Abstract
The generation of CD8+ T-cell memory is a major aim of vaccination. While distinct subsets of CD8+ T-cells are generated following immunization that differ in their ability to confer long-term immunity against infection, the transcriptional profiles of these subsets within endogenous vaccine-induced CD8+ T cell responses have not been resolved. Here, we measure global transcriptional profiles of endogenous effector (TEFF), effector memory (TEM) and central memory (TCM) CD8+ T-cells arising from immunization with three distinct prime-boost vaccine regimens. While a proportion of transcripts were uniquely regulated within distinct CD8+ T cell populations, we observed progressive up- or down-regulation in the expression of a majority of differentially expressed transcripts when subsets were compared in the order TN > TCM > TEM > TEFF. Strikingly, when we compared global differences in gene expression between TN, TCM, TEM and TEFF cells with known transcriptional changes that result when CD8+ T cells repetitively encounter antigen, our analysis overwhelmingly favored a model whereby cumulative antigen stimulation drives differentiation specifically from TN > TCM > TEM > TEFF and this was common to all vaccines tested. These findings provide insight into the molecular basis of immunological memory and identify potential biomarkers for characterization of vaccine-induced responses and prediction of vaccine efficacy.
Keywords: CD8, Memory T cells, T cell memory, Prime-boost vaccination, LCMV vector, Adenovirus vector
1. Introduction
Following immunization or infection, a small number of naïve (TN) CD8+ T-cells bearing specificity for pathogen-associated anti-gens proliferate and differentiate to generate an acute response. Following clearance of antigen there is a contraction in the size of the acute response and only a fraction of cells remain to form long-lived memory cells. Upon infection with a pathogen bearing similar antigens, these memory cells expand to control the infection, often to a better extent than the primary response. Consequently, the generation of CD8+ T-cell memory is an important aim of vaccination.
Phenotypically distinct subsets of CD8+ T cells have been defined with differential capacity for proliferation, lymphoid homing and effector function [1–3]. In particular, naïve (TN), central memory (TCM), effector memory (TEM) and effector (TEFF) CD8+ T cells can be distinguished based upon the expression of surface proteins relating to lymphoid homing and activation status [2,4].Distinct subsets of memory CD8+ T cells confer differential levels of protective immunity in a manner that is pathogen-dependent. For example, adoptive transfer of TCM specific for a recombinant virally encoded model antigen resulted in better protection against vaccinia virus when compared with TEM while a similar number TEM cells provided better protection against lymphocytic choriomeningitis virus infection, respectively, when compared with TCM [5]. Moreover, a persistent recombinant viral vector based on rhesus cytomegalovirus encoding antigens derived from simian immunodeficiency virus (SIV) predominantly generating TEM responses that homed to peripheral tissues exhibited superior protective capacity against subsequent infection with SIV [6]. Resolving the molecular basis for differences in the protective capacity of distinct CD8+ T cell subsets is therefore a priority for the development of better vaccines.
Much has been learnt about the kinetics and composition of CD8+ T-cell responses from the study of antiviral responses in mice [7–10]. Two major models exist to explain the observed predominance of cells with an effector phenotype during acute phases of the response and cells with a memory phenotype at later phases: According to the linear differentiation hypothesis, naive cells uniformly differentiate into effector cells upon encounter with antigen. Upon pathogen clearance, effector cells then either undergo apoptosis or differentiate into central/effector memory T cells [11]. According to the progressive differentiation hypothesis, CD8+ T cells differentiate along a single continuum from TN to TCM to TEM to TEFF. Moreover, the model proposes that T cells acquire irreversible changes while differentiating that result from antigenic or inflammatory signals [12,13]. While there is a predominance of effector cells during the acute phase, central memory cells generated at this time survive while effector cells die upon withdrawal of antigen resulting in a predominance of central memory cells at late time points.
While fate-mapping experiments using artificial Gzmb promoter sequences to induce expression of a heritable marker in effector cells and their progeny indicated that cells that had once been effector cells could contribute to the memory pool [14,15], these studies did not rely on the activity of the endogenous GzmB promoter that may have been subject to further epigenetic or other modulation. Data from adoptive transfer models, on the other hand, support the progressive differentiation model. By performing adoptive transfers of central memory, effector memory and effector T-cell receptor (TCR) transgenic CD8+ T cells arising at an acute phase of an antiviral response into infection-matched recipients, Kaech et al. were able to demonstrate a reduced capacity of effector cells to form memory cells [2]. Thus, there is contradictory evidence in the literature regarding lineage relationship between the CD8+ T cell subsets.
In this study, we have measured global transcriptional profiles of distinct CD8+ T cell subsets arising endogenously from vaccination of mice with three distinct prime-boost vaccine regimens. By using tetrameric peptide/major histocompatibility complex (MHC)-based sorting and highly sensitive microarray platforms, we were able to analyze endogenous vaccine-induced responses, avoiding the need for adoptive T cell transfer and the use of T cell receptor-transgenic model systems. Once fractionated into subsets, transcriptional profiles were remarkably similar between distinct vaccines. This enabled calculation of core gene expression profiles associated with distinct CD8+ T cell subsets independent of vaccine used. While a proportion of transcripts were uniquely regulated within distinct CD8+ T cell subsets, we observed progressive up- or down-regulation in the expression of a majority of differentially expressed transcripts when subsets were compared in the order TN > TCM > TEM > TEFF. When the transcriptional relationships of the CD8+ T-cell subsets were compared in an unbiased fashion with known global transcriptional changes that result when T-cells repeatedly encounter antigen, our results favored a model whereby cumulative antigenic stimulation drives differentiation specifically from TN > TCM > TEM > TEFF. We have established transcriptional profiles of endogenous vaccine-induced CD8+ T-cell responses that provide insight into molecular basis of immunological memory following vaccination and identify potential biomarkers for prediction of vaccine efficacy.
2. Materials and methods
2.1. Animals and immunization protocols
Female BALB/c mice between the ages of 6 and 10 wk (NCI/DCT, Jackson Laboratories or Charles River) were used for our experiments. They were housed in the animal facility of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Bethesda, MD. All animal experiments were reviewed and approved by the Animal Care and Use Committee, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH and were performed in accordance with all relevant federal NIH guidelines and regulations. All immunizations were administered intramuscularly. Recombinant replication-defective adenovirus (rAd5) vectors are replication-defective E1-, E3-, and E4-deleted human adenovirus serotype 5-derived vaccines generated as described previously [16]. Three intramuscular priming immunizations using plasmid DNA vaccines were used as previously described [17]. Recombinant lymphocytic choriomeningitis virus (rLCMV) vectors were generated as described previously [18].
2.2. Flow cytometry and cell sorting
Splenocytes were stained with H2-Dd/PA9-PE tetramers for 15 min at 4°C in PBS prior to staining with fluorescently conjugated antibodies. Tetramers were produced by the NIH tetramer core facility. All other fluorochrome-coupled antibodies were obtained from Becton–Dickinson and used as directed by the manufacturer: CD3 Alexa Fluor 700, CD8a PerCP-Cy5.5, CD127-PECy7, CD62L-APC-Cy7, CD16-PacBlue and CD32-PacBlue. Sorting for microarray analysis was performed using a FACS Aria directly into cold RNALater (Ambion, Inc.) before freezing at −80°C.
2.3. Intracellular cytokine staining
Briefly, spleens were harvested from mice 3 wk after the final immunization, and dissociated over nylon gauze. Single-cell suspensions were washed twice in PBS and 2 million cells per well were distributed into 96-well conical-bottomed plates, in RPMI medium supplemented with 10% FCS containing Brefeldin A (10 μg/mL) and PA9 or an irrelevant peptide (0.1 μg/100 μL). Cells were incubated at 37°C for 5 h before being washed and stained with VIVID dye (Invitrogen) and fluorescently conjugated antibodies against CD3, CD8, and CD4. Cells were then fixed and permeabilized using BD Cytofix/Cytoperm (BD Pharmingen) and stained with antibodies against IFN-γ, TNF-α, and IL-2. Cells were analyzed using an LSRII flow cytometer (Becton–Dickinson) and resultant data were analyzed using FlowJo software (Treestar, Inc.).
2.4. Microarray cDNA hybridization analysis
Quantification was performed using a spectrophotometer (NanoDrop Technologies) and RNA quality was assessed using the Experion automated electrophoresis system (Bio-Rad Laboratories). Total RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification kit, which is based on the Eberwine amplification protocol. This protocol involves a first cDNA synthesis step followed by in vitro transcription for cRNA synthesis. The biotinylated cRNA was hybridized onto Illumina Mouse Chips at 58°C for 20 h and quantified using an Illumina BeadStation 500G× scanner and Illumina BeadStudio v3 software. Illumina probe data were exported from BeadStudio as raw data and screened for quality. Samples failing chip visual inspection and control examination were removed.
Analysis of the GenomeStudio output data was conducted using R (R Development Core Team) and software packages from Bioconductor [19]. Quantile normalization was applied followed by a log transformation. Microarray data are available through the National Center for Biotechnology Information Gene Expression Omnibus (GEO), accession number GSE42459. The LIMMA package [20] was used to fit a linear model to each probe and to perform (moderated) t-tests on various differences of interest. Specifically, contrast formulas were defined as differences between the vaccine averages of CD8+ T cells subsets. The expected proportions of false positives (FDR) were estimated using the Benjamini and Hochberg method, and statistical significance for differential expression was reached at 5% FDR coupled with a minimal difference of one on the log 2 scale (|FC|>2). A CD8+ T cell subset discriminating gene signature was defined as the union of all genes that reached statistical significance for at least one pairwise subset comparison. Probes were collapsed to genes by retaining only the probe with the highest F-statistic over all pairwise comparisons. CD8+ T cell subset fractions for each of the samples from the repetitive antigen stimulation data set [21] were deconvoluted by solving a linear latent model for the least square solution constrained by inequalities (bounded by 0 and 1) [22]. Quadratic programming implemented by the pcls function of the mgcv R package was used for this purpose. The deconvolution algorithm was applied to a subset of the full discriminating gene signature limited to the top 100 genes within each pairwise comparison. Confirmation of microarray gene expression data was performed using Fluidigm Biomark gene expression analysis (Supplemental Fig. 2).
3. Results
3.1. CD8+ T cell responses in diverse states of differentiation are found at a single time point following immunization
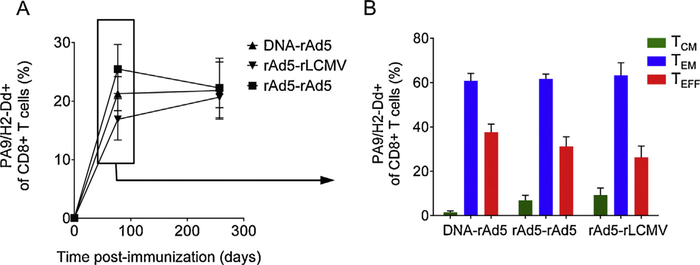
To generate endogenous CD8+ T-cell responses to a single epitope, we immunized mice with three distinct prime-boost vaccine regimens encoding the model antigen HIV-1 Env [16,23]: DNA prime/recombinant adenovirus serotype 5 (rAd5) boost (DNA-rAd5), rAd5 prime/rAd5 boost (rAd5-rAd5) and rAd5 prime/recombinant lymphocytic choriomeningitis virus (rLCMV) boost (rAd5-rLCMV). The prime and boost immunizations were administered 8 weeks apart. HIV-1 Env contains a H-2Dd-restricted immunodominant epitope (PA9) capable of eliciting T-cell responses that can be identified by binding of PA9/H-2Dd tetramers [24]. Thus, we were able to analyze the magnitude and phenotype of vaccine-elicited CD8+ T cell responses. We noted that the three vaccines (DNA-rAd5, rAd5-rAd5 and rAd5-rLCMV) elicited PA9-specific immune responses of similar magnitude at 3 weeks following boost immunization that remained stable over time (Fig. 1A). No significant differences in the ratios of TCM, TEM and TEFF cells were observed between immunization groups (at p < 0.05; Fig. 1B). Importantly, however, at this single time point, we were able to observe PA9-specific CD8+ T cells within the three major antigen-experienced differentiation states (TCM, TEM and TEFF) allowing their analysis at a single contemporaneous time point following immunization.
Fig. 1.
Immunization schema and characterization of endogenous immune reponses. (A) Animals were immunized intramuscularly with DNA-rAd, rAd-rAd or rAd-LCMV vaccine regimens encoding the HIV-1 Env gene insert. H2-Dd/PA9 tetramer staining of splenocytes isolated at indicated time points following primary immunization showing the magnitude of the endogenous immune responses against the PA9 epitope generated by the three vaccine regimens. (B) Phenotypic characterization of splenic Env-specific immune responses (CM: CD62L+ IL7R+; EM: CD62L− IL7R+; EFF: CD62L− IL7R−) at the indicated time point. No significant differences in the frequency of TCM, TEM and TEFF cells were observed between immunization groups at the indicated time point (at p < 0.05; Student’s t-test). Data are representative of two repeated experiments with five mice per group per time point.
3.2. Global transcriptional analysis of endogenous vaccine-elicited CD8+ T cells
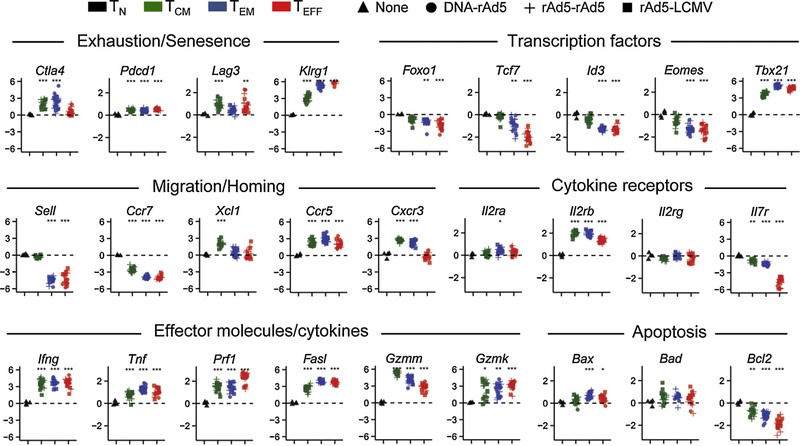
For analysis of gene transcription in the four major CD8+ T cell subsets, we isolated PA9/H-2Dd tetramer-binding TCM, TEM and TEFF cells elicited by each of the three vaccine regimens by fluorescence-activated cell sorting (Supplemental Fig. 1A). Additionally, we isolated naïve CD44−CD62L+ CD8+ T cells from pre-immune mice (Supplemental Fig. 1B). Isolated RNA was subjected to global gene expression analysis using the Illumina BeadChip platform with greater than four biological replicate measurements per cellular subset and per vaccine regimen used. Consistent with our sorting strategy, we observed downregulation of Sell (encoding CD62L) specifically in TEM and TEFF and down-regulation of Il7r expression in TEFF cells alone (Fig. 2). Further, there were dynamic changes in the expression of individual genes between distinct CD8+ T-cell subsets that were remarkably similar between all three vaccination protocols (Fig. 2). In particular, we observed changes in the expression of genes known to regulate T-cell migration (Sell, Ccr7, Xcl1, Ccr5 and Cxcr3), cytokine responsiveness (Il2ra, Il2rb and Il7r) and effector function (Ifng, Tnf, Prf1, Fasl, Gzmm and Gzmk) consistent with previous reports [3,13]. Additionally, we observed the induction of the canonical transcription factor of effector differentiation in CD8+ T cells, Tbet (Tbx21) [25] and downregulation of transcription factors associated with naïve or memory T cells (Foxo1, Tcf7, Id3 and Eomes) [26–29]. Additionally, there was acquisition of genes encoding inhibitory receptors and markers of exhaustion/senescence (Ctla4, Pdcd1, Lag3, Klrg1 and Tim3) as has been previously described upon differentiation of naïve CD8+ T cells into effector cells [30].
Fig. 2.
Dynamic changes in gene expression between endogenous TN, T CM, TEM and TEFF Env-specific CD8+ T cell responses to vaccination. Expression of selected mRNA transcripts in indicated CD8+ T-cell subsets arising endogenously following immunization with given vaccines. Expression values for each transcript are provided as a relative ratio to expression in naïve cells. Gene expression measurements represent pooled results from two independently conducted experiments and results from ≥4 mice per vaccine group are shown. Asterisks p-values; * p < 0.05; ** p < 0.01; *** p < 0.005; ****p < 0.001; two-tailed t-test.
Given the similarity in transcriptional profiles of fractionated populations of TN, TCM, TEM and TEFF cells elicited by the three vaccines, as has been previously described [23], we used the three datasets to derive core gene expression signatures of TCM, TEM and TEFF cells that were independent of the vaccine protocol used. From this dataset, we were able to identify numerous gene expression changes consistent with previous reports from microarray analysis of adoptively transferred TCR-transgenic CD8+ T cells [2,3]. However, we also observed numerous changes in the expression of genes with known or unknown function in CD8+ T cells that were not previously reported (Table 1).
Table 1. Identity and expression of known and novel genes dynamically expressed within endogenously generated CD8+ T cell subsets.
Following the calculation of a core geneset enriched for differences in gene expression independent of vaccine used, differentially expressed genes (p < 0.000001) were ranked in order of greatest differential gene expression compared with naïve cells. Fold change gene expression relative to naïve is presented in the table.
| CM | EM | EFF | CM | EM | EFF | CM | EM | EFF | CM | EM | EFF | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apoptosis | Killer-cell Lectin-like receptors | Transcription (cont’d) | Other/Unknown function (cont’d) | ||||||||||||
| Dapl1 | −2.40 | −14.62 | −15.21 | Klrk1 | 9.07 | 19.19 | 13.87 | Zfp238 | −2.91 | −3.72 | −5.63 | C030011O14Rik | −2.99 | −5.62 | −4.98 |
| Pdcd4 | −4.28 | −7.19 | −2.10 | Klre1 | 3.09 | 16.31 | 39.44 | Tcf7 | −2.39 | −5.38 | Ttc14 | −3.53 | −6.46 | −4.88 | |
| Gadd45b | 2.29 | 3.81 | 6.12 | Klrg1 | 7.43 | 42.99 | 64.41 | Srebf2 | −3.62 | −3.59 | −5.29 | Whrn | −2.16 | −5.13 | −4.81 |
| Dapk2 | 19.08 | 38.17 | 49.61 | Tbl1x | −4.98 | −7.35 | −5.03 | Psmd7 | −4.73 | −5.66 | −4.32 | ||||
| Casp1 | 41.64 | 44.85 | 51.91 | Metabolism | L3mbtl3 | −4.32 | −6.25 | −4.99 | Heatr5a | −4.25 | −5.98 | −4.25 | |||
| Igfbp4 | −2.84 | −23.09 | −17.14 | Rexo1 | −8.94 | −4.86 | −3.71 | Zc3h13 | −2.97 | −10.31 | −4.05 | ||||
| Cell cycle | Ampd1 | −7.61 | −16.02 | −14.63 | Bach1 | −3.11 | −9.34 | −3.04 | Vapa | −6.27 | −4.21 | −4.03 | |||
| Actn1 | −3.53 | −15.79 | −35.35 | Slc2a9 | −8.70 | −10.47 | −9.46 | Zmynd11 | −2.95 | −13.84 | −2.79 | Rab5c | −5.92 | −5.08 | −3.94 |
| 1190002H23Rik | −4.96 | −11.76 | −11.59 | Bdh1 | −3.63 | −4.97 | −8.33 | Gabpb2 | −3.41 | −9.48 | −2.61 | Lmnb1 | −6.64 | −5.09 | −3.81 |
| Pard6g | −6.18 | −10.27 | −8.70 | Slc16a6 | −8.65 | −8.12 | −8.02 | Cep110 | −3.64 | −6.49 | −2.17 | Timp2 | −3.86 | −5.07 | −3.71 |
| Zc3h12d | −4.07 | −5.39 | −8.17 | Osbpl6 | −4.59 | −7.25 | −7.63 | Zbtb7a | −5.32 | 1.69 | LOC100048583 | −4.70 | −5.68 | −3.70 | |
| Trp53 | −3.17 | −5.14 | −6.95 | Art2b | −4.38 | −4.65 | −6.21 | Smad3 | 3.69 | 5.63 | 4.62 | Snx1 | −5.12 | −3.80 | −3.26 |
| Ptp4a2 | −4.48 | −4.51 | −5.33 | Adk | −4.06 | −5.48 | Polr1d | 6.38 | 4.03 | 4.88 | Tmem23 | −2.23 | −7.97 | −2.99 | |
| Rbm38 | −2.82 | −4.12 | −5.20 | Ctps | −2.24 | −3.50 | −5.34 | Tceb2 | 4.74 | 4.18 | 5.16 | Nol5a | −3.23 | −8.58 | −2.93 |
| Cdkn1b | −5.31 | −19.16 | −3.88 | Faah | −2.09 | −2.83 | −5.16 | Rora | 1.66 | 3.03 | 5.33 | Rsrc2 | −4.30 | −8.63 | −2.65 |
| Hsd3b2 | −7.12 | −5.60 | −4.06 | E2f2 | 2.94 | 4.50 | 5.42 | C330021F23Rik | −6.79 | −5.39 | −2.23 | ||||
| Cellular homing and adhesion | Ndufa6 | 6.36 | 4.88 | 4.65 | Scand1 | 2.79 | 3.08 | 6.59 | Rad23a | −1.96 | −5.44 | −1.97 | |||
| Sell | −21.73 | −18.66 | Dcxr | 2.57 | 3.03 | 5.03 | Bhlhb2 | 6.02 | 17.09 | 20.79 | D1Bwg0212e | −3.23 | −6.11 | −1.96 | |
| Itgb1 | 3.22 | 6.12 | 4.15 | Cox6b1 | 4.27 | 3.60 | 5.51 | Tbx21 | 11.64 | 35.19 | 26.24 | Oas1 g | 6.40 | 5.91 | 2.88 |
| Pcdh21 | 2.87 | 6.91 | Dpm3 | 4.33 | 3.35 | 5.66 | Zeb2 | 11.48 | 52.67 | 68.32 | Rps13 | 5.86 | 2.52 | 2.91 | |
| Ndufb10 | 4.19 | 5.44 | 6.89 | 5830482F20Rik | 4.51 | 5.63 | 4.12 | ||||||||
| Chemokines | Tktl1 | 5.70 | 7.82 | Other/Unknown function | Sytl2 | 2.52 | 5.66 | 4.32 | |||||||
| Ccl3 | 2.88 | 8.53 | 10.82 | Alad | 6.24 | 5.64 | 8.30 | St6gal1 | −3.97 | −36.84 | −37.06 | Gabarapl1 | 2.04 | 5.53 | 4.93 |
| Ccl5 | 52.52 | 67.71 | 68.50 | Alas2 | 8.01 | 11.69 | 14.34 | Ppic | −6.37 | −30.36 | −30.87 | Lhfpl2 | 2.99 | 5.10 | |
| Ccl4 | 37.06 | 87.57 | 95.56 | Soat2 | 8.36 | 18.71 | 26.95 | Atp1b1 | −6.82 | −27.99 | −25.59 | 1700020L24Rik | 2.36 | 2.95 | 5.18 |
| Tmie | −6.90 | −20.98 | −20.93 | Clic1 | 3.54 | 4.34 | 5.41 | ||||||||
| Chemokine receptors | Signal transduction | LOC100041103 | −2.79 | −12.81 | −20.31 | Insl6 | 3.71 | 4.22 | 5.54 | ||||||
| Ccr7 | −5.35 | −14.59 | −16.05 | Socs3 | −4.03 | −24.35 | Anp32a | 19.04 | −17.73 | −17.23 | Ahnak | 1.98 | 4.47 | 5.55 | |
| Cxcr3 | 6.46 | 4.77 | Ramp1 | −4.67 | −20.74 | −20.58 | Rasl11b | −6.85 | −6.90 | −17.02 | Sdcbp2 | 4.82 | 6.29 | 5.65 | |
| Ccr5 | 5.32 | 7.59 | 4.07 | Gpr114 | −3.19 | −10.48 | −18.77 | Tpm3 | 12.46 | −17.60 | −13.93 | Dnajc15 | 5.80 | 5.93 | 5.73 |
| Ccl9 | 2.04 | 5.84 | 8.25 | Lrp12 | −8.29 | −14.94 | −17.17 | Fam134b | −3.41 | −7.28 | −10.57 | 1700025G04Rik | 5.17 | 3.97 | 5.83 |
| Cx3cr1 | 4.00 | 18.49 | 29.36 | Rgs10 | −3.67 | −17.22 | −14.62 | Lypd6b | −3.89 | −6.28 | −10.49 | 6330503C03Rik | 2.37 | 4.75 | 5.88 |
| Trib2 | −2.93 | −15.50 | −14.57 | Raly | 15.11 | −11.52 | −10.46 | Plxdc1 | 5.30 | 6.67 | |||||
| Chromatin structure and modification | Pacsin1 | −3.55 | −12.74 | −13.02 | Pscd3 | −4.04 | −10.91 | −10.12 | Snx8 | 4.48 | 3.65 | 6.67 | |||
| Chmp4b | −11.19 | −10.34 | −10.20 | Usp10 | −2.46 | −6.23 | −11.78 | Pitpnm2 | −5.34 | −8.23 | −9.48 | Crip2 | 2.64 | 5.65 | 6.95 |
| Jmjd3 | −5.50 | −3.28 | −1.99 | Ppm1l | −4.13 | −10.76 | −11.52 | Clec2i | −3.40 | −8.01 | −9.14 | Nrp1 | 3.51 | 7.70 | 7.39 |
| Hist1h2bf | 2.47 | 3.15 | 5.59 | Csnk1g1 | −6.83 | −9.11 | −8.90 | Arl6ip5 | 11.51 | −12.34 | −9.09 | Plek | 3.58 | 5.81 | 7.72 |
| Hist2h2ab | 3.62 | 3.86 | 5.88 | Mycbp2 | −1.64 | −4.30 | −7.89 | Cnr2 | −8.51 | −8.88 | LOC641240 | 7.90 | 3.45 | 7.74 | |
| Hist1h2bj | 3.10 | 3.76 | 5.88 | Rras2 | −7.71 | −6.36 | −7.33 | Fntb | −2.51 | −6.66 | −8.70 | 6330403K07Rik | 2.77 | 5.67 | 7.97 |
| Hist1h4m | 4.31 | 7.67 | 6.29 | Amigo2 | −1.95 | −5.99 | −7.30 | Trub1 | −2.16 | −3.50 | −8.69 | Fcgr2b | 4.10 | 9.06 | 8.19 |
| Hist1h2bk | 3.74 | 4.90 | 6.56 | Trat1 | −2.98 | −6.39 | −7.16 | Kif23 | −4.39 | −5.88 | −8.52 | Bloc1s1 | 8.23 | 5.06 | 8.31 |
| Hist2h2ac | 5.30 | 4.86 | 6.65 | Ppp3r1 | −7.59 | −3.32 | −6.71 | Slc11a2 | −2.41 | −3.57 | −8.43 | Trex1 | 3.10 | 4.66 | 8.63 |
| Hist1h4k | 4.66 | 8.07 | 6.76 | Limk1 | −6.98 | −5.46 | −6.52 | 2410066E13Rik | −5.94 | −7.08 | −7.94 | H2-Ea | 9.38 | 4.36 | 8.65 |
| Hist1h3a | 4.70 | 5.04 | 7.93 | Rnf167 | −6.74 | −5.50 | −5.69 | Smc6 | −4.57 | −5.37 | −7.78 | H2-Eb1 | 7.42 | 3.75 | 9.51 |
| Hist1h2ag | 4.79 | 4.52 | 8.41 | 4631426J05Rik | −5.39 | −5.18 | −5.66 | Xkrx | −4.32 | −7.61 | −7.56 | Nt5e | 5.97 | 6.81 | 9.53 |
| Hist1h4i | 5.10 | 7.63 | 8.42 | Dusp10 | −4.68 | −5.13 | −5.11 | Cep68 | −6.39 | −7.39 | −7.33 | Myadm | 5.23 | 14.61 | 9.72 |
| Hist1h2ao | 7.07 | 6.22 | 9.80 | Cd69 | −2.33 | −3.64 | −5.07 | Spnb2 | −4.07 | −3.09 | −7.16 | Kcnk6 | 8.20 | 8.56 | 9.85 |
| Hist1h3d | 6.53 | 6.84 | 9.97 | Usp1 | −4.25 | −7.02 | −5.03 | Psme3 | −6.88 | −8.50 | −7.16 | Hbb−b2 | 3.76 | 6.85 | 9.87 |
| Hist1h4j | 7.57 | 10.17 | 11.41 | Rgs1 | 8.42 | 8.52 | 5.32 | Tatdn2 | −6.49 | −5.14 | −6.80 | Arsb | 4.22 | 9.66 | 10.13 |
| Hist1h3e | 8.46 | 7.83 | 12.04 | Gng2 | 6.02 | 6.37 | 5.88 | Ephx1 | −4.96 | −9.08 | −6.57 | Plekhg3 | 4.58 | 10.05 | 10.21 |
| Hist1h2af | 7.76 | 7.46 | 12.91 | Slamf7 | 3.56 | 7.86 | 6.64 | LOC100047606 | −2.41 | −5.15 | −6.12 | Lmna | 3.90 | 7.97 | 10.36 |
| Hist1h2an | 7.88 | 7.93 | 13.44 | Cish | 5.06 | 10.69 | 7.48 | Crlf3 | −3.84 | −3.89 | −5.84 | Clip4 | 3.34 | 10.60 | |
| Hist1h2ad | 11.04 | 10.89 | 20.17 | Prdx4 | 3.10 | 6.33 | 8.47 | Tmem9 | −2.87 | −3.78 | −5.66 | Stard10 | 2.69 | 7.16 | 14.85 |
| Hist1h2ak | 12.21 | 11.37 | 21.35 | Ier3 | 4.56 | 8.10 | 9.64 | Rcl1 | −2.79 | −4.07 | −5.53 | Fam129b | 3.11 | 15.99 | 15.61 |
| Hist1h2ah | 12.89 | 12.44 | 21.99 | S100a13 | 3.94 | 5.94 | 10.10 | Tspan14 | −3.17 | −3.35 | −5.52 | Anxa2 | 7.74 | 12.32 | 15.75 |
| Socs2 | 5.29 | 7.56 | 10.49 | Spsb1 | −2.81 | −4.85 | −5.48 | Endod1 | 7.98 | 11.87 | 16.27 | ||||
| Cytokines | Ncald | 4.09 | 9.56 | 10.57 | Armcx2 | −2.47 | −5.36 | Mt1 | 5.59 | 9.14 | 16.53 | ||||
| Tnfsf14 | 4.85 | 7.60 | 5.76 | Lrrk1 | 6.19 | 11.45 | 20.39 | Nsg2 | 2.26 | −2.10 | −5.30 | Kcnj8 | 2.86 | 13.71 | 23.98 |
| Ifng | 51.34 | 75.28 | 60.60 | Edg8 | 3.34 | 30.46 | 52.53 | Cd55 | −1.60 | −3.28 | −5.30 | Emp1 | 9.77 | 28.48 | 24.41 |
| 5830457O10Rik | −5.45 | −4.49 | −5.30 | Garnl4 | 6.70 | 29.08 | 28.81 | ||||||||
| Cytokine receptors | Transcription | Emb | −2.56 | −3.29 | −5.22 | Sytl3 | 15.71 | 31.72 | 29.32 | ||||||
| Il7r | −1.93 | −2.67 | −25.83 | Bach2 | −7.42 | −21.29 | −29.62 | Ivns1abp | −3.15 | −2.80 | −5.21 | Esm1 | 8.17 | 24.22 | 33.91 |
| Il18rap | 2.15 | 3.87 | 6.34 | Smad1 | −3.22 | −4.90 | −10.34 | 5530601119Rik | −3.32 | −2.76 | −5.20 | S100a6 | 29.85 | 30.96 | 36.16 |
| Egr2 | −4.22 | −8.41 | −10.14 | Luc7l2 | −4.01 | −4.15 | −5.19 | Lgals1 | 14.29 | 34.21 | 40.32 | ||||
| Cellular cytotoxicity | Egr1 | −4.28 | −6.53 | −7.51 | Golim4 | −7.78 | −6.48 | −5.19 | Kcnk5 | 26.54 | 40.75 | 45.29 | |||
| Prf1 | 2.52 | 2.71 | 5.07 | Taf15 | −6.88 | −4.58 | −7.06 | Agfg1 | −1.84 | −3.12 | −5.13 | Hba-a1 | 31.57 | 36.16 | 48.95 |
| Gzmm | 43.06 | 16.76 | 6.99 | Epc1 | −5.93 | −6.39 | −6.73 | Gbp5 | −4.32 | −6.76 | −5.11 | Smpdl3b | 19.68 | 42.79 | 50.58 |
| Fasl | 6.01 | 15.35 | 12.55 | Eif4a1 | −5.21 | −2.94 | −6.06 | Ddx6 | −4.74 | −12.18 | −5.08 | Osbpl3 | 25.32 | 59.82 | 65.74 |
| Lgals3 | 23.93 | 97.00 | 123.44 | ||||||||||||
3.3. TCM cells exhibit a higher degree of transcriptional relatedness to TN cells than TEM or TEFF cells
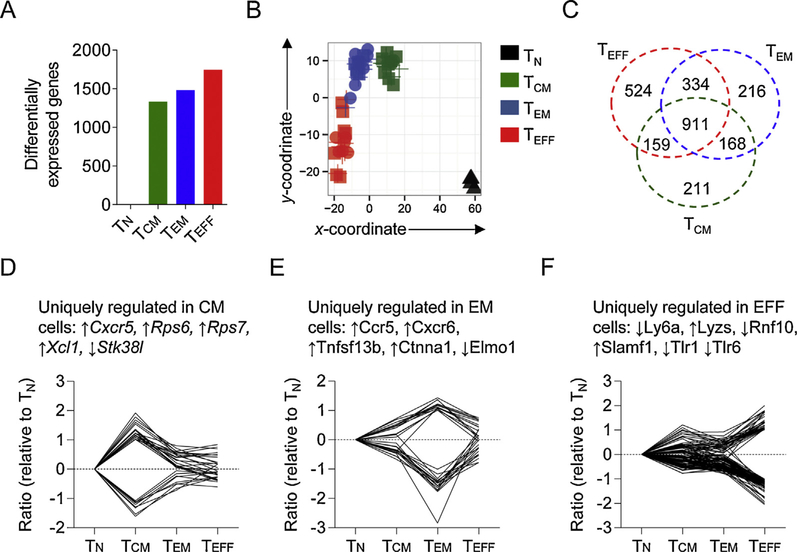
Using derived core signatures, we were able to compare the transcriptional proximity of each antigen-experienced subset to TN cells by analysing the number of differentially expressed genes (Fig. 3A). The number of genes significantly differentially expressed as compared to TN cells was greatest in TEFF cells, less in TEM cells and the lowest in TCM cells. This indicates that at a transcriptional level, TCM cells share greater transcriptional relatedness with TN cells than either TEM or TEFF cells. This transcriptional proximity relationship was also reflected in a multidimensional scaling analysis (Fig. 3B). This analysis reduces the dimensionality of gene expression datasets to allow the transcriptional distance between distinct whole transcriptome measurements to be presented in a lower-dimensional representation while preserving relative between-object distances as well as possible [31]. Our analysis placed TN cells closer to TCM than TEM or TEFF cells. It was also apparent that TCM is closer to TEM than to TEFF. Moreover, the vaccine protocol does not appear to affect substantially affect positioning within clusters, indicating that once fractionated into subsets, the immunogen appeared to play a minor role in determining population-level gene expression.
Fig. 3.
TCM cells exhibit greater transcriptional relatedness to TN cells than either TEM or TEFF cells. (A) Number of differentially expressed genes between the antigen-experienced CD8+ T-cell subsets (TCM, TEM and TEFF) and TN cells from the core signature. (B) Multidimensional scaling analysis of global gene expression data from the CD8+ T-cell subsets separated by vaccine protocol indicating transcriptional proximity of endogenously derived CD8+ T-cell subsets. (C) Venn diagram indicating uniquely up- or down-regulated transcripts within distinct CD8+ T cell subsets as compared with TN. Members of the Venn diagram are provided in Supplemental Tables 1–7. (D–F) Uniquely regulated transcripts in TCM (D), TEM (E) and TEFF (F) cells; selected transcripts and the direction in which they are either up- or down-regulated are identified.
Uniquely regulated transcripts specific to particular cellular subsets are of interest since such genes could be required for lineage-specific maintenance or function. We therefore examined uniquely up- or downregulated genes in TCM, TEM and TEFF cell compared to TN cells (Fig. 3C–F and Supplemental Tables 1–7). Consistent with previous reports, we observed enrichment of the gene encoding Cxcr5 in TCM cells, which guides CD8+ T cells to lymph nodes by binding the ligand Cxcl13, expressed in secondary lymphoid organs. We also observed unique upregulation of the gene encoding Xcl1 in TCM cells which is required for optimal interaction ofCD8+ TcellswithCD8+ dendritic cells [32] and the genes encoding ribosomal proteins Rps6 and Rps7 which play a critical role in ribosome biogenesis [33]. We also observed high levels of expression of the chemokine receptors Ccr5 and Cxcr6 in TEM cells. In TEFF cells we observed specific upregulation of the gene encoding Slamf1, a leukocyte cell-surface glycoprotein involved in TCR signaling and activation-induced cell death [34] and the gene encoding the cytotoxic molecule lysozyme. This indicated that certain transcripts are uniquely expressed in specific subsets of CD8+ T cells.
3.4. Progressive up- or down-regulation in gene expression between TN > TCM > TEM > TEFF account for the majority of transcriptional differences between T-cell subsets
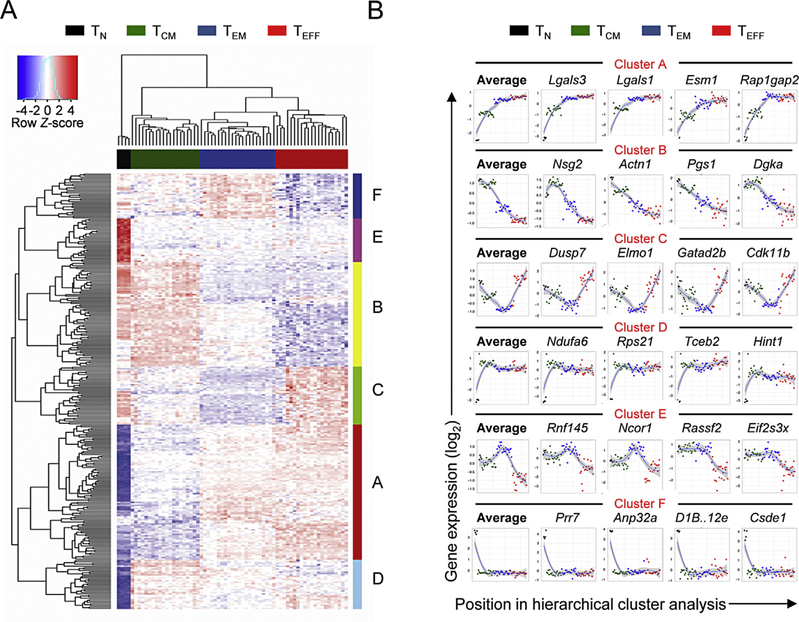
Our results indicated the presence of a small fraction of transcripts that were uniquely regulated in specific CD8+ T-cell subsets. This led us to ask whether there were broader patterns in the regulation of genes that were not uniquely enriched in specific subsets. Thus, we subjected differentially expressed genes from the core gene set to unsupervised hierarchical cluster analysis (Fig. 4A; Supplemental Table 8). Six clusters of transcripts with distinct patterns of fluctuation in expression could be identified. Average gene expression and profiles of the four most representative genes in each cluster are shown (Fig. 4B). Interestingly, the two largest clusters (A and B), showed a pattern of fluctuation in gene expression that was sequential, with progressive up- or down-regulation in gene expression from TN > TCM > TEM > TEFF. Within cluster A (the largest cluster, containing genes progressively upregulated from TN > TCM > TEM > TEFF) we observed genes encoding the surface lectin associated with T cell senescence KLRG1 [35], the effector-associated transcription factors Tbet and Id2, and the chemokine Ccl4 (Supplemental Table 9). Within cluster B (the second largest cluster, containing transcripts progressively downregulated from TN to TCM to TEM to TEFF), we observed genes encoding the transcription factor Tcf7, the lymphoid homing molecule CD62L, the receptor for homeostatic cytokine IL-7 (IL-7R) and a marker of T cell longevity, CD27 (Supplemental Table 10)[36–38]. Smaller clusters of genes with distinct patterns of fluctuation in gene expression could also be identified (Clusters C–E; Supplemental Tables 11–14). Cluster C contained genes whose expression was shared by TN and TEFF cells while cluster E contained genes predominantly expressed in TEM cells. Other clusters could be identified mainly comprising genes specifically up- or down-regulated in naïve cells (clusters F and D, respectively). A majority of genes were assigned to Clusters A and B, indicating that most differentially expressed genes between CD8+ T cell subsets undergo progressive up- or down-regulation in mRNA expression in a pattern that is sequential and in the order TN to TCM to TEM to TEFF.
Fig. 4.
Progressive up- or down-regulation of gene expression accounts for the majority of transcriptional changes occurring in the order TN > TCM > TEM > TEFF. (A) Hierarchical clustering analysis of differentially expressed genes between subsets. Clusters [A-F] of genes exhibiting distinct patterns of regulation between subsets are highlighted along the right hand side corresponding to the dendrogram on the left side of the heatmap. Clustering of CD8+ T-cell subsets based on global transcriptional proximity is highlighted above the heatmap and corresponds to the dendrogram. (B) Average and individual patterns of gene expression within the six identified clusters. Samples are distributed along the x-axis of plots based upon their position in the hierarchical clustering analysis in A and y-axes represent standardized gene expression levels. The top four ranked members of each cluster are displayed to the right of the average gene expression plot for each cluster.
Progressive transcriptional differences between CD8+ T cell subsets are similar to global changes in gene expression when CD8+ T cells progressively encounter antigen
Our data indicated that CD8+ T cells exist along a continuum of transcriptional relatedness with progressive up- or downregulation of a majority of differentially expressed transcripts between subsets in the order TN, TCM, TEM and TEFF. According to the progressive differentiation model, CD8+ T cells differentiate along a unidirectional continuum from TN to TCM to TEM to TEFF as a result of antigenic or inflammatory signals and exhibit progressive acquisition of effector function and concomitant loss of capacity for long-term survival [39].
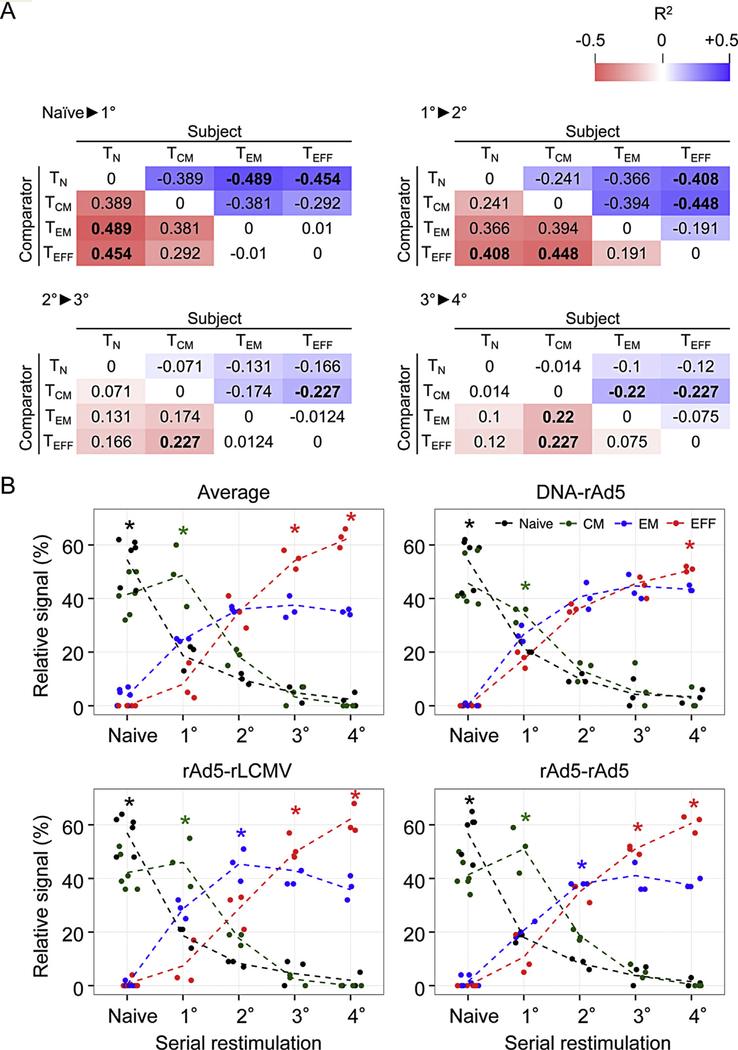
A recent study by Wirth et al. established global transcriptional changes that occur when CD8+ T-cells undergo either primary (1°), secondary (2°), tertiary (3°) or quaternary(4°) stimulation in vivo [21]. We therefore set about comparing the global transcriptional differences we observed between CD8+ T-cell subsets with transcriptional changes that occur when CD8+ T cells repeatedly encounter antigen as defined by Wirth et al. Global transcriptional differences between pairwise combinatorial comparisons of CD8+ T cell subsets from our analysis (e.g. TN vs. TCM or TEFF vs. TCM) were correlated with transcriptional changes accompanying each progressive round of stimulation from the dataset of Wirth et al. (e.g. Naïve ▶ 1°, 1° ▶ 2°, 2° ▶ 3° or 3° ▶ 4°)We observed that transcriptional changes accompanying either Naïve ▶ 1°, 1° ▶ 2°, 2° ▶ 3° or 3° ▶ 4° stimulation were often positively correlated, but never negatively correlated, with transcriptional differences between CD8+ T-cell subsets in the direction TN > TCM > TEM > TEFF (Fig. 5A). The directionality of these correlations add support for a model whereby cumulative exposure to antigen promotes progressive changes in gene expression resulting in differentiation from TN to TCM to TEM to TEFF.
Fig. 5.
Differences in gene expression between CD8+ T-cell subsets isolated at a single time point following immunization resemble known transcriptional changes that occur when T cells repetitively encounter antigen. (A) Differences in gene expression between subject and comparator CD8+ T cell subsets were evaluated in the context of known transcriptional changes that occur when CD8+ T cells repetitively encounter antigen [21]. Pearson’s correlation coefficient between the fold-changes (log2) of a given pairwise CD8+ T cell subset comparison (subject and comparator subsets are indicated as column and row titles) and the fold-changes of a given repetitive antigen stimulation (red boxes) comparison are shown. Statistically significant values (p < 0.05) are indicated in bold text. Naïve to primary (top left), primary to secondary (bottom left), secondary to tertiary (top right) and tertiary to quaternary (bottom right) gene expression changes from the dataset of Wirth et al. were evaluated independently. Only genes that reached statistical significance in the subset comparisons were considered for the correlation calculations. (B) Deconvolution analysis showing relative similarity of TN, TCM, TEM and TEFF cell fractions within cell populations generated following repetitive antigen stimulations of Wirth et al. (naïve, primary (I), secondary (II), tertiary (III) and quaternary (IV)). Colored asterisks represent p-values (* p <0.05; Wilcoxon rank sum test) and indicate statistical significance of enrichment of the indicated CD8+ T-cell subset (as indicated by the color of the asterisk) in populations resulting from indicated repetitive antigen stimulation in vivo.
Given the directionality of these changes, we hypothesised that TCM, TEM and TEFF cells bear greatest transcriptional similarity to distinct populations of repetitively stimulated cells. We therefore compared the transcriptional similarity of TCM, TEM and TEFF cells from our analysis to cells that had undergone either 1°, 2°, 3° or 4° stimulation in the study of Wirth et al. To compare transcriptional similarity of each subset to 1°, 2°, 3° or 4° stimulated cells we performed a deconvolution analysis between the two datasets (Fig. 5B) [22]. From this analysis, we found that cells that had undergone 1° stimulation had the greatest relative transcriptional signal of TCM cells. Likewise, we found that the relative transcriptional signal of TEM cells was most represented in 2° cells while the relative transcriptional signal of TEFF cells was most represented in 4° cells. What is remarkable from this observation is that at a single contemporaneous time point following vaccination, cells are isolated bearing a transcriptional imprint of differential exposure to antigen, and that this imprint coincides with their cellular phenotype.
4. Discussion
In this study we have established global transcriptional profiles of endogenous vaccine-induced effector (TEFF), effector memory (TEM) and central memory (TCM) CD8+ T cells. Previous studies have derived transcriptional profiles of CD8+ T cell subsets arising following adoptive transfer of T-cell receptor (TCR) transgenic cells specific for model antigens expressed by infectious pathogens. While these studies have provided a substantial mechanistic insights into the molecular basis of immunological memory, lymphocyte migration and effector function, the high affinity of transgenic TCRs for cognate model antigens, and imbalances between endogenous immune cells and antigen-specific cells caused by adoptive transfer may leave artifactual imprints in derived transcriptional profiles. By using tetramer-based identification of cellular subsets with defined antigen-specificity and highly sensitive microarray platforms, we were able to circumvent these problems and use prime-boost vaccine vectors to generate endogenous populations of CD8+ T cells specific for a single epitope within a vaccine antigen. Using this approach, we have been able to derive transcriptional profiles from endogenous vaccine-elicited CD8+ T cell responses, leading to identification of novel differentially expressed transcripts between subsets and a number of genes that are uniquely expressed in distinct cell populations.
A new direction in the field of vaccine development is the application of systems biology to define molecular predictors of individual responses to vaccination, potentially enabling rapid screening of promising vaccine candidates [40]. While selected or global gene expression profiles from peripheral blood samples following vaccination may enable resolution of distinct cellular subsets comprising vaccine-induced immune responses using decomposition analysis, such attempts will require gene expression profiles of defined cellular subsets that are reflective of endogenous vaccine-induced responses. By providing such data, we anticipate that this study will substantially enable these attempts.
What was remarkable to us was the observation that the majority of genes that were differentially expressed between CD8+ T cell subsets underwent either progressive up- or down-regulation when comparing subsets in the order TN, TCM, TEM and TEFF. As a result, TCM cells appeared to share greater transcriptional relatedness with TN cells than did either TEM or TEFF cells. An important aspect of the linear differentiation model, whereby effector cells are formed uniformly following antigen stimulation regardless of antigenic signal strength, is that we might expect effector cells to most resemble cells that had undergone few encounters with antigen, and that subsequent differentiation into central or effector memory cells would not result in any decrease in the imprint of antigen signaling. By comparing cells in distinct subsets arising contemporaneously following immunization with known changes that occur when CD8+ T cells undergo cumulative encounters with antigen however, we observed that effector cells exhibited the transcriptional imprint of high antigen exposure, while the analysis placed central and then effector memory cells as intermediates in this spectrum. These results are more consistent with the progressive differentiation hypothesis whereby cumulative encounters with antigen signals results in irreversible differentiation along a continuum defined by a transition from TN to TCM to TEM to TEFF states. While these results are merely correlative and do not definitively establish lineage relationships, our conclusions are in agreement with a recent study which utilized a single-cell fate-mapping strategy to define lineage relationships between antigen-experienced cell populations arising following infection [41]. Importantly, our results do not preclude that a subset of cells with a central memory phenotype revert their transcriptional profiles to a state resembling cells with low antigen exposure history despite arising from effector cells with transcriptional profiles resembling high exposure to antigen.
The relationship between repetitive antigenic signaling and progressive differentiation to TEM and TEFF states is of importance, since rhesus CMV-based vaccine vectors capable of chronic persistence generated effector memory CD8+ T cell responses that afforded robust protection against SIV challenge [6]. Given advantageous peripheral and mucosal homing properties effector memory cells to sites where the majority of HIV infections are acquired in humans, understanding factors that lead to generation of these cells following vaccination, and their transcriptional program is a priority to the field.
Finally, our analysis of endogenous responses identifies novel transcriptional differences between distinct CD8+ T cell subsets. Functional characterization of these transcripts will enable better understanding of the biology of CD8+ T cells and provide molecular targets and signaling pathways relevant to the manipulation of CD8+ T cell responses during vaccination.
Supplementary Material
Acknowledgements
We thank S. Perfetto, E. Lugli, M. Vernez, A. Bergthaler and G. Fabozzi, D.C. Macallan and G.E. Griffin for helpful insights and discussion. We also thank R. Nguyen and D. Ambrozak for their expertise with FACS sorting. This research was supported by the Intramural Research Programs of the US National Institutes of Health, National Institute of Allergy and Infectious Diseases and National Cancer Institute. L.F. is supported by the Stiftung Propter Homines, Vaduz, Principality of Liechtenstein, the Fondation Leenaards and the Medic Foundation. A.N.H. was supported by a European Molecular Biology Organization (EMBO) long-term fellowship (ALTF 116–2012) and a Marie Curie fellowship (FP7-PEOPLE-2012-IEF, Proposal 330621). R.R. is supported by a Fellowship from the Wellcome Trust.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2014.10.007.
Accession codes
GEO: microarray data, GSE42459.
Conflict of interest statement The authors declare no competing financial interests.
References
- [1].Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–12. [DOI] [PubMed] [Google Scholar]
- [2].Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 2003;4:1191–8. [DOI] [PubMed] [Google Scholar]
- [3].Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med 2008;205:625–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 2004;22:745–63. [DOI] [PubMed] [Google Scholar]
- [5].Bachmann MF, Wolint P, Schwarz K, Jager P, Oxenius A. Functional properties and lineage relationship of CD8+ T cell subsets identified by expression of IL-7 receptor alpha and CD62L. J Immunol 2005;175:4686–96. [DOI] [PubMed] [Google Scholar]
- [6].Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011;473:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev 2010;236:151–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol 2001;2:415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8(+) T cells after infection. Nat Immunol 2002;3:619–26. [DOI] [PubMed] [Google Scholar]
- [10].Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature 2009;458:211–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Opferman JT, Ober BT, Ashton-Rickardt PG. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science 1999;283:1745–8. [DOI] [PubMed] [Google Scholar]
- [12].Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 2002;111:837–51. [DOI] [PubMed] [Google Scholar]
- [13].Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2002;2:251–62. [DOI] [PubMed] [Google Scholar]
- [14].Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science 2009;323:505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature 1999;399:593–7. [DOI] [PubMed] [Google Scholar]
- [16].Flatz L, Cheng C, Wang L, Foulds KE, Ko SY, Kong WP, et al. Gene-based vaccination with a mismatched envelope protects against simian immunodeficiency virus infection in nonhuman primates. J Virol 2012;86:7760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, et al. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 2007;25:4085–92. [DOI] [PubMed] [Google Scholar]
- [18].Flatz L, Hegazy AN, Bergthaler A, Verschoor A, Claus C, Fernandez M, et al. Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat Med 2010;16:339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:1544–6115 (Article3). [DOI] [PubMed] [Google Scholar]
- [21].Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity 2010;33:128–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gong T, Hartmann N, Kohane IS, Brinkmann V, Staedtler F, Letzkus M, et al. Optimal deconvolution of transcriptional profiling data using quadratic programming with application to complex clinical blood samples. PLoS One 2011;6:e27156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Flatz L, Roychoudhuri R, Honda M, Filali-Mouhim A, Goulet JP, Kettaf N, et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc Natl Acad Sci USA 2011;108:5724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Honda M, Wang R, Kong WP, Kanekiyo M, Akahata W, Xu L, et al. Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity. J Immunol 2009;183: 2425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 2002;295:338–42. [DOI] [PubMed] [Google Scholar]
- [26].Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol 2011;12:1230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol 2009;10:176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou X, Xue HH. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol 2012;189:2722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 2005;6:1236–44. [DOI] [PubMed] [Google Scholar]
- [30].Wherry EJ. T cell exhaustion. Nat Immunol 2011;12:492–9. [DOI] [PubMed] [Google Scholar]
- [31].Borg I, Groenen PJF. Modern multidimensional scaling: theory and applications. New York, NY: Springer; 1997. [Google Scholar]
- [32].Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Guttler S, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 2009;31: 823–33. [DOI] [PubMed] [Google Scholar]
- [33].Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 2008;14:1918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol 2010;32:157–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 2007;27:281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hendriks J, Gravestein LA, Tesselaar K, van Lier RA, Schumacher TN, Borst J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat Immunol 2000;1:433–40. [DOI] [PubMed] [Google Scholar]
- [37].Ochsenbein AF, Riddell SR, Brown M, Corey L, Baerlocher GM, Lansdorp PM, et al. CD27 expression promotes long-term survival of functional effector-memory CD8+ cytotoxic T lymphocytes in HIV-infected patients. J Exp Med 2004;200:1407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peperzak V, Xiao Y, Veraar EA, Borst J. CD27 sustains survival of CTLs in virus-infected nonlymphoid tissue in mice by inducing autocrine IL-2 production. J Clin Invest 2010;120:168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol Rev 2006;211:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Haining WN, Pulendran B. Identifying gnostic predictors of the vaccine response. Curr Opin Immunol 2012;24:332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Graf P, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science 2013;340:630–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.