aggregation of amyloid beta, after which it can take up to 15 years for the dementia syndrome to manifest (Bateman et al., 2012; Jansen et al., 2015). Because amyloid becomes abnormal very early in the disease when cognition is still normal, it is difficult to estimate when the disease has started and where individuals are in their disease trajectory. As such, when modeling disease progression solely based on time from first assessment, individuals' positions in their disease trajectory are not taken into account. Models that account for disease severity might more accurately estimate disease specific outcome measures within individuals, such as grey matter (GM) atrophy rates.
GM atrophy is thought of as a close biological substrate of decline in cognitive functioning (Jack Jr. et al., 2010; Terry et al., 1991). Still, most studies model changes in GM atrophy and cognitive decline separately, and not within individuals. Although changes over time in both variables are correlated with each other (Jack Jr. et al., 2009; McDonald et al., 2012; Sluimer et al., 2008), these associations do not account for inter-individual differences in disease severity. Modeling change in both GM atrophy and cognition within individuals, Jack Jr. et al. (2012) used Mini-Mental State Examination (MMSE) at baseline as a proxy of disease severity to align individuals according to their initial disease stage and investigated the associations of rates of hippocampal volume and annualized cognitive decline in the MMSE. Results were in line with previous findings of increasing hippocampal atrophy with advancing disease stages, supporting tests of cognitive performance as a continuous approximation for disease severity. Other brain areas in addition to the hippocampus are, however, likely related to disease progression (ten Kate et al., 2017; Vemuri et al., 2011; Zeifman et al., 2015). Another complicating factor is that GM volume declines with age (Bakkour et al., 2013; Fjell et al., 2014; Good et al., 2001; Lee et al., 2018). It remains unclear, however, whether atrophy in more widespread cortical areas is related to decline in MMSE within individuals, and to which extent such associations are specific for AD and influenced by effects of time and/or aging.
In this study we therefore analyzed longitudinal changes in GM volumes using MMSE as an anchor point for individual disease severity and investigated how these associations differed from those using follow-up time or age in individuals with abnormal amyloid across the clinical spectrum. We further studied whether such associations were specific for distinct anatomical regions and for amyloid pathology.
2. Material and methods
2.1. Participants
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). ADNI was approved by the institutional review board of all participating institutions and written informed consent was obtained from all participants at each site.
Details of clinical diagnoses have been previously described elsewhere (Petersen et al., 2010; for a general procedures manual see https://adni.loni.usc.edu/wp-content/uploads/2010/09/ADNI_GeneralProceduresManual.pdf). Briefly, classification into the clinical stages, cognitively normal (CN), and MCI was based on screening with the CDR, MMSE and delayed recall on Logical Memory II subscale of the Wechsler Memory Scale-Revised (adjusted for education). Additionally, MCI and dementia participants had to have a memory complaint. Dementia patients had to have a clinical diagnosis of probable AD according to the NINCDS-ADRDA criteria (McKhann et al., 1984).
We selected participants across the different clinical stages of the AD continuum (preclinical AD, prodromal AD, AD dementia) from ADNI-1/GO/-2 with abnormal amyloid biomarkers at baseline (PiB, AV45-PET or amyloid β 1–42 CSF) and ≥0.9 years of repeated MRI available. A total of 523 patients met inclusion criteria and had MRI scans of sufficient quality available.
We used repeated MMSE as a measure for general cognitive functioning to align individuals according to their disease severity. Over a median follow-up time of 2 years (interquartile range (IQR): 1.6–4), a total of 2281 MMSE assessments were available with a median number of neuropsychological follow-up of 4 (IQR: 3–5).
Additionally, we repeated analyses in a sample of individuals that fulfilled the same inclusion criteria but with normal amyloid markers at baseline (n = 387). For these individuals, a total of 1710 MMSE assessments were available (median (IQR) follow-up time: 2.6 (2–4); median (IQR) number of assessments: 4 (3.8–5)).
2.2. Image acquisition and pre-processing
Image acquisition details and initial preprocessing are described elsewhere (http://adni.loni.usc.edu/methods/mri-analysis/;Jack Jr. et al., 2008). All 3D T1-weighted structural scans available for the selected participants were downloaded from the ADNI LONI Image & Data Archive (IDA) [date of last access: 29.03.2017]. Scans that were preprocessed with gradwarping, B1 correction and/or N3 scaling were downloaded for the present analyses. If available and of sufficient quality, we chose the original sequence over the repeated scan acquisition and the scans with the most preprocessing steps for each acquisition date. In total, 2590 scans met inclusion criteria (n = 1988 scans for individuals with normal baseline amyloid). Within individuals, scans were included when they had the same field strength. First, all images were reoriented with FSL (v5.0.6). Next, a subject-specific median template image from all follow-up scans was created with Freesurfer (v5.3.0) to reduce bias in longitudinal registration (Reuter et al., 2012). Further preprocessing was performed in SPM12. For each subject, scans were co-registered to the subject-specific median template image, and then segmented into GM, white matter and cerebrospinal fluid with the Markov Random Fields (MRF) parameter set to 2 and default settings for all other parameters. Next, the automated anatomical labeling atlas (AAL; Tzourio-Mazoyer et al., 2002) was warped from standard space to subject space using the subject specific inversed normalization parameters. For each of the 90 cortical and subcortical AAL areas we calculated regional GM volumes. Total intracranial volume (TIV) was computed as the sum of GM, white matter and cerebrospinal fluid volumes in cm3. All GM segmentations and subject-specific atlases were visually inspected for quality.
2.3. PET/CSF analysis
Participants were classified as having normal or abnormal amyloid based on amyloid PET results (for processing details see http://adni.loni.usc.edu/methods/pet-analysis/;Jagust et al., 2010; Jagust et al., 2015), and when PET was unavailable, on CSF amyloid β 1–42 results (Shaw et al., 2009). The threshold for abnormal amyloid was for PiB SUVRs >1.5 (available for n = 5, 0.6%), for AV45 SUVR >1.1 (available for n = 631, 69%) (both SUVR calculated in reference to the cerebellum; Jagust et al., 2010; Jagust et al., 2015) and for CSF amyloid β 1–42 levels <192 pg/ml (available for n = 850, 94%) (Shaw et al., 2009).
2.4. Statistical analyses
Comparisons of baseline characteristics between baseline clinical stages (preclinical AD, prodromal AD, AD dementia) were performed with one-way ANOVA, Kruskal tests or chi-squared tests when appropriate. If significant differences were found, we performed post-hoc comparisons with Tukey's tests or Dunn's tests with p values adjusted for multiple comparisons with the Hochberg procedure.
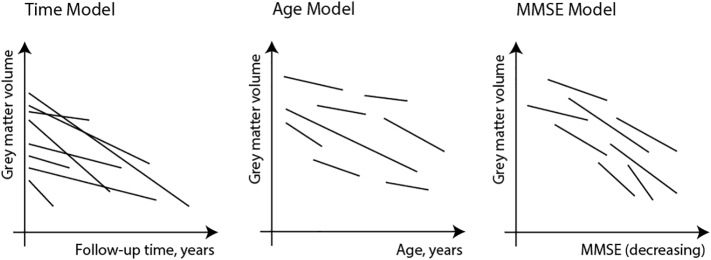
We tested three models taking time, age or MMSE as predictor for change in GM volume (outcome) with linear mixed effects using the R package ‘lme4’ (Bates et al., 2015) (see also Fig. 1): Model 1 included time as the predictor and subject-specific random intercepts and random slopes for time:
Fig. 1.
Hypothesized difference between taking time, age versus MMSE as a measure for disease progression. For the time model, we included follow-up time (in years) as the predictor, a random intercept for subjects and subject-specific random slopes for follow-up time. For the age model, we included age as the predictor, a random intercept for subjects and subject-specific random slopes for age. For the MMSE model, we included MMSE as a predictor, a random intercept for subjects and subject-specific random slopes for MMSE. We additionally included an interaction effect with baseline clinical stage (i.e., CN, MCI, dementia) to estimate cross-sectional and longitudinal differences between the different clinical stages. Models were corrected for age at baseline (time and MMSE model), sex, education and field strength.
Model 2 included age (longitudinal, i.e., baseline age + follow-up time in years) as the predictor and subject-specific random intercepts and random slopes for age:
Model 3 included MMSE as the predictor and subject-specific random intercepts and random slopes for MMSE:
Analyses were adjusted for the potential influence of age at baseline C1 (model 1 and model 3), gender C2, education C3, field strength C4 and baseline total intracranial volume C5. Because it is conceivable that baseline clinical stage (i.e., CN, MCI, AD dementia) may influence the results and in order to assess potential non-linear effects for MMSE, we repeated the models with an additional interaction term for time × diagnosis, age × diagnosis and MMSE × diagnosis, respectively. We only considered paired MRI and MMSE data for these analyses (n = 2281 observations for individuals with abnormal amyloid, n = 1710 observations for individuals with normal amyloid). For all models we used an unstructured covariance matrix. Clinical stage differences in estimated marginal means and trends are reported with the emmeans package (Lenth, 2018). Raw scores of MMSE and GM volumes were used to aid interpretation. Models were repeated for all 90 AAL regions. In order to standardize effects across the different regions, local volumes were normalized to the mean values of cognitively normal individuals with normal amyloid at baseline. Type III analysis of variance with Satterthwaite's method were used to estimate main effects. For each model, local analyses were corrected for multiple testing with the false discovery rate (FDR) procedure (Benjamini and Hochberg, 1995). To investigate whether longitudinal changes in MMSE or grey matter volumes were different depending on amyloid abnormality we refit all models for the total sample including baseline amyloid status as a main term and interaction effect with the respective predictor. Analyses were then repeated for individuals with normal amyloid markers to investigate whether observed effects were specific for the presence of amyloid pathology. We also investigated whether differences in field strength impacted our results and repeated all main models stratified by field strength. Finally, we performed model fit comparisons for non-nested linear mixed models based on differences in AIC and the likelihood ratio test (Vuong, 1989) with the package ‘nonnest2’. R2 for fixed and random effects were estimated with the ‘MuMIn’ package (Johnson, 2014; Nakagawa and Schielzeth, 2013). Statistical analyses were performed in R (version 3.4.4, 2018-03-15) and Surf Ice (version 2017-08-08) was used to visualize regional results.
3. Results
3.1. Sample description
In total 523 individuals with abnormal amyloid were included: 100 preclinical AD participants, 288 individuals with prodromal AD and 135 patients with AD-type dementia (Table 1). Participants were on average 74 ± 7 years old and 48% were female.
Table 1.
Demographic and clinical characteristics of the included sample.
| Total group (n = 523) | Preclinical AD (n = 100, 19%) | Prodromal AD (n = 288, 55%) | AD dementia (n = 135, 26%) | Pairwise comparisons | |
|---|---|---|---|---|---|
| Female | 253 (48%) | 64 (64%) | 124 (43%) | 65 (48%) | CN > MCI; CN > AD |
| Age, years | 73.83 (6.84) | 75.44 (5.57) | 73.19 (6.91) | 73.99 (7.37) | CN > MCI |
| Education, years | 16 (14–18) | 16 (14–18) | 16 (14–18) | 16 (13–18) | n.s. |
| Progression to MCI | 19 (4%) | 19 (19%) | n.a. | n.a. | n.a. |
| Progression to dementia | 154 (29%) | 7 (7%) | 147 (51%) | n.a. | n.a. |
| APOE4 allele (0/≥1) | 177 (34%)/346 (66%) | 53 (53%)/47 (47%) | 96 (33%)/192 (67%) | 28 (21%)/107 (79%) | CN < MCI; CN < AD; MCI < AD c |
| AV45 PET SUVR a | 1.39 (0.17) | 1.34 (0.18) | 1.38 (0.16) | 1.45 (0.16) | CN < AD; MCI < AD |
| Abnormal AV45 PET >1.11 SUVR a | 333 (100%) | 70 (100%) | 189 (100%) | 74 (100%) | n.a. |
| CSF Aβ1–42, pg/ml b | 139.78 (28.66) | 153.33 (35.36) | 139.95 (27.17) | 129.34 (21.28) | CN > MCI; CN > AD; MCI > AD |
| Abnormal Aβ1–42 < 192 pg/ml b | 469 (95%) | 81 (87%) | 263 (96%) | 125 (100%) | CN < MCI; CN < AD; MCI < AD |
| CSF total tau, pg/ml b | 110.17 (55.31) | 77.71 (36.49) | 110.47 (52.7) | 133.65 (60.54) | CN < MCI; CN < AD; MCI < AD |
| Abnormal total tau >93 pg/ml b | 273 (55%) | 27 (29%) | 153 (56%) | 93 (74%) | CN < MCI; CN < AD; MCI < AD |
| Follow up time, years (ADNI) | 2 (1.6–4) | 2.1 (2–4) | 3 (2–4) | 1.5 (1–2) | CN > AD; MCI > AD |
| Number of assessments (MRI and MMSE) | 4 (3–5) | 4 (3–5) | 5 (4–5.8) | 3 (3–4) | CN < MCI; CN > AD; MCI > AD |
| Field strength (3 T) | 1509 (65%) | 260 (61%) | 819 (59%) | 256 (55%) | n.s. |
| Total intracranial volume, cm3 | 1451.67 (148.18) | 1435.76 (140.14) | 1463.19 (145.29) | 1438.89 (158.68) | n.s. |
| Grey matter volume, cm3 | 582.91 (73.39) | 597.74 (66.4) | 593.53 (71.32) | 549.24 (72.67) | CN > AD; MCI > AD |
| Hippocampal volume, cm3 | 7.53 (1.42) | 8.36 (0.96) | 7.71 (1.3) | 6.54 (1.42) | CN > MCI; CN > AD; MCI > AD |
Data are presented as N (%), mean (SD), or median (Q1-Q3), where appropriate. n.a., not applicable; n.s. not significant after adjustment for multiple comparisons.
Available for n = 333.
Available for n = 493.
For ApoE ε4 carriers.
At baseline, AD-type dementia patients performed worst on the MMSE and had the lowest GM and hippocampal volumes, followed by prodromal AD and then preclinical AD (Table 2). At a regional level, AD dementia patients had the lowest volumes in the medial temporal lobes, followed by prodromal AD and then preclinical AD participants (see Inline Supplementary Fig. 1a).
Table 2.
Cross-sectional and longitudinal estimates for MMSE, grey matter and hippocampal volumes over follow-up time.
| Longitudinal estimates |
Cross-sectional estimates |
Cross-sectional differences |
Longitudinal estimates |
Longitudinal differences |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total group | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | |
| MMSE | −1.29 ± 0.08⁎⁎⁎ | 28.75 ± 0.3⁎⁎⁎ | 25.92 ± 0.18⁎⁎⁎ | 20.19 ± 0.27⁎⁎⁎ | −2.82⁎⁎⁎ | −5.74⁎⁎⁎ | −0.27 ± 0.16 | −1.08 ± 0.1⁎⁎⁎ | −2.38 ± 0.18⁎⁎⁎ | −0.8⁎⁎⁎ | −1.3⁎⁎⁎ |
| [−16.17] | [95.09] | [146.55] | [73.78] | [−1.71] | [−11.33] | [−13.46] | |||||
| Grey matter volume in cm3 | −12.23 ± 0.47⁎⁎⁎ | 596.63 ± 5.43⁎⁎⁎ | 570.16 ± 3.21⁎⁎⁎ | 530.72 ± 4.6⁎⁎⁎ | −26.47⁎⁎⁎ | −39.44⁎⁎⁎ | −6.99 ± 0.95⁎⁎⁎ | −12.52 ± 0.57⁎⁎⁎ | −16.53 ± 1.15⁎⁎⁎ | −5.53⁎⁎⁎ | −4.01⁎⁎ |
| [−26.18] | [109.85] | [177.64] | [115.4] | [−7.33] | [−22.07] | [−14.37] | |||||
| Atrophy rate (GM) | −2.1% (−2.26%, −1.94%) | −1.17% (−1.48%, −0.86%) | −2.11% (−2.3%, −1.92%) | −3.01% (−3.42%, −2.6%) | |||||||
| Hippocampal volume in cm3 | −0.32 ± 0.01⁎⁎⁎ | 8.26 ± 0.12⁎⁎⁎ | 7.15 ± 0.07⁎⁎⁎ | 5.98 ± 0.1⁎⁎⁎ | −1.11⁎⁎⁎ | −1.18⁎⁎⁎ | −0.17 ± 0.02⁎⁎⁎ | −0.32 ± 0.01⁎⁎⁎ | −0.45 ± 0.02⁎⁎⁎ | −0.15⁎⁎⁎ | −0.12⁎⁎⁎ |
| [−34.06] | [69.25] | [101.46] | [59.22] | [−9] | [−28.71] | [−22.56] | |||||
| Atrophy rate (Hippocampus) | −4.28% (−4.53%, −4.04%) | −2.03% (−2.47%, −1.59%) | −4.18% (−4.46%, −3.89%) | −6.81% (−7.41%, −6.22%) | |||||||
Data are presented as β ± SE [t ratio] and in percentages (95% CI) for annual atrophy rates. Estimates are based on raw values/scores and were estimated with linear mixed models. The models included the terms age, sex, education, field strength, total intracranial volume (for grey matter volumes) and follow-up time in years for estimates of the total group. We additionally included baseline diagnosis and the interaction term follow-up time × diagnosis for effects per clinical stage. Annual atrophy rates are based on estimates of annual decline divided by mean volumes at baseline for the total group or respective diagnosis for results per clinical stage. GM, grey matter.
p < .01.
p < .001.
3.2. Model 1 results: changes in grey matter volumes and MMSE as a function of time
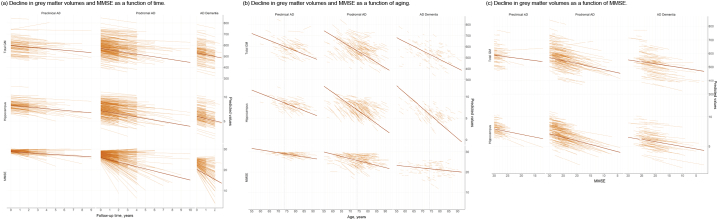
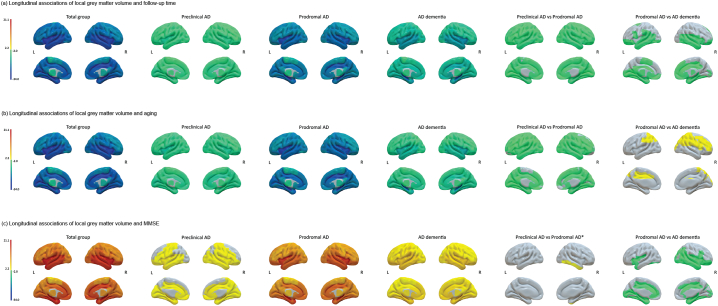
We first modeled changes in GM volumes and MMSE separately over time. Fig. 2a shows the change over time in MMSE and GM volume according to baseline clinical stage. Over time, the total sample showed declines of −1.29 ± 0.08 points per year on the MMSE and −12.23 ± 0.47 cm3 or −2.1% (95%CI; −2.26%, −1.94%) per year in GM volume (all p < .001; Table 2). Patients with AD-type dementia showed the fastest decline in the MMSE and GM volume (β ± SE; −2.38 ± 0.18 points per year and −16.53 ± 1.15 cm3 or −3.01% (95%CI; −3.42%, −2.6%) per year; all p < .001), followed by prodromal AD (β ± SE; −1.08 ± 0.1 points per year and −12.52 ± 0.57 cm3 or −2.11% (95%CI; −2.3%, −1.92%) per year; all p < .001) and then preclinical AD (β ± SE, −0.27 ± 0.16 points per year; p > .05; −6.99 ± 0.95 cm3 or −1.17% (95%CI; −1.48%, −0.86%) per year; p < .001). For annual atrophy rates at a regional level, we found a widespread atrophy pattern with the strongest associations for decline over time for the bilateral hippocampi, insulae and Rolandic opercula (all pFDR < 0.001; Fig. 3a). AD dementia participants showed faster atrophy over time mostly in temporal regions as compared to prodromal AD participants, while the atrophy pattern was similar between prodromal and preclinical AD individuals with faster atrophy for prodromal AD (all pFDR < 0.05).
Fig. 2.
Predicted changes in grey matter volumes and cognitive functioning (a) as a function of follow-up time, (b) as a function of aging and (c) as a function of MMSE for the baseline clinical stages in individuals with abnormal amyloid markers. Predicted values are based on raw values and were estimated with linear mixed models. The models included the terms age (for the time and MMSE models), sex, education, field strength and total intracranial volume (for grey matter volumes), and time, age or MMSE as predictor, and the interaction term predictor × diagnosis. Regression lines are based on cross-sectional (intercepts) and longitudinal (slopes) estimates of the respective model and are based on estimated marginal means for the clinical stages. Vertical line in (b) indicates mean age at baseline for the total sample.
GM, grey matter.
Fig. 3.
Surface plots of longitudinal associations of local grey matter volumes with (a) follow-up time, (b) aging and (c) decline in MMSE over time for the total groups and per baseline clinical stage. The color bar indicates the effect sizes as t ratios based on local GM volumes standardized to the mean values of cognitively normal individuals with normal amyloid at baseline (for descriptive data see Inline Supplementary Table 7) and were obtained with linear mixed models. Analyses were adjusted for age (for time and MMSE models), sex, education, field strength and total intracranial volume. Note that t ratios indicate the strength of the effect and do not correspond to betas. For associations in the total group and baseline clinical stages, negative values indicate steeper grey matter atrophy with increasing time or age and positive values indicate steeper grey matter atrophy with worsening MMSE. For comparison of clinical stages, negative values indicate steeper atrophy rates for e.g. prodromal AD as compared to preclinical AD and positive values indicate less steep atrophy rates for e.g. prodromal AD as compared to preclinical AD. Subcortical structures are plotted in ventricular areas as approximation. The model for the association with MMSE for the total group did not converge for the left supramarginal gyrus.
L, left hemisphere; R, right hemisphere; ⁎puncorrected < 0.05.
3.3. Model 2 results: changes in grey matter volumes and MMSE as a function of aging
When aligning individuals according to their age, we observed smaller estimates as compared to modeling change as a function of time, with −0.28 ± 0.03 points per year of age on the MMSE and −8.87 ± 0.34 cm3 or 1.52% (95%CI; −1.65%, −1.4%) per year of age in GM volume (all p < .001: Table 3 and Fig. 2b). Across clinical stages, individuals with prodromal AD showed the fastest decline over aging for MMSE (β ± SE; −0.22 ± 0.03 points per year of age) and GM volume (−9.6 ± 0.42 cm3 or −1.62% (95%CI; −1.76%, −1.48%) per year of age) compared to AD-type dementia (MMSE: −0.09 ± 0.04 per year of age, p > .05; GM: -7.94 ± 0.77 cm3 or −1.45% (95%CI; −1.72%, −1.17%) per year of age, p < .001) and preclinical AD participants (MMSE: −0.14 ± 0.05 per year of age, p < .05; GM: -6.3 ± 0.74 cm3 or −1.05% (95%CI; −1.3%, −0.81%) per year of age; p < .001). Similar to the time model, we found widespread regional effects for decline in GM volumes with age, with the strongest effects found for the bilateral hippocampi, insulae and Rolandic opercula (all pFDR < 0.001; Fig. 3b). Compared to AD dementia, prodromal AD participants showed faster atrophy in mostly frontoparietal areas, while preclinical and prodromal AD showed a largely similar atrophy pattern albeit with faster rates for prodromal AD (all pFDR < 0.05).
Table 3.
Cross-sectional and longitudinal estimates for MMSE, grey matter and hippocampal volumes over aging.
| Longitudinal estimates |
Cross-sectional estimates |
Cross-sectional differences |
Longitudinal estimates |
Longitudinal differences |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total group | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | |
| MMSE | −0.28 ± 0.03⁎⁎⁎ | 29.03 ± 0.31⁎⁎⁎ | 25.75 ± 0.18⁎⁎⁎ | 21.62 ± 0.27⁎⁎⁎ | −3.28⁎⁎⁎ | −4.13⁎⁎⁎ | −0.14 ± 0.05⁎ | −0.22 ± 0.03⁎⁎⁎ | −0.09 ± 0.04 | −0.08 | 0.13⁎ |
| [−9.41] | [95.08] | [141.56] | [79.02] | [−2.69] | [−8.29] | [−2.23] | |||||
| Grey matter volume in cm3 | −8.87 ± 0.34⁎⁎⁎ | 600.75 ± 6.78⁎⁎⁎ | 563.06 ± 4.1⁎⁎⁎ | 532.19 ± 6⁎⁎⁎ | −37.7⁎⁎⁎ | −30.87⁎⁎⁎ | −6.3 ± 0.74⁎⁎⁎ | −9.6 ± 0.42⁎⁎⁎ | −7.94 ± 0.77⁎⁎⁎ | −3.3⁎⁎⁎ | 1.66 |
| [−25.78] | [88.55] | [137.37] | [88.68] | [−8.56] | [−22.71] | [−10.32] | |||||
| Atrophy rate (GM) | −1.52% (−1.65%, −1.4%) | −1.05% (−1.3%, −0.81%) | −1.62% (−1.76%, −1.48%) | −1.45% (−1.72%, −1.17%) | |||||||
| Hippocampal volume in cm3 | −0.28 ± 0.01⁎⁎⁎ | 8.53 ± 0.2⁎⁎⁎ | 6.96 ± 0.12⁎⁎⁎ | 5.96 ± 0.18⁎⁎⁎ | −1.56⁎⁎⁎ | −1⁎⁎⁎ | −0.16 ± 0.02⁎⁎⁎ | −0.29 ± 0.01⁎⁎⁎ | −0.35 ± 0.02⁎⁎⁎ | −0.13⁎⁎⁎ | −0.06⁎⁎ |
| [−33.58] | [42.95] | [58.37] | [34.04] | [−9.38] | [−28.42] | [−20.16] | |||||
| Atrophy rate (Hippocampus) | −3.73% (−3.96%, −3.51%) | −1.95% (−2.36%, −1.54%) | −3.79% (−4.05%, −3.52%) | −5.43% (−5.95%, −4.9%) | |||||||
Data are presented as β ± SE [t ratio] and in percentages (95% CI) for annual atrophy rates. Estimates are based on raw values/scores and were estimated with linear mixed models. The models included the terms sex, education, field strength, total intracranial volume (for grey matter volumes) and age in years for estimates of the total group. We additionally included baseline diagnosis and the interaction term age × diagnosis for effects per clinical stage. Annual atrophy rates are based on estimates of annual decline divided by mean volumes at baseline for the total group or respective diagnosis for results per clinical stage. GM, grey matter.
p < .05
p < .01
p < .001.
3.4. Model 3 results: changes in grey matter volumes as a function of MMSE
When modeling disease progression as a function of MMSE, we observed that steeper decline on the MMSE was correlated with steeper GM volume loss (β ± SE = 4.1 ± 0.25 cm3 or 0.7% (95%CI; 0.62%, 0.79%) per MMSE point, p < .001; Table 4 and Fig. 2c). Compared to modeling atrophy as a function of time, where individuals showed an atrophy rate of 2.1% and a 1.29 point reduction on MMSE per year (i.e., atrophy rate of 1.63% with 1 point decline on MMSE), when change in both variables was modeled within individuals, atrophy decreased much less with 0.71% decrease in GM volume per 1 point decline in MMSE score. When using age as a proxy for disease progression we, similarly to the model based on time, found steeper slopes for atrophy of 1.54% and a 0.27 point decline on MMSE per year (i.e., atrophy rate of 5.7% with 1 point decline on MMSE). This suggests that using time or age as a proxy for disease progression may result in overestimation of disease-related atrophy, which may not reflect individual decline on MMSE, as a proxy for disease progression. Across clinical stages, prodromal AD participants showed the strongest associations of MMSE and GM volume (β ± SE = 4.46 ± 0.33 cm3 per MMSE point, p < .001) followed by AD dementia patients (β ± SE = 3.04 ± 0.48 cm3 per MMSE point, p < .001) and preclinical AD (β ± SE = 2.64 ± 0.84 cm3 per MMSE point, p < .01). Associations of GM volumes with MMSE did not depend on educational level (see Inline Supplementary Table 1).
Table 4.
Cross-sectional and longitudinal estimates for grey matter and hippocampal volumes over MMSE.
| Longitudinal estimates |
Cross-sectional estimates |
Cross-sectional differences |
Longitudinal estimates |
Longitudinal differences |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total group | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | Preclinical AD | Prodromal AD | AD dementia | Preclinical AD - Prodromal AD | Prodromal AD - AD dementia | |
| Grey matter volume in cm3 | 4.1 ± 0.25⁎⁎⁎ | 588.05 ± 5.53⁎⁎⁎ | 570.02 ± 2.94⁎⁎⁎ | 553.56 ± 4.34⁎⁎⁎ | −18.03⁎ | −16.46⁎⁎ | 2.64 ± 0.84⁎⁎ | 4.46 ± 0.33⁎⁎⁎ | 3.04 ± 0.48⁎⁎⁎ | 1.82 | −1.42⁎ |
| [16.23] | [106.37] | [194.14] | [127.58] | [3.16] | [13.71] | [6.34] | |||||
| Atrophy rate (GM) | 0.7% (0.62%, 0.79%) | 0.44% (0.17%, 0.72%) | 0.75% (0.64%, 0.86%) | 0.55% (0.38%, 0.72%) | |||||||
| Hippocampal volume in cm3 | 0.11 ± 0.01⁎⁎⁎ | 7.94 ± 0.12⁎⁎⁎ | 7.12 ± 0.06⁎⁎⁎ | 6.56 ± 0.09⁎⁎⁎ | −0.82⁎⁎⁎ | −0.56⁎⁎⁎ | 0.09 ± 0.02⁎⁎⁎ | 0.12 ± 0.01⁎⁎⁎ | 0.08 ± 0.01⁎⁎⁎ | 0.03 | −0.04⁎⁎ |
| [21.37] | [67.23] | [112.68] | [70.65] | [5.16] | [18.16] | [8.05] | |||||
| Atrophy rate (Hippocampus) | 1.49% (1.34%, 1.63%) | 1.06% (0.66%, 1.46%) | 1.55% (1.39%, 1.72%) | 1.19% (0.9%, 1.48%) | |||||||
Data are presented as β ± SE [t ratio] and in percentages (95% CI) for annual atrophy rates. Estimates are based on raw values/scores and were estimated with linear mixed models. The models included the terms age, sex, education, field strength, total intracranial volume and MMSE for estimates of the total group. We additionally included baseline diagnosis and the interaction term MMSE × diagnosis for effects per clinical stage. Atrophy rates per 1 score decline on the MMSE are based on longitudinal estimates divided by mean volumes at baseline for the total group or respective diagnosis for results per clinical stage. GM, grey matter.
p < .05.
p < .01.
p < .001.
At a regional level, when modeling atrophy as a function of decline in MMSE, the strongest associations were observed in the bilateral hippocampi, superior and middle temporal poles and insulae (all pFDR < 0.001; Fig. 3c). The anatomical pattern of associations between decline in MMSE and atrophy was similar for AD dementia and prodromal AD patients, albeit with stronger associations for prodromal AD compared to AD dementia patients especially in the left middle cingulate, Rolandic operculum, hippocampus and right inferior occipital gyrus (pFDR < 0.05). Compared to preclinical AD, prodromal AD patients showed stronger associations of decreasing MMSE and atrophy in the right inferior temporal gyrus (puncorrected = 0.04).
We further investigated whether the observed association with MMSE, as a proxy for disease severity, could be explained by the effect of follow-up time or aging. When we repeated the model additionally accounting for time or age, associations of MMSE with GM volumes remained significant for the total group and for individuals with prodromal AD and AD dementia specifically (see Inline Supplementary Tables 2 and 3), suggesting that modeling atrophy as a function of MMSE in more advanced disease stages can explain variance in atrophy in addition to time or aging. Similarly, refitting time and age models with longitudinal measures of MMSE resulted in similar effects as compared to the models that were not adjusted for within-individual cognitive decline (see Inline Supplementary Tables 4 and 5), suggesting that all measures can explain part of the variance in grey matter atrophy. Model fit for all refitted models was significantly improved by additionally adding time, age or MMSE, respectively.
3.5. Changes in grey matter volumes as a function of time, aging and MMSE in individuals with normal amyloid
In order to investigate whether slopes in outcome measures differed for individuals with abnormal and normal amyloid, we included baseline amyloid abnormality as interaction effect with the respective predictor. In all models, longitudinal slopes for MMSE, grey matter atrophy rates were steeper for individuals with abnormal amyloid as compared to those with normal amyloid (see Inline Supplementary Table 6). We subsequently repeated analyses in a sample of 387 individuals with normal amyloid markers (190 CN, 180 MCI, 17 individuals with dementia; see Inline Supplementary Table 7 for demographic and clinical characteristics).
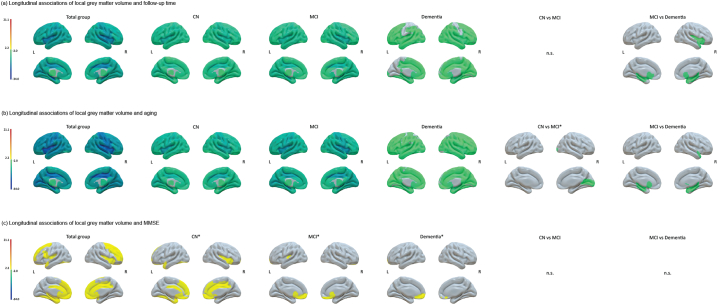
For these participants we found a slower decline in MMSE (β ± SE; −0.09 ± 0.03 points per year; p < .01) and a slower decline in GM volume over time (β ± SE; −5.32 ± 0.33 cm3 or −0.86% (95%CI; −0.97%, −0.76%) per year; p < .001) with 1.6 times weaker effect strength compared to individuals with abnormal amyloid markers (t ratio (abnormal amyloid): −26.18; t ratio (normal amyloid): −16.13; see Inline Supplementary Table 8 also for separate estimates for clinical stages). Using aging as a predictor we similarly found slower decline in MMSE (β ± SE; −0.04 ± 0.01 points per year; p < .001) and GM volume (β ± SE; −4.85 ± 0.25 cm3 or −0.79% (95%CI; −0.87%, −0.71%) per year; p < .001) with 1.3 times weaker effect strength for individuals with normal amyloid compared to those with abnormal amyloid (t ratio (abnormal amyloid): −25.78; t ratio (normal amyloid): −19.68; see Inline Supplementary Table 9). The anatomical pattern for regional volume loss over time or aging was similarly widespread compared to those observed for abnormal amyloid individuals, while we found reduced differences between the clinical stages (Fig. 4a–b; see Inline Supplementary Fig. 2 for cross-sectional associations). When modeling GM atrophy as a function of MMSE, we found that an atrophy rate of only 0.13% (95%CI; 0.01%, 0.25%) was directly related to decline on the MMSE (β ± SE; 0.83 ± 0.37 cm3 per MMSE point; p < .05). Importantly, this association was of 7.2 times weaker effect strength compared to individuals with abnormal amyloid (t ratio (abnormal amyloid): 16.23; t ratio (normal amyloid): 2.24; Table 5), indicating that associations with MMSE were more specific for amyloid pathology than associations with time or age. At a regional level, we only found few associations between decreasing MMSE and atrophy and mean effects were of 7 times weaker strength compared to the abnormal amyloid group (Fig. 4c). The difference in the anatomical patterns and weaker associations further suggest that observed associations between decline in GM volume and MMSE are specific for individuals with abnormal amyloid.
Fig. 4.
Surface plots of longitudinal associations of local grey matter volumes with (a) follow-up time, (b) aging and (c) decline in MMSE over time for the total groups and per baseline clinical stage in individuals with normal amyloid. The color bar indicates the effect sizes as t ratios based on local GM volumes standardized to the mean values of cognitively normal individuals with normal amyloid at baseline (for descriptive data see Inline Supplementary Table 7) and were obtained with linear mixed models. Analyses were adjusted for age (for time and MMSE models), sex, education, field strength and total intracranial volume. Note that t ratios indicate the strength of the effect and do not correspond to betas. For associations in the total group and baseline clinical stages, negative values indicate steeper grey matter atrophy with increasing time or age and positive values indicate steeper grey matter atrophy with worsening MMSE. For comparison of clinical stages, negative values indicate steeper atrophy rates for e.g. prodromal AD as compared to preclinical AD and positive values indicate less steep atrophy rates for e.g. prodromal AD as compared to preclinical AD. Subcortical structures are plotted in ventricular areas as approximation.
L, left hemisphere; R, right hemisphere; ⁎puncorrected < 0.05.
Table 5.
Cross-sectional and longitudinal estimates for grey matter and hippocampal volumes over MMSE in individuals with normal amyloid.
| Longitudinal estimates |
Cross-sectional estimates |
Cross-sectional differences |
Longitudinal estimates |
Longitudinal differences |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total group | CN | MCI | Dementia | CN-MCI | MCI-Dementia | CN | MCI | Dementia | CN-MCI | MCI-Dementia | |
| Grey matter volume in cm3 | 0.83 ± 0.37⁎ | 609.72 ± 3.65⁎⁎⁎ | 597.59 ± 3.89⁎⁎⁎ | 597.6 ± 12.63⁎⁎⁎ | −12.13⁎ | 0.01 | 0.99 ± 0.62 | 0.66 ± 0.5 | 0.8 ± 1.4 | −0.33 | 0.15 |
| [2.24] | [166.93] | [153.64] | [47.3] | [1.6] | [1.31] | [0.58] | |||||
| Atrophy rate (GM) | 0.13% (0.01%, 0.25%) | 0.16% (−0.04%, 0.36%) | 0.11% (−0.05%, 0.27%) | 0.14% (−0.34%, 0.62%) | |||||||
| Hippocampal volume in cm3 | 0.02 ± 0.01⁎⁎ | 8.6 ± 0.08⁎⁎⁎ | 7.95 ± 0.08⁎⁎⁎ | 7.1 ± 0.26⁎⁎⁎ | −0.66⁎⁎⁎ | −0.85⁎⁎ | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.03 | −0.01 | 0.01 |
| [2.67] | [109.26] | [94.5] | [27.1] | [1.7] | [1.41] | [0.77] | |||||
| Atrophy rate (Hippocampus) | 0.22% (0.05%, 0.38%) | 0.22% (−0.04%, 0.47%) | 0.16% (−0.06%, 0.38%) | 0.31% (−0.49%, 1.1%) | |||||||
Data are presented as β ± SE [t ratio] and in percentages (95% CI) for annual atrophy rates. Estimates are based on raw values/scores and were estimated with linear mixed models. The models included the terms age, sex, education, field strength, total intracranial volume and MMSE for estimates of the total group. We additionally included baseline diagnosis and the interaction term MMSE × diagnosis for effects per clinical stage. Atrophy rates per 1 score decline on the MMSE are based on longitudinal estimates divided by mean volumes at baseline for the total group or respective diagnosis for results per clinical stage. GM, grey matter.
p < .05.
p < .01.
p < .001.
For each model and both samples (i.e., amyloid abnormal and normal) we then compared model fit statistics as based on differences in AIC and likelihood ratio tests, calculated explained variance R2 and estimated variances in random effects. For both samples, best model, indicated by lowest AIC and/or highest log-likelihood, was observed for models based on time, followed by models based on aging and then MMSE (see Inline Supplementary Table 10). Aging models explained most variance in declining grey matter volume, as indicated by marginal R2, followed by time and then MMSE. Variance in subject slopes of decline on MMSE was greatest for time models as compared to aging models. Variance in random slope effects was lowest for MMSE models, followed by aging and then time models, suggesting that modeling grey matter atrophy as a function of cognitive decline leads to less inter-individual variance in atrophy slopes, which may reflect that individuals are better aligned according to their disease severity.
4. Discussion
In this study, we compared three different approaches to investigate longitudinal changes in GM volume: by using follow-up time, aging or MMSE, as an anchor point to align individuals according to their disease severity. Compared to modeling atrophy as a function of time or aging we observed less steep slopes when modeling disease progression based on change in MMSE. Associations with time, aging and MMSE generally remained after additionally correcting for the other predictors, suggesting that all measures explain part of the variance in GM atrophy. For the MMSE, associations were specific for individuals with amyloid pathology, suggesting that modeling longitudinal GM atrophy with MMSE as a proxy for disease progression might be specific for cognitive decline in individuals with evidence of AD pathology.
Previous studies taking time between first visit and follow-up to model disease progression found whole-brain atrophy rates of 0.4–0.7% for cognitively normal older individuals and of 0.6–2.2% for AD patients (see Frisoni et al., 2010 for review), which are similar to our estimates of annual atrophy rates. Faster decline in annualized whole-brain atrophy rates were furthermore found to correlate with faster annualized change on the MMSE across individuals (Jack Jr. et al., 2009; Sluimer et al., 2008). Using the MMSE at baseline to align individuals according to their initial disease stage, Jack Jr. et al. (2012) found increasing hippocampal atrophy with advancing disease progression within individuals. Additionally, Donohue et al. (2014) and Lorenzi et al. (2017) used self-modeling regression and Bayesian Gaussian process regression respectively to place individuals according to their disease stage and model declines in whole brain and hippocampal volume. Using the MMSE to align individuals according to their disease severity as a simple approach, our findings are in line with the results of those previous studies and we further show that other regions besides the hippocampus are associated with cognitive decline as a proxy for disease severity within individuals. Moreover, these associations were only observed in individuals with aggregated amyloid.
The most pronounced effect of modeling disease progression by decreasing MMSE compared to follow-up time or age was in the spatial pattern of associations observed in individuals with normal and with abnormal amyloid: Using time or age abnormal and normal amyloid participants showed, albeit with smaller atrophy rates, similar anatomical patterns; but when we used MMSE to model disease progression, we found widespread regional associations in individuals with abnormal amyloid across the clinical stages, whereas only few associations were found in amyloid normal individuals, most of which for cognitively normal individuals. In individuals with abnormal amyloid, associations with cognitive decline were strongest for regions including the hippocampus, superior and middle temporal pole and insula, which have been associated with cognitive dysfunction in AD (Chang et al., 2013; Dupont, 2002). In individuals with normal amyloid, we observed most associations of regional atrophy rates and cognitive decline for cognitively normal individuals in frontal and temporal regions (although these effects did not survive correction for multiple testing). These regions have previously been associated with cognitive performance in ‘normal’ aging in the absence of amyloid pathology (Bakkour et al., 2013). However, when additionally correcting for age the results remained unchanged in regions that included the left superior temporal pole, olfactory gyrus and right Heschl's gyrus (see Inline Supplementary Fig. 3), areas that have been reported to be affected in frontotemporal dementia (Landin-Romero et al., 2017; Moller et al., 2016).This suggests that these alterations are not due to ‘normal aging’, but rather may reflect that some of these individuals have non-AD pathology.
By modeling atrophy as a function of MMSE, we observed a much less steep atrophy rate of 0.7% per 1 point decline on the MMSE compared to 2.1% with 1.29 point decline on the MMSE per year or 1.52% with 0.28 point decline on the MMSE per year of age when modeling atrophy as a function of time or aging, respectively. Possibly, when individuals are not aligned according to their initial disease severity, the steeper slopes may reflect differences between individuals who were in early and those who were in more advanced stages of the disease at their first visit. Our results suggest that modeling GM atrophy as a function of MMSE shows involvement of anatomical areas that seem to be specifically related to cognitive decline in AD. Still, the slopes for the MMSE model were less steep as compared to the other models, and so we cannot exclude the possibility that GM atrophy in other anatomical areas as observed in the time and age models may have yet to affect the MMSE, or that those areas may be related to decline in more specific neuropsychological domains. Future research should further investigate this question by studying how GM atrophy relates to decline on specific neuropsychological tests scores.
Compared to AD dementia, individuals with prodromal AD showed steeper atrophy slopes in age and MMSE models, but not in time models, suggesting that GM atrophy most closely relates with increasing age and decline on MMSE within prodromal AD individuals. Possibly, these findings can be explained due to increased variance in this group of individuals, while participants with dementia might start to show plateau or floor effects in either MMSE and/or atrophy rates. At a regional level, we found similar atrophy patterns between preclinical AD and prodromal AD for models based on time and age in individuals with abnormal amyloid. Compared to prodromal AD participants with AD dementia showed steeper atrophy slopes mostly in temporal regions for time and less steep atrophy slopes mostly in frontoparietal regions for age models. These results suggest that while GM volumes in the temporal lobes continue to decline over time in the dementia stage, atrophy is most associated with increasing age within individuals in the prodromal stage of the disease. Previous studies found higher atrophy rates for younger ages especially in dementia (Fiford et al., 2018; Holland et al., 2012). However, these studies used baseline age to align individuals while we model the effect of aging within individuals. Regional atrophy as modeled over time and aging were also widespread in both samples with steeper slopes for individuals with abnormal amyloid and most pronounced in the medial temporal lobes, especially in the hippocampus. These results provide further support for the notion that amyloid aggregation is not part of normal aging and suggest that atrophy of the hippocampus is not specific for AD per se (Fjell et al., 2014; ten Kate et al., 2017), but rather the rate of hippocampal atrophy.
Finally, model fit comparisons showed that the time model showed the best model fit. However, when additionally including time or age to the MMSE model (and similarly MMSE to the age and time model) effects remained comparable and these refitted models significantly improved model fit compared to the original models, suggesting that adding information on where individuals are in their disease trajectory explains additional variance GM atrophy. These findings further suggest that time, aging and decline on MMSE all explained part of the variance in GM atrophy within individuals. Future research should further investigate the possibility to develop composite measures that combine time or age with decline on MMSE, and should test whether this could improve estimates of GM atrophy over time.
Strengths of our study are the large number of individuals, which we were able to include from ADNI, and patients with different levels of cognitive impairment with and without biomarker evidence of AD over a long follow-up period of up to 10 years. A potential limitation of our approach is the use of MMSE as a measure for general cognitive functioning in this study. The MMSE is a cognitive screening tool that might not adequately capture subtle cognitive impairment and cognitive decline (Tombaugh and McIntyre, 1992), and this may have contributed to the less steep atrophy slopes we observed. Although the MMSE is widely used and readily available for most studies, future research should further investigate how atrophy correlates with other measures that are more sensitive to subtle cognitive decline and to specific cognitive domains, which might relate to distinct, regional atrophy patterns. Furthermore, our sample of individuals with normal amyloid included only few dementia cases, which might have contributed to the weaker associations of MMSE and GM atrophy. Future research should further investigate the association of GM atrophy with time, age and cognitive decline in larger samples of individuals with non-AD dementia. Another possible limitation of our study might be in amyloid status classification. Here, we used PET when available, and otherwise CSF results were used to classify participants. Although these measures correlate (Zwan et al., 2014), they may reflect different aspects of amyloid pathology (Landau et al., 2013; Mattsson et al., 2014; Palmqvist et al., 2016), since PET tracers directly bind to amyloid fibrils, while in CSF amyloid accumulation is measured indirectly by decreasing levels of soluble Aβ1–42. By using PET as first criterion for amyloid abnormality we classified some participants as amyloid normal although they had abnormal levels of CSF Aβ1–42 (n = 76, 8.4%) and vice versa (n = 38, 4.2%). Given that this discordance occurred in <10% of individuals, however, it is unlikely to have influenced the results. Finally, scans included in the present study were obtained with different field strengths. Although we ensured that, within subjects, the same field strength was used, and we adjusted for this variable in all the models, these field strengths may still have influenced our estimates on grey matter atrophy. Additional post-hoc analyses stratifying analyses for field strength showed that for abnormal amyloid individuals, estimates of grey matter and hippocampal volumes for 1.5 Tesla scans were slightly higher, those for 3 Tesla scans slightly lower as compared to the original estimates, which were based on all individuals with abnormal amyloid (see Inline Supplementary Table 11). Importantly, individuals with 1.5 Tesla scans (who were most often included at the start of the study) also showed steeper decline on the MMSE, while individuals with 3 Tesla scans showed less steep decline on the MMSE over time. For individuals with normal amyloid, stratifying by field strength resulted in similar estimates for all outcome measures in all models. These results suggest that differences in subsample characteristics might have driven these differences, but not necessarily field strength per se.
5. Conclusion
One challenge in AD research is the estimation of where an individual is along their disease trajectory. Here, we modeled GM decline as a function of MMSE, in this way aligning individuals according to their disease severity and compared associations to those using follow-up time or aging. Our results suggest that modeling GM with MMSE decline within individuals, as a proxy for disease progression, provides smaller atrophy rates than those based on follow-up time or aging and are specific for cognitive decline in individuals with evidence of amyloid pathology.
Disclosures
E. Dicks and L. Vermunt report no disclosures. W. M. van der Flier's research programs have been funded by ZonMW, the Netherlands Organization of Scientific Research, Seventh European Framework Programme, Alzheimer Nederland, Cardiovascular Onderzoek Nederland, Stichting Dioraphte, Gieskes-Strijbis fonds, Boehringer Ingelheim, Piramal Imaging, Roche BV, Janssen Stellar, Biogen MA and Combinostics. All funding is paid to her institution. PJ Visser reports grants from Innovative Medicine Initiative, grants from ZonMw; non-financial support from GE Healthcare, other from Eli, Lilly, other from Janssen Pharmaceutica, grants from Biogen, outside the submitted work. F. Barkhof is a consultant for Biogen-Idec, Janssen Alzheimer Immunotherapy, Bayer-Schering, Merck-Serono, Roche, Novartis, Genzume, and Sanofi-Aventis; has received sponsoring from European Commission–Horizon 2020, National Institute for Health Research–University College London Hospitals Biomedical Research Centre, Scottish Multiple Sclerosis Register, TEVA, Novartis, and Toshiba; is supported by the University College London Hospitals NHS Foundation Trust Biomedical Research Center; and serves on the editorial boards of Radiology, Brain, Neuroradiology, Multiple Sclerosis Journal, and Neurology. P. Scheltens has acquired grant support (for the institution) from BiogenGE Healthcare, Danone Research, Piramal, and Merck. In the past 2 years, he has received consultancy/speaker fees (paid to the institution) from Lilly, GE Healthcare, Novartis, Sanofi, Nutricia, Probiodrug, Biogen, Roche, Avraham, and EIP Pharma, Merck AG. B. M. Tijms received grant support from ZonMW Memorabel (grant number 73305056).
Author contributions
Ellen Dicks, analysis and interpretation of data, drafting of manuscript.
Lisa Vermunt, critical revision of manuscript.
Wiesje M. van der Flier, interpretation of data, critical revision of manuscript.
Pieter Jelle Visser, critical revision of manuscript.
Frederik Barkhof, critical revision of manuscript.
Philip Scheltens, critical revision of manuscript.
Betty M. Tijms, study concept and design, interpretation of data, critical revision of manuscript.
Acknowledgments
Acknowledgements
This work has been supported by ZonMw Memorabel grant programme (BMT; grant number 73305056). Research of the Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. The Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc Fonds.
ADNI acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101786.
Contributor Information
Ellen Dicks, Email: e.dicks@vumc.nl, ellendicks.ac@gmail.com.
Lisa Vermunt, Email: l.vermunt@vumc.nl.
Wiesje M. van der Flier, Email: wm.vdflier@vumc.nl.
Pieter Jelle Visser, Email: PJ.Visser@vumc.nl.
Frederik Barkhof, Email: f.barkhof@vumc.nl.
Philip Scheltens, Email: p.scheltens@vumc.nl.
Betty M. Tijms, Email: b.tijms@vumc.nl.
Appendix A. Supplementary data
Supplementary material
References
- Bakkour A., Morris J.C., Wolk D.A., Dickerson B.C. The effects of aging and Alzheimer's disease on cerebral cortical anatomy: specificity and differential relationships with cognition. Neuroimage. 2013;76:332–344. doi: 10.1016/j.neuroimage.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., Blazey T.M., Holtzman D.M., Santacruz A., Buckles V., Oliver A., Moulder K., Aisen P.S., Ghetti B., Klunk W.E., McDade E., Martins R.N., Masters C.L., Mayeux R., Ringman J.M., Rossor M.N., Schofield P.R., Sperling R.A., Salloway S., Morris J.C., Dominantly Inherited Alzheimer N. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N. Engl. J. Med. 2012;367(9):795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Machler M., Bolker B.M., Walker S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67(1):1–48. [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57(1):289–300. [Google Scholar]
- Chang L.J., Yarkoni T., Khaw M.W., Sanfey A.G. Decoding the role of the insula in human cognition: functional parcellation and large-scale reverse inference. Cereb. Cortex. 2013;23(3):739–749. doi: 10.1093/cercor/bhs065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue, M.C., Jacqmin-Gadda, H., Le Goff, M., Thomas, R.G., Raman, R., Gamst, A.C., Beckett, L.A., Jack, C.R., Jr., Weiner, M.W., Dartigues, J.F., Aisen, P.S., Alzheimer's Disease Neuroimaging, I. 2014. Estimating long-term multivariate progression from short-term data. Alzheimers Dement. 10(5 Suppl), S400–10. doi: 10.1016/j.jalz.2013.10.003. [DOI] [PMC free article] [PubMed]
- Dupont S. Investigating temporal pole function by functional imaging. Epileptic Disord. 2002;4(Suppl. 1):S17–S22. [PubMed] [Google Scholar]
- Fiford C.M., Ridgway G.R., Cash D.M., Modat M., Nicholas J., Manning E.N., Malone I.B., Biessels G.J., Ourselin S., Carmichael O.T., Cardoso M.J., Barnes J., Alzheimer's Disease Neuroimaging I. Patterns of progressive atrophy vary with age in Alzheimer's disease patients. Neurobiol. Aging. 2018;63:22–32. doi: 10.1016/j.neurobiolaging.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A.M., Westlye L.T., Grydeland H., Amlien I., Espeseth T., Reinvang I., Raz N., Dale A.M., Walhovd K.B., Alzheimer Disease Neuroimaging I. Accelerating cortical thinning: unique to dementia or universal in aging? Cereb. Cortex. 2014;24(4):919–934. doi: 10.1093/cercor/bhs379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Jr., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010;6(2):67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C.D., Johnsrude I.S., Ashburner J., Henson R.N., Friston K.J., Frackowiak R.S. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1):21–36. doi: 10.1006/nimg.2001.0786. Pt 1. [DOI] [PubMed] [Google Scholar]
- Holland D., Desikan R.S., Dale A.M., McEvoy L.K., Alzheimer's Disease Neuroimaging I. Rates of decline in Alzheimer disease decrease with age. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D., Borowski B., Britson P.J., J L.W., Ward C., Dale A.M., Felmlee J.P., Gunter J.L., Hill D.L., Killiany R., Schuff N., Fox-Bosetti S., Lin C., Studholme C., DeCarli C.S., Krueger G., Ward H.A., Metzger G.J., Scott K.T., Mallozzi R., Blezek D., Levy J., Debbins J.P., Fleisher A.S., Albert M., Green R., Bartzokis G., Glover G., Mugler J., Weiner M.W. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W., Petersen R.C., Trojanowski J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9(1):119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Vemuri P., Wiste H.J., Weigand S.D., Lesnick T.G., Lowe V., Kantarci K., Bernstein M.A., Senjem M.L., Gunter J.L., Boeve B.F., Trojanowski J.Q., Shaw L.M., Aisen P.S., Weiner M.W., Petersen R.C., Knopman D.S., Alzheimer's Disease Neuroimaging I. Shapes of the trajectories of 5 major biomarkers of Alzheimer disease. Arch. Neurol. 2012;69(7):856–867. doi: 10.1001/archneurol.2011.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Lowe V.J., Weigand S.D., Wiste H.J., Senjem M.L., Knopman D.S., Shiung M.M., Gunter J.L., Boeve B.F., Kemp B.J., Weiner M., Petersen R.C., Alzheimer's Disease Neuroimaging I. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: implications for sequence of pathological events in Alzheimer's disease. Brain. 2009;132(Pt 5):1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust W.J., Bandy D., Chen K., Foster N.L., Landau S.M., Mathis C.A., Price J.C., Reiman E.M., Skovronsky D., Koeppe R.A. The Alzheimer's disease neuroimaging initiative positron emission tomography core. Alzheimers Dement. 2010;vol. 6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust W.J., Landau S.M., Koeppe R.A., Reiman E.M., Chen K., Mathis C.A., Price J.C., Foster N.L., Wang A.Y. The Alzheimer's disease neuroimaging initiative 2 PET core: 2015. Alzheimers Dement. 2015;11(7):757–771. doi: 10.1016/j.jalz.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen W.J., Ossenkoppele R., Knol D.L., Tijms B.M., Scheltens P., Verhey F.R., Visser P.J., Amyloid Biomarker Study G., Aalten P., Aarsland D., Alcolea D., Alexander M., Almdahl I.S., Arnold S.E., Baldeiras I., Barthel H., van Berckel B.N., Bibeau K., Blennow K., Brooks D.J., van Buchem M.A., Camus V., Cavedo E., Chen K., Chetelat G., Cohen A.D., Drzezga A., Engelborghs S., Fagan A.M., Fladby T., Fleisher A.S., van der Flier W.M., Ford L., Forster S., Fortea J., Foskett N., Frederiksen K.S., Freund-Levi Y., Frisoni G.B., Froelich L., Gabryelewicz T., Gill K.D., Gkatzima O., Gomez-Tortosa E., Gordon M.F., Grimmer T., Hampel H., Hausner L., Hellwig S., Herukka S.K., Hildebrandt H., Ishihara L., Ivanoiu A., Jagust W.J., Johannsen P., Kandimalla R., Kapaki E., Klimkowicz-Mrowiec A., Klunk W.E., Kohler S., Koglin N., Kornhuber J., Kramberger M.G., Van Laere K., Landau S.M., Lee D.Y., de Leon M., Lisetti V., Lleo A., Madsen K., Maier W., Marcusson J., Mattsson N., de Mendonca A., Meulenbroek O., Meyer P.T., Mintun M.A., Mok V., Molinuevo J.L., Mollergard H.M., Morris J.C., Mroczko B., Van der Mussele S., Na D.L., Newberg A., Nordberg A., Nordlund A., Novak G.P., Paraskevas G.P., Parnetti L., Perera G., Peters O., Popp J., Prabhakar S., Rabinovici G.D., Ramakers I.H., Rami L., Resende de Oliveira C., Rinne J.O., Rodrigue K.M., Rodriguez-Rodriguez E., Roe C.M., Rot U., Rowe C.C., Ruther E., Sabri O., Sanchez-Juan P., Santana I., Sarazin M., Schroder J., Schutte C., Seo S.W., Soetewey F., Soininen H., Spiru L., Struyfs H., Teunissen C.E., Tsolaki M., Vandenberghe R., Verbeek M.M., Villemagne V.L., Vos S.J., van Waalwijk van Doorn L.J., Waldemar G., Wallin A., Wallin A.K., Wiltfang J., Wolk D.A., Zboch M., Zetterberg H. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA. 2015;313(19):1924–1938. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.C.D. Extension of Nakagawa & Schielzeth's R-GLMM(2) to random slopes models. Methods Ecol. Evol. 2014;5(9):944–946. doi: 10.1111/2041-210X.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau S.M., Lu M., Joshi A.D., Pontecorvo M., Mintun M.A., Trojanowski J.Q., Shaw L.M., Jagust W.J., Alzheimer's Disease Neuroimaging I. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of beta-amyloid. Ann. Neurol. 2013;74(6):826–836. doi: 10.1002/ana.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landin-Romero R., Kumfor F., Leyton C.E., Irish M., Hodges J.R., Piguet O. Disease-specific patterns of cortical and subcortical degeneration in a longitudinal study of Alzheimer's disease and behavioural-variant frontotemporal dementia. Neuroimage. 2017;151:72–80. doi: 10.1016/j.neuroimage.2016.03.032. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Kim S., Yoo H., Park S., Jang Y.K., Kim H.J., Kim K.W., Kim Y., Jang H., Park K.C., Yaffe K., Yang J.J., Lee J.M., Na D.L., Seo S.W. Trajectories of physiological brain aging and related factors in people aged from 20 to over-80. J. Alzheimers Dis. 2018;65(4):1237–1246. doi: 10.3233/JAD-170537. [DOI] [PubMed] [Google Scholar]
- Lenth R. 2018. Emmeans: Estimated Marginal Means, aka Least-Squares Means. [Google Scholar]
- Lobo A., Launer L.J., Fratiglioni L., Andersen K., Di Carlo A., Breteler M.M., Copeland J.R., Dartigues J.F., Jagger C., Martinez-Lage J., Soininen H., Hofman A. Prevalence of dementia and major subtypes in Europe: a collaborative study of population-based cohorts. Neurologic diseases in the elderly research group. Neurology. 2000;54(11 Suppl 5):S4–S9. [PubMed] [Google Scholar]
- Lorenzi M., Filippone M., Frisoni G.B., Alexander D.C., Ourselin S., Alzheimer's Disease Neuroimaging I. Probabilistic disease progression modeling to characterize diagnostic uncertainty: application to staging and prediction in Alzheimer's disease. Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.08.059. [DOI] [PubMed] [Google Scholar]
- Mattsson N., Insel P.S., Landau S., Jagust W., Donohue M., Shaw L.M., Trojanowski J.Q., Zetterberg H., Blennow K., Weiner M., Alzheimer's Disease Neuroimaging I. Diagnostic accuracy of CSF Ab42 and florbetapir PET for Alzheimer's disease. Ann. Clin. Transl. Neurol. 2014;1(8):534–543. doi: 10.1002/acn3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C.R., Gharapetian L., McEvoy L.K., Fennema-Notestine C., Hagler D.J., Jr., Holland D., Dale A.M., Alzheimer's Disease Neuroimaging I. Relationship between regional atrophy rates and cognitive decline in mild cognitive impairment. Neurobiol. Aging. 2012;33(2):242–253. doi: 10.1016/j.neurobiolaging.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Moller C., Hafkemeijer A., Pijnenburg Y.A.L., Rombouts S., van der Grond J., Dopper E., van Swieten J., Versteeg A., Steenwijk M.D., Barkhof F., Scheltens P., Vrenken H., van der Flier W.M. Different patterns of cortical gray matter loss over time in behavioral variant frontotemporal dementia and Alzheimer's disease. Neurobiol. Aging. 2016;38:21–31. doi: 10.1016/j.neurobiolaging.2015.10.020. [DOI] [PubMed] [Google Scholar]
- ten Kate M., Barkhof F., Visser P.J., Teunissen C.E., Scheltens P., van der Flier W.M., Tijms B.M. Amyloid-independent atrophy patterns predict time to progression to dementia in mild cognitive impairment. Alzheimers Res. Ther. 2017;9(1):73. doi: 10.1186/s13195-017-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013;4(2):133–142. [Google Scholar]
- Palmqvist S., Mattsson N., Hansson O., Alzheimer's Disease Neuroimaging I. Cerebrospinal fluid analysis detects cerebral amyloid-beta accumulation earlier than positron emission tomography. Brain. 2016;139(Pt 4):1226–1236. doi: 10.1093/brain/aww015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R.C., Aisen P.S., Beckett L.A., Donohue M.C., Gamst A.C., Harvey D.J., Jack C.R., Jr., Jagust W.J., Shaw L.M., Toga A.W., Trojanowski J.Q., Weiner M.W. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassman B.L., Langa K.M., Fisher G.G., Heeringa S.G., Weir D.R., Ofstedal M.B., Burke J.R., Hurd M.D., Potter G.G., Rodgers W.L., Steffens D.C., Willis R.J., Wallace R.B. Prevalence of dementia in the United States: the aging, demographics, and memory study. Neuroepidemiology. 2007;29(1–2):125–132. doi: 10.1159/000109998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmansky N.J., Rosas H.D., Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61(4):1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M., Trojanowski J.Q., Alzheimer's Disease Neuroimaging I. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer J.D., van der Flier W.M., Karas G.B., Fox N.C., Scheltens P., Barkhof F., Vrenken H. Whole-brain atrophy rate and cognitive decline: longitudinal MR study of memory clinic patients. Radiology. 2008;248(2):590–598. doi: 10.1148/radiol.2482070938. [DOI] [PubMed] [Google Scholar]
- Terry R.D., Masliah E., Salmon D.P., Butters N., DeTeresa R., Hill R., Hansen L.A., Katzman R. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- Tombaugh T.N., McIntyre N.J. The mini-mental state examination: a comprehensive review. J. Am. Geriatr. Soc. 1992;40(9):922–935. doi: 10.1111/j.1532-5415.1992.tb01992.x. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vemuri P., Weigand S.D., Knopman D.S., Kantarci K., Boeve B.F., Petersen R.C., Jack C.R., Jr. Time-to-event voxel-based techniques to assess regional atrophy associated with MCI risk of progression to AD. Neuroimage. 2011;54(2):985–991. doi: 10.1016/j.neuroimage.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong Q.H. Likelihood ratio tests for model selection and non-nested hypotheses. Econometrica. 1989;57(2):307. [Google Scholar]
- Zeifman L.E., Eddy W.F., Lopez O.L., Kuller L.H., Raji C., Thompson P.M., Becker J.T. Voxel level survival analysis of Grey matter volume and incident mild cognitive impairment or Alzheimer's disease. J. Alzheimers Dis. 2015;46(1):167–178. doi: 10.3233/JAD-150047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwan M., van Harten A., Ossenkoppele R., Bouwman F., Teunissen C., Adriaanse S., Lammertsma A., Scheltens P., van Berckel B., van der Flier W. Concordance between cerebrospinal fluid biomarkers and [11C]PIB PET in a memory clinic cohort. J. Alzheimers Dis. 2014;41(3):801–807. doi: 10.3233/JAD-132561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material