Abstract
The smallest arenaviral protein is the zinc-finger protein (Z) that belongs to the RING finger protein family. Z serves as a main component required for virus budding from the membrane of the infected cells through self-oligomerization, a process that can be aided by the viral nucleoprotein (NP) to form the viral matrix of progeny virus particles. Z has also been shown to be essential for mediating viral transcriptional repression activity by locking the L polymerase onto the viral promoter in a catalytically inactive state, thus limiting viral replication. The Z protein has also recently been shown to inhibit the type I interferon-induction pathway by directly binding to the intracellular pathogen-sensor proteins RIG-I and MDA5, and thus inhibiting their normal functions. This chapter describes several assays used to examine the important roles of the arenaviral Z protein in mediating virus budding (i.e., either Z-self budding or NP-Z budding activities), viral transcriptional inhibition in a viral minigenome (MG) assay, and type I IFN suppression in an IFNb-promoter mediated luciferase reporter assay.
Keywords: arenavirus, Lassa virus, Pichinde virus, matrix protein, RING protein, virus budding, interferon suppression, transcription inhibition
1. Introduction
The Arenaviridae consists of a large group of bi-segmented single-stranded ambisense RNA viruses that can cause significant morbidity and mortality in humans. Lassa virus and Lujo virus (LUJV), found in Africa, and several other arenaviruses found in South America can cause severe hemorrhagic fever (HF). There are currently limited prevention and treatment modalities against these pathogenic arenaviruses. The only available vaccine (Candid #1) has been developed and used extensively to prevent Argentine hemorrhagic fever (AHF) caused by Junín virus (JUNV) (1). Ribavirin, a guanosine analog, has been used in treating arenaviral HF with mixed success and significant toxicity (2).
The arenaviral genome is composed of two segments: the L (large) segment encodes the Z matrix protein and the L polymerase protein, while the S (small) segment encodes the glycoprotein (GPC) and nucleoprotein (NP). The smallest arenaviral protein of 90–99 amino acids in size, depending on the virus species, is the zinc-finger protein (Z) that belongs to the RING finger protein family (3). Like other known RING finger proteins (4), the arenavirus Z protein has been shown to interact with many cellular proteins, including but not necessarily limited to the promyelocytic leukemia protein (PML), eukaryotic elongation factor 4E (eIF4E) or components of the endosomal sorting complexes required for transport (ESCRT) (4, 5). In addition to interacting with cellular proteins, Z also interacts with the arenaviral L protein to latch the protein onto the genome in order to ensure that the L polymerase is incorporated into virion particles (6). The interaction between Z and the L polymerase of the Tacaribe virus (TCRV) appears to be essential for mediating viral transcriptional repression activity (7). Z appears to lock the L polymerase onto the viral promoter in a catalytically inactive state, thus limiting viral replication (6, 8, 9).
Z also serves as a main component required for viral budding by oligomerizing and forming the viral matrix of progeny virus particles (4). The Z homo-oligomerization process is likely important for its function in virion budding (10). A crystal structure of a dodecamer form of the LASV Z protein has recently been solved (11). It shows a ring-like structure of Z with highly basic residues, including a Lys-Trp-Lys triad in the center, as important for Z oligomerization. It is important to note that arenaviral NP protein appears to be required for efficient Z-mediated budding activity of the Tacaribe virus (TCRV) (12) and Pichinde virus (PICV) (13) possibly through specific NP-Z interactions (14, 15).
Arenaviral Z protein has also recently been shown to inhibit the type I interferon-induction pathway by directly binding to the N-terminal CARD domain of the intracellular pathogen-sensor proteins RIG-I and MDA5 (16). Z proteins from all known pathogenic arenaviruses [e.g., LASV, LUJV, JUNV, Machupo (MACV), Sabia (SABV), Chapare (CHPV), Guanarito (GTOV), and Dandenong (DANV)] as well as the relatively low pathogenic lymphocytic choriomeningitis virus (LCMV) are able to bind to RIG-I as well as MDA5 and thus, suppress the production of interferon-beta (IFNβ). In contrast, Z proteins of many known non-pathogenic arenaviruses do not bind to RIG-I or MDA5 and fail to inhibit the IFN-induction pathway (16). The fact that this small Z protein plays multiple roles in arenaviral replication and in modulating host immune responses to viral infection highlights the importance of this viral protein as a potential target for antiviral drug development. This chapter describes several assays used to examine the important roles of the arenaviral Z protein in mediating virus budding (i.e., either Z-self budding or NP-Z budding activities), viral transcriptional inhibition in a viral minigenome (MG) assay, and type I IFN suppression in an IFN-β-promoter-mediated luciferase reporter assay.
2. Materials
Cells: Human kidney epithelial 293T cells were grown in DMEM supplemented with 10% FBS and 50 μg of penicillin-streptomycin/ml.
Plasmids: For PICV Z protein and NP expression constructs, genes was amplified from the respective plasmid of the full-length PICV L or S segment using primers containing hemagglutinin (HA) or Myc tag at the C terminus, and cloned into the pCAGGS expression vector. IFN-β promoter-directed LUC plasmid was used for Z induced interferon suppression assay. MGLF7–5Rluc3 plasmid was used for Mini-genome assay. pSV-β-Galactosidase Control Vector was purchased from promega. Human RIG-I N terminal domain (RIG-iN) and LASV Z gene (with HA tag) were cloned into the pCAGGS vector as previously described (16).
Assay kits for Beta-galactosidase, Firefly luciferase, and Renilla luciferase.
Promega cell culture lysis 5X reagent
Lipofectamine 2000.
PCR thermal cycler.
Microcentrifuge.
Ultracentrifuge.
Waterbath.
Enhanced chemiluminescence (ECL) kit to detect bands in Western blots.
ECL detection device (Thermo).
Polyethylene glycol (PEG).
Plate reader.
2X HBS: 50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.10.
2.5 M calcium chloride stock solution.
RIPA buffer: 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl.
3. Methods
3.1. Z-self budding assay
293T cells were seeded in 10 cm tissue culture dish at 3×106/dish with complete DMEM.
On the morning of the day of transfection, replace cell-culture media with an equal aliquot of fresh complete DMEM.
-
Prepare plasmids and calcium chloride mix fresh as follows:
10 μl pCAGGS-PICV-Z-HA plasmid (1 μg/μl)
50 μl 2.5 M calcium chloride
440 μl water
Add 500 μl 2X HBS to above mixture while vortexing at the lowest speed setting for 10 s. The mixture was added dropwise using a pipetman into dish. Media were replaced with same amount of complete DMEM 6 h post-transfection.
48 h post-transfection, supernatants were collected and centrifuged at 15,000 × g for 10 min at 4°C. The budded VLP in the cleared supernatants were precipitated with 8% PEG 8000, 0.5 M sodium chloride, collected after centrifugation at 15,000 × g for 15 min, and lysed in RIPA buffer.
Cell lysates and the budded VLP were analyzed on a 15% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and Z proteins were detected by Western blotting with the anti-HA antibody.
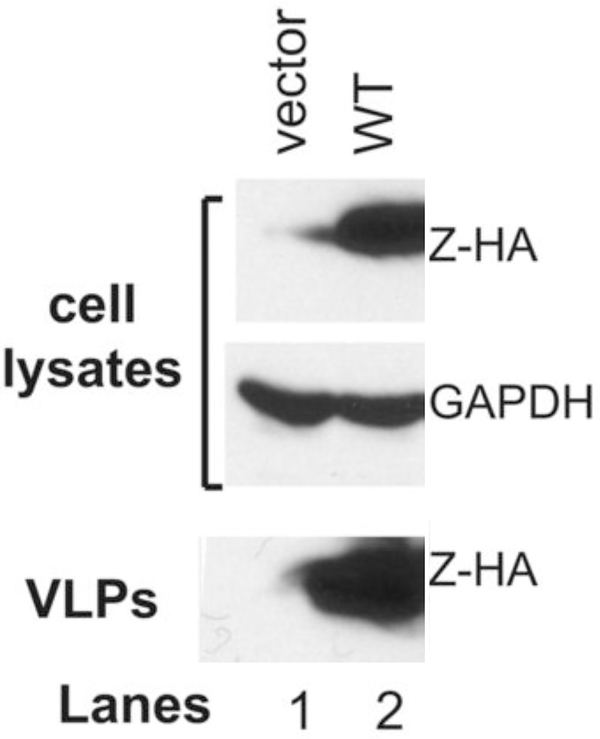
Figure 1 shows results of the wildtype PICV Z-self budding as VLP.
Figure 1. Analysis of the self-budding activity of the wildtype PICV Z protein.
293T cell were transfected with an empty vector (lane 1) or the WT PICV Z expression vector (lane 2). Expressions of HA-tagged Z proteins (Z-HA) along with GAPDH as loading control in the cell lysates were analyzed by Western blot. Virus supernatants (VLP) were collected, precipitated by PEG-8000, separated on 15% SDS-PAGE, and analyzed by Western blot.
3.2. NP-Z budding assay
293T cells were seeded in a 10 cm tissue culture dish at 3×106/dish with complete DMEM.
On the morning of the day of transfection, replace culture media with an equal aliquot of fresh complete DMEM.
-
Prepare plasmids and calcium chloride mix fresh as follows:
10 μl pCAGGS-PICV-NP plasmid (1 μg/μl)
10 μl pCAGGS-PICV-Z-HA plasmid (1 μg/μl)
50 μl 2.5 M calcium chloride
440 μl water
Add 500 μl 2X HBS into above mixture by vortexing using the lowest speed setting for 10 s. The mixture was added dropwise using a pipetman into dish. Media were replaced with same amount of complete DMEM 6 h post transfection.
48 h post-transfection, supernatants were collected and centrifuged at 15,000 × g for 10 min at 4°C. The budded VLP in the cleared supernatants were precipitated with 8% PEG-8000, 0.5 M sodium chloride, collected after centrifugation at 15,000 × g for 15 min, and lysed by RIPA buffer. Cell lysates and the budded VLP were analyzed on a 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel and detected for Z and NP proteins by Western blotting with the anti-HA antibody and rabbit anti-PICV NP sera, respectively (see Note 1).
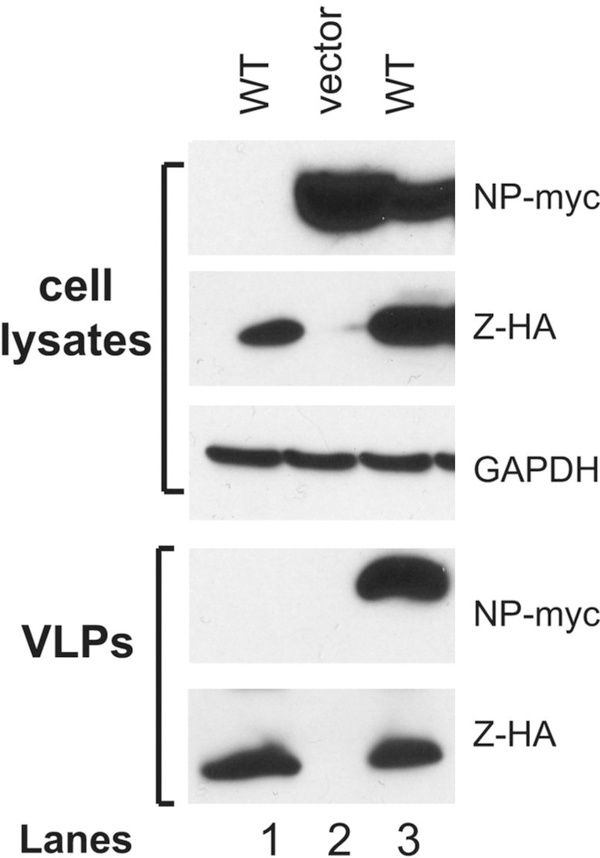
Figure 2 shows results of NP incorporation into the Z-mediated VLP.
Figure 2. NP incorporation into Z-mediated VLP formations.
293T cells were transfected with WT PICV Z protein expression vector (lanes 1 and 3), empty vector (lane 2), and with the NP expression vector (lanes 2 and 3). The amounts of VLP released into the supernatants were analyzed by Western blot against Z and NP proteins on 15% SDS-PAGE along with GAPDH as loading control in the cell lysates.
3.3. IFN β promoter-driven luciferase reporter assay
293T cells were seeded into 12 well-plate at 3×105/well with complete DMEM.
In the morning of the day of transfection, culture media were replaced by an equal aliquot of fresh media.
-
Prepare DNA and calcium phosphate mix as follow:
1 μl pSV-β-Galactosidase plasmid (50 ng/μl)
1 μl IFNβ promoter reporter plasmid (100 ng/μl)
1 μl pCAGGS-PICV-Z-HA or pCAGGS-LASV-Z-HA plasmid (1 μg/μl)
1 μl pCAGGS-RIG-iN plasmid (50 ng/μl)
5 μl 2.5M calcium chloride
47 μl water
To prepare triplicates for each test condition, mix as follows:
3 μl pSV-β-Galactosidase plasmid (50 ng/μl)
3 μl IFN β promoter reporter plasmid (100 ng/μl)
3 μl pCAGGS-PICV-Z-HA plasmid or pCAGGS-LASV-Z-HA plasmid (1 μg/μl)
15 μl 2.5M calcium chloride
141 μl water
Into each tube of DNA/calcium phosphate mixture, add 75 μl 2xHBS, vortex using very low speed to mix, and add the mixture to 3 wells of cells (98 μl per well)
24 h post transfection, the supernatants were aspirated and cells were lysed by 100 μl of Luciferase Cell Culture Lysis 1X Reagent. 20 μl of cell lysate was used for Firefly luciferase assay and another 50 μl of cell lysate was used for β-gal assay according to manufacturer’s instructions.
The Firefly luciferase activity is normalized by dividing the Firefly luciferase reading by the β-gal reading. The IFNβ promoter activity is represented by the folds of the normalized Firefly luciferase activity of the test to that the control (see Note 2).
Figure 3 shows results of the LASV Z-mediated suppression of the IFNβ promoter activity.
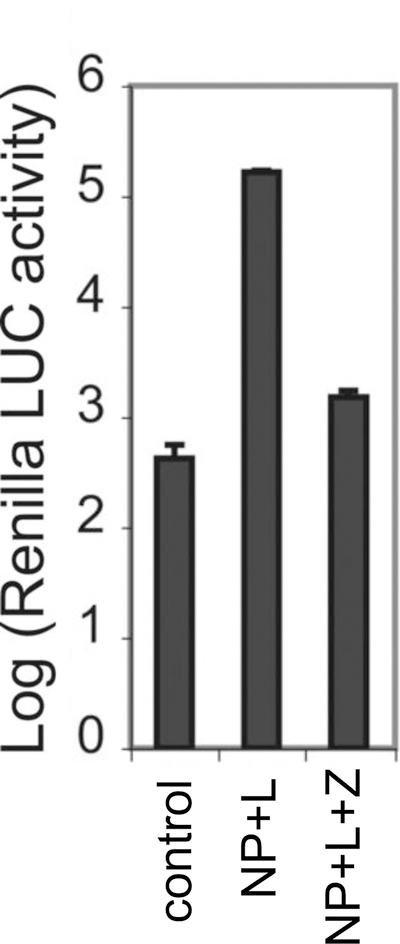
Figure 3. Z-mediated inhibition of viral RNA synthesis.
PICV Z protein was examined for its ability to inhibit viral RNA transcription in the mini-genome assay. Control reaction contains the minigenomic RNA and the NP-expressing plasmid only, whereas other reactions also contain other protein-expressing plasmids (i.e., either L-expressing plasmid or L- and Z-expressing plasmids). Results shown are the average of at least three independent experiments with error bars representing standard deviations.
3.4. Z-mediated viral mini-genome (MG) replication and transcription assay
3.4.1. In vitro RNA transcription
The plasmid MGLF7–5Rluc3 was linearized by digestion with Nhe I restriction enzyme.
DNA was extracted by standard phenol/chloroform and isopropanol precipitation.
Transcription reaction was assembled at room temperature according to Ambion MEGAscript® Kit manual. Calculate the volume of nuclease-free water to bring the total volume to 20 μL, and to this amount add 2 μL ATP, 2 μL CTP, 2 μL GTP, 2 μL UTP, 2 μL 10X reaction buffer, 1 μg linearized template DNA, and 2 μL enzyme mix.
Sample was incubated at 37°C for 4 h.
115 μL nuclease-free water and 15 μL ammonium acetate stop solution were added.
RNA was extracted by phenol/chloroform and isopropanol precipitated using the standard technique.
Carefully discard the precipitating solution and the RNA was resuspended in 80μl of DEPC-treated water.
RNA was quantified and adjusted to a concentration of 1 μg/μl (see Note 3)
3.4.2. Plasmid transfection:
293T cells were seeded into 24-well plates at 1×105 cells/well that contains 0.5 ml of DMEM with 10% FBS and 50 μg/ml of penicillin and streptomycin.
On the morning of the day of transfection, a fresh aliquot of the cell media was added to the 293T cell culture.
-
DNA and calcium phosphate mixture was prepared at room temperature as follows:
Each of the 25-μl of DNA mix (for one 24-well transfection) contains:
1 μl Beta-gal reporter plasmid (50 ng/μl)
1 μlpCAGGS-PICV-Z-HA plasmid (1 μg/μl)
1 μl pCAGGS-NP plasmid (250 ng/μl)
1 μl pCAGGS-L plasmid (500ng/μl)
2.5 μl 2.5 M calcium chloride
18.5 μl water
To prepare triplicates for each test condition, make a mixture as follows:
3 μl Beta-gal reporter plasmid (50ng/μl)
3 μl pCAGGS-PICV-Z-HA plasmid (1μg/μl)
3 μl pCAGGS-NP plasmid (250ng/μl)
3 μl pCAGGS-L plasmid (500ng/μl)
7.5 μl 2.5 M calcium chloride
55.5 μl water
Into each tube of DNA/calcium phosphate mixture (25 × 3 = 75 μl), add 75 μl of 2xHBS. Sample was gently mixed by vortexing using the lowest speed setting. The mixture was added dropwise using a pipetman into each well of cells (48μl of sample into each well of the 24-well plate).
Cells were incubated in a 37°C CO2 incubator for 24 hours.
3.4.3. RNA Transfection at 24 h after plasmids transfection:
4 h before transfection, an aliquot of fresh antibiotics-free DMEM was used to replace the overnight cell culture media.
For each well, 1 μg of in vitro-transcribed RNA (see Note 3) was diluted into 25 μl OptiMEM® I Medium.
For each well, 1 μl of Lipofectamine™ 2000 was diluted into 25 μl OptiMEM® I Medium and incubated for 5 min at room temperature.
The diluted RNA was added to diluted Lipofectamine™ and incubated at room temperature for no more than 10 min.
RNA-Lipofectamine™ 2000 complexes were directly added to each well containing cells and mixed gently by rocking the plate back and forth. The cells were incubated at 37°C in a CO2 incubator.
The cell medium was replaced by fresh DMEM medium (10% FBS) at 6 h post transfection.
24 h post RNA transfection, cells were washed once by PBS. Then cells were lysed by lysis buffer for Renilla luciferase assay.
3.4.4. Renilla luciferase assay:
20 μl of cell lysate was used for Renilla luciferase assay and another 50 μl of cell lysate was used for β-gal assay according to manufacturer’s instructions.
Renilla luciferase activity is normalized by dividing the Renilla luciferase reading by the β-gal reading. The minigenome replication activity is represented by the folds of the normalized Renilla luciferase activity of the test to that of the control.
Figure 4 shows results of Z-mediated viral mini-genome inhibition.
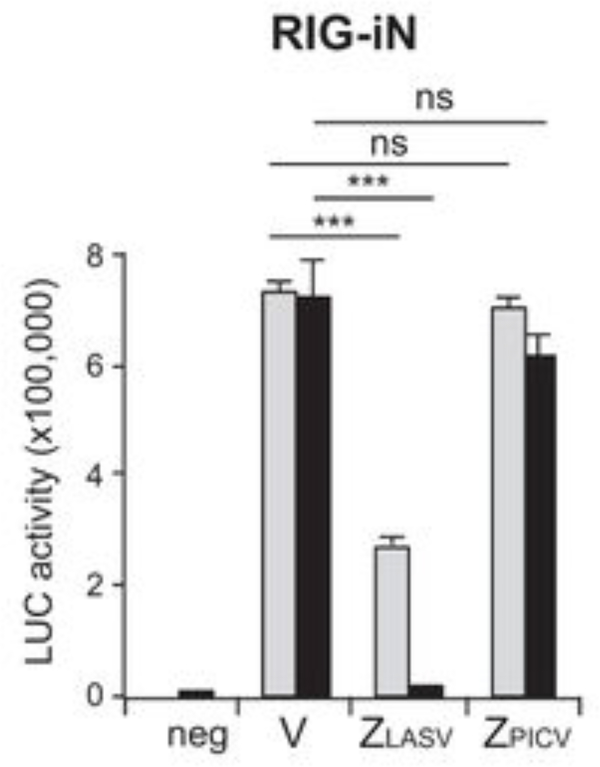
Figure 4. Z-mediated suppression of the IFNβ promoter-driven luciferase reporter activity.
LASV Z, but not PICV Z, inhibits RIG-iN-induced IFN-β activation in a luciferase (LUC)-based promoter assay. 293T cells were transfected with two different concentrations [100 ng (gray bars) and 1,000 ng black bars)] of either an empty vector (V) or Z plasmids (LASV or PICV) together with IFN-β–LUC, β-Gal, and RIG-iN plasmids. neg, negative control (no RIG-iN transfection).
Acknowledgements
This work was supported in part by the NIAID/NIH through the new-direction awards mechanism of the SERCEB grant (U54-AI057157) to YL and HL, by the NIAID/NIH R01 AI083409 to YL, and R01 AI093580 and R56 AI091805 to HL.
Footnotes
Western blot analyses were conducted using rabbit sera raised against specific short peptides within the NP protein as previously described (17). Peptide sequences and limited aliquots of the rabbit sera are available upon request.
When performing the IFNβ promoter assay, the amount of RIG-iN and Z plasmid is critical. Low Z and RIG-iN protein expression will lead to unreliable IFNβ readings.
The integrity of the RNA needs to be validated by agarose gel electrophoresis prior to use in the transfection reaction.
References
- 1.Maiztegui JI, McKee KT Jr., Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, Enria DA, Briggiler AM, Levis SC, Ambrosio AM, Halsey NA, Peters CJ. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis 177:277–283. [DOI] [PubMed] [Google Scholar]
- 2.Gunther S, Lenz O. 2004. Lassa virus. Crit Rev Clin Lab Sci 41:339–390. [DOI] [PubMed] [Google Scholar]
- 3.Salvato MS, Shimomaye EM. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1–10. [DOI] [PubMed] [Google Scholar]
- 4.Fehling SK, Lennartz F, Strecker T. 2012. Multifunctional nature of the arenavirus RING finger protein Z. Viruses 4:2973–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwyer EJC, Lai HK, MacDonald RC, Salvato MS, Borden KLB. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol 74:3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kranzusch PJ, Whelan SP. 2011. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc Natl Acad Sci U S A 108:19743–19748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilda M, Lopez N, Casabona JC, Franze-Fernandez MT. 2008. Mapping of the tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J Virol 82:11454–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornu TI, de la Torre JC. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol 75:9415–9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez N, Jacamo R, Franze-Fernandez MT. 2001. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol 75:12241–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kentsis A, Gordon RE, Borden KL. 2002. Self-assembly properties of a model RING domain. Proc Natl Acad Sci U S A 99:667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie KM, Zandonatti M, Liu T, Li S, Woods VL Jr., Saphire EO. 2016. Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z. J Virol 90:4556–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groseth A, Wolff S, Strecker T, Hoenen T, Becker S. 2010. Efficient budding of the tacaribe virus matrix protein z requires the nucleoprotein. J Virol 84:3603–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Danzy S, Kumar N, Ly H, Liang Y. 2012. Biological roles and functional mechanisms of arenavirus Z protein in viral replication. J Virol 86:9794–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capul AA, de la Torre JC, Buchmeier MJ. 2011. Conserved residues in Lassa fever virus Z protein modulate viral infectivity at the level of the ribonucleoprotein. J Virol 85:3172–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J Virol 83:7029–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing J, Ly H, Liang Y. 2015. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-I-like receptor-dependent interferon production. J Virol 89:2944–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Lan S, Ly H. 2009. Molecular determinants of Pichinde virus infection of guinea pigs--a small animal model system for arenaviral hemorrhagic fevers. Ann N Y Acad Sci 1171 Suppl 1:E65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]