Abstract
Biological membranes are predominantly composed of structural glycerophospholipids such as phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol. Of the membrane glycerophospholipids, phosphatidylinositol (PtdIns) and its phosphorylated derivatives (phosphoinositides) constitute a minor fraction yet exert a wide variety of regulatory functions in eukaryotic cells. Phosphoinositides include PtdIns, three PtdIns monophosphates, three PtdIns bisphosphates, and one PtdIns triphosphate, in which the hydroxy groups of the inositol head group of PtdIns are phosphorylated by specific lipid kinases. Of all the phosphoinositides in eukaryotic cells, phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2] constitutes the smallest fraction, yet it is a crucial lipid in animal and yeast membrane trafficking systems. Here, we review the recent findings on the physiological functions of PtdIns(3,5)P2 and its enzyme—formation of aploid and binucleate cells (FAB1)—along with the regulatory proteins of FAB1 and the downstream effector proteins of PtdIns(3,5)P2 in Arabidopsis.
Keywords: Arabidopsis; phosphoinositides; biological membranes; PtdIns(3,5)P2; PtdIns-binding proteins
Introduction
Biological membranes are physical barriers that regulate cellular biological reactions through dedicated permeable zones. They are composed of major structural glycerophospholipids such as phosphatidylcholine and phosphatidylethanolamine alongside minor regulatory phospholipids such as phosphatidylinositol (PtdIns) and its phosphorylated derivatives (phosphoinositides), which exert a wide variety of regulatory functions in all eukaryotic cells. Phosphoinositides spatiotemporally regulate diverse downstream cellular pathways via the recruitment of various effector proteins through specific membrane domains (Balla, 2013). Phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2] is the least abundant phosphoinositide in eukaryotic cells, comprising approximately 0.05–0.1% of the total phospholipids. Studies have shown that various mammalian physiological signals such as hormones and growth factors, and osmotic or oxidative stress signals in yeast and plant cells, cause a rapid elevation in PtdIns(3,5)P2 levels (Dove et al., 1997; Meijer et al., 1999; Sbrissa et al., 1999; Bridges et al., 2012; Hirano et al., 2015). PtdIns(3,5)P2 levels are regulated by a synchronized mechanism consisting of PtdIns 3-phosphate 5-kinase, formation of aploid and binucleate cells/FYVE finger-containing phosphoinositide kinase (FAB1/PIKfyve), and a PtdIns(3,5)P2-phosphatase (SAC3/FIG 4) (Hasegawa et al., 2017). FAB1 and PIKfyve are localized in the vacuoles and endolysosomes, respectively, and carry out essential roles in endosomal membrane trafficking, including vacuolar sorting, endocytosis of membrane proteins, ion transport, cytoskeleton dynamics, and retrograde transport in animals and yeasts (Efe et al., 2005; Shisheva, 2008; Ikonomov et al., 2011). PtdIns(3,5)P2 is the product of FAB1, and has crucial roles in maintaining membrane trafficking, autophagy, and signaling mediation in response to various cellular stresses (Shisheva, 2008). It has been demonstrated that loss of FAB1/PIKfyve function causes severe defects during embryogenesis, resulting in embryonic lethality in Drosophila spp., Caenorhabditis elegans, and Mus musculus (Nicot et al., 2006; Rusten et al., 2006; Ikonomov et al., 2011; Takasuga et al., 2012).
The majority of eukaryotes contain a single copy of the FAB1-encoding gene; however, Arabidopsis has four distinct FAB1-related genes (FAB1A, FAB1B, FAB1C, and FAB1D) of which only FAB1A and FAB1B contain a conserved FYVE (FAB1, YOTB, Vac1, and EEA1) domain (Mueller-Roeber and Pical, 2002). The diversity of FAB1 genes indicates the vast array of functions that FAB1 and PtdIns(3,5)P2 have in Arabidopsis and higher plants. This review summarizes the current findings on the physiological roles of PtdIns(3,5)P2, its catalyst enzyme FAB1, and its regulating and effector proteins in Arabidopsis.
Structures of FAB1/PIKfyve Complexes in Yeast, Mammals, and Arabidopsis
FAB1/PIKfyve forms a large protein complex with several regulatory proteins to simultaneously catalyze PtdIns 3-phosphate kinase and PtdIns(3,5)P2 phosphatase reactions, thereby regulating phosphoinositide 3,5-bisphosphate levels.
In yeast, the FAB1/PIKfyve complex is localized to the vacuole membrane and is composed of Fab1p, Fig 4p (Gary et al., 2002), an adaptor-like protein (Vac14p) (Bonangelino et al., 2002), and a FAB1 regulatory protein (Vac7p) (Bonangelino et al., 1997). FIG 4 is a phosphoinositide phosphatase enzyme that specifically catalyzes the production of PtdIns3P from PtdIns(3,5)P2 (Gary et al., 2002; Rudge et al., 2004; Duex et al., 2006a,b). The loss of FIG 4 function impairs the phosphatase and PtdIns3P 5-kinase activity of the FAB1 complex (Rudge et al., 2004; Duex et al., 2006b; Botelho et al., 2008), suggesting that FIG 4 may have an additional regulatory role in PtdIns(3,5)P2 synthesis. The adaptor-like protein Vac14p consists of multiple HEAT repeat arrays (Andrade and Bork, 1995) over almost its entire sequence (Jin et al., 2008), and forms oligomers via its C-terminal region (Dove et al., 2002; Jin et al., 2008; Alghamdi et al., 2013). Vac14p associates with all of the FAB1 complex proteins, acting as a scaffold protein, and it has been proven that the association of Vac14p with Fig 4p is necessary for the positive regulation of PtdIns(3,5)P2 synthesis (Duex et al., 2006a,b; Botelho et al., 2008; Jin et al., 2008). The FAB1 regulatory protein Vac7p is the only transmembrane protein with no conserved motif and no known metazoan homologs (Gary et al., 2002). Vac7p and Vac14p have been demonstrated to independently regulate PtdIns(3,5)P2 levels in yeast (Duex et al., 2006b).
In metazoan cells, FAB1 (also called PIKfyve in mammals) (Sbrissa et al., 1999) is localized in the early and late endosomes where it forms a complex with VAC14 (ArPIKfyve in mammals) (Sbrissa et al., 2004) and SAC3 (FIG 4) (Sbrissa et al., 2007; Ikonomov et al., 2013). The triple complex composed of PIKfyve (FAB1), ArPIKfyve (Vac14), and SAC3 (FIG 4) is known as the PAS complex, and regulates the synthesis and turnover of PtdIns(3,5)P2 (Sbrissa et al., 2007, 2008; Ikonomov et al., 2009).
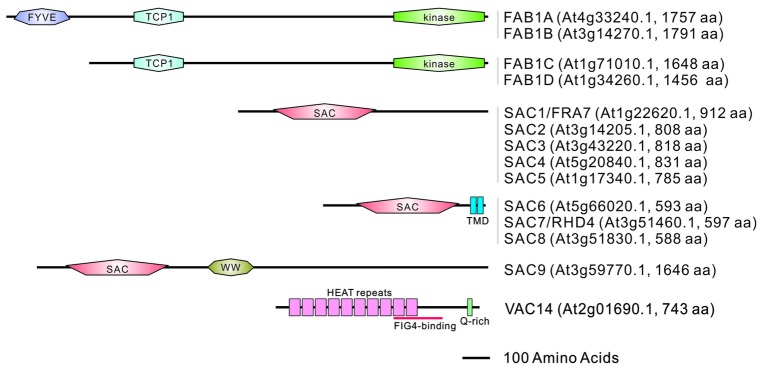
The Arabidopsis genome encodes four types of FAB1 (FAB1A–FAB1D), named based on their similarity to the yeast FAB1 (Whitley et al., 2009). Yeast FAB1 and mammalian PIKfyve have a conserved N-terminal FYVE (FAB1p, YOTB, Vac1p, and EEA1)-finger domain, the central part of which comprises a Cpn_TCP1 (HSP chaperonin T-complex protein 1) homology domain and the C-terminal of which comprises a kinase catalytic domain. The FYVE-finger domain is responsible for binding PtdIns3P and localizing it to the endosomes (Shisheva, 2008). Arabidopsis FAB1A and FAB1B also possess a conserved FYVE domain, and the fab1afab1b double mutant has a male gametophyte-lethal phenotype suggesting functional redundancy between FAB1A and FAB1B in Arabidopsis. Phylogenic analysis has indicated that FAB1 orthologs without the FYVE domain are clustered in the higher plant lineage (Whitley et al., 2009). Since the expression of FAB1C and FAB1D has been detected in various tissues (Bak et al., 2013; Serrazina et al., 2014), these proteins are likely to have diversified during the evolution of higher plants where they may have acquired novel functions beyond the canonical FAB1 (FAB1A and FAB1B) proteins. FAB1B and FAB1D have complementary roles in the regulation of membrane recycling, vacuolar pH, and the homeostatic ROS control in pollen tube growth, despite the cytosolic localization of FAB1D in the pollen tube (Serrazina et al., 2014). Unlike the gene arrangement of FAB1, Arabidopsis has a single VAC14 gene (Zhang et al., 2018). Bimolecular fluorescence complementation has revealed that FAB1A/B and VAC14 physically interact to form a functional protein complex in Arabidopsis (Whitley et al., 2009; Hirano et al., 2011; Zhang et al., 2018). The Arabidopsis genome includes 9 SAC-domain containing phosphatase (SAC1-SAC9) genes (Zhong and Ye, 2003) which can be separated into three different classes based on their homology; SAC1-SAC5 are the most similar to yeast FIG4p, SAC6-SAC8 have two C-terminal transmembrane domains and are the most similar to yeast SAC1p, and SAC9 is the largest protein containing a unique WW domain (Zhong and Ye, 2003) (Figure 1).
Figure 1.
Domain structures of FAB1, SAC family proteins, and VAC14 in Arabidopsis. Domain structures were identified using the PROSITE program (https://prosite.expasy.org/). FYVE; FAB1, YOTB, VAC1, EEA1 zinc finger domain, TCP1; Tailless Complex Polypeptide 1 domain found in chaperone proteins, SAC; suppressor of actin domain, TMD; transmembrane domain, WW; a domain containing two conserved tryptophans, HEAT repeats; a tandem repeat structural motif composed of two alpha helices linked by a short loop found in Huntingtin, Elongation factor 3, protein phosphatase 2A, TOR1, Q-rich; a glutamine rich domain.
Subcellular Localization FAB1 and PtdIns(3,5)P2 in Arabidopsis
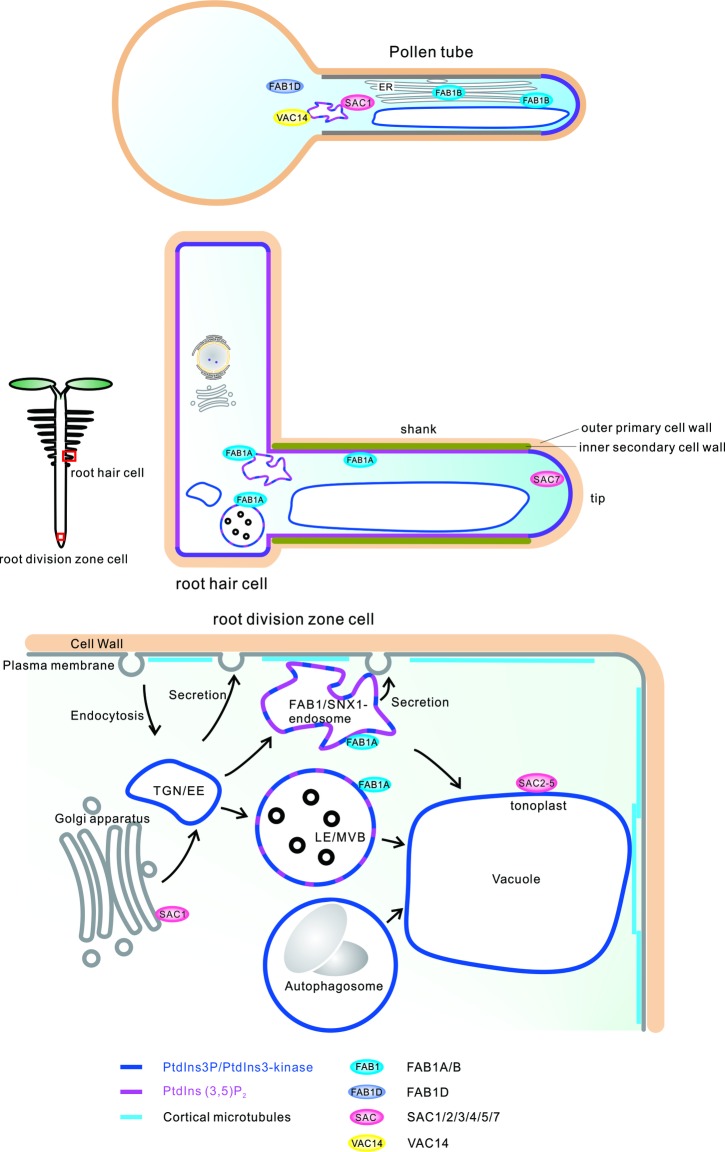
In yeast and mammalian cells, the FAB1/PAS complex (comprising of FAB1, VAC14, & FIG 4) is localized in the vacuoles, endosomes, and lysosomes (Gary et al., 1998; Shisheva et al., 2001; Rudge et al., 2004; Botelho et al., 2008; Jin et al., 2008; Zhang et al., 2012). In Arabidopsis, FAB1A is predominantly localized in the SORTING NEXIN-1 (SNX-1)-residing late endosomes of the developmental cell division, transition, and elongation zones of epidermal and cortical cells (Hirano et al., 2015), whereas FAB1B localizes to the endomembrane compartments including endoplasmic reticulum (ER)-like reticulate structures and vacuoles of the pollen tubes. However FAB1D, the plant-specific FAB1 ortholog with no FYVE-domain, is mainly localized in the cytosol indicating that the N-terminal FYVE-domain of FAB1 is necessary for its endosomal localization in Arabidopsis (Serrazina et al., 2014).
The fluorescent PtdIns(3,5)P2-specific probe, based on tandem repeats of the cytosolic PtdIns(3,5)P2-interacting domain (ML1N) of the mammalian lysosomal transient receptor potential cation channel, Mucolipin 1 (TRPML1), was developed to label mammalian (Li et al., 2013) and Arabidopsis cells. Using this probe, PtdIns(3,5)P2 was predominantly observed on late endosomes in root cells. Unlike the yeast and mammalian cells, the vacuolar localization of the PI(3,5)P2-specific probe was never observed in Arabidopsis (Hirano et al., 2017b).
Intriguingly, FAB1A and PtdIns(3,5)P2 have been shown to be localized in the plasma membrane on the shank of growing root hairs, hardening the region by constructing a secondary cell wall and cortical microtubule structures. This suggests that FAB1 and PtdIns(3,5)P2 have acquired a novel function whereby they harden the root hair cell wall to regulate the organization of cortical microtubules and secretion of secondary cell wall structural components in higher plants (Hirano et al., 2018) (Figure 2).
Figure 2.
Subcellular localization of PtdIns3P, PtdIns(3,5)P2, FAB1s, SACs, and VAC14 in developing and mature Arabidopsis cells.
Physiological Functions of FAB1 and PtsIns(3,5)P2 in Arabidopsis
The most striking feature of PtdIns(3,5)P2 deficiency or FAB1(PIKfyve) dysfunction in many eukaryotic cells is the enlargement of vacuoles, endosomes, or lysosomes (Yamamoto et al., 1995; Ikonomov et al., 2001; Rutherford et al., 2006; Jefferies et al., 2008; Whitley et al., 2009; Hirano et al., 2011; Takasuga et al., 2012). In mammalian cells, impaired FAB1(PIKfyve) function has been reported to cause severe defects during embryogenesis, resulting in embryonic lethality in animals (Nicot et al., 2006; Rusten et al., 2006; Ikonomov et al., 2011; Takasuga et al., 2012). Furthermore, the fab1afab1b revealed a lethal male gametophyte phenotype in Arabidopsis (Whitley et al., 2009), with mutant pollen showing severe vacuolar reorganization and vacuolar acidification defects following the first mitotic division (Whitley et al., 2009). The inhibition of PtdIns(3,5)P2 production reduced vacuolar acidification and convolution, and delayed stomatal closure in response to ABA. Since vacuolar H+-pyrophosphatase has been shown to bind PtdIns(3,5)P2 in vitro, the authors hypothesized that PtdIns(3,5)P2 may stimulate the H+-pumping activity of vacuolar H+-pyrophosphatase in ABA-dependent stomatal closure (Bak et al., 2013). However, a patch-clamp study of the Arabidopsis vacuole demonstrated that nanomolar levels of PI(3,5)P2 regulate chloride channel a (CLC-a), a member of the anion/H+ exchanger family, which is implicated in stomatal movements in Arabidopsis, but not H+-pyrophosphatase (Carpaneto et al., 2017). These observations suggest that PtdIns(3,5)P2 is localized in the vacuolar membrane where it exerts various vacuolar functions to regulate PtdIns(3,5)P2 effector proteins, however the presence of PtdIns(3,5)P2 on the vacuolar membrane could not be detected by ML1N-based fluorescent probably due to the detection limit of the probe (Kd of GFP-ML1N*2 was calculated to be 5.6 μM (Li et al., 2013)).
The conditional down-regulation of FAB1A and FAB1B expression has been shown to cause various abnormal phenotypes including growth inhibition, hypo-sensitivity to exogenous auxin, disturbance of root gravitropism, and floral organ abnormalities. These pleiotropic developmental phenotypes are likely related to auxin signaling in Arabidopsis (Hirano et al., 2011). Auxin signaling defects in fab1afab1b knockdown are caused by the abnormal localization of the PIN2 and AUX1 auxin transporters, likely due to the inhibition of the late endosomal maturation process (Hirano and Sato, 2011; Hirano et al., 2015, 2017a). It has also been reported that conditional knockdown of FAB1A and FAB1B, or inhibition of PtdIns(3,5)P2 production using a FAB1 inhibitor (YM201636), induces the release of late endosomes from cortical microtubules and disturbs cortical microtubule organization, highlighting the importance of late endosome association in proper cortical microtubule construction (Hirano et al., 2015). Studies have shown although FAB1C lacks the conserved N-terminal FYVE domain required to bind PtdIns3P, it still successfully produces PtdIns(3,5)P2 from PtdIns3P in vitro (Bak et al., 2013). In addition, fab1b and fab1c T-DNA mutants exhibit stomatal closure defects, implying that FAB1B and FAB1C have overlapping functions in stomatal closure to control PtdIns(3,5)P2 levels in Arabidopsis guard cells (Bak et al., 2013). In fab1b and fab1d single mutants, pollen viability, germination, and tube morphology were not significantly affected, although the pollen tubes of these mutants were found to exhibit altered membrane recycling, vacuolar acidification, and decreased reactive oxygen species (ROS) production (Serrazina et al., 2014). Lack of the N-terminal FYVE-domain in FAB1C and FAB1D may confer different subcellular localization patterns of these proteins in plant cells. In fact, a subcellular localization prediction program (SUBA4) predicts the nuclear localization of FAB1D (Hooper et al., 2017), suggesting the PtdIns(3,5)P2 synthesis role of FAB1D in nucleus. Future studies are necessary to determine the precise subcellular localization of FAB1C and FAB1D in plant cells.
Function of FAB1- and PtdIns(3,5)P2-Associated Proteins
Loss of VAC14 function leads to a lethal male gametophyte phenotype caused by vacuolar reorganization defects during pollen development (Zhang et al., 2018). A similar male gametophyte phenotype is observed in vac14 and fab1afab1b mutants, and bimolecular fluorescence complementation suggests that FAB1A/B and VAC14 physically interact to form a crucial functional protein complex in Arabidopsis (Whitley et al., 2009; Hirano et al., 2011; Zhang et al., 2018).
The SAC1 protein has been shown to have PtdIns(3,5)P2 phosphatase activity (Zhong et al., 2005), with root hair elongation defects observed in sac1–2 homozygous T-DNA mutants. In contrast, gain of function SAC1 mutants have longer root hairs than the wild-type, indicating that SAC1 is essential for elongation during root hair morphogenesis (Vijayakumar et al., 2016). Although a direct interaction between FAB1 and SAC1 has not yet been reported, SAC1 and FAB1B have been found to co-localize in the wortmannin-sensitive vesicles of pollen and pollen tubes (Zhang et al., 2018). These data suggest the formation of a complex between SAC1 and FAB1 in Arabidopsis.
The data also suggests the presence of PtdIns(3,5)P2 phosphatase on the vacuolar membranes of Arabidopsis. In fact, yeast FIG 4 orthologs such as SAC2-SAC5 are localized in the vacuolar membrane to catalyze the conversion of PtdIns(3,5)P2 to PtdIns3P, thereby controlling the balance between these phosphoinositides and maintaining the morphology of storage and lytic vacuoles (Vermeer et al., 2006; Nováková et al., 2014). SAC6/SAC1b-, SAC7/SAC1c/RHD4- and SAC8/SAC1a-encoded proteins have been found to be similar to yeast Sac1p and can rescue yeast from Sac1p mutations (Despres et al., 2003), whereas SAC7 and SAC8 are expressed broadly and SAC6 is expressed only in pollen (Despres et al., 2003; Zhong and Ye, 2003). Mutations in the SAC7/RHD4 protein have been associated with aberrant root hairs, while mutant rhd4–1 roots accumulated higher levels of PtdIns4P in vivo, indicating that SAC7/RHD4 has a role in the regulation of PtdIns4P accumulation on the membrane compartments of growing root hair tips (Thole et al., 2008). sac9 mutants have been shown to accumulate significant levels of PtdIns(4,5)P2 and PtdIns4P, and have a constitutively stressed phenotype with shorter roots and extreme cell wall and membrane structure abnormalities (Williams et al., 2005; Vollmer et al., 2011) (Figure 2).
PtdIns(3,5)P2 Effector Molecules
PtdIns(3,5)P2 is the least abundant phosphoinositide in eukaryotes (Balla, 2013), and constitutes 0.02% of the total phospholipids in Arabidopsis (Hirano et al., 2015). Although low in abundance, PtdIns(3,5)P2 has various physiological functions in Arabidopsis as a signaling molecule, which cannot be carried out without the help of a range of effector proteins. Recently, a wide variety of effector proteins have been reported to associate with PtdIns(3,5)P2. In yeast, it has been demonstrated that autophagy related protein 18 (ATG18) is necessary for autophagy, the cytoplasm-to-vacuole targeting (Cvt) pathway, the delivery of the hydrolase aminopeptidase I to the vacuole, and the early steps of autophagosome formation (Barth et al., 2001; Guan et al., 2001). ATG18 has both PtdIns3P- and PtdIns(3,5)P2-binding sites, which are essential for its localization on vacuole membranes and its function as a negative regulator of PtdIns(3,5)P2 synthesis (Hasegawa et al., 2017). Eight orthologous ATG18 proteins (ATG18a-ATG18h) have been found in the Arabidopsis genome, forming three major subgroups: ATG18a, ATG18c, ATG18d, and ATG18e form a cluster similar with the yeast protein Ygr223c; ATG18b is similar to yeast ATG18, whilst ATG18f, ATG18g, and ATG18h are more divergent forms (Xiong et al., 2005). Among these, ATG18a is required for autophagosome formation during nutrient stress and senescence conditions (Xiong et al., 2005) in coordination with ATG9 (Zhuang et al., 2017).
The plant immunophilin ROF1 containing a FK506 binding domain (FKBP), plays a significant role in the osmotic/salt stress responses of germinating seeds, and interacts directly with PI3P and PtdIns(3,5)P2 (Karali et al., 2012). The hydrophilic cation-binding proteins, PCaP1 and PCaP2, are involved in stomatal closure (Nagata et al., 2016) and root hair morphogenesis (Kato et al., 2013). They were also found to preferentially interact with PI(3,5)P2 and PtdIns(3,4,5)P3, however these interactions were inhibited by association with calmodulin in a Ca2+-dependent manner (Nagasaki et al., 2008; Kato et al., 2013). The endosomal protein SNX1 binds to PI3P as well as PI(3,5)P2, and is involved in the auxin pathway, regulates endosome maturation (Hirano et al., 2015), PIN2 trafficking (Jaillais et al., 2006), and cortical microtubule organization along with CLIP-associated protein (CLASP) (Ambrose et al., 2013). Vacuolar membrane-localized H+-translocating pyrophosphatase (V-PPase) binds to PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 suggesting that PtdIns(3,5)P2 may regulate vacuole acidification (Bak et al., 2013). Type-II Rho-related GTPase from plants 10 (ROP10) binds FAB1 and various phosphoinositides [PtdIns3P, PtdIns(3,5)P2, PtdIns4P and PtdIns(4,5)P2], resulting in localization to and hardening of the root hair shank (Hirano et al., 2018).
Conclusions and Future Prospects
In plants, the establishment of cell polarity is important for patterning processes. It has been reported that PtdIns(4,5)P2 and PtdIns4P 5-kinase, which mediates their interconversion, are specifically enriched in the apical and/or basal polar plasma membrane domains, thereby controlling the polar localization of apical and basal cargoes in specialized cells such as root hairs, pollen tubes, and root cells (Ischebeck et al., 2008; Kusano et al., 2008; Stenzel et al., 2008; Tejos et al., 2014). A recent study showed that FAB1B and PtdIns(3,5)P2 are predominantly localized in the plasma membrane of the root hair shank to control cortical microtubule organization and cell wall construction, thereby mediating root hair shank hardening in Arabidopsis. These results suggest that PtdIns(3,5)P2 and PtdIns(4,5)P2 have crucial roles in establishing cell polarity in specialized cells like root hairs. However, the function and significance of the majority of molecules involved in the PtdIns(3,5)P2-mediated regulation of cellular processes remains largely unknown. Future studies are required to determine the roles of PtdIns(3,5)P2 and its unique regulatory mechanisms in higher plants.
Data Availability
The datasets for this manuscript are not publicly available. Requests to access the datasets should be directed to mhsato@kpu.ac.jp.
Author Contributions
TH and MS wrote the review. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.jp) for English language editing.
Footnotes
Funding. This work was supported by the Japan Society for the Promotion of Science, a grant-in-aid for Scientific Research (16H05068 to MS and 17K08200 to TH).
References
- Alghamdi T. A., Ho C. Y., Mrakovic A., Taylor D., Mao D., Botelho R. J. (2013). Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J. Biol. Chem. 288, 9363–9372. 10.1074/jbc.M113.453712, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L. M., Catching A., Kirik V., et al. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24, 649–659. 10.1016/j.devcel.2013.02.007, PMID: [DOI] [PubMed] [Google Scholar]
- Andrade M., Bork P. (1995). HEAT repeats in the Huntington’s disease protein. Nat. Genet. 11, 115–116. 10.1038/ng1095-115, PMID: [DOI] [PubMed] [Google Scholar]
- Bak G., Lee E.-J., Lee Y., Kato M., Segami S., Sze H., et al. (2013). Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell 25, 2202–2216. 10.1105/tpc.113.110411, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. 10.1152/physrev.00028.2012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H., Meiling-Wesse K., Epple U. D., Thumm M. (2001). Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508, 23–28. 10.1016/S0014-5793(01)03016-2, PMID: [DOI] [PubMed] [Google Scholar]
- Bonangelino C. J., Catlett N. L., Weisman L. S. (1997). Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 17, 6847–6858. 10.1128/MCB.17.12.6847, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C. J., Nau J. J., Duex J. E., Brinkman M., Wurmser A. E., Gary J. D., et al. (2002). Osmotic stress-induced increase of phophatidylinositol 3,5-bisphosphate requreis Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 18, 1015–1028. 10.1083/jcb.200201002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho R. J., Efe J. A., Teis D., Emr S. D. (2008). Assembly of a Fab1 phosphoinositide kinase Ssignaling complex requires the Fig 4 phosphoinositide phosphatase. Mol. Biol. Cell 19, 4273–4286. 10.1091/mbc.e08-04-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D., Ma J.-T., Park S., Inoki K., Weisman L. S., Saltiel A. R. (2012). Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 23, 2955–2962. 10.1091/mbc.e11-12-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpaneto A., Boccaccio A., Lagostena L., Zanni E. D., Scholz-Starke J. (2017). The signaling lipid phosphatidylinositol-3,5-bisphophate targets plants CLC-a anion/H+ exchange activity. EMBO Rep. 18, 1100–1107. 10.15252/embr.201643814, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres B., Bouissonnié F., Wu H. J., Gomord V., Guilleminot J., Grellet F., et al. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. 10.1046/j.1365-313X.2003.01720.x, PMID: [DOI] [PubMed] [Google Scholar]
- Dove S. K., Cooke F. T., Douglas M. R., Sayers L. G., Parker P. J., Michell R. H. (1997). Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390, 187–192. 10.1038/36613, PMID: [DOI] [PubMed] [Google Scholar]
- Dove S. K., Mcewen R. K., Mayes A., Hughes D. C., Beggs J. D., Michell R. H. (2002). Vac14 controls PtdIns(3,5)P2 synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12, 885–893. 10.1016/S0960-9822(02)00891-6, PMID: [DOI] [PubMed] [Google Scholar]
- Duex J. E., Nau J. J., Kauffman E. J., Weisman L. S. (2006a). Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukyaryot. Cell 5, 723–731. 10.1128/EC.5.4.723-731.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex J. E., Tang F., Weisman L. S. (2006b). The Vac14p-Fig 4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 172, 693–704. 10.1083/jcb.200512105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J. A., Botelho R. J., Emr S. D. (2005). The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Curr. Opin. Cell Biol. 17, 402–408. 10.1016/j.ceb.2005.06.002, PMID: [DOI] [PubMed] [Google Scholar]
- Gary J. D., Sato T. K., Stefan C. J., Bonangelino C. J., Weisman L. S., Emr S. D. (2002). Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig 4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell 13, 1238–1251. 10.1091/mbc.01-10-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J. D., Wurmser A. E., Bonangelino C. J., Weisman L. S., Emr S. D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65–79. 10.1083/jcb.143.1.65, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J., Stromhaug P. E., George M. D., Habibzadegah-Tari P., Bevan A., Dunn W. A., Jr., et al. (2001). Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell 12, 3821–3838. 10.1091/mbc.12.12.3821, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J., Strunk B. S., Weisman L. S. (2017). PI5P and PI (3,5)P2: minor, but essential phosphoinositides. Cell Struct. Funct. 60, 49–60. 10.1247/csf.17003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Konno H., Takeda S., Dolan L., Kato M., Aoyama T., et al. (2018). PtdIns(3,5)P2 mediates root hair shank hardening in Arabidopsis. Nat. Plants 4, 888–897. 10.1038/s41477-018-0277-8, PMID: [DOI] [PubMed] [Google Scholar]
- Hirano T., Matsuzawa T., Takegawa K., Sato M. H. (2011). Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol. 155, 797–807. 10.1104/pp.110.167981, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Munnik T., Sato M. H. (2015). Phosphatidylinositol 3-phosphate 5-kinase, FAB1/PIKfyve kinase mediates endosome maturation to establish endosome-cortical microtubule interaction in Arabidopsis. Plant Physiol. 169, 1961–1974. 10.1104/pp.15.01368, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Munnik T., Sato M. H. (2017a). Inhibition of phosphatidylinositol 3,5-bisphosphate production has pleiotropic effects on various membrane trafficking routes in Arabidopsis. Plant Cell Physiol. 58, 120–129. 10.1093/pcp/pcw164 [DOI] [PubMed] [Google Scholar]
- Hirano T., Sato M. H. (2011). Arabidopsis FAB1A/B is possibly involved in the recycling of auxin transporters. Plant Signal. Behav. 6, 583–585. 10.4161/psb.6.4.15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Stecker K., Munnik T., Xu H., Sato M. H. (2017b). Visualization of phosphatidylinositol 3,5-bisphosphate dynamics by a tandem ML1N-based fluorescent protein probe in Arabidopsis. Plant Cell Physiol. 58, 1185–1195. 10.1093/pcp/pcx011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C. M., Castleden I. R., Tanz S. K., Aryamanesh A., Millar A. H. (2017). SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acid Res. 45, D1064–D1074. 10.1093/narlgkw1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Heilmann I. (2008). Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotina tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 20, 3312–3330. 10.1105/tpc.108.059568, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Filios C., Sbrissa D., Chen X., Shisheva A. (2013). The PIKfyve-ArPIKfyve-Sac3 triad in human breast cancer: functional link between elevated Sac3 phosphatase and enhanced proliferation of triple negative cell lines. Biochem. Biophys. Res. Commun. 440, 342–347. 10.1016/j.bbrc.2013.09.080, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D., Delvecchio K., Xie Y., Jin J.-P., Rappolee D., et al. (2011). The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J. Biol. Chem. 286, 13404–13413. 10.1074/jbc.M111.222364, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D., Fenner H., Shisheva A. (2009). PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J. Biol. Chem. 284, 35794–35806. 10.1074/jbc.M109.037515, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D., Shisheva A. (2001). Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 276, 26141–26147. 10.1074/jbc.M101722200, PMID: [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miege C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443, 106–109. 10.1038/nature05046, PMID: [DOI] [PubMed] [Google Scholar]
- Jefferies H. B. J., Cooke F. T., Jat P., Boucheron C., Koizumi T., Hayakawa M., et al. (2008). A selective PIKfyve inhibitor blocks PtdIns(3,5)P2 production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 9, 164–170. 10.1038/sj.embor.7401155, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Chow C. Y., Liu L., Zolov S. N., Bronson R., Davisson M., et al. (2008). VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 27, 3221–3234. 10.1038/emboj.2008.248, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali D., Oxley D., Runions J., Ktistakis N., Farmaki T. (2012). The Arabidopsis thaliana immunophilin ROF1 directly interacts with PI(3)P and PI(3,5)P2 and affects germination under osmotic stress. PLoS One 7:e48241. 10.1371/journal.pone.0048241, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Aoyama T., Maeshima M. (2013). The Ca2+ binding protein PCaP2 located on the plasma membrane is involved in root hair development as a possible signal transducer. Plant J. 74, 690–700. 10.1111/tpj.12155, PMID: [DOI] [PubMed] [Google Scholar]
- Kusano H., Testerink C. H., Verneer J. E. M., Tsuge T., Shimada H., Oka A., et al. (2008). The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K2 is a key regulator of root hair tip growth. Plant Cell 20, 367–380. 10.1105/tpc.107.056119, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Zhang X., Zhao M., Tsang W. L., Zhang Y., et al. (2013). Genetically encoded fluorescent probe to bisualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. U. S. A. 110, 21165–21170. 10.1073/pnas.1311864110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer H. J. G., Divecha N., van den Ende H., Musgrave A., Munnik T. (1999). Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta 208, 294–298. 10.1007/s004250050 [DOI] [Google Scholar]
- Mueller-Roeber B., Pical C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46. 10.1104/pp.004770, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki N., Tomioka R., Maeshima M. (2008). A hydrophilic cation-binding protein of Arabidopsis thaliana, AtPCaP1, is localized to plasma membrane via N-myristoylation and interacts with calmodulin and the phosphatidylinositol phosphates PtdIns(3,4,5)P3 and PtdIns(3,5)P2. FEBS J. 275, 2267–2282. 10.1111/j.1742-4658.2008.06379.x, PMID: [DOI] [PubMed] [Google Scholar]
- Nagata C., Miwa C., Tanaka N., Kato M., Suito M., Tsuchihira A., et al. (2016). A novel-type phosphatidylinositol phosphate-interactive, Ca-binding protein PCaP1 in Arabidopsis thaliana: stable association with plasma membrane and partial involvement in stomata closure. J. Plant Res. 129, 539–550. 10.1007/s10265-016-0787-2, PMID: [DOI] [PubMed] [Google Scholar]
- Nicot A., Fares H., Payrastre B., Chisholm A. D., Labouesse M., Laporte J. (2006). The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol. Biol. Cell 17, 3062–3074. 10.1091/mbc.e05-12-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková P., Hirsch S., Feraru E., Tejos R., van Wijk R., Viaene T., et al. (2014). SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 2818–2823. 10.1073/pnas.1324264111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Anderson D. M., Emr S. D. (2004). Vacuole size control: regulation of PtdIns (3,5)P2 levels by the vacuole-associated Vac14-Fig 4 complex, a PtdIns (3,5)P2-specific phosphatase. Mol. Biol. Cell 15, 24–36. 10.1091/mbc.e03-05-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T. E., Rodahl L. M. W., Pattni K., Englund C., Samakovlis C., Dove S., et al. (2006). Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol. Biol. Cell 17, 3989–4001. 10.1091/mbc.e06-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A. C., Traer C., Wassmer T., Pattni K., Bujny M. V., Carlton J. G., et al. (2006). The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 119, 3944–3957. 10.1242/jcs.03153, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Fenner H., Shisheva A. (2008). ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J. Mol. Biol. 384, 766–779. 10.1016/j.jmb.2008.10.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., et al. (2007). Core protein machinery for mammalian phosphatidylinositol 3, 5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. J. Biol. Chem. 282, 23878–23891. 10.1074/jbc.M611678200, PMID: [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Shisheva A. (1999). PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. J. Biol. Chem. 274, 21589–21597. 10.1074/jbc.274.31.21589, PMID: [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Strakova J., Dondapati R., Mlak K., Deeb R., et al. (2004). A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol. Cell. Biol. 24, 10437–10447. 10.1128/MCB.24.23.10437.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrazina S., Dias F. V., Malhó R. (2014). Characterization of FAB1 phosphatidylinositol kinases in Arabidopsis pollen tube growth and fertilization. New Phytol. 203, 784–793. 10.1111/nph.12836, PMID: [DOI] [PubMed] [Google Scholar]
- Shisheva A. (2008). PIKfyve: partners, significance, debates and paradoxes. Cell Biol. Int. 32, 591–604. 10.1016/j.cellbi.2008.01.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisheva A., Rusin B., Ikonomov O. C., DeMarco C., Sbrissa D. (2001). Localization and Insulin-regulated relocation of phosphoinositide 5-kinase PIKfyve in 3T3-L1 adipocytes. J. Biol. Chem. 276, 11859–11869. 10.1074/jbc.M008437200, PMID: [DOI] [PubMed] [Google Scholar]
- Stenzel I., Ischebeck I., König S., Holubowska A., Sporysz M., Hause B., et al. (2008). The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20, 124–141. 10.1105/tpc.107.052852, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasuga S., Horie Y., Sasaki J., Sun-wada G., Kawamura N., Iizuka R. (2012). Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc. Natl. Acad. Sci. U. S. A. 110, 1726–1731. 10.1073/pnas.1213212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejos R., Sauer M., Vanneste S., Palacios-Gomez M., Li H., Heilmann M., et al. (2014). Bipolar plasma membrane distribution of phosphoinosities and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26, 2114–2128. 10.1105/tpc.114.126185, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole J. M., Vermeer J. E. M., Zhang Y., Gadella T. W. J., Nielsen E. (2008). ROOT HAIR DEFECTIVE4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20, 381–395. 10.1105/tpc.107.054304, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J. E. M., van Leeuwen W., Tobeña-Santamaria R., Laxalt A. M., Jones D. R., Divecha N., et al. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700. 10.1111/j.1365-313X.2006.02830.x, PMID: [DOI] [PubMed] [Google Scholar]
- Vijayakumar P., Datta S., Dolan L. (2016). ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 212, 944–953. 10.1111/nph.14095, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer A. H., Youssef N. N., DeWald D. B. (2011). Unique cell wall abnormalities in the putative phosphoinositide phosphatase mutant AtSAC9. Planta 234, 993–1005. 10.1007/s00425-011-1454-4, PMID: [DOI] [PubMed] [Google Scholar]
- Whitley P., Hinz S., Doughty J. (2009). Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol. 151, 1812–1822. 10.1104/pp.109.146159, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. E., Torabinejad J., Cohick E., Parker K., Drake E. J., Thompson J. E., et al. (2005). Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 138, 686–700. 10.1104/pp.105.061317, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Bassham D. C., Biology C., Plant I., Program P. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546. 10.1111/j.1365-313X.2005.02397.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Dewald D. B., Boronenkov I. V., Anderson R. A., Emr S. D., Koshland D. (1995). Novel PI (4) P5-kinase homologue, Fablp, essential for normal vacuole function and morphology in yeast. Mol. Cell. Biol. 6, 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li E., Guo Y., Yu S., Wan Z., Ma T., et al. (2018). Arabidopsis VAC14 is critical for pollen development mediating vacuolar organization. Plant Physiol. 177, 1529–1538. 10.1104/pp.18.00495, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCartney A. J., Zolov S. N., Ferguson C. J., Meisler M. H., Sutton M. A., et al. (2012). Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P2 and PI(5)P. EMBO J. 31, 3442–3456. 10.1038/emboj.2012.200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Burk D. H., Nairn C. J., Wood-Jones A., Morrison W. H., Ye Z.-H. (2005). Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17, 1449–1466. 10.1105/tpc.105.031377, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z. (2003). The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 132, 544–555. 10.1104/pp.103.021444, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Chung K. P., Cui Y., Lin W., Gao C., Kang B.-H., et al. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. 114, E426–E435. 10.1073/pnas.1616299114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets for this manuscript are not publicly available. Requests to access the datasets should be directed to mhsato@kpu.ac.jp.