Abstract
The human Respiratory Syncytial Virus (hRSV) is the major causative agent of lower respiratory tract diseases in infants, young children and elderly. The membrane protein G is embedded in the viral lipid envelope and plays an adhesion function of the virus to host cells. The present study reports the production of the group A hRSV recombinant G protein ectodomain (edG) and its characterization of secondary structure and thermal unfolding by circular dichroism (CD), as well as the binding investigation of flavonoids quercetin and morin to this protein by fluorescent quenching. CD data reveal that edG is composed mostly of β-structure and its melting temperature is of 325 K. Fluorescence quenching experiments of hRSV edG show that the dissociation constants for the flavonoids binding are micromolar and the binding affinity for the edG/quercetin complex is inversely dependent on rising temperature while is directly dependent for the edG/morin interaction. The thermodynamic parameters suggest that hydrophobic contacts are important for the edG/morin association while van der Waals forces and hydrogen bonds contribute to the stabilization of the edG/quercetin complex. Thus, data reported herein may contribute to the development of new treatment strategies that prevent the viral infection by hRSV.
Keywords: Biochemistry, Biophysics, Molecular biology
1. Introduction
The human Respiratory Syncytial Virus (hRSV) is the major causative agent of chronic lower respiratory tract diseases in infants, young children, elderly, and people who have cardiopulmonary problems and are immunocompromised [1, 2, 3, 4, 5, 6, 7]. Most patients exhibit symptoms of bronchiolitis or pneumonia that in severe cases require hospitalization [8]. Although previous infections by hRSV enhance the development of significant levels of serum neutralizing antibodies, they do not prevent reinfections [9]. The vaccine trials were unsuccessful and the currently available treatments and prophylaxis against hRSV present restrictions such as high costs, low efficacy, and several side effects [10].
RSV belongs to the family Pneumoviridae, species Human orthopneumovirus [11], and has a non-segmented, single-stranded negative sense RNA genome of 15,200 bases that encodes eleven proteins [12, 13]. The viral nucleocapsid is surrounded by an outer lipid envelope where the membrane proteins F and G are embedded and play a key role during virus attachment, fusion, and entry. They are closely related to the humoral response, being the main antigenic groups and antiviral targets [12, 14].
The hRSV G protein is classified as a glycosylated type II integral membrane protein which has an adhesion function between the virus and the host cell [15, 16]. This protein contains 289 to 299 residues, depending on the strain of group A or B which diverge in nucleotide and amino acid residue sequences both between and within the two groups [17]. In its ectodomain (from residue 67 to 298), there is a highly conserved non-glycosylated region which is composed of the partial overlap of the cysteine noose domain, with four cysteine residues forming two disulfide bonds, and highly basic heparin-binding domain (HBD) [18, 19, 20]. Studies indicate that the HBD of the G protein functions as a primary site of virus-host cell interaction; however, mutations in this domain do not preclude infection [18]. In addition, the G protein has a highly conserved chemokine-like structural motif (CX3C) located in the cysteine noose domain between amino acid residues 182 and 186, whose function is binding to fractalkine receptor (CX3CR1) which is mainly expressed in cells of the immune system such as dendritic, monocytes, macrophages, T lymphocytes and natural killer (NK). The interaction of secreted G protein with CX3CR1 receptor is associated with modulation of immune system responses and consequent aggravation of the viral infection in the host. Therefore, the G protein is capable of suppressing the inflammation process, innate and memory immune responses, production of β-interferon, proliferation of T lymphocytes, and response of phagocytic immune system cells [21, 22, 23, 24, 25, 26]. Lee et al. (2017) and Jones et al. (2018) suggest that antibodies against the G protein conserved non-glycosylated region (mentioned above) might be used as a prophylactic treatment against hRSV diseases, since the studies in vivo demonstrated that these immunoglobulins promote a decrease in the viral titer and infection [27, 28]. Thus, the hRSV G membrane protein can be considered as an important antiviral target, not just for antibodies but also for small molecule as it can be inferred from the studies of Kaul et al. (1985), which showed a potential virucidal effect promoted by the flavonoid quercetin that reduced the infectivity of RSV by 90% [29].
Flavonoids are a class of natural polyphenolic compounds widely found in plants that are part of the daily human diet. Literature shows that flavonoids have potential biological and pharmacological activities such as antioxidant, anticarcinogenic, antibacterial, anti-inflammatory, and antiviral [30]. This paper presents the production of the recombinant ectodomain of group A hRSV G protein and its secondary structure and thermal unfolding characterization by circular dichroism spectroscopy, as well as the interaction studies of this protein with the flavonoids quercetin and morin (Fig. 1) investigated by the fluorescent quenching method. The data reported herein may contribute to the development of new treatment strategies that prevent the viral infection by hRSV.
Fig. 1.
Molecular structures of the flavonoids quercetin (top) and morin (bottom).
2. Materials and methods
2.1. Expression and purification of the recombinant G protein ectodomain
E. coli BL21 C41 bacteria were transformed by the thermal shock method with the recombinant plasmid pJexpress401 (DNA2.0, USA) containing the ectodomain G cDNA from A2 genotype of group A hRSV. A colony was chosen and grown in LB medium with 50 μg mL-1 kanamycin at 37 °C until it reached an optical density of 0.6 absorbance units. Next, the temperature was lowered to 30 °C and the cells were induced with 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 18 h. After the induction time, the suspension was centrifuged at 3500×g for 25 min at 4 °C and the pellet was resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 1.0 mM β-mercaptoethanol). The cells were lysed by sonication with pulses of 15 s and intervals of 45 s in a total time of 10 min. After this procedure, the crude cell extract was centrifuged at 25,000×g for 60 min at 4 °C. The supernatant was collected and filtered on a 0.2 μm diameter filter (Minisart) before being loaded on a Ni-NTA column. The affinity chromatography column was pre-equilibrated with lysis buffer and the bound protein was washed extensively with loading buffer plus gradients of 5–30 mM imidazole. The edG protein was eluted with a 60–500 mM imidazole gradient. The eluted fractions were concentrated using Amicon Ultra-15 centrifugal filter (MWCO: 3.0 kDa) and injected onto a Superdex 75 10/300 GL (GE Healthcare) size exclusion column, pre-equilibrated in sample buffer (50 mM NaH2PO4/Na2HPO4 pH 7.5, 150 mM NaCl, 1.0 mM β-mercaptoethanol). The fractions containing pure edG protein were pooled and concentrated. The sample purity after each purification step was assessed by 15% SDS-PAGE and Western Blotting gels. For immunodetection, the monoclonal anti-polyhistidine primary antibody (Sigma Aldrich, USA) and polyclonal anti-mouse IgG (Fab specific)-peroxidase secondary antibody were used. The protein concentration was determined spectrophotometrically (UV-Visible Spectrophotometer, BioMATE 3S, Thermo Scientific, USA) at 280 nm using the molar extinction coefficient of 6,990 M-1 cm-1 [31].
2.2. Preparation of the stock solutions of flavonoid quercetin and morin
The stock solutions of flavonoid quercetin (302.24 Da) and morin (302.24 Da) were prepared in absolute ethanol at concentrations of 0.86 and 0.33 mM, respectively. The concentrations were determined spectrophotometrically (UV-Visible Spectrophotometer, BioMATE 3S, Thermo Scientific, USA) using the molar extinction coefficient of 21,880 [32] and 15,900 M-1 cm-1 [33] at 376 and 354 nm for quercetin and morin, respectively.
2.3. Circular dichroism spectroscopy
The secondary structure analysis and thermal unfolding thermodynamics of the edG protein were performed by the circular dichroism (CD) technique using a Jasco 815 spectropolarimeter (Jasco, USA) equipped with a Peltier-type temperature control system and a 0.5 mm path-length quartz cuvette which was coupled a metal spacer block. The far UV-CD spectrum of 11.8 μM edG protein was collected at 293 K (20 °C) in a spectral range of 240–200 nm with a resolution of 0.2 nm at the scan speed of 50 nm min-1 and 1.0 nm spectral bandwidth. The signal was averaged over 10 scans. The baseline was corrected by subtracting the spectrum of the obtained buffer solution (50 mM NaH2PO4/Na2HPO4 pH 7.5, 150 mM NaCl, 1.0 mM β-mercaptoethanol). The CD spectrum was taken as millidegrees (θ) and expressed in terms of mean residues ellipticity (MRE or [Θ]) in deg·cm2·dmol−1 using the following equation:
| (1) |
where [P] is the molar concentration of the edG protein, l is the optical path length (cm), and nr is the quantity of amino acid residues. Percentages of secondary structures of the edG protein in solution were calculated with the CONTINLL software of the CDPro package, using the reference set of proteins SMP56 [34].
In the thermal denaturation experiments, the protein sample was heated from 293 to 353 K (20–80 °C, respectively) at a scan rate of 1.0 K/min, recording a spectral region of 226–218 nm every step of 5 K with the equipment adjusted in the same conditions described above. The average value of θ (mdeg) at 220, 221, 222, 223, and 224 nm was used for the denaturation analysis. The thermal unfolding data were adjusted by Eq. (2) assuming a two states reaction, native and denatured [35].
| (2) |
where, θ is the temperature-dependent signal, θN and θU are signals of the protein in the native and denatured state, respectively, T is the absolute temperature, Tm is the melting temperature of the edG protein, and ΔHu is the enthalpy change of the unfolding process.
2.4. Fluorescence spectroscopy
The fluorescence quenching experiments were performed using the Fluorescent Spectrometer Lumina (Thermo Fisher Scientific, USA) equipped with a thermostat bath and quartz cell of 10 mm of optical path length. The fluorescence spectra of the edG protein in absence and presence of flavonoids were collected in the range of 290–450 nm with the excitation at 280 nm with the increment of 0.1 nm. Both excitation and emission spectral bandwidths were set at 5.0 nm. Each point in the emission spectrum of the protein is the average of 10 accumulations. The spectra were corrected for the background fluorescence of the buffer and for inner filter effects by flavonoids using the following equation [36]:
| (3) |
where Fcorr is the corrected fluorescence intensity of edG in presence of the flavonoids, Fobs is the observed intensity from spectrofluorimeter, and ODex and ODem is the optical density attenuated at excitation and emission wavelength of the fluorescence spectrum of the protein, respectively.
The titration experiments were performed by adding small aliquots from flavonoid stock solutions to protein solution (1.8 mL) with a constant concentration of 4.0 μM at 292 and 310 K (19 and 37 °C, respectively). The concentration of the flavonoids quercetin and morin varied from 0 to 6.4 μM. In all measurements, the final volume of ethanol in buffer was less than 1%.
The edG/flavonoid binding process was described by the following site-binding model [37, 38]:
| (4) |
where P is the protein, L is the ligand (flavonoids), and PL is the molecular complex formed with a dissociation constant (Kd) given by
| (5) |
The observed fluorescence intensity of the protein in the titration experiments with flavonoids can be described by a linear combination of the emission of its free and bound species, as follow:
| (6) |
where F0 and F is the fluorescence intensity in the absence and presence of quencher (flavonoids), respectively. fF and fB is the molar fluorescence intensity of the free and bound protein, respectively.
Considering the law of conservation of mass for the total concentration of protein and ligand as being the sum of concentrations of the free and bound species, and site-binding model in Eq. (4), the concentration of molecular complex (bound protein) can be calculated using the following equation:
| (7) |
The values of Kd for the flavonoid-binding experiments were adjusted over the fitting process by nonlinear least-squares optimization using Levenberg–Marquardt interactions.
2.5. Thermodynamic analysis
The driving interactions responsible for the association and stabilization of the protein-ligand complex involve the formation of hydrogen bonds, van der Waals forces, electrostatic interactions, and hydrophobic contacts. In order to elucidate the interactions between edG protein and flavonoids quercetin and morin, the enthalpy change (ΔH) was calculated from the van't Hoff equation:
| (8) |
where Kd1 and Kd2 are dissociation constants at the absolute temperature T1 and T2, which were 292 and 310 K, respectively. R is the ideal gas constant. The Gibbs free energy change (ΔG) and entropy change (ΔS) for the edG/flavonoid complexes were calculated from the relations:
| (9) |
| (10) |
3. Results and discussions
3.1. Expression and purification of group A hRSV edG
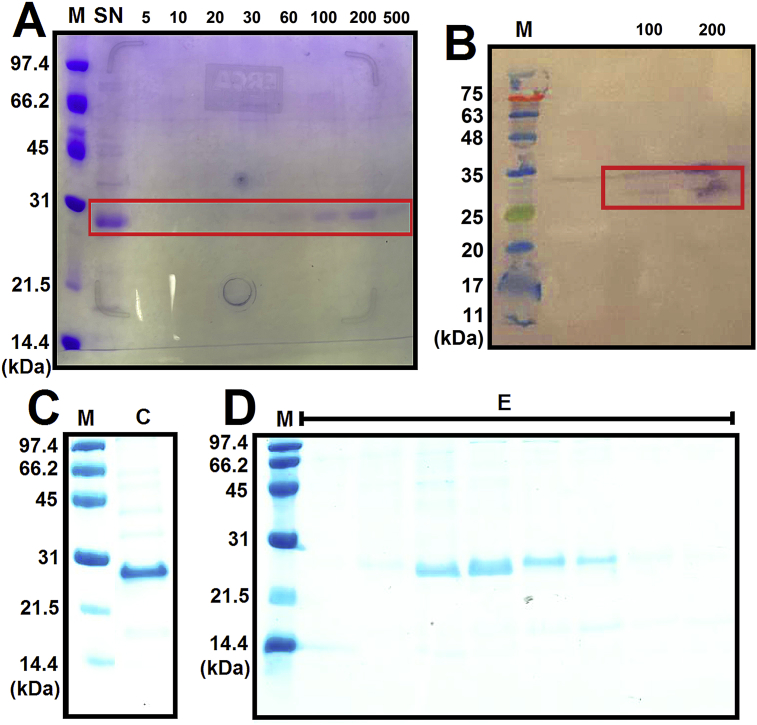
The gene that encodes the edG protein of group A hRSV was fused to a hexahistidine affinity tag and cloned into plasmid pJexpress401 by DNA2.0. The vector was transformed to and expressed in E. coli BL21 C41 bacterium. The highest level of expression along with solubility was obtained by induction with 0.4 mM IPTG at 30 °C for 18 h. The lysed cell extract was purified using a Ni-NTA column and presented a band on the 15% SDS-PAGE gel corresponding to ∼26.4 kDa edG protein eluted with imidazole gradient of 60–500 mM (Fig. 2A). At elution fractions of 100 and 200 mM imidazole, this band was confirmed as A-hRSV edG protein by immunodetection (Western Blotting, Fig. 2B). After that, the fractions containing the protein of interest were concentrated (Fig. 2C) and injected onto a size exclusion column for further purification and exchange the buffer, 50 mM NaH2PO4/Na2HPO4 pH 7.5, 150 mM NaCl, and 1.0 mM β-mercaptoethanol. Next, the eluted edG protein at volume fractions of 9–11 mL was assessed by 15% SDS-PAGE gel (Fig. 2D, Lanes E). The pure protein fractions were pooled and concentrated (yield of ∼0.5 mg/mL) for performing the experiments of biophysical and flavonoids-binding characterization.
Fig. 2.
(A) Coomassie-stained 15% SDS-PAGE gel of the purification process of the A-hRSV edG protein by Ni-NTA affinity chromatography. Lane M: molecular weight marker; Lane SN: supernatant; and Lanes 5–500 indicate the imidazole gradient concentrations (mM). The red box denotes the molecular weight corresponding to the edG protein (∼26.4 kDa). (B) Immunodetection of the edG protein of group A hRSV. Lane M: molecular weight marker; and Lane 100 and 200 denote the elution fraction where the protein was detected (red box). (C) Coomassie-stained 15% SDS-PAGE gel of the concentrated pool (Lane C) of the protein fractions eluted with 60–500 mM imidazole, before injected onto a size exclusion column. (D) Coomassie-stained 15% SDS-PAGE of the purification process of the edG protein by size exclusion chromatography. Lanes E shows the fractions in which the pure protein was eluted.
3.2. Secondary structure and thermal unfolding characterization of the edG protein
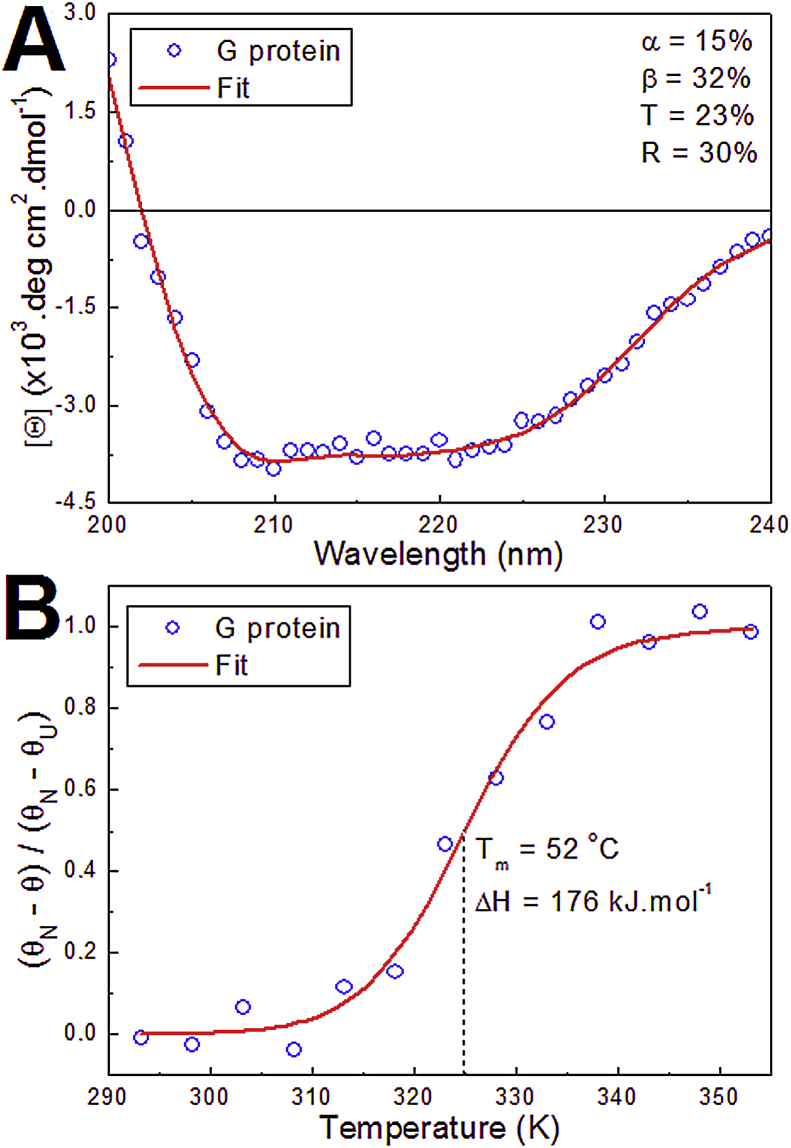
The circular dichroism (CD) spectroscopy was applied to study the secondary structure profile and thermal unfolding process of the edG protein of group A hRSV (Fig. 3). The far UV-CD spectrum of the edG presented characteristics of a structured protein, which shows minimum values of [Θ] at 221–208 nm range and a positive ellipticity at 200 nm (Fig. 3A). The secondary structure analysis suggests that the edG protein has approximately 32% of β-structure, 15% of α-helix, 23% of turn, and 30% of random coil (insert in Fig. 3A). Khan et al. (2016) characterized by using CD spectroscopy the secondary structures of the G protein ectodomain of group B hRSV. The CD spectrum of B-hRSV edG protein presented a single pronounced ellipticity minimum at ∼206 nm, being determined 75 % of α-helix and 4% of β-structure [39]. In addition to the methodology aspects, the different primary sequences of the G proteins of groups A and B hRSV can be responsible for the discrepancy between the CD results present herein and by Khan et al (2016).
Fig. 3.
(A) Far UV–CD spectrum of A-hRSV edG protein (11.8 μM, unfilled blue circles) at 298 K and 50 mM NaH2PO4/Na2HPO4 (pH 7.5) containing 150 mM NaCl and 1.0 mM β-mercaptoethanol. The red line represents the best result for curve fit performed by CONTINLL program. (B) Temperature dependence of the average value of θ (mdeg) between 220 and 224 nm for thermal unfolding process of the edG protein obtained with a scan rate of 1.0 K/min. The solid red line represents the best curve adjusted using Eq. (2) and the dotted line indicates the melting temperature (Tm) and the correspondent enthalpy change of the unfolding process (ΔHu).
The evaluation of the thermal unfolding data of the A-hRSV edG reveals that the protein denaturation is a cooperative process starting at 313 K (40 °C) and ending at 338 K (65 °C). The fitting of this experimental data using Eq. (2) presents a melting temperature of 325 K (52 °C) and van't Hoff enthalpy change of 176 kJ mol-1 (Fig. 3B). This result indicates that the edG protein is thermally stable considering the temperature of physiologic processes (∼310 K) and also regard the flavonoids-binding experiments performed at 292 and 310 K using the fluorescence quenching method.
The literature shows that proteins in its glycosylated and non-glycosylated form present similar CD spectra, indicating that its amount of secondary structures is preserved. However, the thermal unfolding processes of these proteins show different melting temperatures, being the Tm value of the glycosylated state larger than non-glycosylated protein which suggests higher thermal stability of the protein containing the glycosylation [40, 41, 42]. Therefore, despite the ectodomain of hRSV edG protein to be glycosylated in vivo, it is probable that this glycosylation does not significantly change the percentage of secondary structures herein determined to its non-glycosylated form but likely it promote an increment of its melting temperature (higher than 325 K) regard to the unfolding process of the recombinant protein, characterizing an increase in the thermal stability.
3.3. Binding aspects of the edG/flavonoid complexes determined by fluorescence quenching
The fluorescence intensity of the protein is quenched upon titration with the flavonoids (Figs. 4A and 4B) and therefore suggests that the microenvironment of the fluorophores of edG (one Tyr and one Trp in the protein sequence) is affected at a molecular level by the presence of the quercetin and morin. The fluorescence quenching data were analyzed using the site-binding model of Eq. (4). The adjusted curves to the experimental data are in agreement with the relations described in Eqs. (6) and (7), as it can be seen in Figs. 4C and 4D. The dissociation constants (Kd) values obtained for the binding of quercetin and morin to the edG are on order of micromolar (μM) and their magnitudes present a different behavior with increasing temperature (Table 1). For the edG/quercetin complex, Kd values increase with the increment of the temperature while decrease for the interaction with morin. This fact suggests that the affinity of the edG/quercetin association decreases with rising temperature while the edG/morin becomes more stable. The flavonoids quercetin and morin just present different positions of hydroxyl groups on the B-ring (Fig. 1), indicating that the different arrangement of these hydroxyls affects the thermal stability of the complexes.
Fig. 4.
Corrected and normalized emission spectra of A-hRSV edG protein in absence and presence of increments of quercetin (A) and morin (B) concentrations (pH 7.5, T = 292 K (19 °C), λex = 280 nm). [edG] = 4.0 μM; [flavonoids] = 0–6.4 μM. The dotted lines correspond to the emission spectrum of quercetin and morin in phosphate buffer at the highest concentration (6.4 μM). Analysis of the fluorescence quenching data of the edG protein by the flavonoids quercetin (C) and morin (D) at 292 and 310 K. The continuous lines represent the curves adjusted to the experimental data (red square and blue circle) according to Eqs. (6) and (7).
Table 1.
Dissociation constant (Kd), enthalpy change (ΔH), Gibbs free energy change (ΔG), and entropy change (ΔS) of the edG/flavonoid interaction determined using fluorescence quenching experiments at 292 and 310 K.
| Flavonoid | |||||
|---|---|---|---|---|---|
| Quercetin | 292 | 1.4 ± 0.2 | –43 ± 6 | –32.71 ± 0.03 | –10 ± 6 |
| 310 | 3.9 ± 0.1 | –32.08 ± 0.01 | –10 ± 6 | ||
| Morin | 292 | 2.3 ± 0.2 | 44 ± 16 | –31.50 ± 0.02 | 76 ± 16 |
| 310 | 0.8 ± 0.3 | –36.2 ± 0.1 | 80 ± 16 |
All correlations coefficients are ≥0.998.
The dissociation constants (Kd) obtained at varying temperatures were used to evaluate the thermodynamic properties of flavonoids binding. values were analyzed using the van't Hoff equation (Eq. 8) at 292 and 310 K. The values of enthalpy changes (ΔH) for the binding of quercetin and morin to the edG protein are –43 ± 6 and 44 ± 16 kJ mol-1 (Table 1), indicating that the binding reactions are exothermic and endothermic, respectively. The negative values of ΔG indicate that the flavonoids binding processes are spontaneous. For the edG/quercetin complex, ΔH < 0 and T·ΔS < 0 values suggest that the interaction is enthalpically favorable and entropically unfavorable, being the enthalpic term the major contribution to ΔG and therefore characterizing an enthalpy-driven binding reaction. The positive values of ΔH and T·ΔS for the edG/morin interaction characterize an enthalpically unfavorable and entropically favorable binding with an entropic term contributing mostly to ΔG and thus describing an entropy-driven binding reaction. Therefore, the thermodynamic parameters suggest that hydrophobic contacts play a key role in the association of the edG/morin complex, while van der Waals forces and hydrogen bonds are important contributions to the stabilization of the edG/quercetin complex [43]. Regarding the association of the edG/morin complex, the large positive values of may be interpreted as a release process of ordered water molecules (desolvation) from the binding pocket of the flavonoid in the protein [44]. On the other hand, the arrangement of the hydroxyl groups on the B-ring of the flavonoid quercetin may be considered as more favorable than of the morin for the formation of hydrogen bonds with the binding site of the protein.
4. Conclusions
The expression and purification process of the hRSV recombinant edG was successful and this soluble protein was characterized regarding the composition of secondary structure, thermal unfolding, and binding to flavonoids. The CD results reveal that the recombinant edG has 32% of β-structure and 15% of α-helix and a melting temperature of 325 K for its thermal unfolding process, characterizing the production of a structured and thermally stable protein. The analysis of fluorescence quenching data indicates that the Kd values of the edG/flavonoid complexes are on order of micromolar (10−6 M) and the dissociation constants for the edG/quercetin interaction are directly dependent on increasing temperature while are inversely dependent for the edG/morin complex. The thermodynamic parameters analysis indicates that the hydrophobic effect plays a role key in the association of the edG/morin complex, while the van der Waals forces and hydrogen bonds are important contributions to the stabilization of the edG/quercetin complex. The outcomes described herein suggest that the flavonoids quercetin and morin may be considered as virucidal and antiviral agents against hRSV, corroborating with the infectivity essays of Kaul et al. (1985) and also pointing out the use of these polyphenols as a new strategy of treatment that prevents the viral infection. However, it is fair to note that specific biological assays should be made to confirm the potential virucidal and antiviral activity of the quercetin and morin against RSV.
Declarations
Author contribution statement
Vitor Brassolatti Machado, Jéssica Maróstica de Sá: Performed the experiments; Analyzed and interpreted the data.
Ana Karla Miranda Prado: Performed the experiments.
Karina Alves de Toledo: Conceived and designed the experiments.
Luis Octávio Regasini: Contributed reagents, materials, analysis tools or data.
Fátima Pereira de Souza: Conceived and designed the experiments; Analyzed and interpreted the data.
Ícaro Putinhon Caruso: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Marcelo Andres Fossey: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2016/01749-3 and 2015/09261-7).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Prof. Dr. Marcelo de Freitas Lima for the access to the Fluorescent Spectrometer Lumina located in the Bio-organic Environmental Laboratory of the Department of Chemistry and Environmental Sciences.
Contributor Information
Ícaro Putinhon Caruso, Email: icaro.caruso@unesp.com.
Marcelo Andres Fossey, Email: marcelo.fossey@unesp.com.
References
- 1.Hall C.B. Prospects for a respiratory syncytial virus vaccine. Science. 1994;265:1393–1394. doi: 10.1126/science.7915433. [DOI] [PubMed] [Google Scholar]
- 2.Mufson M.A., Levine H.D., Wasil R.E., Mocega-Gonzalez H.E., Krause H.E. Epidemiology of respiratory syncytial virus infection among infants and children in Chicago. Am. J. Epidemiol. 1973;98:88–95. doi: 10.1093/oxfordjournals.aje.a121542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrott R.H., Kim H.W., Arrobio J.O., Hodes D.S., Murphy B.R., Brandt C.D., Camargo E., Chanock R.M. Epidemiology of respiratory syncytial virus infection in Washington, D.C. Am. J. Epidemiol. 1973;98:289–300. doi: 10.1093/oxfordjournals.aje.a121558. [DOI] [PubMed] [Google Scholar]
- 4.Dowell S.F., Anderson L.J., Gary H.E., Erdman D.D., Plouffe J.F., File T.M.J., Marston B.J., Breiman R.F. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 1996;174:456–462. doi: 10.1093/infdis/174.3.456. [DOI] [PubMed] [Google Scholar]
- 5.Falsey A.R., Cunningham C.K., Barker W.H., Kouides R.W., Yuen J.B., Menegus M., Weiner L.B., Bonville C.A., Betts R.F. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 1995;172:389–394. doi: 10.1093/infdis/172.2.389. [DOI] [PubMed] [Google Scholar]
- 6.Hertz M.I., Englund J.A., Snover D., Bitterman P.B., McGlave P.B. Respiratory syncytial virus-induced acute lung injury in adult patients with bone marrow transplants: a clinical approach and review of the literature. Medicine (Baltim.) 1989;68:269–281. doi: 10.1097/00005792-198909000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Whimbey E., Couch R.B., Englund J.A., Andreeff M., Goodrich J.M., Raad I.I., Lewis V., Mirza N., Luna M.A., Baxter B., Tarrand J.J., Bodey G.P. Respiratory syncytial virus pneumonia in hospitalized adult patients with leukemia. Clin. Infect. Dis. 1995;21:376–379. doi: 10.1093/clinids/21.2.376. [DOI] [PubMed] [Google Scholar]
- 8.Hall C.B. Respiratory syncytial virus. In: Feigin R.D., Cherry J.D., editors. Textbook of Pediatric Infectious Diseases. second ed. Saunders; Philadelphia: 1987. [Google Scholar]
- 9.Walsh E.E., Peterson D.R., Falsey A.R. Risk factors for severe respiratory syncytial virus infection in elderly persons. J. Infect. Dis. 2004;189:233–238. doi: 10.1086/380907. [DOI] [PubMed] [Google Scholar]
- 10.Sidwell R.W., Barnard D.L. Respiratory syncytial virus infections: recent prospects for control. Antivir. Res. 2006;71:379–390. doi: 10.1016/j.antiviral.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses. Arch. Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins P.L., Huang Y.T., Wertz G.W. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J. Virol. 1984;49:572–578. doi: 10.1128/jvi.49.2.572-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins P.L. The molecular biology of human respiratory syncytial viruses (RSV) of the genus Pneumovirus. In: Kingsbury D.W., editor. The Paramyxoviruses. Plenum Publishing; New York: 1991. [Google Scholar]
- 14.Berthiaume L., Joncas J., Pavilanis V. Comparative structure, morphogenesis and biological characteristics of the respiratory syncytial (RS) virus and the pneumonia virus of mice (PVM) Archiv. f. Virusforschung. 1974;45:39–51. doi: 10.1007/BF01240540. [DOI] [PubMed] [Google Scholar]
- 15.Walsh E.E., Schlesinger J.J., Brandriss M.W. Purification and characterization of GP90, one of the envelope glycoproteins of respiratory syncytial virus. J. Gen. Virol. 1984;65:761–767. doi: 10.1099/0022-1317-65-4-761. [DOI] [PubMed] [Google Scholar]
- 16.Wertz G.W., Collins P.L., Huang Y., Gruber C., Levine S., Ball L.A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan J.S., Ray W.C., Peeples M.E. Structure and function of RSV surface glycoproteins. Corr. Top. Microbiol. Immunol. 2013;372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman S.A., Hendry R.M., Beeler J.A. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 1999;73:6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorman J.J., Ferguson B.L., Speelman D., Mills J. Determination of the disulfide bond arrangement of human respiratory syncytial virus attachment (G) protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sci. 1997;6:1308–1315. doi: 10.1002/pro.5560060619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langedijk J.P., Schaaper W.M., Meloen R.H., van Oirschot J.T. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J. Gen. Virol. 1996;77:1249–1257. doi: 10.1099/0022-1317-77-6-1249. [DOI] [PubMed] [Google Scholar]
- 21.Harcourt J., Alvarez R., Jones L.P., Henderson C., Anderson L.J., Tripp R.A. Respiratory syncytial virus G protein and G protein CX3C motif adversely affect CX3CR1+ T cell responses. J. Immunol. 2006;176:1600–1608. doi: 10.4049/jimmunol.176.3.1600. [DOI] [PubMed] [Google Scholar]
- 22.Boyoglu-Barnum S., Todd S.O., Meng J., Barnum T.R., Chirkova T., Haynes L.M., Jadhao S.J., Tripp R.A., Oomens A.G., Moore M.L., Anderson L.J. Mutating the CX3C motif in the G protein should make a live respiratory syncytial virus vaccine safer and more effective. J. Virol. 2017;91 doi: 10.1128/JVI.02059-16. e02059–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernie B.F., Gerin J.L. Immunochemical identification of viral and non-viral proteins of the respiratory syncytial virus virion. Infect. Immun. 1982;37:243–249. doi: 10.1128/iai.37.1.243-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendricks D.A., Baradaran K., Mcintosh K., Patterson J.L. Appearance of a soluble form of the G protein of respiratory syncytial virus in fluids of infected cells. J. Gen. Virol. 1987;68:1705–1714. doi: 10.1099/0022-1317-68-6-1705. [DOI] [PubMed] [Google Scholar]
- 25.Tripp R.A., Jones L.P., Haynes L.M., Zheng H., Murphy P.M., Anderson L.J. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2001;2:732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- 26.Chirkova T., Lin S., Oomens A.G., Gaston K.A., Boyoglu-Barnum S., Meng J., Stobart C.C., Cotton C.U., Hartert T.V., Moore M.L., Ziady A.G., Anderson L.J. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J. Gen. Virol. 2015;96:2543–2556. doi: 10.1099/vir.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H.J., Lee J.Y., Park M.H., Kim J.Y., Chang J. Monoclonal antibody against G glycoprotein increases respiratory syncytial virus clearance in vivo and prevents vaccine-enhanced diseases. PLoS One. 2017;12:e0169139. doi: 10.1371/journal.pone.0169139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones H.G., Ritschel T., Pascual G., Brakenhoff J.P.J., Keogh E., Furmanova-Hollenstein P., Lanckacker E., Wadia J.S., Gilman M.S.A., Williamson R.A., Roymans D., van 't Wout A.B., Langedijk J.P., McLellan J.S. Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies. PLoS Pathog. 2018;14:e1006935. doi: 10.1371/journal.ppat.1006935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaul T.N., Jr Middleton E., Ogra P.L. Antiviral effect of flavonoids on human viruses. J. Med. Virol. 1985;15:71–79. doi: 10.1002/jmv.1890150110. [DOI] [PubMed] [Google Scholar]
- 30.Grotewold E. Springer; New York: 2006. The Science of Flavonoids. [Google Scholar]
- 31.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker John M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 32.The Merck Index. eleventh ed. 1989. Entry# 8044. [Google Scholar]
- 33.Yoshimura E., Liu Z.D., Khodr H., Hider R.C. Purity of commercial morin products and their spectrophotometric characterization. Bunseki Kagaku. 2002;51:979–982. [Google Scholar]
- 34.Sreerama N., Woody R.W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 35.Olsson U., Wolf-Watz M. Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nat. Commun. 2010;1:111–119. doi: 10.1038/ncomms1106. [DOI] [PubMed] [Google Scholar]
- 36.Lakowicz J.R. second ed. Kluwer Academic Publishers/Plenum Press; New York: 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 37.Cantor C.R., Schimel P.R. W. H. Freeman and Company; New York: 1980. Biophysical Chemistry, Part III the Behavior of Biological Macromolecules. [Google Scholar]
- 38.Gianotti A.R., Ferreyra R.G., Ermácora M.R. Binding properties of sterol carrier protein 2 (SCP2) characterized using Laurdan. BBA - Proteins Proteom. 2018;1866:1143–1152. doi: 10.1016/j.bbapap.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Khan W.H., Srungaram V.L.N.R., Islam A., Beg I., Haider Md.S. H., Ahmad F., Brood S., Parveen S. Biophysical characterization of G protein ectodomain of group B human respiratory syncytial virus from E. coli, Prep. Biochem. Biotechnol. 2016;46:483–488. doi: 10.1080/10826068.2015.1084512. [DOI] [PubMed] [Google Scholar]
- 40.Sinha S., Surolia A. Attributes of glycosylation in the establishment of the unfolding pathway of soybean agglutinin. Biophys. J. 2007;92:208–216. doi: 10.1529/biophysj.106.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kar B., Verma P., den Haan R., Sharma A.K. Effect of N-linked glycosylation on the activity and stability of a β-glucosidase from Putranjiva roxburghii. Int. J. Biol. Macromol. 2018;112:490–498. doi: 10.1016/j.ijbiomac.2018.01.201. [DOI] [PubMed] [Google Scholar]
- 42.Wang C., Eufemi M., Turano C., Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 43.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 44.Hatsumi A., Magobei Y. Thermodynamic characterization of drug binding to human serum albumin by isothermal titration microcalorimetry. J. Pharm. Sci. 1994;83:1712–1716. doi: 10.1002/jps.2600831213. [DOI] [PubMed] [Google Scholar]