Mishra et al. present a composite extreme-phenotype strategy for gene mapping of cerebral small vessel disease in population cohorts of older persons. They identify associations with variants in HTRA1 and NOTCH3, and describe two participants heterozygous for known pathogenic variants for familial small vessel disease in NOTCH3 and HTRA1.
Keywords: cerebral small vessel disease, white matter hyperintensity, lacunes of presumed vascular origin, extreme phenotype, exome sequencing study
Abstract
We report a composite extreme phenotype design using distribution of white matter hyperintensities and brain infarcts in a population-based cohort of older persons for gene-mapping of cerebral small vessel disease. We demonstrate its application in the 3C-Dijon whole exome sequencing (WES) study (n = 1924, nWESextremes = 512), with both single variant and gene-based association tests. We used other population-based cohort studies participating in the CHARGE consortium for replication, using whole exome sequencing (nWES = 2,868, nWESextremes = 956) and genome-wide genotypes (nGW = 9924, nGWextremes = 3308). We restricted our study to candidate genes known to harbour mutations for Mendelian small vessel disease: NOTCH3, HTRA1, COL4A1, COL4A2 and TREX1. We identified significant associations of a common intronic variant in HTRA1, rs2293871 using single variant association testing (Pdiscovery = 8.21 × 10−5, Preplication = 5.25 × 10−3, Pcombined = 4.72 × 10−5) and of NOTCH3 using gene-based tests (Pdiscovery = 1.61 × 10−2, Preplication = 3.99 × 10−2, Pcombined = 5.31 × 10−3). Follow-up analysis identified significant association of rs2293871 with small vessel ischaemic stroke, and two blood expression quantitative trait loci of HTRA1 in linkage disequilibrium. Additionally, we identified two participants in the 3C-Dijon cohort (0.4%) carrying heterozygote genotypes at known pathogenic variants for familial small vessel disease within NOTCH3 and HTRA1. In conclusion, our proof-of-concept study provides strong evidence that using a novel composite MRI-derived phenotype for extremes of small vessel disease can facilitate the identification of genetic variants underlying small vessel disease, both common variants and those with rare and low frequency. The findings demonstrate shared mechanisms and a continuum between genes underlying Mendelian small vessel disease and those contributing to the common, multifactorial form of the disease.
Introduction
Cerebral small vessel disease (SVD) encompasses a group of pathological processes affecting small arteries, arterioles, capillaries and small veins in the brain. It is associated with cognitive impairment, mood disorders, dysfunction of gait and balance, and with increased risk of stroke, dementia and death (Pantoni, 2010). Specific mechanistic treatments for SVD are yet to be identified. Identifying genes underlying SVD may provide important insight on pathways driving this disease and accelerate the discovery of novel drug targets. SVD is driven by a complex mix of environmental and genetic risk factors (Longstreth, 2005) and both familial and sporadic conditions of the disease have been reported. Mutations in the NOTCH3, HTRA1, COL4A1, COL4A2 and TREX1 genes are known to cause rare familial forms of SVD (Joutel et al., 1997; Richards et al., 2007; Vahedi et al., 2007; Hara et al., 2009; Gunda et al., 2014) but endeavours to detect genetic risk factors for the common multifactorial form of SVD are still at a preliminary stage. Studies on multiple complex disorders including a few reports on SVD have suggested that genes harbouring mutations leading to the Mendelian form of the disease may also harbour polymorphisms leading to the sporadic condition (Schmidt et al., 2011; Rannikmae et al., 2015; Stitziel et al., 2015; Fuchsberger et al., 2016).
MRI markers of vascular brain injury, including burden of white matter hyperintensities (WMH) and small subcortical infarcts, namely lacunes of presumed vascular origin (hereafter referred to as lacunes), have been shown to reflect primarily SVD and are commonly used for its diagnosis and assessment of severity (Wardlaw et al., 2013). These MRI markers are heritable with reported heritability estimates ranging between 49% and 80% for WMH burden, and ∼29% for lacunes (Turner et al., 2004; DeStefano et al., 2009; Sachdev et al., 2013). To date, genome-wide association studies (GWAS) reported five WMH burden risk loci that explain only a small proportion of its heritable component (Fornage et al., 2011; Verhaaren et al., 2015), whereas no robust genetic association with lacunes has been described. The extreme-phenotype design was shown to be a more powerful strategy to identify rare risk alleles underlying complex traits (Peloso et al., 2016). Moreover, using a composite extreme phenotype derived from two key MRI markers of SVD may increase the phenotype specificity and reduce misclassification bias that may arise when studying individual MRI-markers, as a ‘control’ without lacunes may for instance well have extensive WMH burden reflecting underlying SVD (Traylor et al., 2015).
Whole exome sequencing (WES) allows a comprehensive survey of both rare and common variants in coding regions and has been helpful in deciphering the genetic architecture of complex diseases (Cruchaga et al., 2014; Lange et al., 2014). Here, we report the first WES study on MRI markers of SVD using a composite extreme phenotype study design, and focus our exploration on genes harbouring mutations causing Mendelian forms of SVD.
Materials and methods
Study population
The Three City Dijon (3C-Dijon) study is a population-based cohort of 4931 French non-institutionalized individuals aged 65 years and older (3C Study Group, 2003). A total of 2763 individuals aged ≤80 years were invited to undergo a brain MRI between June 1999 and September 2000. Participation rate was high (83%, n = 2285) but because of financial restrictions, only 1924 MRI scans were performed. Among the 1924 participants with MRI data, 1683 had also undergone genome-wide genotyping. After exclusion of individuals with brain tumours (n = 8), stroke (n = 71), or dementia (n = 7) at baseline, the remaining sample comprised 1497 participants with automated WMH volume measurement.
The Ethical Committee of the University Hospital of Kremlin-Bicêtre approved the study protocol. All participants signed an informed consent to participate in the study.
Brain MRI
MRI acquisition was performed with a 1.5 T Magnetom scanner (Siemens,) using T1-weighted, T2-weighted, and proton density-weighted sequences, according to the same protocol at both baseline and follow-up (Kaffashian et al., 2014). Fully automated image processing software was developed to detect and localize WMH and to measure WMH volume (Maillard et al., 2008). Infarcts were rated on T1-, T2- and proton density-weighted images by the same examiner (Y.-C.Z.), using a standardized assessment grid, to visually review all brain scans. Lacunes were defined as infarcts 3–15 mm in diameter having the same signal characteristics as cerebrospinal fluid on all sequences, located in basal ganglia, brainstem or cerebral white matter. Characteristics of lesions were visualized simultaneously in axial, coronal, and sagittal planes to discriminate them from dilated perivascular spaces. Lesions with a typical vascular shape and following the orientation of perforating vessels were regarded as dilated perivascular spaces (Zhu et al., 2010). The nomenclature of MRI markers of SVD in our study is consistent with the recently proposed neuroimaging standards for research into SVD (STRIVE) (Wardlaw et al., 2013; Kaffashian et al., 2014).
Definition of extreme cerebral small vessel disease
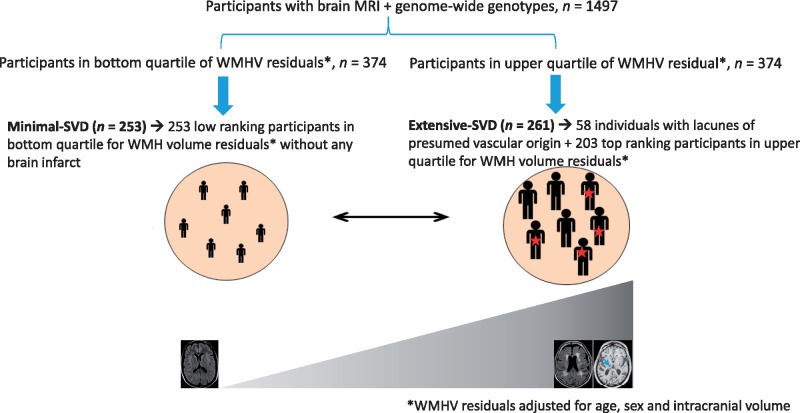
The composite extreme phenotype was defined based on the distribution of WMH volume and presence or absence of lacunes in the 3C-Dijon study using 1497 participants from the 3C-Dijon study who had both MRI scan and GWAS data. The objective was to define a group with extensive SVD severity (individuals in the upper quartile of WMH distribution and having one or more brain infarcts) and a group with minimal SVD severity (lower quartile of WMH distribution and without any brain infarcts). We log-transformed WMH volume (natural log of [WMH volume in cm3 + 1]) and extracted residuals adjusted for age, gender, and white matter mask volume, hereafter referred to as WMH-burden residuals. The first and fourth quartiles of these residuals were taken to represent small and large WMH volume, respectively. The 261 participants with extensive SVD were defined from 374 participants within the fourth quartile of WMH-burden residuals distribution by including all participants who also had at least one lacune (n = 58) and by selecting additionally 203 participants with the highest WMH-burden residuals within the fourth quartile. Similarly, the 253 participants with minimal SVD were defined from 374 participants within the first quartile of WMH-burden residuals distribution by the absence of MRI-defined brain infarcts and having WMH burden residuals at the bottom tail of the WMH burden residual distribution. The design used for defining extreme SVD is summarized in Fig. 1.
Figure 1.
Schematic representation of the extreme SVD design in the 3C-Dijon study.
Covariates and clinical events
At baseline, socio-demographics, medical history, and drug use data were collected at home during an interview by trained psychologists. Centralized measurements of fasting plasma glucose, serum total cholesterol, high density lipoprotein cholesterol, and triglycerides were performed using enzymatic methods by the Biochemistry Laboratory of the University Hospital of Dijon. Low density lipoprotein (LDL) cholesterol was calculated with the Friedewald formula (Friedewald et al., 1972). Body mass index (BMI) was defined as the ratio of weight (kg) to the square of height (m). Smoking status was categorized as never, former, and current. Diabetes mellitus was defined as intake of antidiabetic drugs or fasting blood glucose ≥ 7 mmol/l. Hypertension was defined by systolic blood pressure (BP) ≥140 mm Hg, or diastolic BP ≥90 mm Hg, or antihypertensive drug intake. History of cardiovascular disease was defined by history of myocardial infarction, bypass cardiac surgery, angioplasty, or peripheral vascular disease. Hypercholesterolaemia was defined as fasting total cholesterol ≥6.2 mmol/l or use of any lipid-lowering drug. Information concerning stroke occurrence over time was collected at each follow-up. Incident stroke was defined as a new focal neurological deficit of sudden or rapid onset, of presumed vascular origin, that persisted for >24 h, or leading to death. An expert panel of neurologists adjudicated diagnosis of stroke based on criteria of the WHO (1988). Dementia status was evaluated prospectively by an expert panel using a three-step procedure (Schilling et al., 2017): (i) participants underwent neuropsychological evaluations carried out by trained psychologists; (ii) an examination by a neurologist for those who screened positive at step 1 based on the MMSE and the Isaacs’ Set Test; and (iii) an independent committee of neurologists and geriatricians reviewed all suspected prevalent and incident dementia cases to reach consensus on the diagnosis and aetiology according to the DSM-IV criteria, using all available information (e.g. cognitive functioning, severity of cognitive disorders, hospitalization records when possible, computed tomography scans, MRI, functional assessments).
Exome sequencing and quality control
The DNA samples of 514 participants (261 with extensive SVD and 253 with minimal SVD) with the extremes of SVD severity underwent high depth WES. The majority of samples (n = 508) were sequenced at the McGill Genome Center, Montreal, Canada, and remaining six participants were sequenced at the Centre National de Génotypage, Paris, France. The Agilent SureSelect Human All Exome V5 exome capture kit was used for exome capture except for five samples for which the Agilent SureSelect Human All Exome V4 or V5+UTR exome capture kits were used. The Illumina HiSeq2000 instrument was used to perform paired-end sequencing (2 × 100 bp). The reads were aligned to the GRCh37 human reference genome sequence using the software Burrows-Wheeler Aligner and duplicate reads were tagged with Picard MarkDuplicates (Li and Durbin, 2009). The Genome Analysis Toolkit (GATK) software was used to perform realignment around InDels and base quality score recalibration (BQSR) (McKenna et al., 2010). Single-sample calling was performed using HaplotypeCaller from GATK 3.3 in GVCF mode with base-pair resolution, except 15 samples, whose calling was generated with default band definition as part of the Alzheimer Disease Exome Sequencing-France (ADSP-FR) project (Bellenguez et al., 2017). Calling was done on the target intervals of each exome kit using a padding of 100 bp. Multi-sample calling was performed with the GenotypeGVCFs tool implemented in GATK 3.4, together with other samples from the ADES-FR project. Our whole exome sequence data covered 17 649 RefSeq genes with an average depth of coverage of ∼80× (Supplementary Fig. 1). We filtered out samples with missingness >20%, and individuals with >6 standard deviations (SD) for number of singletons, heterozygote to homozygote ratio, mean depth, and transition to transversion (Ti/Tv) ratio. This protocol resulted in filtering out two participants with extensive SVD because of high number of singletons and low mean depth coverage. We filtered out genotypes with Phred-scaled confidence for genotype call <20 or average depth of coverage <8×. In our study, we included only biallelic variants [(single nucleotide polymorphism (SNPs) and insertions/deletions (Indels)]. Additionally, we filtered out variants with mean depth higher than 500-fold, missingness >20%, and Hardy Weinberg equilibrium P-value < 5 × 10−6. Overall, we achieved high quality WES data for 259 extensive SVD and 253 minimal SVD participants (n = 512).
Description of the study sample
Baseline characteristics of participants with extensive SVD and participants with minimal SVD were compared using analysis of covariance for continuous variables and chi-square test for categorical variables. After verifying the proportional hazard assumption through Schoenfeld residuals, we examined the association of extreme SVD and 12-year incident dementia using Cox proportional regression with age as the time scale, adjusted for sex and education status. For incident stroke, we used the Cox model with age as a timescale and adjusted for sex, BMI, smoking status, diabetes mellitus, hypertension, history of cardiovascular disease, and hypercholesterolaemia.
Genetic association tests
We performed single-variant and gene-based tests using the R package SeqMeta (https://cran.r-project.org/web/packages/seqMeta/index.html). The primary association models were adjusted for age, sex and the first four principal components of population stratification. In secondary analyses, we additionally adjusted for hypertension status.
Single variant association tests
We performed single variant association tests considering common and low frequency variants [minor allele frequency (MAF) >0.01] located within 100 kb of the 5′ and 3′ UTR of five candidate genes: NOTCH3, HTRA1, COL4A1, COL4A2 and TREX1. The 100 kb arbitrary boundary was considered to capture cis regulatory variants that might be localized within neighbouring genes. We used a permutation approach to derive the significance threshold correcting for multiple association tests for 389 common and low frequency variants that might be in linkage disequilibrium (Supplementary material, part A). Additionally we performed the top-SNP association tests implemented in the VEGAS2 software (Mishra and Macgregor, 2015) to account for number of variants and linkage disequilibrium structure in the locus.
Gene-based analysis
To increase power for association testing of rare and low frequency variants (MAF < 0.05), we also performed gene-based association tests focusing on five candidate genes: NOTCH3, HTRA1, COL4A1, COL4A2 and TREX1. We used the variant effect predictor (v90) software (McLaren et al., 2016) to annotate functional consequences of genetic variants localized within five candidate genes considering the default ‘GRCh37’ ensemble annotation database. We used ‘filter_vep’ module to extract variants with the following functional consequences: splice acceptor variant, splice donor variant, start lost, stop lost, stop gained, frameshift variant, inframe insertion, inframe deletion, and missense variant, to perform gene-based association tests on protein-modifying variants only. We used the SKAT-O approach (Lee et al., 2012) for gene-based analyses of protein-modifying rare and low frequency variants. We considered genes with a cumulative MAF of rare or low frequency protein-modifying variants higher than 1%. We performed power calculations for SKAT-O test using the R package SKAT (Wu et al., 2011).
Replication of significant associations
We sought replication of non-exonic significant findings in genome-wide genotyped subsets imputed to the Haplotype Reference Consortium (HRC) panel and of exonic variants in WES subsets of the Atherosclerosis Risk in Communities (ARIC) study, the Cardiovascular Health study (CHS), the Framingham Heart study (FHS) and the Rotterdam study, all participating in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. The ARIC study and the CHS analysed European and African ancestry samples separately; the FHS and the Rotterdam study analysed only European ancestry samples. The MRI measurements of WMH burden and lacunes in these cohorts are described in the Supplementary material, part B, which also provides details on quality control of genotyped and WES datasets of these studies. The extreme SVD phenotype in the replication cohorts was defined using the same strategy as described above for the 3C-Dijon cohort. We defined one-third of the total sample with phenotype and genotype information as having extreme-SVD (extensive SVD for one-sixth of the sample, with extensive WMH burden with or without lacunes, and minimal SVD for another sixth of the sample with minimal WMH burden and no brain infarcts). In the FHS and Rotterdam study, WMH burden residuals were computed using automated quantitative WMH volume measures adjusting for age, gender, and intracranial volume, whereas in the ARIC study and CHS WMH burden residuals were derived from visual semi-quantitative WMH burden measures adjusting for age and gender, as intracranial volume was accounted for in WMH burden assessment. In the FHS, WMH burden residuals were additionally adjusted for family structure. We separately defined extreme SVD in genotyped and WES subsets of individual studies (see Supplementary Tables 2 and 3, respectively, for population characteristics of extreme SVD cohorts of genotyped and WES subsets of the ARIC, CHS, FHS and Rotterdam studies). Of those participants with MRI SVD phenotype data, the total sample size with genome-wide genotypes was 9924 for European ancestry participants, of whom 3308 had extreme SVD (n = 1654 with extensive SVD and n = 1654 with minimal SVD) and 1170 for African ancestry participants, of whom 390 had extreme SVD (n = 195 with extensive SVD and n = 195 with minimal SVD). The total sample size with WES was 2877 for European ancestry participants, of whom 956 had extreme SVD (n = 480 with extensive SVD and n = 477 with minimal SVD) and 726 for African ancestry participants, of whom 242 had extreme SVD (n = 121 with extensive SVD and n = 121 with minimal SVD).
In the ARIC, CHS and Rotterdam studies, the single variant association tests were performed with an additive model in R using logistic regression, whereas in the FHS, a generalized estimation equation was used to account for family structure. Analyses were adjusted for age, sex and the first four principal components of population stratification. We used METAL software (Willer et al., 2010) to perform an inverse variance weighted meta-analysis of association statistics across replication cohorts, and with the discovery study.
We used seqMeta software to perform the SKAT-O gene-based analysis across all replication cohorts. We then meta-analysed the SKAT-O P-values from discovery and replication cohorts using Stouffer’s method for sample size weighted combination of p-values.
Association of extreme small vessel disease risk variants with related phenotypes
We also tested for association of extreme SVD associated common variants with stroke and continuous measures of WMH, in previously reported GWASs of small vessel ischaemic stroke [NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC), 2016] [defined using the Causative Classification of Stroke (CCS) system] by the National Institute of Neurological Disorders and Stroke and the Stroke Genetics Network (NINDS-SiGN) and of WMH burden (Verhaaren et al., 2015). The GWAS summary statistics on small vessel ischaemic stroke by the NINDS-SiGN consortium were accessed using the Cerebrovascular Disease Knowledge Portal (Crawford et al., 2018).
We additionally performed SKAT-O gene-based analysis of protein-modifying rare and low frequency variants observed within a NOTCH3 targeted Sanger sequenced subsample of the Austrian Stroke Prevention Study (ASPS) (n = 277) cohort. Of these 24 participants were filtered out due to missing information on principal components of population stratification, leaving 171 participants with either coalescent white matter lesions or lacunes and 82 randomly selected participants with no focal changes on magnetic resonance images (Schmidt et al., 2011) for the follow-up analysis. The SKAT-O gene-based analysis was adjusted for age, sex and the first four principal components of population stratification.
In silico functional exploration of non-exonic variants
We used the HaploReg (Ward and Kellis, 2012) (version 4.1) software to perform functional annotation of non-exonic variants that are in linkage disequilibrium (r2 > 0.6 in the 1000 Genomes European panel) with the lead SNP associated with extreme SVD. We also manually explored expression quantitative trait locus (eQTL) databases: the GTeX database (Mele et al., 2015) and the blood eQTL resource (Westra et al., 2013).
NOTCH3 glycosylation site prediction
NOTCH3 functions are regulated by different types of O-glycosylation of the EGF repeat (EGFr) domain including O-fucose (Moloney et al., 2000), O-glucose (Moloney et al., 2000), O-GlcNAc (N-acetylglucosamine) (Matsuura et al., 2008), O-xylose (Takeuchi et al., 2011) and mucin-type O-GalNAc (Boskovski et al., 2013). O-fucosylation is mediated by proteins O-fucosyltransferase 1 and Fringe. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL)-causing mutations were reported to affect carbohydrate chain elongation of NOTCH3 by Fringe proteins (Arboleda-Velasquez et al., 2005). We investigated whether rare and low frequency missense variants in the NOTCH3 EGFr domain observed in the 3C-Dijon cohort localized at these computationally predicted mucin-type O-GalNAc glycosylation sites, using the publicly available software for mucin-type O-GalNAc sites prediction (Steentoft et al., 2013).
Survey of pathogenic variants
We manually surveyed the ClinVar database (Landrum et al., 2016) (accessed on 27 February 2017, Supplementary Table 7) to identify participants in the 3C-Dijon cohort carrying a rare allele at SVD causing pathogenic or likely pathogenic mutations in the following genes: NOTCH3 (Joutel et al., 1997) [causing CADASIL (OMIM:125310)], HTRA1 (Hara et al., 2009) [causing cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL, OMIM:600142)], COL4A1 (Vahedi et al., 2007) [causing COL4A1-related familial vascular leukoencephalopathy (OMIM:607595); and pontine autosomal dominant microangiopathy with leukoencephalopathy (PADMAL), porencephaly-1 (OMIM:175780)], COL4A2 (Gunda et al., 2014) [causing porencephaly-2 (OMIM: 614483)], and TREX1 (Richards et al., 2007) [causing retinal vasculopathy and cerebral leukodystrophy (RVCL, OMIM:192315)].
In addition to the pathogenic and likely pathogenic variants classified in the ClinVar database, we systematically searched for NOTCH3 EGFr domain cysteine-modifying missense variants, the typical type of mutation causing CADASIL (Rutten et al., 2016b), and for variants recently reported to cause HTRA1 autosomal dominant forms of SVD (Verdura et al., 2015).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
The approach for defining the composite extreme phenotype of SVD (extreme SVD) is schematically presented in Fig. 1. From a total sample of 1497 participants with MRI and genome-wide genotype information within the 3C-Dijon study, 514 participants (261 with extensive SVD and 253 with minimal SVD) were identified. Characteristics of 3C-Dijon participants with extreme SVD are described in Table 1. Participants with extensive and minimal SVD had similar age and gender distributions. Participants with extensive SVD had more vascular risk factors than those with minimal SVD, the most significant association being observed for hypertension. Compared to participants with minimal SVD, those with extensive SVD were more often current smokers, and had more frequently a history of cardiovascular disease, as well as higher fasting plasma glucose, triglycerides, and BMI, but lower LDL-cholesterol (Table 1). Over the mean follow-up period of 9.2 ± 2.7 years, 40 participants were diagnosed with dementia, and 20 with stroke. Compared to participants with minimal SVD, those with extensive SVD showed a significantly increased risk of developing incident dementia [hazard ratio (HR) (95% confidence interval, CI) = 1.94 (1.01–3.73), P = 0.05] and a trend towards an increased risk of incident stroke [HR (95%CI) = 2.54 (0.95–6.74), P = 0.06]. Characteristics of replication studies with extreme-SVD are described in Supplementary Tables 1 and 2.
Table 1.
Baseline characteristics of 3C-Dijon participants with extreme cerebral SVD
| Characteristics | Extensive SVD | Minimal SVD | P-value* |
|---|---|---|---|
| Participants, n | 259 | 253 | NA |
| WMH volume, ml, mean ± SD | 13.18 ± 7.07 | 2.05 ± 0.63 | <0.0001 |
| Presence of lacunes, n (%) | 58 (22.4) | 0 | NA |
| Age at MRI, years, mean ± SD | 73.5 ± 4.01 | 73.19 ± 4.45 | 0.4 |
| Female, n (%) | 150 (58.1) | 155 (61) | 0.51 |
| Hypertension, n (%)a | 223 (86.4) | 184 (72.4) | <0.0001 |
| Systolic BP, mmHg, mean ± SD | 152.05 ± 22.51 | 147.07 ± 21.85 | 0.011 |
| Antihypertensive drug intake, n (%) | 146 (56.6) | 93 (36.6) | <0.0001 |
| Fasting plasma glucose, mmol/l, mean ± SD | 5.18 ± 1.51 | 4.95 ± 0.67 | 0.026 |
| Diabetes mellitus, n (%)b | 25 (9.7) | 14 (5.5) | 0.07 |
| HDL cholesterol, mmol/l, mean ± SD | 1.64 ± 0.39 | 1.68 ± 0.41 | 0.23 |
| LDL cholesterol, mmol/l, mean ± SD | 3.53 ± 0.89 | 3.68 ± 0.84 | 0.046 |
| TG, mmol/l, mean ± SD | 1.26 ± 0.56 | 1.15 ± 0.52 | 0.031 |
| Lipid lowering drug, n (%) | 96 (37.2) | 71 (28) | 0.026 |
| BMI, kg/m2, mean ± SD | 25.84 ± 3.92 | 24.86 ± 3.71 | 0.004 |
| Current smoker, n (%) | 22 (8.5) | 8 (3.1) | 0.012 |
| History of CVD at MRI, n (%)c | 15 (5.8) | 5 (2) | 0.025 |
| Hypercholesterolaemia, n (%)d | 142 (55) | 140 (55.3) | 0.95 |
*Significant differences across SVD status obtained from analysis of covariance (continuous variables) or chi-square tests (categorical variables). Models with WMH volume as the dependent variable are adjusted for intracranial volume.
aSystolic blood pressure ≥140 mmHg, or diastolic blood pressure ≥90 mmHg, or use of antihypertensive drugs.
bFasting blood glucose ≥7 mmol/l or antidiabetic drug intake.
cHistory of myocardial infarction, bypass cardiac surgery, angioplasty, or peripheral artery disease.
dHypercholesterolaemia was defined as fasting total cholesterol ≥6.2 mmol/l or use of any lipid-lowering drug (fibrates, statins or bile acid sequestrant).
BMI = body mass index; BP = blood pressure; CVD = cardiovascular diseases; HDL = high-density lipoprotein; LDL = low-density lipoprotein; TG = triglycerides.
Single variant association analyses identified a significant association of an intronic variant in HTRA1 (rs2293871-T, frequency = 0.19) with extreme SVD (Table 2). This association was significant after correcting for multiple testing (permutation derived 95% empirical significance threshold P < 2.89 × 10−4, Supplementary material, part A) and remained significant after additionally adjusting for hypertension status (Table 2). Moreover the top-SNP locus-based test implemented in the VEGAS2 software (Mishra and Macgregor, 2015) confirmed that the association of rs2293871 with extreme SVD is independent of the regional properties of the HTRA1 locus: the linkage disequilibrium structure or number of variants in the region (Table 2). The effect estimates of rs2293871-T appeared larger when comparing the 58 extensive SVD participants with lacunes to minimal SVD participants [OR (95%CI) = 3.04 (1.67–5.50), P = 2.56 × 10−4] than when comparing the 203 participants with extensive SVD without lacunes to minimal SVD participants [OR (95%CI) = 1.80 (1.27–2.56), P = 9.60 × 10−4]. We replicated the association of rs2293871 in independent cohorts of European ancestry (n extreme SVD = 3308) using genome-wide genotype data for this common intronic variant (Table 2). The association of rs2293871 was not significant in the only African ancestry sample (rs2293871-T frequency = 0.14, Supplementary Table 3). The inverse variance weighted meta-analysis of discovery and replication cohorts of European ancestry showed an association of rs2293871-T with extensive SVD at an OR (95%CI) of 1.29 (1.14–1.46), P = 4.72 × 10−5 (Table 2). The same allele at rs2293871 was also associated with increased risk of small vessel ischaemic stroke defined using the CCS system in 16 851 cases and 31 259 controls in the NINDS-SiGN study [NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC), 2016]: OR (95%CI) = 1.12 (1.03–1.22), P = 6.14 × 10−3 for causative CCS and OR (95%CI) = 1.12 (1.04–1.22), P = 4.68 × 10−3 for phenotypic CCS. The rs2293871 variant showed nominal association with continuous WMH burden (n = 17 936, P-value = 0.03) in a previously reported GWAS meta-analysis (Verhaaren et al., 2015). Functional explorations using HaploReg (Ward and Kellis, 2012) suggest that rs2293871 lies in the H3K9ac promoter and H3K4me1, H3K4me3 and H3K27ac enhancer histone marks (Supplementary Table 4). Two proxies of rs2293871 (rs876790 and rs2736928, r2 = 0.75 with rs2293871) are eQTL for HTRA1 in blood (Westra et al., 2013), with C alleles at rs876790 and rs2736928 (in phase with rs2293871-T) showing significant association with lower HTRA1 transcript levels (false discovery rate-corrected P-value = 0.03 and 0.04, respectively).
Table 2.
Top common or low frequency variant at individual candidate locus
| Gene | Top variant (rsID) | Hg19_chr:bp | RA/OA | RA Freq. | 3C-Dijon (Discovery, n = 512) | ARIC, CHS, FHS and RS1–3 (Replication, n = 3308) | Joint analysis (n = 3802) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (CI 95%) | P-value* | Variants, n | Top-SNP P-value*** | OR (95% CI) | P-value | OR (95% CI) | P-value | |||||
| HTRA1# | rs2293871 | 10:124273671 | T/C | 0.19 | 1.92 (1.39–2.65) | 8.21 × 10−5 | 49 | 1.77 × 10−3 | 1.21 (1.06–1.38) | 5.25 × 10−3 | 1.29 (1.14–1.46) | 4.72 × 10−5 |
| COL4A1 | rs2275842 | 13:110813523 | T/C | 0.17 | 1.52 (1.09–2.11) | 0.01 | 89 | 0.38 | NA | NA | NA | NA |
| COL4A2 | rs2275842 | 13:110813523 | T/C | 0.17 | 1.52 (1.09–2.11) | 0.01 | 154 | 0.55 | NA | NA | NA | NA |
| NOTCH3 | rs1043997 | 19:15300136 | G/A | 0.05 | 1.69 (0.96–2.96) | 0.07 | 60 | 0.65 | NA | NA | NA | NA |
| TREX1 | rs78159609 | 3:48419898 | A/G | 0.06 | 0.62 (0.36–1.08) | 0.09 | 29 | 0.59 | NA | NA | NA | NA |
*Significance threshold for discovery is P-value < 2.89 × 10−4 correcting for 389 common and low frequency variants tested.
***Significance threshold for VEGAS2 top-SNP test is P-value < 0.01 correcting for five loci tested.
#After adjustment for hypertension status the association of rs2293871 with extreme SVD was OR = 1.85 (95%CI: 1.33- 2.58), P = 2.39 × 10−4 in the 3C-Dijon Study.
ARIC = Atherosclerosis Risk In Communities; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; OA = other allele; RA = risk allele; RS1–3 = Rotterdam Studies 1, 2 and 3.
We analysed the association of protein-modifying (splice acceptor variant, splice donor variant, start lost, stop lost, stop gained, frameshift variant, inframe insertion, inframe deletion, and missense variants only) rare and low frequency variants (MAF < 0.05) in candidate genes using the SKAT-O approach (Lee et al., 2012). Only three genes NOTCH3, COL4A1, and COL4A2, satisfied the criteria of cumulative MAF of protein-modifying rare or low frequency variants of >1% (Supplementary Table 5), thus qualifying for gene-based analyses. The SKAT-O gene-based analysis identified a significant association of protein-modifying rare and low frequency variants in the NOTCH3 gene with extreme SVD (SKAT-O P = 1.61 × 10−2, n extreme SVD = 512, Table 3), which remain associated after additionally adjusting for hypertension status (SKAT-O P = 1.58 × 10−2, Table 3). We successfully replicated the gene-based association of protein-modifying rare and low-frequency variants in NOTCH3 with extreme SVD in four independent cohorts of European ancestry (n extreme SVD = 956): SKAT-O P = 3.99 × 10−2 for the replication set and SKAT-O P = 5.31 × 10−3 for the combined discovery and replication samples (Table 3). The NOTCH3 association was not significant in the African ancestry sample of 242 extreme-SVD participants (SKAT-O P = 0.78). Follow-up in a previously described Sanger sequencing subset of the ASPS (Schmidt et al., 2011) did not show any significant association in a cohort of 171 participants with either coalescent white matter lesions or lacunes compared with 82 randomly selected participants with no focal changes on magnetic resonance images (SKAT-O P = 0.53), possibly due to limited sample size.
Table 3.
Gene-based association of rare and low frequency protein-modifying variants in NOTCH3, COL4A1, and COL4A2 genes with extreme cerebral SVD
| Genea (Transcript) | 3C-Dijon (Discovery, n = 512) | ARIC, CHS, FHS and RS1 (Replication, n = 956) | Combined (n = 1467) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Variants, n | Cumulative MAF | P (SKAT-O)* | P (SKAT-O) additional adjusted for HT status | Variants, n | Cumulative MAF | P (SKAT-O) | P (SKAT-O) additional adjusted of HT status | P (SKAT-O) | |
| NOTCH3 (ENST00000263388) | 31 | 0.10 | 1.61 × 10−2 | 1.58 × 10−2 | 36 | 0.11 | 3.99 × 10−2 | 4.60 × 10−2 | 5.31 × 10−3 |
| COL4A2 (ENST00000360467) | 29 | 0.09 | 0.23 | 0.19 | NA | NA | NA | NA | NA |
| COL4A1 (ENST00000375820) | 13 | 0.02 | 0.40 | 0.48 | NA | NA | NA | NA | NA |
aSorted by SKAT-O P-value in discovery cohort.
*Significance threshold for discovery is SKAT-O P-value < 1.67 × 10−2 correcting for three tested genes.
ARIC = Atherosclerosis Risk In Communities; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; HT = hypertension; MAF = minor allele frequency; RS1 = Rotterdam Study 1.
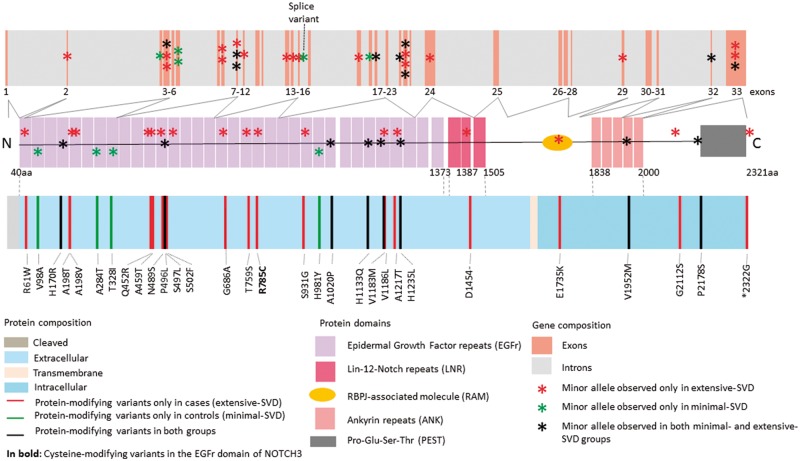
Further exploratory protein-domain specific SKAT-O analyses showed significant association of extreme SVD with the EGFr domain determining region of the NOTCH3 gene, which is known to preferentially harbour mutations causing CADASIL (SKAT-O P = 2.14 × 10−2 for 3C-Dijon, SKAT-O P = 3.30 × 10−2 for the replication samples, and SKAT-O P = 4.98 × 10−3 for the combined discovery and replication). We also observed, in the 3C-Dijon extreme SVD sample, that five of the missense variants in the EGFr determining region (T328I, S497L, S502F, T759S, and S931G) were predicted mucin type GalNAc O-glycosylation sites with predication scores ranging between 0.18 and 0.82 (Supplementary Table 6). The S502F, T759S and S931G variants were observed exclusively in the 3C-Dijon extensive-SVD sample (Fig. 2).
Figure 2.
NOTCH3 protein modifying rare and low frequency variants in the 3C-Dijon extreme SVD cohort.
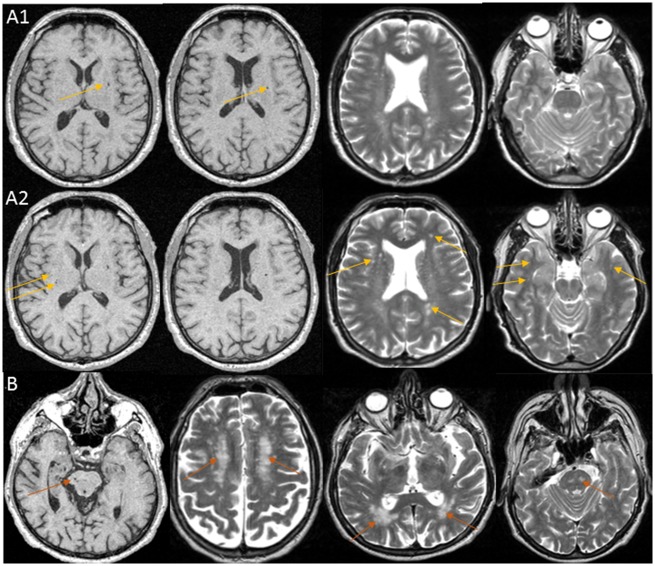
Screening of 3C-Dijon extreme SVD participants for rare alleles at pathogenic or likely pathogenic variants in five candidate genes harbouring mutations causing monogenic SVD, identified two such alleles in the NOTCH3 and HTRA1 genes. One extensive SVD participant carried a heterozygote genotype at a NOTCH3 EGFr domain cysteine altering variant: NM_000435.2 (NOTCH3):c.C2353T:p.R785C (Fig. 2) (participant level depth coverage = 33× and Phred scaled genotype quality = 99, note: Phred score vary from 0 to 99 and 99 represents the highest Phred scaled confidence for genotype quality). This variant leads to addition of a seventh cysteine residue in EGF repeat 20 of the NOTCH3 N-terminus, typical of CADASIL, and was previously described in one Italian CADASIL family with an autosomal dominant inheritance pattern (Mosca et al., 2014). Another extensive SVD participant carried a heterozygote genotype at the CARASIL-causing variant: NM_002775.4 (HTRA1):c.1108C > T (p.Arg370Ter), a nonsense variant resulting in a stop codon at amino acid position 370 (participant level depth coverage = 96× and Phred scaled genotype quality = 99). Only the homozygous TT genotype at this variant has been reported to cause CARASIL in the literature, in Asian populations (Hara et al., 2009). This variant was not included in the list of variants reported to cause a dominant HTRA1-related SVD phenotype in Europeans (Verdura et al., 2015). Brain imaging characteristics of both participants are shown in Fig. 3. Neither of them had typical imaging features of CADASIL or CARASIL at baseline, but the participant with a NOTCH3 EGFr domain cysteine altering genotype developed WMH in the anterior temporal lobe, a location typical for CADASIL, on a 4-year follow-up MRI scan. Both participants were free of stroke and dementia.
Figure 3.
MRI images of participants carrying heterozygote genotypes at CADASIL and CARASIL causing mutations. (A) Baseline (1) and 4-year follow-up (2) MRI scans of a 65-year old female participant with extensive SVD, in whom a NOTCH3 EGFr domain cysteine-modifying mutation was found: NM_000435.2 (NOTCH3):c.C2353T:p.R785C. Images show lacunar infarcts and dilated perivascular spaces in basal ganglia and white matter, and WMH in the periventricular region and deep white matter; on the follow-up MRI scan WMH and dilated perivascular spaces burden had increased and WMH became visible in the anterior temporal lobes (yellow arrows), a typical location for CADASIL. This participant remained free of stroke and dementia until the end of her follow-up at age 77. Her MMSE score was 28 at baseline and 26 at 12 years follow-up (secondary school education but no high school). (B) Baseline MRI scan of a 74-year-old female participant with extensive SVD, in whom a heterozygous CARASIL causing mutation was found: NM_002775.4 (HTRA1):c.1108C > T (p.Arg370Ter). Images show WMH and lacunes in the pons and extensive WMH in the deep white matter and periventricular region (magenta arrows). This participant was free of stroke and dementia at baseline but was lost to follow-up. Her baseline MMSE score was 27 (primary school education).
We also observed two minimal-SVD participants carrying a heterozygote genotype at one glycine residue altering missense variant in COL4A1 [NM_001845.4 (COL4A1): c.3158G > A (p.Gly1053Asp), participant level depth coverage = 125× and Phred scaled genotype quality = 99], and one nonsense (stop gained) variant in COL4A2 [NM_001846.2 (COL4A2) c.3766C > T (p.Arg1256Ter), participant level depth coverage = 77× and Phred scaled genotype quality = 99]. Heterozygous glycine residue changes and nonsense mutations in COL4A1 and COL4A2 are typically described in SVD families with cerebral bleedings, although, to our knowledge, these specific variants have not been described previously in any SVD family. All protein-modifying variants observed in 3C-Dijon participants with extreme-SVD within the five candidate genes are displayed in Fig. 2 (NOTCH3) and Supplementary Fig. 2–5 (HTRA1, COL4A1, COL4A2 and TREX1).
Discussion
We report a novel gene-mapping strategy for SVD in population-based cohorts of older person with MRI-defined extremes of SVD severity. We explored the association with extreme SVD in the general population of common and rare variants in five genes known to harbour mutations causing Mendelian SVD, with a discovery sample of 512 participants and a follow-up sample of 3698 participants for common variants and n = 1198 for rare and low frequency variants. We report significant association of a common intronic variant in the HTRA1 gene with extreme SVD, with evidence suggesting that the risk allele is lowering HTRA1 expression. We also found a significant association with extreme SVD of rare and low frequency NOTCH3 protein-modifying variants using a gene-based approach. Finally, in 512 participants from the 3C-Dijon discovery population-based sample, we also screened for pathogenic variants causing Mendelian SVD and identified two participants with extensive SVD harbouring heterozygote genotypes for such variants in NOTCH3 (CADASIL-causing missense variant modifying a cysteine-residue of the EGFR domain) and in HTRA1 (heterozygous carrier of a CARASIL causing mutation).
Our novel approach complements the gene-mapping strategies traditionally being used to identify SVD risk loci in population-based cohorts, consisting of studying each MRI-marker of SVD individually: presence or absence of brain infarct (Debette et al., 2010) and, quantitative measure of WMH burden (Verhaaren et al., 2015), and of efforts to reveal genetic determinants of the clinically defined small vessel ischaemic stroke subtype [NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC), 2016]. The extreme composite phenotype of SVD presented here is likely to be more specific for underlying SVD pathology and provides a better contrast by excluding participants with either lacunes or moderate to extensive WMH burden from the control group. Extreme phenotype association studies have been reported to be better powered for identifying rare risk variants associated with disease by reducing the phenotypic heterogeneity (Peloso et al., 2016), which we demonstrate through association of rare and low frequency NOTCH3 protein-modifying variants with extreme-SVD. Notably, we also demonstrate that our study design is more powerful to identify some common SVD risk variants, as the common intronic variant rs2293871 in HTRA1 has greater significance in relation with extreme SVD compared to associations observed with small vessel ischaemic stroke [NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC), 2016] and continuous WMH burden (Verhaaren et al., 2015), which had comparatively more number of participants than the former study. This observation is in line with simulation studies demonstrating that extreme sample phenotyping might identify additional common risk variants (MAF > 0.05) associated with complex diseases by reducing the impact of phenotype misclassification on observed genetic effect size estimates (van der Sluis et al., 2010; Manchia et al., 2013).
We report and replicate an association of a common intronic variant (rs2293871) in HTRA1 with extreme-SVD in European ancestry cohorts. HTRA1 encodes a secretory protein of the serine protease family, which regulates transforming growth factor (TGF) signalling (Beaufort et al., 2014). Disruption in HTRA1 activity causing cell death by modulating TGF signalling has been suggested as a possible causal mechanism underlying CARASIL (Beaufort et al., 2014). We identified two blood eQTLs in linkage disequilibrium (r2 = 0.75) with rs2293871, suggesting that the allele associated with increased risk of extensive SVD is associated with lower HTRA1 expression in blood. Although limited to blood, and based on proxies in moderate linkage disequilibrium, this observation is in line with suggested mechanisms of reduced HTRA1 activity in CARASIL (Hara et al., 2009; Beaufort et al., 2014), and also the recent description of heterozygote loss-of-function variants in HTRA1 causing SVD phenotypes in European populations (Verdura et al., 2015).
Using a gene-based approach we also demonstrate significant association with extreme SVD of NOTCH3 protein-modifying rare and low frequency variants, which were replicated in independent cohorts. This association is primarily driven by variants located in the EGFr domain, known to preferentially harbour CADASIL causing mutations. CADASIL, the most common of all known Mendelian forms of SVD, is an autosomal dominant disease typically caused by cysteine residue altering variants in NOTCH3 resulting in an uneven number of cysteines in the EGFr domain of NOTCH3, disrupting disulphide bridge formation, causing misfolding of EGFr, and increasing NOTCH3 multimerization (Monet-Lepretre et al., 2013). Some cysteine-modifying mutational hotspots (R91C, R170C or C213S) were reported to cause aberrant dimerization of NOTCH3 fragments by reducing Fringe-mediated elongation of O-fucose glycosylation (Arboleda-Velasquez et al., 2005). In rare instances, cysteine-sparing variants were also reported to cause CADASIL in some families, but their pathogenicity is still debated. Five missense variants in EGFr determining region observed in the 3C-Dijon extreme SVD sample are predicted mucin type GalNAc O-glycosylation sites, of which three variants were exclusively observed in participants with extensive SVD. Because of the lack of publicly available resources to computationally predict other types of O-glycosylation than mucin (O-fucose, O-glucose, O-GlcNAc, and O-xylose) we have only partially captured the impact of observed NOTCH3 missense variants on glycosylation disruption in the EGFr domain. Further functional studies are essential to understand the impact of glycosylation disruption in the EGFr domain of NOTCH3 by genetic variants in complex SVD pathophysiology.
Interestingly, two participants with extensive SVD, representing 0.4% of our discovery population-based sample, and 0.8% of participants with extensive SVD, carried heterozygous mutations in NOTCH3 or HTRA1 described previously as pathogenic and causing CADASIL or CARASIL, two Mendelian forms of SVD. Of note, this observation is based on high quality WES data but lacks technical validation using targeted Sanger sequencing. The frequency of known pathogenic variants in our community sample is higher than expected, but in line with a recent analysis of NOTCH3 likely pathogenic variants described in 0.3% of the 60,706 exomes of the publicly available exome aggregation consortium (ExAC) database (Rutten et al., 2016a). The main shortcoming of the ExAC database is the limited clinical information on participants included in the database and the lack of data on covert, MRI-defined SVD phenotypes. Our study provides further evidence that pathogenic variants known to cause rare Mendelian forms of SVD (CADASIL and CARASIL) are less exceptional than previously suspected in the general population. Although they were observed in persons with extensive SVD on brain imaging, they appeared to have mild clinical expression in this population-based setting. The CADASIL causing variant reported in our cohort modifies the cysteine residue of the EGF repeat 20 of the NOTCH3 N-terminus. Interestingly, cysteine modifying mutations at EGF repeats 7–34 may have a milder CADASIL phenotype than those affecting EGFr domain 1–6 at the C-terminal end, because of lower likelihood of interaction of unpaired cysteine with other proteins (Rutten et al., 2016a). Our results further add to the debate around returning results on incidental findings from next generation sequencing considering that the penetrance of likely pathogenic variants may be highly variable (Hehir-Kwa et al., 2015; Hofmann, 2016).
Intriguingly we observed two missense variants in COL4A1 and COL4A2 in 3C-Dijon participants with minimal SVD, with typical characteristics of SVD causing mutations, although they have not been described previously in SVD families. This may reflect low penetrance. Indeed, clinical studies have shown that a significant proportion of mutation carriers do not develop intracerebral bleedings (Meuwissen et al., 2015). This may also be explained by the fact that our brain imaging protocol did not include gradient echo images to detect previous microbleeds or intracerebral haemorrhages, as the most common manifestations of COL4A1/2 related SVD are brain haemorrhages (Lanfranconi and Markus, 2010).
Our proof-of-concept gene-mapping study focused on genetic variants within five candidate genes observed using the WES technique. One notable limitation of this work is that it did not report on association of some common risk variants relevant to SVD pathology that were identified using the GWAS approach, as these were not captured by WES, particularly COL4A2 intronic variants, respectively, rs9515201, rs9521732, rs9521733, and rs9515199, which were recently reported to be associated with WMH volume (Traylor et al., 2016) and deep intracerebral haemorrhage (Rannikmae et al., 2015). Furthermore, as we focused only on SVD candidate genes, our study did not explore the impact on extreme SVD of rare and common variants in other candidate loci, such as those previously associated with continuous WMH burden (Verhaaren et al., 2015; Traylor et al., 2016), stroke (Malik et al., 2018) or Alzheimer’s disease (Lambert et al., 2013). These limitations will need to be addressed through a large multi-cohort gene-mapping study using GWAS and possibly whole genome sequencing approaches.
In summary, our proof-of-concept study provides strong evidence that using a novel composite MRI-derived phenotype for extremes of SVD can facilitate the identification of genetic variants underlying SVD, both common variants and those with rare and low frequency. The findings demonstrate shared mechanisms and a continuum between genes underlying Mendelian SVD and those contributing to the common, multifactorial form of the disease. Future studies exploring rare and common genetic variants associated with this composite extreme SVD phenotype at a genome-wide or whole genome level are warranted. Indeed, SVD is a major contributor to stroke and dementia risk worldwide with no specific therapy available to date, and efforts to decipher underlying biological pathways to accelerate the discovery of novel treatment strategies represent a public health priority.
Supplementary Material
Acknowledgements
We thank Dr Anne Boland (CNG) for her technical help in preparing the DNA samples for analyses. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, and Carolina Medina-Gomez, for their help in creating the GWAS database, and Karol Estrada, Yurii Aulchenko, and Carolina Medina-Gomez, for the creation and analysis of imputed data. The generation and management of the exome sequencing data for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We thank Pascal Arp, Mila Jhamai, Jeroen van Rooij, Marijn Verkerk, and Robert Kraaij for their help in creating the RS-Exome Sequencing database. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Glossary
Abbreviations
- CADISIL
cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
- CARASIL
cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy
- GWAS
genome-wide association studies
- SVD
small vessel disease
- WES
whole-exome sequencing
- WMH
white matter hyperintensity
Funding
This project is supported by the Fondation Leducq (Transatlantic Network of Excellence on the Pathogenesis of SVD of the Brain) and is an EU Joint Programme -Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND www.jpnd.eu: Australia, National Health and Medical Research Council, Austria, Federal Ministry of Science, Research and Economy; Canada, Canadian Institutes of Health Research; France, French National Research Agency; Germany, Federal Ministry of Education and Research; Netherlands, The Netherlands Organisation for Health Research and Development; United Kingdom, Medical Research Council. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 643417. This project has also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No 640643. This project has received funding from European Union’s Horizon 2020 research and innovation programme under grant agreement No 667375. Computations were performed on the Bordeaux Bioinformatics Center (CBiB) computer resources, Université de Bordeaux. Funding support for additional computer resources has been provided to S.D. by the Fondation Claude Pompidou.
The Three City Study: The Three City (3C) Study is conducted under a partnership agreement among the Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Bordeaux, and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l’Education Nationale (MGEN), Institut de la Longévité, Conseils Régionaux of Aquitaine and Bourgogne, Fondation de France, and Ministry of Research–INSERM Programme ‘Cohortes et collections de données biologiques.’ C.T. and S.D. have received investigator-initiated research funding from the French National Research Agency (OPE-2016-0500) and from the Fondation Leducq (12CVD01). This work was supported by the National Foundation for Alzheimer’s disease and related disorders, the Institut Pasteur de Lille, the Centre National de Génotypage, the French government’s LABEX (laboratory of excellence program investment for the future) DISTALZ grant (Development of Innovative Strategies for a Transdisciplinary approach to Alzheimer’s disease), and the GENMED labex.
The Atherosclerosis Risk in Communities study: The Atherosclerosis Risk in Communities study (ARIC) was performed as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HSN268201100006C, HSN26 8201100007C, HHSN268201100008C, HHSN2682011 00009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL70825, R01HL087 641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health (NIH) contract HHSN268200625226C. Infrastructure was partly supported by grant No. UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. This project was also supported by NIH R01 grant NS087541 to M.F.
The Cardiovascular Health Study: The Cardiovascular Health study (CHS) research was supported by contracts HHSN268201200036C, HHSN268200800007C, HHSN 268201800001C, N01HC55222, N01HC85079, N01HC 85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, N01HC15103, and HHSN2682009 60009C and grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, R01HL 085251, and U01HL130114 from the National Heart, Lung, and Blood Institute (NHLBI) with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 and R01AG033193 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at chs-nhlbi.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding support for ‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium’ was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for ‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium’ were provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. Sequencing was carried out at the Baylor Genome Center (U54 HG003273).
The Framingham Heart Study (FHS): This work was supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contracts No. N01-HC-25195 and No. HHSN268201500001I), and its contract with Affymetrix, Inc. for genotyping services (Contract No. N02-HL-6–4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study was also supported by grants from the National Institute of Aging (R01s AG033040, AG033193, AG054076, AG049607, AG008122, and U01-AG049505) and the National Institute of Neurological Disorders and Stroke (R01-NS017950, UH2 NS100605).
The Rotterdam Study: The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911–03–012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014–93–015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050–060–810. The Exome Sequencing dataset was funded by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) sponsored Netherlands Consortium for Healthy Aging (NCHA; project nr. 050–060–810), by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by the and by a Complementation Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl; project number CP2010–41). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
The Austrian Stroke Prevention Study (ASPS): The research reported in this article was funded by the Austrian Science Fund (FWF) grant number P20545-P05, P13180 and P20545-B05, by the Austrian National Bank Anniversary Fund, P15435, and the Austrian Ministry of Science under the aegis of the EU Joint Programme - Neurodegenerative Disease Research (JPND) www.jpnd.eu received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 643417.
Competing interests
The authors report no competing interests.
References
- C Study Group. Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology 2003; 22: 316–25. [DOI] [PubMed] [Google Scholar]
- Arboleda-Velasquez JF, Rampal R, Fung E, Darland DC, Liu M, Martinez MC, et al. CADASIL mutations impair Notch3 glycosylation by Fringe. Hum Mol Genet 2005; 14: 1631–9. [DOI] [PubMed] [Google Scholar]
- Beaufort N, Scharrer E, Kremmer E, Lux V, Ehrmann M, Huber R, et al. Cerebral small vessel disease-related protease HtrA1 processes latent TGF-beta binding protein 1 and facilitates TGF-beta signaling. Proc Natl Acad Sci USA 2014; 111: 16496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenguez C, Charbonnier C, Grenier-Boley B, Quenez O, Le Guennec K, Nicolas G, et al. Contribution to Alzheimer’s disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol Aging 2017; 59: 220.e1–e9. [DOI] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, et al. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 2013; 504: 456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford KM, Gallego-Fabrega C, Kourkoulis C, Miyares L, Marini S, Flannick J, et al. Cerebrovascular disease knowledge portal: an open-access data resource to accelerate genomic discoveries in stroke. Stroke 2018; 49: 470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 2014; 505: 550–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debette S, Bis JC, Fornage M, Schmidt H, Ikram MA, Sigurdsson S, et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke 2010; 41: 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano AL, Seshadri S, Beiser A, Atwood LD, Massaro JM, Au R, et al. Bivariate heritability of total and regional brain volumes: the Framingham Study. Alzheimer Dis Assoc Disord 2009; 23: 218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornage M, Debette S, Bis JC, Schmidt H, Ikram MA, Dufouil C, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol 2011; 69: 928–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972; 18: 499–502. [PubMed] [Google Scholar]
- Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The genetic architecture of type 2 diabetes. Nature 2016; 536: 41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunda B, Mine M, Kovacs T, Hornyak C, Bereczki D, Varallyay G, et al. COL4A2 mutation causing adult onset recurrent intracerebral hemorrhage and leukoencephalopathy. J Neurol 2014; 261: 500–3. [DOI] [PubMed] [Google Scholar]
- Hara K, Shiga A, Fukutake T, Nozaki H, Miyashita A, Yokoseki A, et al. Association of HTRA1 mutations and familial ischaemic cerebral small-vessel disease. N Engl J Med 2009; 360: 1729–39. [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Claustres M, Hastings RJ, van Ravenswaaij-Arts C, Christenhusz G, Genuardi M, et al. Towards a European consensus for reporting incidental findings during clinical NGS testing. Eur J Hum Genet 2015; 23: 1601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann B. Incidental findings of uncertain significance: to know or not to know–that is not the question. BMC Med Ethics 2016; 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet 1997; 350: 1511–5. [DOI] [PubMed] [Google Scholar]
- Kaffashian S, Tzourio C, Soumare A, Dufouil C, Zhu Y, Crivello F, et al. Plasma beta-amyloid and MRI markers of cerebral small vessel disease: three-city Dijon study. Neurology 2014; 83: 2038–45. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016; 44: D862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranconi S, Markus HS. COL4A1 mutations as a monogenic cause of cerebral small vessel disease: a systematic review. Stroke 2010; 41: e513–8. [DOI] [PubMed] [Google Scholar]
- Lange LA, Hu Y, Zhang H, Xue C, Schmidt EM, Tang ZZ, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet 2014; 94: 233–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012; 13: 762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longstreth WT., Jr Brain vascular disease overt and covert. Stroke 2005; 36: 2062–3. [DOI] [PubMed] [Google Scholar]
- Maillard P, Delcroix N, Crivello F, Dufouil C, Gicquel S, Joliot M, et al. An automated procedure for the assessment of white matter hyperintensities by multispectral (T1, T2, PD) MRI and an evaluation of its between-centre reproducibility based on two large community databases. Neuroradiology 2008; 50: 31–42. [DOI] [PubMed] [Google Scholar]
- Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018; 50: 524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One 2013; 8: e76295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, et al. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem 2008; 283: 35486–95. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015; 348: 660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen ME, Halley DJ, Smit LS, Lequin MH, Cobben JM, de Coo R, et al. The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature. Genet Med 2015; 17: 843–53. [DOI] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: Software for more flexible gene-based testing. Twin Res Hum Genet 2015; 18: 86–91. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem 2000; 275: 9604–11. [DOI] [PubMed] [Google Scholar]
- Monet-Lepretre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V, et al. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain 2013; 136 (Pt 6): 1830–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca L, Rivieri F, Tanel R, Bonfante A, Burlina A, Manfredini E, et al. Mutational screening of NOTCH3 gene reveals two novel mutations: complexity of CADASIL diagnosis. J Mol Neurosci 2014; 54: 723–9. [DOI] [PubMed] [Google Scholar]
- NINDS Stroke Genetics Network (SiGN), International Stroke Genetics Consortium (ISGC).Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol 2016; 15: 174–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol 2010; 9: 689–701. [DOI] [PubMed] [Google Scholar]
- Peloso GM, Rader DJ, Gabriel S, Kathiresan S, Daly MJ, Neale BM. Phenotypic extremes in rare variant study designs. Eur J Hum Genet 2016; 24: 924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannikmae K, Davies G, Thomson PA, Bevan S, Devan WJ, Falcone GJ, et al. Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology 2015; 84: 918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, et al. C-terminal truncations in human 3’-5’ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet 2007; 39: 1068–70. [DOI] [PubMed] [Google Scholar]
- Rutten JW, Dauwerse HG, Gravesteijn G, van Belzen MJ, van der Grond J, Polke JM, et al. Archetypal NOTCH3 mutations frequent in public exome: implications for CADASIL. Ann Clin Transl Neurol 2016a; 3: 844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten JW, Dauwerse HG, Peters DJ, Goldfarb A, Venselaar H, Haffner C, et al. Therapeutic NOTCH3 cysteine correction in CADASIL using exon skipping: in vitro proof of concept. Brain 2016b; 139 (Pt 4): 1123–35. [DOI] [PubMed] [Google Scholar]
- Sachdev PS, Lee T, Wen W, Ames D, Batouli AH, Bowden J, et al. The contribution of twins to the study of cognitive ageing and dementia: the Older Australian Twins Study. Int Rev Psychiatry 2013; 25: 738–47. [DOI] [PubMed] [Google Scholar]
- Schilling S, Tzourio C, Soumare A, Kaffashian S, Dartigues JF, Ancelin ML, et al. Differential associations of plasma lipids with incident dementia and dementia subtypes in the 3C Study: a longitudinal, population-based prospective cohort study. PLoS Med 2017; 14: e1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Zeginigg M, Wiltgen M, Freudenberger P, Petrovic K, Cavalieri M, et al. Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebral small vessel disease. Brain 2011; 134 (Pt 11): 3384–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 2013; 32: 1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitziel NO, Peloso GM, Abifadel M, Cefalu AB, Fouchier S, Motazacker MM, et al. Exome sequencing in suspected monogenic dyslipidemias. Circ Cardiovasc Genet 2015; 8: 343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Fernandez-Valdivia RC, Caswell DS, Nita-Lazar A, Rana NA, Garner TP, et al. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc Natl Acad Sci USA 2011; 108: 16600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The World Health Organization MONICA Project (monitoring trends and determinants in cardiovascular disease): a major international collaboration. WHO MONICA Project Principal Investigators. J Clin Epidemiol 1988; 41: 105–14. [DOI] [PubMed] [Google Scholar]
- Traylor M, Bevan S, Baron JC, Hassan A, Lewis CM, Markus HS. Genetic architecture of lacunar stroke. Stroke 2015; 46: 2407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor M, Zhang CR, Adib-Samii P, Devan WJ, Parsons OE, Lanfranconi S, et al. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology 2016; 86: 146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner ST, Jack CR, Fornage M, Mosley TH, Boerwinkle E, de Andrade M. Heritability of leukoaraiosis in hypertensive sibships. Hypertension 2004; 43: 483–7. [DOI] [PubMed] [Google Scholar]
- Vahedi K, Boukobza M, Massin P, Gould DB, Tournier-Lasserve E, Bousser MG. Clinical and brain MRI follow-up study of a family with COL4A1 mutation. Neurology 2007; 69: 1564–8. [DOI] [PubMed] [Google Scholar]
- van der Sluis S, Verhage M, Posthuma D, Dolan CV. Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS One 2010; 5: e13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdura E, Herve D, Scharrer E, Amador Mdel M, Guyant-Marechal L, Philippi A, et al. Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain 2015; 138 (Pt 8): 2347–58. [DOI] [PubMed] [Google Scholar]
- Verhaaren BF, Debette S, Bis JC, Smith JA, Ikram MK, Adams HH, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet 2015; 8: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward linkage disequilibrium, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013; 12: 822–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45: 1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 2011; 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YC, Tzourio C, Soumare A, Mazoyer B, Dufouil C, Chabriat H. Severity of dilated Virchow-Robin spaces is associated with age, blood pressure, and MRI markers of small vessel disease: a population-based study. Stroke 2010; 41: 2483–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.