Abstract
Top-down proteomics (TDP) by mass spectrometry (MS) is a technique by which intact proteins are analyzed. It has become increasingly popular in translational research because of the value of characterizing distinct proteoforms of intact proteins. Compared to bottom-up proteomics (BUP) strategies, which measure digested peptide mixtures, TDP provides highly specific molecular information that avoids the bioinformatic challenge of protein inference. However, the technique has been difficult to implement widely because of inherent limitations of existing sample preparation methods and instrumentation. Recent improvements in proteoform pre-fractionation and the availability of high-resolution benchtop mass spectrometers have made it possible to use high-throughput TDP for the analysis of complex clinical samples. Here, we provide a comprehensive protocol for analysis of a common sample type in translational research: human peripheral blood mononuclear cells (PBMCs). The pipeline comprises multiple workflows that can be treated as modular by the reader and used for various applications. First, sample collection and cell preservation are described for two clinical biorepository storage schemes. Cell lysis and proteoform pre-fractionation by gel-eluted liquid fractionation entrapment electrophoresis are then described. Importantly, instrument setup and liquid chromatography–tandem MS are described for TDP analyses, which rely on high-resolution Fourier-transform MS. Finally, data processing and analysis are described using two different, application-dependent software tools: ProSight Lite for targeted analyses of one or a few proteoforms and TDPortal for high-throughput TDP in discovery mode. For a single sample, the minimum completion time of the entire experiment is 72 h.
Introduction
MS serves to identify and characterize proteins within a variety of cellular and disease contexts. Indeed, MS-based proteomics is popular among researchers wishing to study complex proteomes across diverse phenotypes, as current MS instrumentation enables the complicated measurements required to identify peptides or intact proteins within biological or clinical samples1. Furthermore, the relatively strong mechanistic connection of proteins to health and disease has inspired the application of MS-based proteomics to translational research questions, as proteins are expected to yield specific diagnostic biomarkers and therapeutic targets for a wide swath of human diseases. Many translational proteomics initiatives have adopted human blood as a sample type of interest because of the relative ease and non-invasiveness of its acquisition, as well as the proximal location and functional importance of the bloodstream to many organ systems. Human blood contains a host of proteins secreted by many different tissues and cell types, in addition to various hematopoietic cell populations (such as PBMCs) with their respective proteomes that are crucial to processes including oxygen transport, wound healing, and adaptive and innate immunity. A large body of research details the proteomic analysis of blood and its cellular components across various applications2,3.
A notable shortcoming of many proteomic studies, however, is that they require the enzymatic digestion of proteins into peptides, a methodology commonly referred to as BUP4. BUP is technically accessible and widely used for a variety of reasons. However, it has an associated protein inference problem stemming from the fact that identical peptide sequences can be shared among different proteins or protein isoforms, leading to a high degree of ambiguity during data interpretation5. The inference problem renders quite difficult the characterization of myriad post-translational modifications (PTMs), single-nucleotide polymorphisms (SNPs), and mutations, which can have high functional relevance in disease networks6. TDP involves the study of intact, rather than proteolytically digested, proteins and is capable of direct detection and examination of proteoforms, which are the highly specific, modified protein variants that naturally occur and functionally operate in cells7. Top-down measurements can be targeted to the analysis of just a few purified proteins in a sample, or expanded using high-throughput proteomics techniques to characterize hundreds or even thousands of proteoforms in a sample proteome, depending on the sample type and study design. In human samples, we often confidently characterize one to ten proteoforms for each base sequence (unmodified amino acid sequence associated with the parent protein or isoform accession number) identified. This variation is most often due to the presence of PTMs such as acetylation or phosphorylation in varying stoichiometries in the sample, or the presence of proteolytically processed variants demonstrating removal of signal and transit peptide sequences. The protocol presented here is one of the first to describe the application of TDP to human PBMC analysis (Fig. 1) with a high level of technical detail such that proteoform-resolved translational proteomics workflows can be widely disseminated. Although high-throughput analyses by TDP are less widespread than BUP for a multitude of reasons, namely, lack of access to expensive instrumentation and the difficulty of data analysis for more complex intact protein spectra8, recent advances in intact protein separations, instrumentation, and high-throughput data analysis methods have allowed TDP to be established as a viable technique using off-the-shelf tools to study the proteoform content of proteomes9. In addition, the semantics and nomenclature necessary for implementing TDP across a wider foundation of users have been developed to not only describe proteoforms and evaluate their quality of characterization, but also to establish a proteoform repository (http://atlas.topdownproteomics.org/) and a standard nomenclature, ProForma, by which individual characterized proteoforms can be formally annotated10. The importance of this advance cannot be understated: before the formalization of the proteoform as a specific molecular entity, there were various nonstandard and redundant strategies for describing proteoforms, which greatly complicated their annotation and collection into repositories for the benefit of the broader research community.
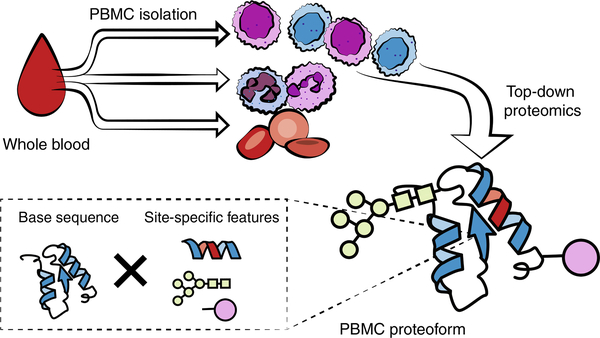
Fig. 1 |. Schematic depicting characterization of peripheral blood mononuclear cell proteoforms from whole blood by top-down proteomics.
The protocol described here has been optimized for the performance of translational top-down proteomics from a single ~8-mL draw of human blood. Whole blood is subsequently fractionated into its component parts by density-gradient centrifugation, and the PBMCs, which are highly relevant to immune-related pathologies, are isolated and prepared for analysis. The key value proposition of top-down proteomics is that it analyzes proteoforms, rather than peptides yielded by tryptic or other enzymatic digestion. This allows the analyst to characterize the specific post-translational modifications and other chemical and genetic sequence variants in a sample of whole proteome that would be lost by bottom-up proteomics, but that may have high mechanistic relevance to the phenotypes of interest. As depicted in the figure inset marked by the dashed box, a single gene product often gives rise to multiple proteoforms after post-translational modification or because of the existence of sequence variants derived from proteolytic processing, multiple isoforms, or even single-nucleotide polymorphisms.
Initial high-throughput TDP studies evaluated the relatively simple proteomes of bacteria11,12, archaea13, or yeast14–17; these studies resulted in the development of best practices for pre-analytical separations, instrumentation requirements, and proteoform characterization. For example, solution-phase strategies for separation of intact proteoforms, such as gel-eluted liquid fraction entrapment electrophoresis (GELFrEE)18,19, helped mitigate some issues of variability and intact protein recovery presented by slab gel separations coupled to proteoform-resolved analyses. Furthermore, the Orbitrap mass analyzer20,21, and particularly its implementation in hybrid ion trap (IT)-Orbitrap instruments22–25, became a robust option for high-throughput TDP studies of the more complex human proteome26. Concurrently, numerous efforts to advance database searching of high-throughput TDP data with software tools have been developed for proteoform characterization and visualization27–34. Recent efforts in translational TDP have applied the aforementioned technological improvements to the proteomic characterization of human tissues35–38, cerebrospinal fluid39, saliva40,41, and plasma and pleural effusions42. However, these studies employ divergent methods for patient sample preparation, separation of denatured proteins, proteoform ionization, characterization of putative proteoform biomarkers, and dataset storage. Our group has been reducing our innovations to better practice and therefore has set out to establish a robust and reproducible pipeline for translational TDP from human clinical samples, from sample collection to data analysis (Fig. 2). Several of our simpler protocols are provided on our website and are designed for ready use or adaptation by others. We elected to focus on human PBMCs for this pipeline, as they represent a sample type highly relevant to a multitude of biomedical applications ranging from cancer to autoimmunity.
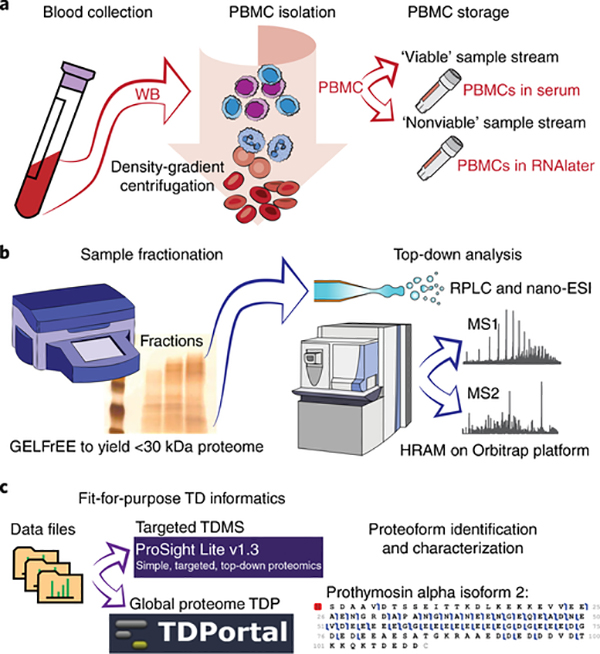
Fig. 2 |. Experimental workflow for top-down proteomic analysis of peripheral blood mononuclear cell proteoforms using either targeted or discovery informatics solutions.
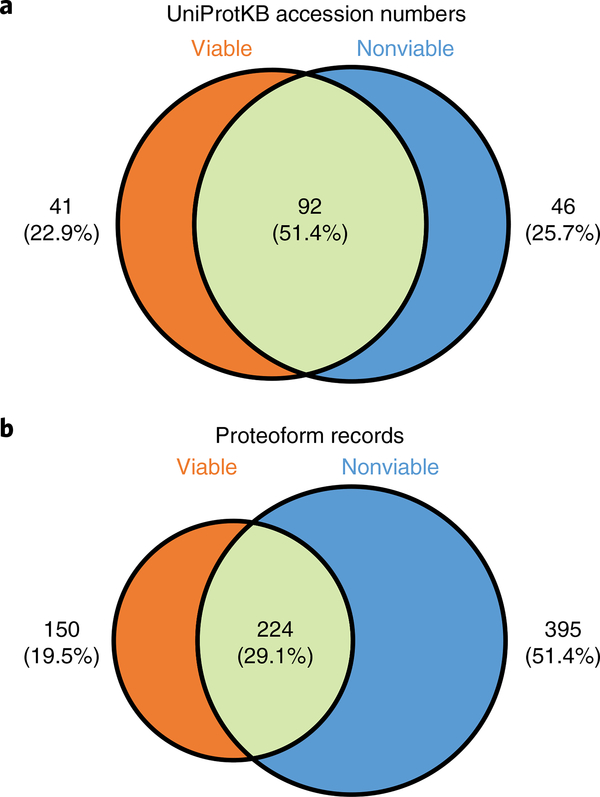
a, After blood collection from a patient, whole blood is separated by density-gradient centrifugation in the CPT tube, and the PBMCs are isolated and stored by two alternative cell processing schema: ‘viable’ cells are frozen in serum, and ‘nonviable’ cells are emulsified in RNA-stabilizing reagents. b, Lysed samples are then fractionated by MW in the liquid phase and submitted to top-down analysis by mass spectrometry after desalting, reversed-phase LC, and ESI. c, Finally, we present two methods for data analysis to identify and characterize proteoforms: targeted top-down mass spectrometry using ProSight Lite, and discovery-mode top-down proteomics using the TDPortal interface. GELFrEE, gel-elution liquid fraction entrapment electrophoresis; ESI, electrospray ionization; HRAM, high-resolution accurate mass measurement; WB, whole blood.
In the following protocol, we describe some best practices for preparing PBMCs collected by two common methodologies used by clinical biorepositories: one for collecting ‘viable’ cells isolated from blood and stored in serum-containing freeze medium before cryogenic preservation, and a second for collecting ‘nonviable’ cells emulsified in RNA-stabilizing reagents (e.g., RNAlater). We have developed TDP workflows adaptable to both types of clinical sample collection, as well as to storage of samples banked with diverse ‘-omics’ strategies in mind43. For both TDP workflows, these protocols have been demonstrated in prior studies from our lab to delineate proteoform-resolved molecular signatures of kidney and liver transplant rejection from biorepository-stored human PBMCs44,45. Here, we expand upon the methods published in these previous studies to describe in high detail alternative clinical sample storage strategies, robust GELFrEE fractionation, and high-resolution measurements, using LC–MS/MS for translational TDP as specifically applied to human PBMCs. Finally, we provide details for multiple data analysis strategies, including targeted analysis of known proteoforms using the freely accessible ProSight Lite software46,47 and high-throughput global proteome analysis in discovery mode using TDPortal (http://nrtdp.northwestern.edu/resource-software/), a high-performance computing environment for TDP data processing available through the National Resource for Translational and Developmental Proteomics (NRTDP). TDPortal harbors the associated Top Down Viewer visualization tool and incorporates the top-down-specific characterization score (C score), a Bayesian metric designed to score the degree of quality achieved in any given proteoform characterization48.
Experimental design
The Procedure below entails human PBMCs as the sample type, GELFrEE as the pre-fractionation method, an LTQ Velos Orbitrap Elite as the mass spectrometer, and ProSight Lite or TDPortal as the data analysis software. Fit-for-purpose variations to the Procedure can be incorporated to accommodate other sample types, instrumentation, and software solutions—for example, one can simply skip cell lysis and move on to fractionation when working with serum or plasma samples. With these latter samples, we have found that the addition of a depletion step to remove highly abundant proteins before fractionation increases the depth of analysis. Other sample types for which we have modified this protocol include human and mouse cell lines and tissues; in some of these cases, adapted homogenization or disruption strategies were used during sample preparation. It is also important to note that the instrumentation and software described here are not the sole solutions for top-down measurements, although options are much more limited in this field than for BUP. Although beyond the scope of this specific protocol, ref. 9 thoroughly reviews the history and continued development of divergent strategies and general instrumentation for intact proteoform fractionation and analysis.
As always, we recommend incorporating the proper controls and degree of replication into any discovery proteomics study design; these will depend on the application at hand. For example, we often include ‘normal’ or ‘healthy’ patients in our translational study designs to compare to ‘abnormal’ patients, or patients with a disease of interest. Importantly, the primary studies from which this protocol was adapted incorporated replication at multiple levels throughout the complicated TDP process as a strategy to assign variation in proteoform signals to discrete levels in the study, and thus control for possible variation introduced at the bench or that is due to natural differences among patients of the relevant population. To begin, we will underscore the importance of quality control (QC) for complex TDP experiments with biological and technical replicates by describing the top-down standard. Then we will address other key components of the Procedure in the order of their practice.
The top-down standard for LC–MS quality control
Before the analysis of clinical or biorepository samples by TDP, it is essential to establish that the parameters selected for LC and tandem MS (LC–MS/MS) are suitable, and that the components required for both are working optimally. The chosen method for QC of TDP at the NRTDP involves the LC–MS/MS analysis of a mixture of purified standard proteins described below and collectively referred to as the ‘top-down standard’ (TD standard). A commercial mixture of intact proteins, the Pierce Intact Protein Standard Mix (cat. no. A33526) is also available from Thermo Fisher Scientific. The TD standard includes bovine carbonic anhydrase (UniProt ID P00921), equine myoglobin (UniProt ID P68082), bovine pancreatic trypsinogen (UniProt ID Q547S4), and bovine ubiquitin (UniProt ID P0CG53). The TD standard is prepared in bulk and stored frozen in individual aliquots, so that reproducible analyses of a single batch can be conducted over the course of 6 months or more. An individual injection of TD standard should contain ~0.6 pmol carbonic anhydrase (29 kDa), 1 pmol myoglobin (17 kDa), 0.5 pmol trypsinogen (26 kDa), and 0.15 pmol ubiquitin (9 kDa). A fifth protein, bovine superoxide dismutase (UniProt ID P00442, 16 kDa), may also be present in variable concentrations as a contaminant of the commercial preparation of carbonic anhydrase.
Upon injection of the TD standard for LC–MS/MS, the quality of LC performance is evaluated by the order of elution, baseline chromatographic resolution, peak width at half height, peak symmetry, peak tailing, and retention time drift over multiple technical replicates exhibited by the five individual protein peaks within the resulting chromatogram. The quality of instrument performance is subsequently evaluated by a series of metrics, including the intensity ratio between intact mass spectra of each protein acquired in the IT and the Orbitrap (FT), the appearance of the charge state distributions contained within each intact mass spectrum, the degree of fragment ion coverage obtained by a single MS2 scan after higher-energy collisional dissociation (HCD), and the resulting probability-based confidence scores (namely, the p score and E value) for identification of each protein after analysis with ProSight Lite47 or a TD standard–specific TDPortal tool available by request through the NRTDP (http://nrtdp.northwestern.edu/resource-software). A dedicated NRTDP TDPortal tool for analysis of the Pierce Intact Protein Standard is also available by request. Additional metrics, including the MS1 injection times (IT and FT), peak full width at half-maximum, and pressure within the Orbitrap during LC–MS/MS analysis can also be incorporated, particularly for longitudinal QC tracking of instrument performance. An example analysis of the TD standard on an LTQ Velos Orbitrap Elite is depicted in Fig. 3.
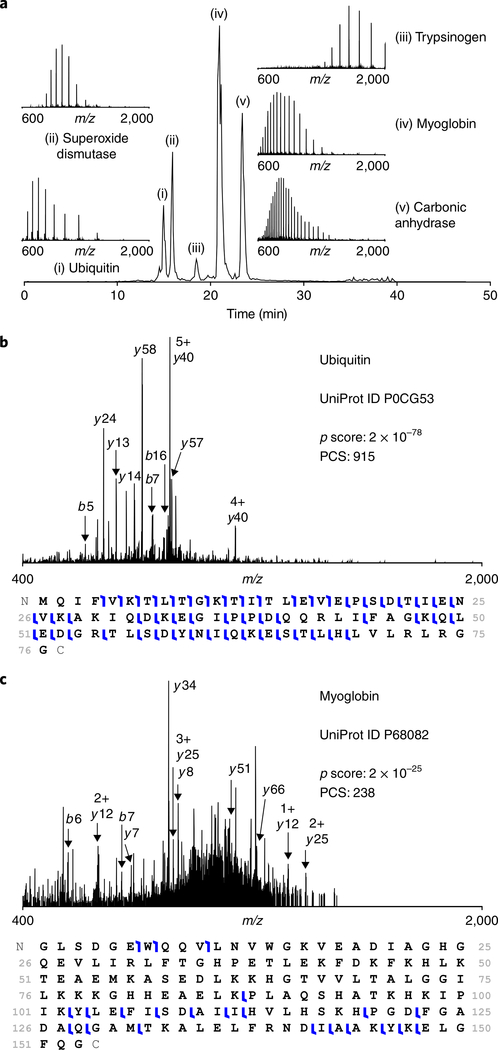
Fig. 3 |. An example of using the top-down standard for quality control of top-down proteomics experiments.
a, The top-down standard is composed of four proteins and one common contaminant that elute in the following order: (i) ubiquitin; (ii) superoxide dismutase; (iii) trypsinogen; (iv) myoglobin; and (v) carbonic anhydrase. Labeled inset spectra correspond by number to the chromatographic elution peak from which they were obtained. b, An example fragmentation spectrum of ubiquitin yielded by higher-energy collisional dissociation, with corresponding scoring metrics and graphical fragment map. c, An example fragmentation spectrum of myoglobin yielded by higherenergy collisional dissociation, with corresponding scoring metrics and graphical fragment map. PCS, protein characterization score.
The NRTDP recommends a minimum of three replicate injections of TD standard to evaluate LC and MS performance before the analysis of clinical or biological samples, at least one injection of TD standard for every 24 h of total experiment time, and a further three replicate injections of TD standard at the end of the experiment period. If data quality and reproducibility are of primary importance, as in the case of label-free top-down quantitation of proteins within clinical samples, the recommended practice is to include an additional TD standard injection between blocks of randomized technical replicate injections and to perform daily analyses of each standard run. The increased frequency of these QC measures will enable on-the-fly detection of issues in LC or MS performance that might affect final data quality or reproducibility and reduce the loss of samples or resulting data due to issues emerging during the course of a multi-day or multi-week experiment.
Sample treatment from whole blood to denatured proteoforms
In our experience, PBMCs collected by researchers for -omics studies are stored in various ways that may affect downstream analysis by MS45. Therefore, we have included methods in the following protocol that branch based on common biorepository cell storage procedures, allowing for greater flexibility in applying TDP to translational studies. In our workflow, 5–8 mL of whole blood is first collected in tubes with built-in technology for cell separation by density-gradient centrifugation. PBMCs are then extracted and stored via two different mechanisms: ‘viable’ cell collection, which involves cryopreserving the PBMCs in medium; and ‘nonviable’ cell collection, which involves emulsifying the cells in RNA-stabilizing reagents. We have included methods for analyzing the latter sample type because we have often found that clinical researchers collecting cells for future -omics studies stored their samples with genomic and transcriptomic applications in mind43. Retrospectively, this was a sensible strategy, considering the proliferation of these techniques in translational research; however, we have found that common RNA-stabilizing reagents, such as RNAlater, interfere with conventional TDP sample preparation methods. Namely, the high salt content of these reagents is predisposed to lyse portions of cell pellets and also to complicate downstream cell lysis, protein quantitation, and protein desalting protocols common to TDP. We therefore describe how we have modified existing centrifugation-based strategies to integrate the use of these reagents for sample storage with BUP49 for translational work using TDP, while streamlining the process for minimal time, cost, and pre-analytical variation. We then detail our existing methods for the final step in the reduction of whole blood from a patient to denatured proteoforms, which involves cytolysis and denaturation of the proteome with detergents as a prerequisite to downstream pre-fractionation50,51. Finally, we describe how to quantify the content of lysates by the bicinchoninic acid (BCA) assay to control the amount of total protein input into GELFrEE by measuring absolute concentration.
Protein pre-fractionation by GELFrEE and nano-UHPLC
In proteolysis-based proteomic approaches, the molecular weight (MW) of peptides subjected to analysis is confined to a rather narrow mass bin (typically 0.6–3 kDa) because of proteolytic digestion at specific residues, such as lysine and/or arginine. Conversely, TDP omits proteolysis, and thus measures much larger proteins, which present analytical challenges in terms of detection and interpretation, especially when measured from complex mixtures. For this reason, successful analysis of complex mixtures at the proteoform level requires some degree of pre-fractionation. In our laboratory, we use a two-tiered fractionation approach. First, we perform offline protein separation in the first dimension by GELFrEE18,19. This methodology allows us to obtain MW-based protein fractions in the liquid phase that are binned according to the type of GELFrEE cartridge and its polyacrylamide concentration. For this protocol, we will describe the analysis of the first eluted fraction from an 8% (wt/vol) GELFrEE cartridge, as this fraction contains all proteins <30 kDa in the sample proteome of interest, which is a MW range that TDP can robustly detect and characterize. Next, collected GELFrEE fractions are cleaned of detergent and subjected to a second dimension of separation via hydrophobicity-based chromatography performed online.
In MS-based proteomics, pre-fractionation and analyte separation enhance dynamic range, minimize ion suppression during ESI, and increase the depth of proteome coverage. The most popular separations technology applied to complex protein mixtures in denaturing TDP is reversed-phase LC (RPLC). For proteomics analyses, high chromatographic resolution is fundamentally important, as it allows for minimization of the number of species, or intact proteoforms in the case of TDP, that are simultaneously directed into the mass spectrometer. This greatly reduces problems due to signal suppression and overlapping charge state envelopes, which spread across a wide m/z range for intact protein ions, and ultimately leads to the detection of more species with high specificity and selectivity. In the language of chromatography, increasing the number of theoretical plates (e.g., by increasing the column length) will result in increased resolution for improved analyte separation. In addition, this will lead to higher peak capacity, or the maximum number of resolvable peaks. In contrast to peptide separations, we have observed that intact proteins often exhibit broader elution peaks over the analytical gradient. From an MS point of view, slight peak broadening can be advantageous for the acquisition of multi-scan MS/MS spectra for polypeptides of >30 kDa; however, MS1-based label-free quantitation can be made cumbersome by the loss of chromatographic resolution for the same number of theoretical plates.
The choice of column is not only of great importance for qualitative TDP, but it is particularly so for quantitative TDP experiments, as robust quantitation depends upon the number of identifiable species per chromatographic elution profile. For the optimization of reproducible, high-throughput TDP, we have heavily tested two columns with the same functional group (cross-linked copolymeric stationary phase based on divinylbenzene and polystyrene) but created with different packing methodologies: either as macroporous polydivinylbenzene particles (PLRP-S) or prepared as a monolithic resin (RP4H). Generally, one of the most pronounced advantages of monolithic columns over their bead-packed counterparts is a faster mass transfer. This ultimately results in faster analyte convection flow and, therefore, higher-resolution separations. By contrast, particle-based columns suffer from peak broadening because of their slower mass transfer, which is largely controlled by diffusion. A secondary benefit of monolithic columns is their reduced backpressure as compared to those of most bead-based stationary phases. The difference in backpressure is particularly pronounced when small-diameter beads (e.g., <2 μm) are used to improve resolution. In our experience, monolithic column performance exceeded that of the porous bead resin when compared using a metric of proteome coverage (number of proteoforms identified per GELFrEE fraction), and was therefore chosen for most future TDP experiments. In addition to the previously mentioned factors, we attribute this difference, which is quantifiable as a ≥10% increase in unique identified UniProt accession numbers and proteoforms, to the difficulties in homogeneously packing long (≥50 cm) porous bead columns, as PLRP-S and monolithic columns show comparable performance, with relatively small differences for shorter lengths (e.g., 25 cm). Moreover, we tested column performance using the same identification metrics by either injecting samples directly onto the analytical column or first concentrating them on a trap column. We found an increased number of identifications and characterized proteoforms when using the latter setup (trap column in line with analytical column), and so elected to incorporate a trap column for future TDP experiments. Specifically, the use of a trap column increased the number of identified UniProt accession numbers per GELFrEE fraction by 30%, and unique proteoforms by 50% or more.
Finally, once the column has been chosen, the LC gradient must be optimized based on the type of analyte. Notably, the chromatographic gradient can be adjusted according to the chemical and physical properties of proteins of interest to obtain the best possible separation of the analytes. For instance, isolated nuclear proteins such as histones, often studied in epigenetic research, may require a more ‘shallow’ gradient (slow increase of mobile phase B over time) because of their inherent hydrophilicity. Conversely, when characterizing membrane proteins or large proteoforms (>30 kDa), a stepped gradient consisting of an initial rapid increase in the organic phase (up to ~20%), followed by a longer resolving gradient leading to a final organic phase concentration of 50–60% would be beneficial. For proteoforms <30 kDa, the optimal separation conditions that we have determined in our laboratory are illustrated in Table 1, whereas other technical details are summarized in Box 1 and Fig. 4 (our LC valve configuration for TDP using monolithic RP-4H columns).
Table 1 |.
LC parameters
| NC pump | ||
|---|---|---|
| Solvents | Mobile phase A: water/acetonitrile/formic acid (94.8:5:0.2 (vol/vol/vol)) | |
| Mobile phase B: water/acetonitrile/formic acid (94.8:5:0.2 (vol/vol/vol)) | ||
| Column(s) | Trap: PepSwift Monolithic, 200 μm i.d. × 5 mm | |
| Analytical: ProSwift RP-4H, 100 μm i.d. × 50 cm | ||
| Column oven temperature | 35 °C (lower limit:upper limit, 20:75 °C) | |
| Injection loop | 20 μL | |
| Injection mode | μlPickUp | |
| Transfer vial | Mobile phase A | |
| Injection volume | 6 μLa | |
| Flow | Multi-step gradient | |
| Flow rate | 1 μL/min | |
| Gradient conditions | ||
| Time (min) | % B | Comment |
| 0 | 5 | See Fig. 4: middle panel (inject; 6-port valve) |
| 3 | 5 | See Fig. 4: right panel (gradient; 10-port valve) |
| 64 | 50 | |
| 65 | 95 | |
| 68 | 95 | |
| 69 | 5 | |
| 83 | 5 | |
| Micro (loading) pump | ||
| Solvent | Mobile phase A: water/acetonitrile/formic acid (94.8:5:0.2 (vol/vol/vol)) | |
| Flow rate | 1 μL/min | |
| Flow | Isocratic (100% (vol/vol/vol) mobile phase A) | |
Refer to Box 1 for details on sample injection.
Box 1 |. LC setup and testing ● Timing 4–5 h.
For our LC separations, we use a Dionex UltiMate 3000 RPLC nano system (Thermo Fisher Scientific). This LC system is equipped with a nanopump (with a flow selector allowing for flow rates between 0.05 and 1 μL/min) and a micropump (operating at >1 μL/min). The former is used for protein separation, whereas in our setup, the latter is used to wash and concentrate proteins on a trap column right after injection and before analytical separation. We use a two-valve setup: a 10-port switch valve for the column oven compartment and a 6-port valve for the injection module. The two valves are synchronized to perform two main operations: loading (switch valve 1_10, 6-port ‘inject’) and delivering the analytical gradient (switch valve 1_2, 6-port ‘load’) (see Fig. 4 for specific LC valve plumbing setups). Before sample analysis, we find it critical to undertake the following steps to reduce bias introduced by temporal LC variance and improve system performance before the LC metrics evaluation via analysis of the TD standard.
Prepare fresh solvents (Reagent setup) and purge both pumps and the flow meter thoroughly (45 min for the blocks of the nanopump and the flow meter, and 15 min for the loading pump). We recommend changing (not topping off) solvents every 2–3 weeks; in doing so, we ensure that the acid content does not greatly reduce over time.
Perform the pressure transducer and viscosity tests.
Disconnect the loop from the autosampler while in ‘load’ position and rinse it well with organic solvent from the micropump. This will ensure that there are no residual samples present within the loop that could interfere with the injection of our sample and subsequent analysis. In addition, by disconnecting the loop from the valve, we can make sure that no contaminants or particles come into the valve, rotor seal, or ultimately the column during this process.
Prime the syringe and wash the needle and fluidics of the autosampler with mobile phase A.
-
Reconnect the system and condition the trap and analytical column. Column specifications can be found in Table 1 and the Equipment section. We perform conditioning with the trap column separate from the analytical column to ensure that all residual polymeric species elute to waste rather than onto the analytical column, where those species might be bound irreversibly in high-organic solvent, reducing binding capacity and therefore column longevity. The recommended parameters for column conditioning are as follows (with run times dependent on column length):
% B Trap column run time (min) Analytical column run time (min) 95 15 45 80 15 45 50 15 45 A minimum of three repetitions of this program is recommended for optimal conditioning before performing QC and sample analysis.
Connect the LC system to the mass spectrometer. We use a custom ESI source in which the voltage is delivered through a zero-dead-volume microtight high-voltage union (depicted in Fig. 4). Connect the analytical column to the high-voltage union with the minimum possible length of fused silica capillary (260 μm o.d., 30 μm i.d.) fitted with a zero-dead-volume union at the column outlet end, so as to minimize total potential dead volume. Finally, connect the third port of the high-voltage union to a packed PicoTip spray emitter (Equipment setup).
-
Perform final column conditioning and a complete saturation step once the setup is connected, by injecting three TD standards for QC before samples are to be run.
For proteoform separation from lysed human PBMCs, we inject via the microliter pickup method, using a 20-μL sample loop. We inject 6 μL (one third of the loop), which corresponds to ~1 μg of sample loaded on the column, and the rest of the loop is filled with buffer from the transfer vial (mobile phase A). Although full loop injection would in theory be the optimal method for injection, to obtain the highest degree of reproducibility between injections, more sample volume is required, even in a 1-μL loop injection, and therefore full loop injection is not sustainable for sample-limited studies. Variability across injections is compensated for by running multiple technical replicates from each biological replicate sample.
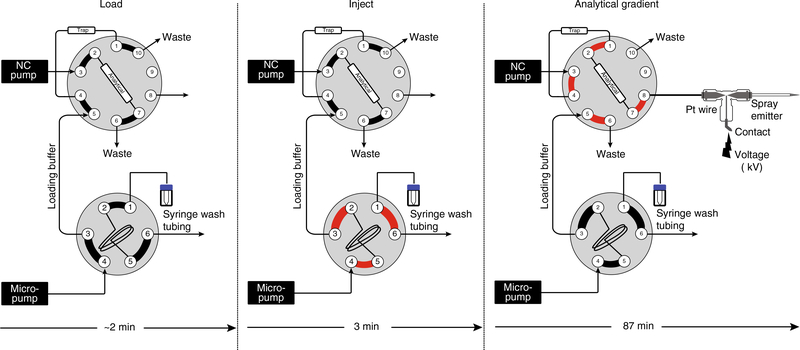
Fig. 4 |. LC valve setup for top-down proteomics used in this protocol.
Depiction of the positions for the 6-port and 10-port valve setup on a Dionex Ultimate 3000 nano-UHPLC system for loading sample from the vial (left panel), injecting sample onto the trap column (middle panel), and delivering the analytical gradient to elute the sample for mass spectrometry analysis (right panel). Note the switch in valve position in the 10-port valve from 10–1 during sample injection onto the trap to 1–2 for delivery of the analytical gradient from the NC pump. In addition, note that connected ports for each valve position are denoted by thick black or red lines, with the path of sample within the valve highlighted in red during the injection and analytical gradient. Here, ‘NC pump’ refers to the high-pressure gradient pump.
High-resolution Fourier-transform mass spectrometry on the Orbitrap platform
Our choice of instrumentation for TDP experiments is the hybrid linear IT (LTQ)-Orbitrap mass spectrometer, equipped with a compact high-field Orbitrap mass analyzer (i.e., D20 cell) and the enhanced signal processing (eFT) algorithm (Orbitrap Elite)52. This mass spectrometer combines the high resolution and mass accuracy afforded by Fourier-transform mass spectrometry (FTMS) required to isotopically resolve multiply charged intact protein cations, and an automatic gain control (AGC) function to accurately estimate the number of charges injected into the high-resolution mass analyzer. The AGC plays a pivotal role in quantitative applications of this workflow, as it ensures that the intensity values of the different detected species (expressed as normalized level (NL)) are not biased by the detection of different numbers of ions from scan to scan, or from one run to another. In our experience, the AGC calculated using the electron multipliers of the IT returned a linear response to different concentrations of selected standard proteins spiked into a complex background (i.e., a GELFrEE fraction from yeast lysate)17. The AGC is based on a short pre-scan in the LTQ that anticipates the survey scan in the Orbitrap, which implies that every MS1 event is based on the same number of charges, independent of the spray stability. However, we do not use the predictive AGC function for MS2 on the Orbitrap Elite when performing TDP experiments.
The ion activation technique we describe here for MS2 is HCD53, which ensures good sequence coverage for proteoforms and higher speed than radical-driven dissociation methods (e.g., electron transfer dissociation), making it the most efficient ion fragmentation technique for large-scale TDP studies. If the TDP experiment is being forwarded for a label-free workflow, the quantitation step is based on MS1 intensities, and therefore a sufficient number of survey scans must be collected throughout the chromatographic elution peak. This limits the number of tandem MS scans that can follow the survey scan in our data-dependent acquisition method such that we select only the two most abundant peaks from MS1 for fragmentation. Even so, we calculated that on average (across an entire gradient) ~75% of the total instrument duty cycle is spent on MS2 events54 (Fig. 5). In the 0–30 kDa range, considering the ~60-s elution time of most proteins, this translates to a minimum of approximately ten MS1 scans per LC peak. Newer instrumentation can further improve this value, owing primarily to faster electronics (Box 2). Finally, we average multiple microscans (i.e., timedomain transients) for each scan to improve the spectral signal-to-noise ratio, in both MS1 and MS2. All of the values for AGC targets, numbers of microscans, instrument resolution, and other instrument parameters are listed in Table 2, whereas further technical details for the analysis of intact proteins are given in Box 2.
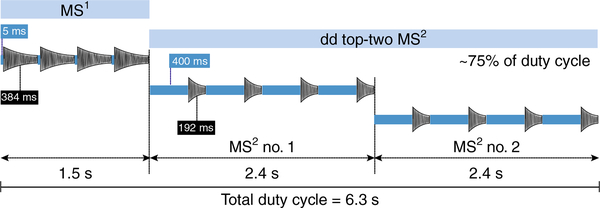
Fig. 5 |. MS duty cycle for the data-dependent, top-two experiment method used in this protocol.
Note that the indicated durations of the single scan events are specifically based on an Orbitrap Elite mass spectrometer. Briefly, the experimental design for intact protein analysis is composed of one survey scan (indicated as ‘MS1’) followed by the acquisition of two data-dependent (dd) tandem mass spectra (or ‘MS2’ scans). Each scan event uses four microscans to improve the spectral signal-to-noise ratio. In calculating the duration of each scan event, it is therefore necessary to account for four repetitions of ion injection (here represented by blue bars), followed by transient recording (indicated by the stylized time-domain transient signal). In the scheme, both ion injection and transient recording icons are represented with lengths proportional to their actual duration. The MS1 and each of the MS2 scan events have, on average, different durations, with the latter lasting longer (2.4 s versus 1.5 s) due to substantially higher injection times (as a high AGC target value is required to obtain quality fragmentation data). During the overall duty cycle of ~6.3 s, the mass spectrometer is working for ~75% of the time in MS2 mode. The increase in ion injection duration, which lasts a maximum of a few milliseconds in MS1 (5 ms in this example) and hundreds of milliseconds for MS2 (400 ms in this example), counterbalances the reduction in time spent recording transients in MS2. The applied resolving power is set at 120,000 (corresponding to time-domain transients of 384 ms) in MS1, whereas it is reduced to 60,000 (equivalent to 192 ms transients) for MS2.
Box 2 |. MS considerations.
Aside from the standard calibration and tuning of the mass spectrometer, the efficient detection of intact proteoform cations in an Orbitrap FTMS instrument requires dedicated settings. In particular, modern instruments such as those based on the so-called Tribrid platform (Orbitrap Fusion and Orbitrap Fusion Lumos), as well as some of the quadrupole-Orbitrap family (e.g., Q Exactive HF), allow the possibility of operating in ‘intact protein mode’. Briefly, this operation mode reduces the gas (i.e., N2) pressure in the HCD cell, and hence in the ultrahigh-vacuum region of the instrument where the Orbitrap is located. In addition, this mode extends the ion path beyond the C trap for MS1, so that intact proteoform ions are cooled in the HCD cell (where they are transferred using low potential to prevent fragmentation) before being back-transferred to the C trap and then to the Orbitrap mass analyzer. The ‘protein mode’ is natively available in modern Orbitrap instruments (including the Orbitrap Elite) as either a standard feature or an option. The proper transfer of ions from the source region to the LTQ and then to the FTMS mass analyzer is evaluated using the TD standard (see above), where unlike our experiments using real samples that rely uniquely on FTMS acquisition, survey scans are recorded both at low resolution in the LTQ ion trap and at high resolution in the Orbitrap. In our experience, we find that the NL values measured for the standard proteins should be similar between the two mass analyzers under the applied settings (Table 2), with the only exception being carbonic anhydrase, whose NL in the Orbitrap drops to approximately one-fifth its value in the IT due to its larger collisional cross-section.
In addition, it is fundamental to choose the proper settings for precursor isolation for TDP. We usually apply a 15 m/z unit-wide isolation window for multiply charged proteoform cations, to avoid space-charge effects during waveform isolation in the LTQ. This width can be reduced in instruments using a quadrupole for isolation, such as the Q Exactive, without penalizing ion transfer efficiency64. The TD standard is also useful for evaluating the isolation and fragmentation efficiency (through the analysis of ion injection times), as well as the optimal normalized collision energy values for HCD (by assessing for each protein the fragmentation quality in terms of p score and sequence coverage), which is generally lower than for peptide ions.
Table 2 |.
MS parameters
| Parameter | Value | Comment |
|---|---|---|
| Source region | ||
| Electrospray voltage (V) | 1.9–2.1 | |
| Inlet capillary temperature (°C) | 320 | Higher than for peptides to favor desolvation |
| S-lens radiofrequency value (%) | 50 | Higher values may moderately help transmission of large proteins, but contaminates the ion optics faster |
| In-source dissociation (V) | 15 | Helps declustering and adduct removal |
| MS1 acquisition parameters | ||
| Resolving power (at 400 m/z) | 120,000 | |
| AGC target value | 1 × 106 | |
| Number of microscans | 4 | |
| Maximum injection time (ms) | 200 | |
| Scan range (m/z) | 500–2,000 | |
| MS2 acquisition parameters | ||
| Resolving power (at 400 m/z) | 60,000 | |
| AGC target value | 1 × 106 | The amount of potential fragmentation channels requires a high value for MS2 |
| Number of microscans | 4 | |
| Maximum injection time (ms) | 1,000 | |
| Isolation window ( m/z) | 15 | |
| HCD-normalized collision energy | 23 | |
| Scan range (m/z) | 300–2,000 | Low m/z ions are generally not informative in TDP and are therefore excluded |
| Predictive AGC | Disabled | |
| Preview mode | Disabled | We rely on the full transient length to generate the list of candidates; it is fundamental to correctly determining the charge state |
| Dynamic exclusion | ||
| Repeat count | 1 | |
| Duration (s) | 60 | This should be based on the chromatographic performance |
| Exclusion window (m/z) | 15 | |
| Exclusion list size | 500 |
Data processing in targeted and discovery modes
The dual data processing modes we describe here differ based on the objective of the analyst. For cases in which the analyst already knows the identity of the protein under study but wishes to better characterize its proteoforms or benchmark new fragmentation techniques and optimize their settings, we recommend the ProSight Lite tool for data analysis (available at http://prosightlite.northwestern.edu)46. For traditional large-scale proteomics experiments, in which the contents of an unknown proteome are released by cytolysis, pre-fractionated, and analyzed by LC–MS/MS, streamlined tools for proteoform identification and characterization, database searching, and false discovery rate (FDR) determination are required. For high-throughput searches, we offer a software package including TDPortal, our Galaxy framework55,56 search system supported by high-performance computing, and Top Down Viewer for data visualization and further interpretation (both tools accessible at http://nrtdp.northwestern.edu/resource-software). In general, ProSight Lite is an application best suited for top-down mass spectrometry (TDMS) experiments in which a few proteoforms of interest are studied, whereas the TDPortal modality is designed for large-scale TDP in discovery mode. For example, we tend to use ProSight Lite when analyzing a single purified proteoform or small collection of proteoforms, such as the subset of three or four major glycoforms present in therapeutic immunoglobulins. TDPortal is best applied to profile all of the proteoforms detectable in a sample of interest for discovery or comparative experiments, and can adeptly identify hundreds or even thousands of well-characterized proteoforms per experiment, depending on the number of samples, replicates, and level of fractionation.
Before visualizing known proteoforms of interest in ProSight Lite for targeted TDMS analysis, separate software tools are required to deconvolute and deisotope mass spectral data. Tools for data visualization and algorithms for this purpose are usually supplied with instruments by vendors, and thus vary in a vendor-specific manner. Here, we use Xtract (which deconvolutes only isotopically resolved spectra) within QualBrowser. Accessible, free options for deconvolution and deisotoping are plentiful on the web, including MS-Deconv57 and YADA58, but the use of these tools will not be detailed in this protocol. Once precursor (MS1) and fragmentation (MS2) scans for a proteoform of interest are located in QualBrowser and their monoisotopic, zero-charge spectra are obtained using Xtract, these values are exported to ProSight Lite for targeted analysis. ProSight Lite relies on both experimental data at the MS1 and MS2 levels and sequence information (which can be downloaded for any protein of interest at http://www.uniprot.org) to match fragments from the exported list to the candidate sequence and display useful graphical fragment maps. Importantly, the software also calculates a p score59, which uses a Poisson model to calculate the specificity of the observed fragmentation data for protein identification. Analysis of proteoforms using ProSight Lite is an iterative technique that is especially suited to localizing unexpected mass shifts (Δm) and characterizing PTMs. Analysts can apply in silico a selection of PTMs or a custom mass to amino acid residues displayed in the candidate sequence as the application updates the fragment map and metrics in real time. Figure 6 depicts a detailed flow diagram documenting the use of ProSight Lite with helpful screenshots. We have also recently published a more in-depth description of how to use ProSight Lite to analyze data from targeted TDMS experiments46.
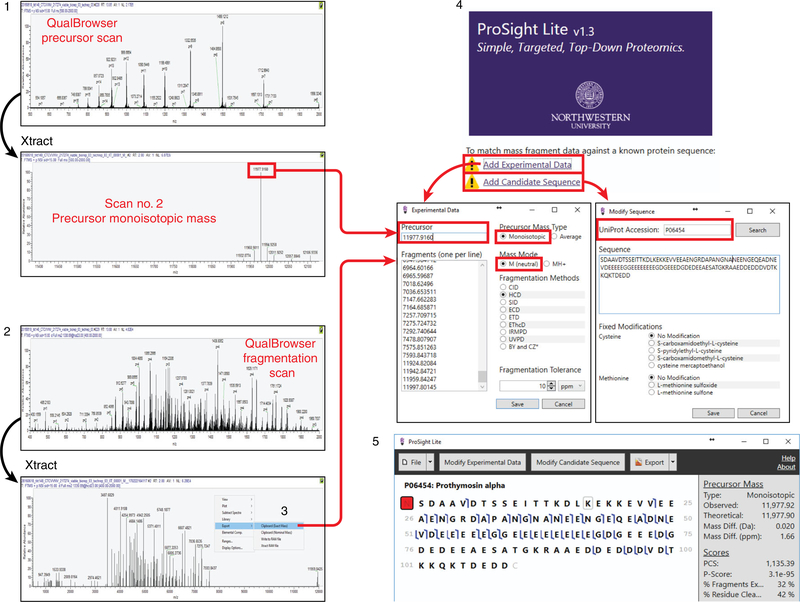
Fig. 6 |. General workflow for analyzing top-down mass spectrometry data using ProSight Lite.
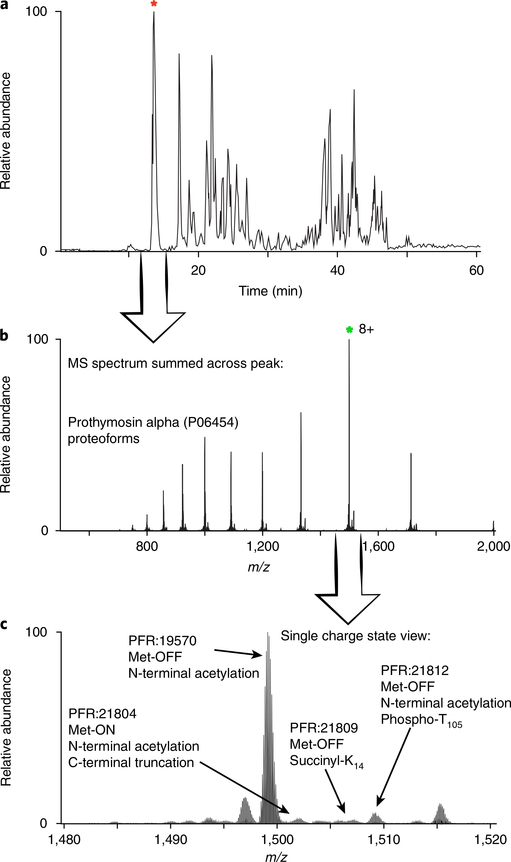
A stepwise flow diagram for analyzing top-down mass spectrometry data targeted to one or a few proteoforms of interest using ProSight Lite. In step 1, the precursor spectrum (from prothymosin alpha, in this example) is Xtracted, and the generated monoisotopic mass value is input into ProSight Lite. In steps 2 and 3, the fragmentation spectrum associated with the precursor is Xtracted, and the list of fragment monoisotopic masses is input into ProSight Lite. Step 4 shows the parameters required for ProSight Lite analysis, and step 5 depicts the final graphical fragment map yielded by this experiment.
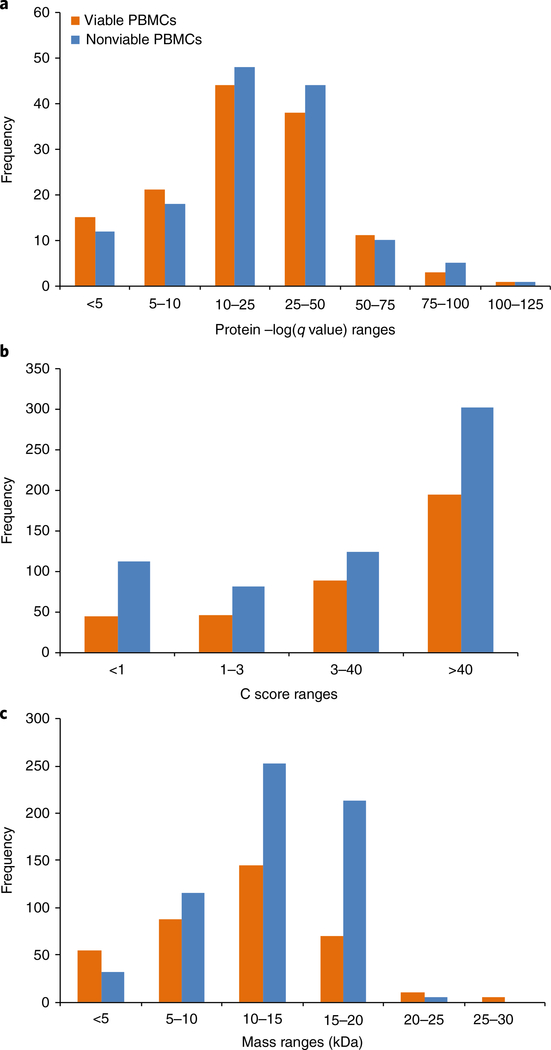
We will also detail the procedures for the use of TDPortal and Top Down Viewer for performing a high-throughput analysis of fractions of PBMC whole proteome44,60. Processing datasets for these experiments with the TDPortal interface is executed via a search environment that runs within a Galaxy framework on Northwestern University’s Quest high-performance computing resource, made available to researchers free of charge by the NRTDP. Thermo RAW files containing the experimental observations are supplied by the user, and are searched against a highly annotated human database from UniProt using an iterative, three-tiered search tree as previously described for discovery TDP experiments44,54. For other spectral data types, software tools such as ProteoWizard61 can be used to convert the data into the .mzML extensible markup language format. The general workflow performed by TDPortal is as follows (Fig. 7): first, RAW files are converted to .mzML format and then interrogated to generate sets of precursor and fragment ion data grouped across scans within a preset retention time window and mass tolerance. Precursor and fragmentation scans for targets generated in this manner are then deconvoluted using Xtract, and the output is submitted for database searching. Protein identification and proteoform characterization are first attempted using a tight absolute mass search with narrow mass tolerances at the MS1 and MS2 levels. A so-called ‘Biomarker’ search is then performed to identify any natural and artificial proteolytic fragments by matching the experimental precursor and fragment masses with any theoretical protein subsequence, using very narrow mass tolerances14. A final search for proteoforms with unknown modifications is then performed with a much higher allowed precursor mass tolerance using Delta M mode, which applies any mass differences that could be due to an unknown PTM to all theoretical fragment ions to compensate for and localize unexpected mass shifts14. Each of the three searches is subjected to a local FDR estimation to control for multiple testing, and a global FDR is then determined via the Higdon method62. Datasets can then be exported to Top Down Viewer, which displays key details for the proteoforms identified and characterized in the study, including p scores, C scores, and visualization of the raw spectra from which they were identified.
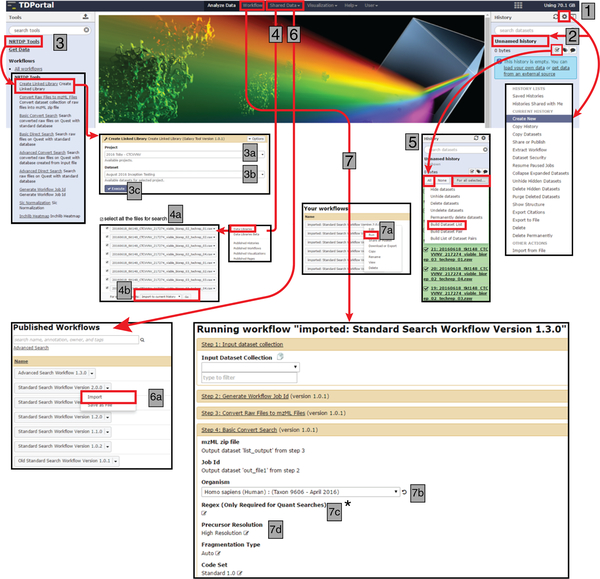
Fig. 7 |. General workflow for searching high-throughput, discovery mode top-down proteomics data using TDPortal.
A stepwise flow diagram for using the TDPortal interface to search high-throughput, discovery top-down proteomics datasets. In steps 1 and 2, a new history is created and named. In step 3, a linked library is created from the uploaded data files. In step 4, the shared data files are then imported to the created history, where they are used to build a dataset list (step 5). In step 6, a published workflow for data searching is imported (we recommend Standard Search Workflow 1.3); and in step 7, the workflow is run for the selected dataset list.
Finally, the Consortium for Top Down Proteomics (http://topdownproteomics.org/) hosts an online storage database to house proteoforms discovered by TDP experiments. The open-access Proteoform Atlas (accessible at http://atlas.topdownproteomics.org/) provides researchers with a central location to browse uniquely observed proteoforms and submit their own datasets for storage. The repository currently stores >260,000 proteoforms (each with specific proteoform identifiers (PFRs)) mapping to >38,000 protein accession numbers from four organisms.
Generally, the total time required to perform the experiment detailed herein, from blood collection to data analysis, is based on the number of samples involved. For a minimal number of samples, the entire protocol takes ~72 h to complete, if the pause points included in the protocol are limited, and only the break after acetone precipitation is allowed to continue overnight (or for ~12 h). We believe that the expertise required to complete the sample preparation protocols is appropriate for a senior graduate student or trained practitioner of MS-based proteomics with experience in protein handling and fractionation. The LC and MS methods detailed here can be routinely deployed by most MS core facilities, as well as research labs that specialize in MS-based proteomics. Blood collection, of course, should be performed only by a certified phlebotomist on participants when informed consent has been obtained and the corresponding institutional ethics committees have approved the study protocol.
Limitations
One of the primary and most well-known limitations of the denatured-mode TDP experiment is the mass range of proteoforms that can be resolved by contemporary, commonly accessible instrumentation. For this reason, only the <30-kDa proteome after GELFrEE fractionation is submitted for analysis here, as, in our experience, that represents the MW range that can be most robustly resolved on Orbitrap platforms. Fortunately, recent advancements in instrumentation and data processing strategies have been reducing this barrier over the past few years. For instance, recent work in targeted TDMS achieved greater sensitivity in the analysis of large proteins by averaging the time-domain transients collected for multiple LC–MS/MS experiments before signal processing by Fourier transformation63 or by using short time-domain transients for improving signal-to-noise ratio at the expense of resolving power64. However, advanced instrumentation options and data processing strategies are not widely available to most researchers and are not yet highly automated, respectively, and therefore serve as examples of the cutting edge in TDP as the field progresses. We believe that advancements along these lines will continue to increase the MW range robustly applicable for TDP and will soon find traction across a broader range of labs.
In addition, top-down analysis is known to suffer from a dynamic range challenge, as multiple charge states for proteoforms after ESI and the longer scan times and higher ion numbers required for TDP make it difficult for experiments to interrogate low-abundance proteoforms. To address this challenge, more intelligent data acquisition strategies are being developed, most notably Autopilot65, which has recently demonstrated increases in proteome coverage by TDP54. Our group continues to address the dynamic range challenge through similar advances, which we hope will become widely available on standard instrumentation platforms of the future. Furthermore, it is key to note that the sample type of interest in this protocol, PBMCs, represents a diverse collection of cell populations with variable frequencies between patients. PBMC proteomics experiments are thus ‘bulk’ measurements and are not suited to assigning signals to specifically phenotyped cell populations and subpopulations. A final limitation of TDP is the substantial ESI interference provided by surfactants necessary for cytolysis and pre-fractionation workflows (e.g., SDS). The methods that we describe here for their removal before analysis are not easily automated and may have the potential to introduce substantial pre-analytical variation if not performed carefully.
The described limitations of TDP make it difficult to match the depth of analysis achieved by BUP, which generally yields more expansive lists of identified proteins from a given sample, across a broader mass range. Although great advancements have been made to narrow the divide between TDP and BUP and mitigate these limitations for intact protein analysis, it is key to note that the information generated by these divergent methods is intrinsically different, and therefore complementary. Proteoform-resolved information generated by TDP on targeted proteins or mass range–defined fractions achieves an unmatched breadth of analysis in regard to visualizing molecular complexity in the context of a protein or protein family, whereas a much deeper snapshot of the sample proteome is generated by BUP, at the expense of molecular specificity
Materials
Reagents
PBMC collection and cell lysis (Reagent setup)
RNAlater RNA stabilization reagent (Qiagen, cat. no. 76104) !CAUTION RNAlater is a suspected human carcinogen. Wear gloves and safety glasses.
FBS (USDA approved, sterile-filtered, suitable for cell culture; Sigma-Aldrich, cat. no. F0926)
HyClone Dulbecco’s PBS (DPBS, without calcium, magnesium; GE Healthcare Life Science, cat. no. SH30028.02)
DMSO for molecular biology (Sigma-Aldrich, cat. no. D8418)
N-Lauroylsarcosine sodium salt (Sigma-Aldrich, cat. no. L9150–50G) !CAUTION Lauroylsarcosine sodium salt causes skin irritation and serious eye damage, and may be fatal if inhaled. Avoid breathing dust or fumes, and wear gloves and safety glasses when handling.
Tris base (Fisher Scientific, cat. no. BP152–1) !CAUTION Tris base may cause mild eye irritation; handle with gloves and wear safety glasses.
Sodium chloride (Sigma-Aldrich, cat. no. S9888)
DL-Dithiothreitol (DTT; Sigma-Aldrich, cat. no. D0632) !CAUTION Reagent is harmful if swallowed and may cause skin, eye, and respiratory irritation. Avoid breathing dust or fumes, and wear gloves and safety glasses when handling.
Magnesium chloride (Sigma-Aldrich, cat. no. M8266) !CAUTION Avoid breathing dust or vapors generated by this product.
Benzonase nuclease (25 kilo-units; Sigma-Aldrich, cat. no. E1014–25KU) !CAUTION Avoid contact with skin or eyes.
Halt protease and phosphatase inhibitor cocktail (EDTA-free; Thermo Fisher Scientific, cat. no. 78443) !CAUTION Halt protease and phosphatase inhibitor cocktail contains sodium fluoride, which is toxic if swallowed and also a skin and eye irritant. Handle with gloves and wear safety glasses.
Acetone (certified ACS; Fisher Scientific, cat. no. A18–4) !CAUTION Acetone is highly flammable, so keep away from heat and open flames. It causes serious eye irritation, so wear gloves and safety glasses when handling. Avoid inhalation and ingestion, as it is toxic to the central nervous system and internal organs.
Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, cat. no. 23225)
GELFrEE and silver staining reagents (Reagent setup)
GELFrEE 8100 HEPES running buffer (Expedeon, cat. no. 42202)
GELFrEE 8100 Tris acetate sample buffer (Expedeon, cat. no. 42302)
Methanol (HPLC grade; Fisher Scientific, cat. no. A452SK-4) !CAUTION Methanol is highly flammable, so keep away from heat and open flames. It is toxic upon inhalation and contact with skin and eyes. Use methanol under a chemical fume hood, and wear gloves and safety glasses when handling.
Formaldehyde (37% by weight, certified ACS; Fisher Scientific, cat. no. F79–500) !CAUTION Formaldehyde is highly poisonous, and may cause blindness or fatality if swallowed. Formaldehyde is toxic upon inhalation, causes burns by all exposure routes, and may affect the central nervous system and cause birth defects. Use this reagent only under a chemical fume hood, and wear gloves and safety glasses when handling.
Silver nitrate (Sigma-Aldrich, cat. no. 209139) !CAUTION Silver nitrate is an oxidizer and may intensify fire, so keep it away from heat as well as clothing and combustible materials. Silver nitrate also causes skin burns and eye damage, so be sure to wear gloves and safety glasses when handling.
Sodium thiosulfate (Sigma-Aldrich, cat. no. 217263) !CAUTION Wear gloves and safety glasses when handling sodium thiosulfate.
Acetic acid (glacial, certified ACS; Fisher Scientific, cat. no. A38–212) !CAUTION Acetic acid is a flammable liquid and vapor, so keep it away from any sources of ignition. Use personal protective equipment and avoid contact with skin, eyes, or respiratory/digestive tract.
Sodium carbonate (Sigma-Aldrich, cat. no. S7795) !CAUTION Sodium carbonate causes serious eye irritation, so wear eye and skin protection when handling.
SDS (Sigma-Aldrich, cat. no. L3771) !CAUTION SDS is a flammable solid, so keep it away from ignition sources. SDS causes eye, skin, and respiratory irritation, so wear protective equipment when handling.
Glycine (Sigma-Aldrich, cat. no. G8898)
2× Laemmli sample buffer (Bio-Rad, cat. no. 1610737)
Precision Plus Protein Dual Color Standards (Bio-Rad, cat. no. 1610374)
Fraction desalting and LC–MS reagents
Methanol (Optima LC/MS grade; Fisher Scientific, cat. no. A456) !CAUTION Methanol is highly flammable, so keep it away from heat and open flames. Methanol is toxic upon inhalation and contact with skin and eyes. Use methanol under a chemical fume hood, and wear gloves and safety glasses when handling it.
Chloroform (Sigma-Aldrich, cat. no. C2432) !CAUTION Chloroform is harmful if swallowed, and causes skin and serious eye irritation. Chloroform is toxic if inhaled and may cause drowsiness or dizziness. Handle reagent only in a chemical fume hood and always wear protective gloves and safety glasses. Be aware that chloroform is a suspected carcinogen and may damage fertility or the unborn child.
Water (Optima LC/MS grade; Fisher Scientific, cat. no. W6)
Acetonitrile (Optima LC/MS grade; Fisher Scientific, cat. no. A955) !CAUTION Acetonitrile is highly flammable, so keep it away from open flames and hot surfaces. Acetonitrile may also cause skin and respiratory tract irritation, central nervous system effects, and internal organ damage, so always wear protective gloves and safety glasses when handling it.
Pierce formic acid ampules (Fisher Scientific, cat. no. PI28905) !CAUTION Formic acid is flammable, so keep it away from open flames and hot surfaces. Formic acid is toxic if swallowed or inhaled, and causes severe skin burns and eye damage. Wear protective gloves and safety glasses when handling it.
Pierce LTQ Velos ESI positive ion calibration solution (Thermo Fisher Scientific, cat. no. 88323) !CAUTION This solution is highly flammable, so keep it away from open flames and hot surfaces. This solution exhibits acute oral, dermal, and inhalation toxicity, as well as causing serious eye irritation, so avoid inhaling it, and wear protective gloves and safety glasses while handling it.
PLRP-S 5-μm particles (1,000-Å pore size; Agilent, cat. no. PL1912–1502) ▲CRITICAL Bulk medium must be collected from a cracked column, owing to lack of availability of bulk PLRP-S from this supplier. Alternative reversed-phase media qualified for protein separations, such as MAbPac (Thermo Fisher Scientific, cat. no. 088644) and Proteomix RP-1000 (Sepax Technologies, cat. no. 465950), can be used to pack spray emitters.
Top-down standard reagents (Reagent setup)
Ubiquitin (Sigma-Aldrich, cat. no. U6253)
Trypsinogen (Sigma-Aldrich, cat. no. T1143) !CAUTION Trypsinogen causes skin and serious eye irritation, so wear protective gloves and safety glasses when handling it. Trypsinogen is also known to cause allergy or asthma symptoms in certain individuals when inhaled, so avoid breathing dust or fumes.
Myoglobin (Sigma-Aldrich, cat. no. M5696)
Carbonic anhydrase (Sigma-Aldrich, cat. no. C2624) !CAUTION Carbonic anhydrase causes serious eye irritation, so wear protective gloves and safety glasses when handling.
Equipment
PBMC collection and sample preparation
BD Vacutainer CPT mononuclear cell preparation tube, sodium citrate (BD Biosciences, cat. no. 362761) !CAUTION Tubes must be stored upright at room temperature (20–25 °C), and protected from direct light. Tube shelf life in these conditions is 1 year.
1.5-mL Protein LoBind microcentrifuge tubes (Fisher Scientific, cat. no. 13–698-794)
Falcon 15-mL conical centrifuge tubes (Fisher Scientific, cat. no. 14–959-70C)
Benchtop mini-centrifuge (Fisher Scientific, cat. no. 13–100-676)
Benchtop vortex mixer (Fisher Scientific, cat. no. 02–215-414)
Benchtop thermal mixer or heating block (Fisher Scientific, cat. no. 13–687-720)
Benchtop lab rotator or orbital shaker (Thermo Fisher Scientific, cat. no. 11–676-066)
Colorimetric 96-well plate reader (Thermo Fisher Scientific, cat. no. BTS1A)
96-Well plate (clear, flat-bottom; Falcon, cat. no. 351172)
Automated cell counter with slides (Thermo Fisher Scientific, cat. no. AMQAX1000)
−86 °C freezer (Thermo Fisher Scientific, cat. no. 90–9)
−120 °C liquid nitrogen storage system (Thermo Fisher Scientific, cat. no. 13–762-353)
Biological safety cabinet (Thermo Fisher Scientific, cat. no. 13–261-334)
Cell-freezing container (Thermo Fisher Scientific, cat. no. 13–350-50)
1-mL Capacity cryovials (Thermo Fisher Scientific, cat. no. 03–337-7X)
pH indicator strips (EMD Millipore, cat. no. 1095350001)
GELFrEE 8100 fractionation system (Expedeon, cat. no. 48100)
GELFrEE 8100 8% tris-acetate cartridge (Expedeon, cat. no. 42103) or other desired cartridge, depending on mass range of interest
Mini-PROTEAN Tetra gel rig assembly, with power supply (Bio-Rad, cat. no. 1658004)
Any kD Mini-PROTEAN TGX precast gels (Bio-Rad, cat. no. 4569036)
Nano-UHPLC platform
Dionex 3000 Ultimate nano-UHPLC (Thermo Fisher Scientific, cat. no. IQLAAAGABHFAPBMBFB)
Monolithic PepSwift trap/guard column (200-μm i.d. × 5 mm; Thermo Fisher Scientific, cat. no. 164558)
Monolithic RP-4H analytical column (100-μm i.d. × 50 cm; Thermo Fisher Scientific, cat. no. 164921)
Nanospray emitters (15-μm i.d. ×125 mm; New Objective, cat. no. FS360–50-15-N-20-C12)
Various viper-fitted LC tubing for waste lines and connection to MS. For example: fused silica nanoviper tubing, 20-μm i.d. × 260-μm o.d. × 1-m length, as line to high-voltage union (Thermo Fisher Scientific, cat. no. 6041.5293); and fused silica nanoviper tubing, 75 μm × 250-mm length, as waste lines (Thermo Fisher Scientific, cat. no. 6041.5730)
High-voltage union (Vanderhulst Associates, cat. no. 97044–60290S)
LC autosampler vials (Thermo Fisher Scientific, cat. no. 03-FIRV)
LC autosampler vial caps (Thermo Fisher Scientific, cat. no. C4011–53G)
High-pressure capillary column packing station (product numbers and assembly instructions available at https://proteomicsresource.washington.edu/docs/protocols05/Packing_Capillary_Columns.pdf).
Alternatively, ref. 65 also provides an in-depth description of column fabrication66.
Hamilton syringe (Thermo Fisher Scientific, cat. no. 14–815-113)
High-resolution FTMS platform
Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, cat. no. IQLAAEGAAPFADBMAZQ)
Ion Max ion source with HESI-II probe (Thermo Fisher Scientific, cat. no. IQLAAEGABBFACTMAJI)
Custom nano-source (product numbers and instructions for assembly available at https://proteomicsresource.washington.edu/docs/protocols05/UWPR_NSI_Source.pdf) ▲CRITICAL Alternatively, the Nanospray Flex Ion Source (Thermo Fisher Scientific, cat. no. ES071) is a commercially available source for nano-ESI
Informatics tools for TDP data analysis
ProSight Lite (freely available for download online at http://prosightlite.northwestern.edu)
TDPortal (accounts available by request at http://nrtdp.northwestern.edu/resource-software)
Top Down Viewer (freely available for download online at http://topdownviewer.northwestern.edu)
Xcalibur Software with QualBrowser (Thermo Fisher Scientific, cat. no. OPTON-30487)
Chromeleon Chromatography Data System (Thermo Fisher Scientific, cat. no. CHROMELEON7)
Microsoft Excel or similar spreadsheet software of choice (Microsoft, https://office.microsoft.com/excel/)
Reagent setup
Cell lysis buffer
Our lysis buffer is composed of 20 mM Tris base (adjusted with 11 N HCl to pH 7.5), 100 mM NaCl, 1% (wt/vol) N-lauroylsarcosine, 1 mM MgCl2, and 1× Halt inhibitor cocktail (EDTA-free) in dH2O. We recommend creating this solution at the bench from the following stored stock solutions: 1 M Tris base (pH 7.5), 5 M NaCl, 10% (wt/vol) N-lauroylsarcosine, and the commercial stock of 100× Halt inhibitor cocktail. We recommend that the cell lysis buffer be freshly prepared, whereas all stock solutions except for the inhibitor cocktail are stable on the bench at room temperature for up to a year. 100× Halt inhibitor cocktail must be stored at 4 °C, and is stable for up to a year in our experience.
SDS–PAGE running buffer
We create a final 1× solution of running buffer from a 10× stock of 248 mM Tris base, 1.92 M glycine, and 1% (wt/vol) SDS. We freshly prepare the 1× solution, whereas the stock solutions are stable at room temperature for up to a year.
Top-down standard
The final TD standard is composed of 0.64 pmol carbonic anhydrase, 1.09 pmol myoglobin, 0.49 pmol trypsinogen, and 0.14 pmol ubiquitin loaded on a column in a 6-μL injection. ▲CRITICAL We recommend preparing 2 mg/mL stocks of these proteins for storage at −80 °C for up to several years. Use these to create a stock pot of 25.7 pmol/μL carbonic anhydrase (40 μL of 2 mg/mL stock), 43.9 pmol/μL myoglobin (40 μL of 2 mg/mL stock), 19.6 pmol/μL trypsinogen (25 μL of 2 mg/mL stock), and 5.5 pmol/μL ubiquitin (2.5 μL of 2 mg/mL stock). This stock pot can then be divided into 2.5-μL aliquots and stored at −80 °C, for reconstitution in 600 μL of mobile phase A (or 100× the intended injection volume) at the time of the experiment. A standard prepared in this way is stable for a few days at 4 °C. A comprehensive protocol for the preparation of TD standard, in addition to analysis of this standard on an LTQ-Velos Orbitrap Elite mass spectrometer is available for download from http://nrtdp.northwestern.edu/protocols.
LC mobile phase A
Prepare 1 L of mobile phase A in a clean glass bottle reserved only for LC solvents, using a clean glass graduated cylinder also reserved only for LC solvents. Prepare a solution of 94.8% (vol/vol) Optima-grade water and 5% (vol/vol) Optima-grade acetonitrile. Using a clean Hamilton syringe reserved only for formic acid, add formic acid to a final concentration of 0.2% (vol/vol). We recommend freshly preparing LC solvents each month in a continuously running mass spectrometry core or facility.
LC mobile phase B
Using the same sample-handling precautions as outlined above, prepare a solution of 4.8% (vol/vol) water, 95% (vol/vol) acetonitrile, and 0.2% (vol/vol) formic acid. We recommend freshly preparing LC solvents each month in a continuously running mass spectrometry core or facility.
Silver staining solutions
Create the following 1× stock solutions in 1 L of dH2O and store at room temperature for up to a month. Only the silver nitrate solution needs to be prepared at the time of staining.
Fixing solution: 50% (vol/vol) methanol and 5% (vol/vol) acetic acid in water
Washing solution: 50% (vol/vol) methanol in water
Sensitizing solution: 0.02% (wt/vol) sodium thiosulfate in water
Silver nitrate solution: 0.1% (wt/vol) silver nitrate in water (0.05 g of silver nitrate per 50-mL conical tube of dH2O is appropriate for one gel)
Developing solution: 0.04% (vol/vol) formaldehyde and 2% (wt/vol) sodium carbonate in water
Terminating solution: 5% (vol/vol) acetic acid in water
Equipment setup
GELFrEE device setup and testing
An Expedeon GELFrEE 8100 device is used to separate proteins by MW. For our analysis, we collect proteins up to ~30 kDa using an 8% GELFrEE cartridge. The GELFrEE device is operated according to manufacturer’s instructions (see Procedure for further information). Importantly, for safety reasons, the device is operational only if the lid is properly covering the cartridge compartment.
UHPLC setup and testing
For the chromatographic separation of intact proteins, we use a Thermo Fisher Scientific Ultimate 3000 LC system. This is equipped with two pumps: a gradient pump (also called an NC pump) fitted with a specific nano-flow selector, and a loading pump, which is configured to transport the sample from the loop to the trap column. Notably, the NC pump delivers the gradient up to 1,000 bar, whereas the loading pump has an upper pressure limit of 500 bar. Further details about the LC configuration are specified in Box 1 and Fig. 4. Details on the UHPLC method for sample analysis are provided in Table 1. Both pumps of the LC system are purged on a weekly basis, and the column performance is tested using the TD standard (see above). Nanospray emitters should be packed with ~2–3 mm of reversed-phase resin (PLRP-S or MAb-Pac), using a high-pressure column packing station as described in the ‘Packing Capillary Columns and Pre-columns (traps)’ protocol found at https://proteomicsresource.washington.edu/docs/protocols05/Packing_Capillary_Columns.pdf.
MS setup and testing
The TDP experiments described herein are performed on an Orbitrap Elite mass spectrometer. This instrument combines two mass analyzers in series, a linear IT (LTQ) and a high-resolution Orbitrap. In addition, this mass spectrometer is equipped with an N2-filled collision chamber that is used to perform HCD, and also to cool intact protein ions when the instrument is operated in ‘protein mode’67. Detailed information about the MS acquisition methods can be found in Box 2 and Table 2. ▲CRITICAL Instrument performance for TDP is assessed using the TD standard, with the data analyzed via TDPortal.
ProSight Lite setup
ProSight Lite can be freely downloaded at http://prosightlite.northwestern.edu. After the installation, the software can be accessed using the Windows ‘Start’ button. Updates are downloaded and installed automatically when available. ProSight Lite can be used to match a selected sequence and related set of fragment masses, or to visualize the graphical fragmentation map of an identified proteoform from a database search performed through TDPortal.
Top Down Viewer setup
Top Down Viewer is freely available at http://topdownviewer.northwestern.edu. The software can be opened from the Windows ‘Start’ button. Updates are downloaded and installed automatically when available. Top Down Viewer is used to visualize .tdReport files generated by the TDPortal environment.
TDPortal setup
An account for TTDPortal can be requested on the Resource Software page of the NRTDP website at http://nrtdp.northwestern.edu/resource-software. Upon completion of the request form, the TDPortal administrator will provide login information by email. In addition, a secure shell file transfer protocol (SFTP) application will need to be downloaded for transferring files to TDPortal. We recommend WinSCP (download at: https://winscp.net/eng/download.php).
Procedure
Blood collection, PBMC isolation, and two application-specific methods for cell storage ● Timing ~2 h
-
Draw 8 mL of blood by venipuncture (do not use a syringe) into the CPT tube and invert the tube ten times immediately after draw. Do not shake the tube; keep it at room temperature.
▲CRITICAL STEP Centrifuge as soon as possible, and no later than 2 h from the time of draw.
▲CRITICAL STEP For all human subjects, informed consent must be obtained and the corresponding institutional ethical committees must approve the study protocol.
Centrifuge the CPT tubes within 2 h of the time of the blood draw in a swinging-bucket rotor for 20 min, at room temperature, at 1,700g.
After centrifugation, using a transfer pipette, in aseptic conditions, carefully and slowly remove the clear plasma from the uppermost layer (either to waste or for storage in tubes at −80 °C for future plasma proteomics), and avoid disturbing the whitish cell layer.
Using a pipette with a wide opening, collect the whitish cell layer and transfer it to a labeled 15-mL conical tube, then pipette delicately up and down the remaining plasma and add it to the same 15-mL conical tube.
Add DPBS to bring the volume to 15 mL, cap the tube, and mix the cells by inverting the tube five times.
Centrifuge for 15 min, at room temperature, at 300g, and aspirate as much supernatant as possible without disturbing the cell pellet, leaving a few microliters of supernatant with the cell pellet.
Resuspend the pellet by gently and briefly vortexing or tapping the tube with a finger. Add DPBS to bring the volume to 2 mL, cap the tube, mix the cells by inverting five times, take 40 μL for cell counting, and record the cell number after counting on an automated cell counter.
Add DPBS to bring the volume to 10 mL, cap the tube, and mix the cells by inverting five times.
Centrifuge for 10 min, at room temperature, at 300g, and aspirate as much supernatant as possible without disturbing the cell pellet, leaving a few microliters of supernatant with the cell pellet.
-
To collect cells for storage, follow either option A, for the viable cell method, which does not use RNA-stabilizing reagent, or option B, for the nonviable cell method, which utilizes RNAlater to stabilize RNA for future -omics studies.
▲CRITICAL The choice of cell storage method depends on institutional biorepository protocols and intended downstream applications. We have found that many PBMC samples retroactively applied to proteomics methods were stored using nonviable protocols, and are thus suitable for parallel transcriptomic assays. Viable cells are more amenable to TDP and can also be used for parallel cell-based functional assays.
-
Viable cell collection
Add a sufficient volume of cold FBS to make a cell concentration of 4–5 × 106 cells/mL.
Immediately add the same volume of 2× cold freezing medium (80% FBS (vol/vol) and20% DMSO (vol/vol)) dropwise to obtain ~2–2.5 × 106 cells/mL and transfer to a 2-mL labeled cryovial.
Place the cryovials in the freezing container and store the container in the −80 °C freezer for a minimum of 24 h.
Transfer the cryovials to −120 °C long-term storage in N2 within 1 week.
-
Nonviable cell collection
Immediately add 1.5 mL of RNAlater solution, resuspend, and then transfer the suspensionto a labeled cryovial.
-
Transfer the cryovial to storage in a −80 °C freezer.
■PAUSE POINT Samples can be stored at −120 °C (viable cells) or −80 °C (nonviable cells) until required. We have kept samples frozen in this manner for >2 years with no substantial changes to the number and quality of detected proteoforms.
-
Cell lysis, protein precipitation, and lysate quantitation ● Timing ~3 h
-
11
Follow option A for viably collected cells and option B for non-viably collected cells.
-
Viable cells
Remove the cryovial containing PBMCs for analysis from the N2, and quickly thaw for 1–2 min in a 30–37 °C water bath.
-
Gently tap the vial multiple times to adequately resuspend the thawed cells, and then transfer the cell suspension to a labeled 1.5-mL Protein LoBind centrifuge tube.
▲CRITICAL STEP Keep the tubes on ice throughout the beginning of the prep protocol (Steps 11 and 12), to minimize protein degradation and loss of labile PTMs.
Centrifuge at 300g for 10 min at 4 °C to pellet the PBMCs.
Gently remove the tubes from the centrifuge, and use a pipette to slowly remove the freezing medium supernatant without disturbing the cell pellet.
-
Add 1 mL of DPBS to resuspend the pellet, and centrifuge again at 300g for 10 min at 4 °C.
▲CRITICAL STEP This is to wash out the remaining serum from the freezing medium; depending on how much medium was left over the pellet; this step can be repeated two to three more times.
Carefully remove the DPBS wash and discard.
-
Nonviable cells
Remove the cryovial containing PBMCs for analysis, and quickly thaw it for 1–2 min in a 30–37 °C water bath.
-
Gently tap the vial multiple times to adequately resuspend the thawed cells, and then transfer the cell and protein suspension to a labeled 1.5-mL Protein LoBind centrifuge tube.
▲CRITICAL STEP Keep the tubes on ice throughout the beginning of the prep protocol (Steps 11 and 12), to minimize protein degradation and loss of labile PTMs.
▲CRITICAL STEP Note that the high salt content of RNAlater will have lysed some of the cells and already precipitated some protein. It is therefore important to pipette carefully during the following steps to avoid any sample loss.
Centrifuge the tubes at maximum centrifugal force (~21,000g) for 25 min at 4 °C.
-
Carefully remove all of the supernatant from the tubes, without disturbing the resulting protein/cell debris pellet, which should adhere to the sides and bottom of the tube after maximum-speed centrifugation.
▲CRITICAL STEP It is important to remove as much RNAlater supernatant as possible, because it crystallizes in downstream protein preparation procedures and complicates lysate quantitation by BCA assay.
-
-
12
To the dry pellets yielded by either of the above options, add 225 μL of cell lysis buffer (including protease/phosphatase inhibitor cocktail), and pipette up and down to completely resuspend the pellet.
▲CRITICAL STEP The solution may become very clumpy and viscous. This is due to DNA released by the PBMCs, which have a high nucleus-to-cytoplasm ratio. Be careful to minimize sample loss in the pipette tip by expunging completely any remaining fluid back into the tube, and dragging the tip up the side of the tube.
-
13
Using a pipette, add 750 units of benzonase to each lysate, vortex the tubes for 10–20 s, and place the tubes in a 37 °C incubator or water bath for 15 min.
▲CRITICAL STEP Keep benzonase nuclease on ice when working with it at the bench.
▲CRITICAL STEP Benzonase is used to enzymatically digest DNA, RNA, and all other nucleotides to make the lysate homogeneous, which is important for protein quantitation and accurate pipetting throughout the rest of the protocol.
? TROUBLESHOOTING
-
14
To desalt and precipitate proteins in the lysate, add 4 volumes (~1 mL) of ice-cold acetone to the lysates, vortex for 10–20 s, and place the tubes in a −80 °C freezer for at least 30 min.
■PAUSE POINT This is a natural stopping point in the protocol, as the protein precipitate is stable in acetone overnight.
-
15
15 Remove the tubes from the freezer and immediately centrifuge at maximum centrifugal force (~21,000g) for 30 min at 4 °C.
-
16
Using a pipette, completely remove as much acetone as possible from the protein pellets without disturbing them. Place open tubes in a tube rack in an operating biological safety cabinet for 5–10 min to evaporate off any excess acetone.
? TROUBLESHOOTING
-
17
Resuspend the protein pellet in 225 μL of 1% (wt/vol) SDS solution, frequently pipetting up and down to break up the pellet and resolvate the proteins until the solution is clear and without precipitates.
▲CRITICAL STEP Resuspension of the protein pellet is often an arduous task for intact proteins, as opposed to peptides. It may take multiple rounds of vigorous pipetting to completely dissolve the precipitates.
? TROUBLESHOOTING
-
18
Transfer 25 μL of the solution to a new 1.5-mL Protein LoBind tube for the BCA assay. The tubes containing the remaining 200 μL of resuspended proteins should be kept on ice.
-
19
A BCA assay should now be performed on the 25-μL aliquots of sample proteome, using a 96-well plate and following the manufacturer’s recommended protocol, which can be downloaded from: https://tools.thermofisher.com/content/sfs/manuals/MAN0011430_Pierce_BCA_Protein_Asy_UG.pdf.
▲CRITICAL STEP We recommend following the 96-well plate protocol for high-throughput analysis for larger collections of samples. In addition, we recommend preparing three dilutions for each 25 μL of sample (e.g., 1:5, 1:10, and 1:20) in order to be sure to stay within the linear range of the assay.
-
20
After collection of colorimetric data for BCA assay on a 96-well-plate reader, export the data containing the absorbance values for each well into Microsoft Excel. Create a standard curve, using the BSA values and linear fitting in Microsoft Excel, and fit the average of the triplicate values for each sample dilution to the standard curve to determine the protein concentration of each sample, after correcting for dilutions.
Protein fractionation by GELFrEE ● Timing ~3 h
-
21
Once the protein concentration of each sample has been determined, calculate the volume of the remaining 200 μL of samples on ice required for GELFrEE analysis. We recommend a GELFrEE input value of 100–300 μg of total protein, depending on the protein content of the lysates. Typically, we use 200 μg of total protein as the input amount, and we will continue with the Procedure using this value, although any input in the range of 50–500 μg can feasibly be used for GELFrEE.
-
22
For each sample, transfer the calculated volume containing 200 μg of protein to a new 1.5-mL Protein LoBind tube. The leftover lysates in 1% (wt/vol) SDS can be stored for several months at −80 °C, in our experience.
▲CRITICAL STEP From this point on, keep the sample tubes on ice until immediately before GELFrEE.
-
23
To each sample, add 8 μL of 1 M DTT, 30 μL of GELFrEE sample loading buffer, and dH2O to a final volume of 150 μL.
▲CRITICAL STEP If the samples contain relatively low amounts of protein, the 150 μL of final input volume may need to be surpassed to reach a baseline of 50–100 μg of total protein input, although we rarely find this to be the case. We do not recommend adding >200 μL of input volume to GELFrEE cartridges, and samples that would surpass this value are either ruled out for our translational TDP experiments, or concentrated in spin filters.
-
24
Vortex each tube for 10–20 s, and heat it on a 95 °C heat block or in a thermal mixer for 5 min. Centrifuge the samples at 1,000g for 2 min at 4 °C to re-collect any condensation at the tube lids after heating.
-
25
Perform fractionation by GELFrEE by following the manufacturer’s protocol for 8% cartridges (the general GELFrEE manual can be found at https://p2v6h7b4.stackpathcdn.com/wp-content/uploads/2018/01/GELFREE-Manual.pdf and the 8% cartridge Quick Reference Card can be found at https://p2v6h7b4.stackpathcdn.com/wp-content/uploads/2018/01/GelFree-8pct-Quick-Ref-Card.pdf).
▲CRITICAL STEP As mentioned previously, we typically use fraction 1 from a GELFrEE 8% cartridge for large-scale TDPs, because it contains the robustly resolvable MW range (<30 kDa) of proteins all within one fraction. If more targeted subsets of MW below the 30-kDa range are required for analysis, other, higher-percentage cartridges will yield multiple fractions that are appropriate for TDMS and TDP. For example, we have analyzed multiple fractions of 10 and 12% cartridges either to detect a more low-abundant proteoform in a less complex fraction (TDMS) or to increase our overall coverage of the <30-kDa proteome with analysis of multiple fractions (TDP).
? TROUBLESHOOTING
-
26
Once fraction 1 of each sample has been collected into a new, labeled 1.5-mL Protein LoBind tube, the analyst must decide whether to continue collecting fractions according to the GELFrEE protocol, or to end the fractionation process if only the first fraction is desired.
▲CRITICAL STEP Often, for high-value patient samples from the clinic, it may be desirable to collect every fraction for each sample, if they may prove useful for future experiments or other proteomics analyses (i.e., BUP or western blots).
■PAUSE POINT This is a natural stopping point in the protocol, as GELFrEE fractions are stable for several months at −80 °C.
Quality control of GELFrEE fraction by SDS–PAGE with silver staining ● Timing ~2 h
▲CRITICAL At this point, we advise that the analyst visualize collected fractions by SDS–PAGE and silver staining as described by Schevchenko et al.68 in order to confirm that the fractionation process worked correctly and that fractions of the correct MW range are submitted for LC–MS/MS analysis.
▲CRITICAL Briefly, the silver staining protocol is as follows (with all steps conducted at room temperature unless otherwise stated).
-
27
Mix 5–10 μL of each fraction of interest with the appropriate amount of Laemmli sample buffer for the gel lane capacity in a new microcentrifuge tube. Boil these gel samples at 95 °C for 5 min and spin down on a benchtop microcentrifuge (5,000g, 20–25 °C, 30–60 s).
-
28
Separate the proteins in each fraction by MW using SDS–PAGE, being careful not to run lower-MW proteins off the gel that may be of interest for TDMS or TDP characterization. Use a commercially available MW standard ladder to gauge the MW range of each protein fraction after staining.
-
29
Remove the gel and submerge it in fixing solution in a clean container (e.g., a 15-cm Petri dish). Place on an orbital shaker for 20 min at low speed to fix the gel.
-
30
Pour the fixing solution out of the staining container and replace it with enough washing solution to submerge the gel. Place on an orbital shaker for 10 min at low speed to fix the gel.
-
31
Pour the washing solution out of the staining container and replace it with sensitizing solution. Place on an orbital shaker for 1 min at low speed to sensitize the gel.
-
32
Pour out the sensitizing solution and rinse the gel with two washes of dH2O for 1 min each.
-
33
Add enough silver nitrate solution to submerge the gel and incubate it for 20 min on an orbital shaker at low speed to stain the gel.
-
34
Pour out the silver nitrate solution and rinse the gel twice in dH2O for 1 min.
!CAUTION We advise collecting silver nitrate waste in a special waste container, to be disposed of according to institutional policies.
-
35
Add enough developing solution to submerge the gel, and develop under rapid shaking.
▲CRTICAL STEP Discard the developing solution and replace it with fresh developing solution, once the liquid turns yellow, to reduce background staining.
-
36
Pour out the developing solution and replace with terminating solution, once the desired staining intensity has been achieved, to terminate the development (Fig. 2b). The gel can now be scanned or photographed.
Desalting and concentrating GELFrEE fractions ● Timing ~2 h
-
37
To each fraction, add 4 volumes of Optima-grade methanol (600 μL), and vortex each tube vigorously for 20 s.
▲CRITICAL STEP In the following steps, the fractions will be desalted and concentrated by methanol–chloroform–water precipitation of proteins69. Our general procedure for this technique is described below.
-
38
Add 1 volume (150 μL) of chloroform to each fraction, and vortex vigorously for 20 s.
!CAUTION Handle chloroform in a biological safety cabinet, to avoid respiratory exposure.
-
39
Add 3 volumes (450 μL) of LC–MS-grade water to each fraction. The liquid should immediately become cloudy and white in color. Vortex vigorously for 20 s.
? TROUBLESHOOTING
-
40
Centrifuge the tubes at 21,000g for 10 min at 4 °C.
-
41
Carefully remove the tubes from the centrifuge. The separated aqueous and organic phases should be clearly observed, with a visible white protein pellet floating at the interface.
▲CRITICAL STEP Be very careful when transporting tubes to avoid shaking them or otherwise disturbing the protein pellet more than is necessary.
-
42
Using a pipette, remove and discard the top layer of solution, using extreme care to not remove or disturb the protein pellet. It may be necessary to leave a few millimeters of volume above the pellet to avoid accidentally pipetting it out.
-
43
Very slowly add 3 volumes (450 μL) of Optima methanol to the tube, to avoid breaking up the protein pellet more than is necessary. Slowly mix the solution by carefully rocking the tube back and forth three to five times, while keeping an eye on the protein pellet and being careful to break it up as little as possible.
▲CRITICAL STEP We try to keep the protein pellet as intact as possible to avoid accidentally removing proteins when removing all solvents later on in this protocol.
-
44
Centrifuge the tubes at 21,000g for 10 min at 4 °C.
-
45
At this point, the protein pellet should now be at the bottom of the tube. Carefully remove and discard as much methanol as possible from the tube without disturbing the pellet, and repeat steps 42–44 two more times.
-
46
After the final methanol wash, remove as much methanol as possible from the tube without accidentally removing protein pellet particles. Place open tubes in a tube rack in an operating biological safety cabinet for 5–10 min to evaporate off any excess methanol.
-
47
Cleaned and desalted protein pellets can now be resuspended in mobile phase A as a prerequisite to nano-UHPLC–MS/MS. Add enough mobile phase A to account for the number of injections planned per sample, with enough left over for an extra injection or two, if needed.
▲CRITICAL STEP We typically prepare enough sample for at least four 6-μL injections. Our normal rule of thumb is therefore to resuspend cleaned pellets resulting from the above preparative procedures in 35 μL of mobile phase A, as this provides enough volume for an extra injection even after slight sample loss in the pipette tip after vigorous pipetting. In addition, this volume typically results in a final total protein amount within the range of 0.5–1 μg of protein per injection, which is a ‘sweet spot’ that allows acceptable sensitivity and subsequent proteome coverage without overloading the LC columns and causing clogs that complicate downstream analysis.
▲CRITICAL STEP Resuspension of intact proteoforms in mobile phase A is more difficult than that of peptide pellets, and may take multiple rounds of vigorous pipetting. In addition, if pellet particles are observed to remain in solution after vigorous pipetting, the tubes can be sonicated on ice for 5 min.
-
48
Centrifuge the samples dissolved in mobile phase A at 21,000g for 5 min at 4 °C, and slowly remove the majority of the solution, pipetting from just below the surface to avoid disturbing any unnoticed particles. Transfer each sample to a separate, labeled, pre-chilled LC vial.
▲CRITICAL STEP We perform this centrifugation and transfer step to avoid moving any possible particulates into the final LC vial; such particulates may lodge in the LC lines and decrease performance. Therefore, it is paramount to ensure that the sample protein is completely dissolved, and that no particulates from the environment were introduced to the vial. It may be necessary to perform these steps in a functioning biological safety cabinet if this is an issue in the analyst’s lab.
-
49
Transport the vials on ice to the nano-UHPLC autosampler.
▲CRITICAL STEP It is critical to analyze the samples by LC–MS as soon as possible after resuspension in mobile phase A. We typically observe sample degradation and an associated decrease in data quality by 48–72 h after sample reconstitution. For certain less stable proteoforms, sample degradation can begin much quicker. In these cases, we recommend reconstituting the protein pellet immediately before injection onto the instrument and initiation of the method.
Ultra-HPLC coupled to tandem mass spectrometry ● Timing QC: ~2–3 h; PBMC samples: ~2 h per sample per replicate
-
50
If necessary, perform MS calibration as described in Box 3.
-
51
Prepare the UHPLC setup for the top-down experiment by performing the testing and optimization steps described in Box 1 after plumbing the LC and installing the column set as depicted in Fig. 4.
!CAUTION Ensure that all the tubing is compatible with all solvents of choice. Proteins have different sensitivities and affinities to various contaminants than do peptides. Dissolved plasticizers readily stick to proteins, creating additional peaks in the mass spectrum, dissipating ion current, and lowering the overall signal intensity of the analyte of interest. In addition, the masses of some of these species are similar to those of PTMs, thus complicating their discovery and characterization.
-
52
Using Chromeleon or another LC method manager, create a new UHPLC method and set up the gradient, following the guidelines set out in Table 1.
▲CRITICAL STEP We tailor the gradient specifically to the sample we plan to investigate, so as to obtain the maximum chromatographic performance. For PMBCs, we use a 90-min gradient (Table 1). For samples with higher protein complexity, the gradient could be prolonged accordingly until a plateau is reached for the given number of theoretical plates, which will in turn vary with different columns and column lengths. Note that by doubling the column length (and, hence, the number of theoretical plates), the efficiency of separation theoretically doubles; however, the final chromatographic resolution depends on the square root of efficiency, and thus will increase by only 1.4 times. Furthermore, longer columns will require an increased duration of the gradient and will produce higher backpressure.
▲CRITICAL STEP When setting the chromatographic method, particularly its total duration, consider that quantitative experiments will require multiple technical replicates of the same samples. Therefore, minimizing the gradient run time is critical for limiting the time that the sample will have to sit in the autosampler, as longer sample wait times may facilitate protein degradation, adsorption, or precipitation.
-
53
Set up the MS method in Xcalibur, following the considerations set out for top-down analysis in Box 2 and using the guidelines delineated in Table 2.
▲CRITICAL STEP Be sure to initiate MS acquisition in the LC software to align with the start of the analytical gradient.
-
54
Before analyzing samples, be sure to perform QC to monitor instrument performance by running three TD standards.
▲CRITICAL STEP The LC–MS parameters for the separate TD standard method are available for download at http://nrtdp.northwestern.edu/protocols.
-
55
Prepare a sequence in Xcalibur for the samples of interest, incorporating the LC–MS methods set up previously in this section.
▲CRITICAL STEP We run blanks in between different biological replicates to minimize carryover. For longer sequences, we run blanks between blocks of three or four injections to minimize the overall time of analysis. In addition, we typically randomize the injection order for any quantitative, discovery-mode experiments.
▲CRITICAL STEP We advise using mobile phase A as the blank sample in a separate vial and avoiding isopropanol blanks, as the latter could affect retention times due to the viscosity discrepancy between isopropanol and the elution buffer used for the gradient, which may be retained in the LC lines.
-
56
Initiate the sequence, and thus inject 0.5–1 μg of each sample in the order created in the previous step.
▲CRITICAL STEP We recommend triplicate injections for each sample of interest, especially if any downstream quantitation methodology is incorporated into the study.
? TROUBLESHOOTING
Box 3 |. MS calibration ● Timing ~1 h.
The correct calibration of the mass spectrometer is fundamental to ensuring high mass accuracy measurements and, thus, correct proteoform identification. The main steps of this procedure are as follows:
Mount the HESI source on the Orbitrap Elite, and fill a glass syringe (e.g., a 500-μL Hamilton syringe) with Pierce LTQ Velos ESI positive ion calibration solution (Calmix). Use an automatic syringe pump to obtain a 5 μL/min flow of Calmix.
Adjust the HESI settings to obtain a stable spray. This is essential for achieving correct calibrations of important devices such as electron multipliers and functions such as enhanced signal processing for FTMS. A typical set of parameters is 4 kV spray voltage, sheath gas flow at 7 arbitrary units, 275° C heated capillary temperature, and 60% S-lens radiofrequency. Note that the stability of the electrospray can be monitored using the specific function that is present in the Tune software under the ‘Diagnostics’ dialog box (i.e., ‘Monitor API Stability’).
Once the spray is stabilized, it is possible to proceed with the calibration, starting from the LTQ. When clean, ion optics of the source region can use default values and do not require special tuning. It is recommended to visualize the progress of the calibration using the ‘Plot Graph’ function offered by the Tune software. For the calibration, we use the ‘Semi-automatic calibration’ function. Importantly, a full calibration, which includes all items in the ‘LTQ – Positive ion’ list (with the exclusion of mass and resolution calibration for the LTQ modes ‘Enhanced’, ‘Zoom’, and ‘Ultra-zoom’), should be performed on a monthly basis. Conversely, the main LTQ items requiring a biweekly calibration check (followed by potential re-calibration) are the (i) transfer lenses and (ii) electron multiplier gain.
With regard to the FT portion of the calibration, we recommend performing monthly a full calibration of all the positive mode items. Weekly recalibration of (i) FT masses and (ii) predictive AGC is highly recommended. Note that on the Orbitrap Elite, the FT mass calibration is necessarily accompanied by the enhanced Fouriertransform (eFT) calibration.
Data processing and analysis ● Timing ProSight Lite: ~1–2 h per sample; TDPortal: ~6–8 h per dataset
-
57
Depending on the experiment, follow either option A for ProSight Lite analysis of TDMS data targeted to one or a few proteoforms of interest, or option B for TDPortal on the Quest platform for high-throughput TDP in discovery mode.
-
ProSight Lite (video tutorial available at the URL mentioned below)
Open the .raw file of interest in QualBrowser and navigate to the precursor and fragmentation scans for the proteoform interest. As an example, we depict the second isoform of prothymosin alpha (PTMA; accession no. P06454) in Fig. 6, which graphically lays out the ProSight Lite workflow.
Acquire the intact mass of the proteoform of interest, as well as a table of its fragment masses, by performing Xtract on the requisite scans to generate deisotoped and deconvoluted spectra for each.
In the new window for the Xtracted precursor scan, navigate to scan 2 to obtain the monoisotopic, zero-charge mass spectrum of the precursor and record this value for input into ProSight Lite.
In the PostXtract scan for fragmentation data, navigate to scan 2, right-click to bring upoptions, and then copy exact masses to the clipboard, using the Export menu. Paste the fragment masses into Microsoft Excel for import into ProSight Lite in the next step.
Open the ProSight Lite software (Equipment setup) and click on ‘Add Experimental Data’.
First, enter the value recorded for the Precursor. Then, copy the fragment mass data(column A) from the Excel Spreadsheet and paste them into the ‘Fragments’ box (these masses should appear as one per line). Set the parameters to those depicted in Fig. 6 (i.e., monoisotopic precursor mass type, M (neutral) mass mode, HCD fragmentation method, and 10 p.p.m. fragmentation tolerance).
Click on ‘Add Candidate Sequence’, and enter the sequence of the protein of interest in the text box. Alternatively, input the UniProt accession number of the protein of interest to import the sequence from the UniProt database.
-
Results are displayed, including the graphical fragment map and key scoring metrics for theproteoform. Results can be saved for future use or exported to Excel, and fragmentation maps can be downloaded in SVG or PNG formats.
▲CRITICAL STEP A video detailing the ProSight Lite workflow step by step as it appears on the computer screen can be accessed at http://prosightlite.northwestern.edu. ref. 45 also provides a more detailed method for how to perform this analysis.
-
TDPortal
▲CRITICAL A detailed guide to TDPortal for external users can be downloaded from http://nrtdp.northwestern.edu/wp-content/uploads/2016/11/ExternalUserNov29.pdf. A video tutorial is also available at https://www.youtube.com/watch?v=1XHpF-W9Zeo.
Obtain access to TDPortal (Equipment setup), and transfer files to the portal using WinSCP by creating a new site. Use the SFTP file protocol, port number 22, and portal.nrtdp.northwestern.edu as the host name. The TDPortal administrator will supply each user with a username and password.
-
Once the above parameters have been set, log in and create a folder containing the .raw files generated by LC–MS that are to be searched.
▲CRITICAL STEP Be sure to place all .raw files within a subfolder titled ‘Samples’, so that the TDPortal software can detect them.
Use the username and password information supplied by the administrator to log into TDPortal at http://portal.nrtdp.northwestern.edu.
-
Perform the TDPortal search (Step 57B(iv–xv)) (refer to Fig. 7 for a detailed workflow with helpful screenshots). Create a new history in the ‘History’ panel on the right side of the screen by clicking on the gear icon and then clicking on ‘Create New’ in the drop-down menu.
▲CRITICAL STEP We recommend renaming the new history for easier identification by clicking on the Unnamed History heading.
Click on ‘NRTDP Tools’ in the ‘Tools’ panel on the left side of the screen, and then click on ‘Create Linked Library’ in the drop-down menu.
In the new window, select the dataset to be searched, and click ‘Execute’.
Navigate to the ‘Shared Data’ subheading in the ‘Galaxy’ toolbar at the top of the screen, and click ‘Data Libraries’ in the drop-down menu to view the linked library.
In the new window, click on the desired dataset library to view all the .raw files in the dataset. Using the checkboxes to the left of each file, select which files to import to the history. Then, navigate to the ‘For Selected Datasets’ drop-down menu at the bottom of the page and select ‘Import to current history’, then click ‘Go’. A new window depicting a green checkbox should confirm that the datasets of interest were successfully imported to the correct history.
Return to the homepage by clicking the ‘Galaxy’ icon at the top left of the screen, and confirm that the ‘History’ panel has been populated with green boxes for every imported .raw file.
Click on the checkbox below the name of the history, and click the ‘All’ button to select all imported files. Create a dataset list by then clicking the ‘For all selected’ button and selecting ‘Build Dataset List’ in the drop-down menu. The new dataset list should appear in the ‘History’ panel.
Navigate to the ‘Shared Data’ subheading again and click on ‘Published Workflows’ in the drop-down menu.
-
Import the ‘Published Standard Search Workflow 1.3’ after clicking on the down arrow of the ‘Standard Search Workflow Version 1.3’ button. A new window depicting a green checkbox should confirm that the workflow has been successfully imported.
▲CRITICAL STEP Use the most updated search workflow available for external users if v.1.3 is no longer available or becomes redundant in the future.
Navigate to the ‘Workflow’ subheading in the top toolbar. In the drop-down menu for ‘Standard Search Workflow Version 1.3’, click ‘Run’.
-
In the next window, select the created dataset list under the ‘Step 1’ heading. Under the ‘Step 3’ heading, select ‘homo sapiens’ in the ‘Organism’ drop-down menu, and ‘High Resolution’ in the ‘Precursor Resolution’ drop-down menu. Finally, click ‘Run Workflow’.
▲CRITICAL STEP The search will be added to the ‘History’ panel on the Galaxy home screen, and the progress of the search can be monitored by clicking on the ‘Check Progress’ button found under ‘mzML Zip file’.
-
To view the ‘Top Down Report’ after the search has finished, click the ‘Download’ button (floppy disk icon) under the ‘Generated Top Down Report’ section in the ‘History’ panel. Downloaded results can then be viewed in Top Down Viewer (Equipment setup).
? TROUBLESHOOTING
-
Troubleshooting
Troubleshooting advice can be found in Table 3.
Table 3 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| Sample preparation | |||
| 13 | Lysate will not homogenize | Benzonase is not working properly | Make sure MgCl2 is present in the lysis buffer, as this is a cofactor for benzonase. If EDTA or other chelators have been added, bring the final concentration of MgCl2 to 10 mM to compensate. Incubation time with benzonase can also be increased to 30 min, with pipetting every 10 min to homogenize the lysate |
| 16 | No discernable pellet noted after acetone precipitation | Low protein yield | Precipitate the sample overnight to increase the yield of salvageable protein |
| 17 | Precipitates are still present after multiple attempts at dissolution | Precipitated proteins are not properly dissolving back into solution | Precipitates in 1% (wt/vol) SDS can be placed in a 95 °C heat block or thermal mixer for no more than 5 min, to aid in dissolution without destroying PTMs or breaking down protein structure |
| 25 | A GELFrEE lane containing sample does not show any measurable current when the procedure is started | There is an air bubble in either the sample or the collection chamber | Use a pipette to gently aspirate sample out of the sample chamber or buffer out of the collection chamber, and carefully replace, being careful to avoid introducing any bubbles into the system |
| The GELFrEE cartridge lane is faulty, due to mechanical error in the production process | Move sample from the sample chamber to a free lane in the GELFrEE cartridge that exhibits a consistent current when activated | ||
| 39 | A high degree of SDS adducts (+265 Da) are noted in spectra across the chromatogram after analysis | SDS was not completely removed during the desalting/washing process | Add an extra methanol wash to the methanol/ chloroform/water precipitation protocol |
| In some cases of persistent adducts, we have found it helpful to halt the GELFrEE procedure and wash the collection chamber with running buffer immediately before the dye front elutes | |||
| LC | |||
| 56 | Peak broadening/peak tailing | Sample overloading | Adjust the injected volume or dilute the sample |
| Presence of dead volume | Check all connections of the fluidics, particularly the post-column ones; make sure that all o.d. and i.d. of lines and unions and/or sleeves match; verify valve functionality and check for potential leaks between the rotor seal and the stator | ||
| Worn trap column | Replace with a new trap column | ||
| Shifts in protein retention times | Presence of air in the pump blocks | Purge the NC pump blocks and the flow meter | |
| The flow meter is not working properly | Purge the flow meter; check the pressure transducer calibration and the viscosity calibration | ||
| Leak in the system | Verify the quality of connection; run a detailed leak test using the LC system software (if available) | ||
| Progressive increase in the backpressure of the trap column over the runs | The trap column is being overloaded | If the increase in backpressure is <15–20%, simply reduce the volume of injected samples; otherwise, replace the trap column | |
| The total ion chromatogram shows that all proteins in the sample elute in a very narrow time window | The flow meter requires a new calibration | Perform a new viscosity measurement calibration | |
| The pressure sensor is out of calibration | Recalibrate the pressure transducer and, eventually, also repeat the viscosity measurement | ||
| The LC solvents are old and their composition is altered | Prepare fresh solvents, purge the system, and proceed to a new pressure transducer and viscosity measurement calibration | ||
| Orbitrap FTMS | |||
| 56 | Poor spray stability | Clogged emitter/clogged high-voltage union | Replace the emitter and/or high-voltage union |
| Wrongly positioned emitter | Position the spray emitter 1–2 mm away from the heated capillary of the mass spectrometer | ||
| Bubbling emitter, causing intermittent spray | Pack a new emitter with ~5 mm of PLRP-S resin, checking with a microscope that it is completely filled to the end and that there are no clogs | ||
| Poor protein MS2 fragmentation | HCD energy is too low/too high | Recalibrate the HCD collision energy using the calibration solution. Check the N2 pressure in the HCD cell | |
| Wrong AGC settings | Switch off the ‘predictive AGC’ function and set the AGC target for MS2 to 1 × 106 | ||
| MS1 spectra of protein show repeated satellite masses not corresponding to known PTMs | Adducts derived from poor sample cleanup (e.g., SDS adducts, +265 Da) | Re-prepare samples from GELFrEE | |
| Adducts are contaminants from the LC solvents | Prepare new solvents and purge the LC system | ||
| Adducts are produced by a worn or contaminated high-voltage union | Replace the high-voltage union and, possibly, the spray emitter | ||
| Proteins in MS1 appear highly oxidized | The terminal surface of either the capillary connecting the analytical column to the high-voltage union or the spray emitter is not even | Recut with a ceramic blade and eventually polish the terminal ends of these capillaries | |
| Malfunctioning high-voltage union | Replace the high-voltage union | ||
| Contaminants such as metallic shavings are present in the packed spray emitter | Replace the spray emitter | ||
| Progressive reduction of the signal intensity over the runs; loss in sensitivity | Charge has accumulated on one or more ion optics elements, such as ion lenses or multipoles | Perform an ‘ion optics charging evaluation’ test, and eventually vent the mass spectrometer and clean the dirty optics (or contact a field engineer) | |
| Overloaded trap column | Replace the trap column and run a top-down standard to verify that the signal intensity is back to normal levels | ||
| Data analysis | |||
| 57B | The number of identified proteoforms/accessions is low, especially considering that the chromatogram seems rich in proteins | There is a problem with mass calibration | Verify this by running a new database search with less strict fragment tolerance (e.g., 20 p.p.m.). If needed, recalibrate the instrument |
| The dynamic exclusion settings do not follow the chromatographic performance | Assess the average peak width and adjust the dynamic exclusion time accordingly; this time cannot be shorter than the time of elution | ||
| Proteoforms are identified, but their C score is low | Adjust fragmentation energy | Repeat the calibration of HCD energy and eventually re-run the whole study | |
Timing
Steps 1–10, blood collection, PBMC isolation, and two application-specific methods for cell storage: ~2 h
Steps 11–20, cell lysis, protein precipitation, and lysate quantitation: ~3 h
Steps 21–49, protein fractionation by GELFrEE, QC by SDS–PAGE with AgNO3 stain, and desalting for downstream nano-UHPLC–MS/MS: ~7 h
Steps 50–56, ultra-HPLC coupled to tandem mass spectrometry: ~2 h per sample per replicate and ~5–7 h for setup and testing and initial QC
Step 57, data processing and analysis: ~1–2 h per sample for targeted TDMS; ~6–8 h per dataset for discovery mode TDP
Box 1, LC setup and testing: ~4–5 h
Box 3, MS calibration: ~1 h
Anticipated results
In this section, we show results from an experiment in which we analyzed human PBMCs from three donors prepared according to both sample streams in the provided protocol: nonviable cells stored in RNAlater and viable cells stored in traditional freezing medium after cell collection and isolation. ForSboth samples, two blood draws of 5–8 mL were collected from three healthy donors. Informed consent was obtained from each participant, and the corresponding institutional ethical committees approved the study protocol. PBMCs were isolated from each sample as described above, and one sample from each donor was treated according to the viable cell sample stream protocol and the other was treated by the nonviable sample stream protocol. For all samples, 100 μg of PBMC protein was the input into the fractionation workflow, and the desalted <30 kDa fractions were analyzed with quadruplicate injections using the LC and MS methods described above. Both data analysis streams were conducted for demonstrative purposes: the entire dataset was analyzed as a high-throughput experiment on TDPortal, and proteoforms of an immune-relevant protein family, PTMA (accession no. P06454) were chosen for targeted TDMS analysis by ProSight Lite (fragment map displayed in Fig. 6).
Data from a representative .raw file from this experiment are displayed in Fig. 8. The HPLC base peak chromatogram in Fig. 8a depicts a complex pattern of elution peaks corresponding to proteoforms from the human PBMC sample fraction as they elute over time and are detected in the mass spectrometer. In our experience, the two broad clusters of peaks showing the majority of proteoforms eluting between 10 and 30 min, or around the 40-min mark, are typical for this sample type and are broadly reflective of the chemical and physical properties of the PBMC proteome in this MW range. The MS spectra acquired over the most abundant elution peak (Fig. 8a, red asterisk), which is primarily composed of PTMA proteoforms, were summed and depicted in Fig. 8b. This parent MS1 spectrum is commonplace in TDMS analysis, in which the partitioning of signal from proteoforms (with many more ionizable residues than tryptic peptides) into multiple charge states is observed after ESI. The value of proteoform-resolved measurements is most apparent in Fig. 8c, where we selected only the m/z window containing a cluster of 8+ charge state proteoforms of PTMA (Fig. 8b, green asterisk). As evidenced by this spectrum, in which each different PTMA proteoform is signified by its distinctive isotopic distribution cluster, several proteoforms of this parent protein exist in the sample and can be identified after database searching. The specificity of analysis afforded by TDP adds great complementary value to the higher depth of coverage achievable by BUP, with which it would be very difficult to determine exactly which proteoforms of PTMA are present in the sample.
Fig. 8 |. Display of top-down data from a representative .raw file.
a, The LC base peak chromatogram for the <30-kDa fraction of a human PBMC sample using the method described in this protocol is displayed. A large number of elution peaks, consisting of proteoform signals as they are detected over the course of the LC gradient, can be observed between the 10- and 60-min marks, which is typical for this sample type. b, The MS spectrum of the most abundant elution peak from a (denoted by the red asterisk) was summed across the peak and is displayed here. Multiple charge states of the proteoforms of prothymosin alpha, an immune-relevant protein family, were detected over this retention time window. c, The m/z window containing the 8+ charge states from b (denoted by the green asterisk) is selectively displayed, showing the diversity and abundance of various proteoforms of prothymosin alpha that were detected, each represented by a different isotopic distribution cluster. Met-OFF, Methionine-off.
After database searching via TDPortal, data were visualized in Top Down Viewer. For the combined study and at a FDR threshold of 1%, 179 protein groups/UniProt KB accession numbers were identified, corresponding to 769 characterized proteoforms. Of these proteoforms, 372 were characterized with a C score >40, which means that 48% of the proteoforms yielded by the entire study were unambiguously characterized. In Figs. 9 and 10, we compare data from the viable and nonviable cell preparations for the three donors, as a way to visualize any differences in the final result due to the diverging cell treatment and storage methods. Interestingly, although both sample treatments yielded a similar number of UniProt KB accession numbers (133 identified in the viable PBMCs and 138 identified in the nonviable PBMCs), a markedly increased number of proteoforms wereRE PROTOCCdiscovered in the nonviable (619 proteoforms) compared to viable (374 proteoforms) PBMCs. This perhaps suggests that this cell lysis and treatment method may yield a broader snapshot of the proteoform-resolved content of the PBMC proteome, or conversely that the high salt concentrations in RNA-stabilizing reagents may induce chemical changes at the bench that create artifactual proteoforms. The application of variation components modeling by ANOVA of proteoform intensity values across the files demonstrated a substantial increase in the sample-level variation due to preanalytical variables, suggesting that the viable cell preparation method may be more appropriate for quantitative contexts.
Fig. 9 |. Qualitative comparison of identified protein accession numbers and characterized proteoforms in viable and nonviable PBMC samples.
Qualitative comparison of identified (a) protein accession numbers and (b) characterized proteoforms in viable and nonviable PBMC samples. Two whole-blood draws each from the same three patients were prepared according to either the viable cell protocol or the nonviable cell protocol. A comparison of the total list of identified gene products (each referring to a unique accession number from the UniProt KnowledgeBase (KB)) and related characterized proteoforms (indicated by unique proteoform record identifiers, or PFRs) shows that the two alternative sample-processing schemes yield substantially different results, especially at the proteoform level. Multiple PFRs can refer to a single accession number, and the characterization of four to five different proteoforms for each identified accession number is common in top-down proteomics. Notably, many more proteoforms were specifically characterized in the nonviable samples, in comparison to the viable samples.
Fig. 10 |. Histogram comparisons of key metrics for protein identification and proteoform characterization in viable and nonviable blood samples.
a, A histogram of protein q values, which score the confidence of identification and subsequent assignment of the UniProt accession number, was created to compare the viable samples (orange bars) to nonviable samples (blue bars). b, A histogram of proteoform C scores, which score the confidence of proteoform characterization, was generated to compare viable (orange) and nonviable (blue) samples. c, Finally, a histogram of proteoform mass values was generated to compare viable (orange) and nonviable (blue) samples. In general, both sample types reflected the same trends in all three metrics, with the primary difference being attributed to the increase in proteoforms detected in the nonviable samples.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary.
Acknowledgements
We thank the following members of the Kelleher Research Group and Proteomics Center of Excellence for helpful discussions and experimental assistance: R. Fellers, J. Greer, P. Compton, and P. Thomas. We also acknowledge the Northwestern Comprehensive Transplant Center Biorepository Core. This work was supported by the Paul G. Allen Family Foundation (grant award 11715 to N.L.K.), and the National Institutes of Health via the National Resource for Translational and Developmental Proteomics under grant P41 GM108569 from the National Institute of General Medical Sciences. T.K.T. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award no. T32GM105538, as well as by an American Chemical Society Division of Analytical Chemistry Fellowship, sponsored by the Society for Analytical Chemists of Pittsburgh.
Footnotes
Competing interests
N.L.K. declares an affiliation with Thermo Fisher Scientific. The remaining authors declare no competing interests.
Data availability
The .raw files converted to .mzML format used for this analysis are accessible at https://scholarworks.iu.edu/dspace/handle/2022/21091. In addition, the well-characterized proteoforms (C score >40) elucidated here are available for public access in the Consortium for Top Down Proteomics proteoform repository (http://atlas.topdownproteomics.org/proteoforms/?DataSetId=15).
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-018-0085-7.
Referenrereces
- 1.Savaryn JP, Toby TK & Kelleher NL A researcher’s guide to mass spectrometry-based proteomics. Proteomics 16, 2435–2443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liumbruno G, D’Alessandro A, Grazzini G & Zolla L Blood-related proteomics. J. Proteomics 73, 483–507 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Zhu P, Bowden P, Zhang D & Marshall JG Mass spectrometry of peptides and proteins from humanblood. Mass Spectrom. Rev. 30, 685–732 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y et al. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 113, 2343–2394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesvizhskii AI & Aebersold R Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Savaryn JP et al. The emergence of top-down proteomics in clinical research. Genome Med. 5, 53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith LM & Kelleher NL Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compton PD, Zamdborg L, Thomas PM & Kelleher NL On the scalability and requirements of whole protein mass spectrometry. Anal. Chem 83, 6868–6874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toby TK, Fornelli L & Kelleher NL Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem 9, 499–519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeDuc RD et al. ProForma: a standard proteoform notation. J. Proteome Res 17, 1321–1325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunger MK et al. Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal. Chem 80, 1459–1467 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Li Y et al. Optimizing capillary electrophoresis for top-down proteomics of 30–80 kDa proteins. Proteomics 14, 1158–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson JT, Wenger CD, Metcalf WW & Kelleher NL Top-down proteomics reveals novel proteinforms expressed in Methanosarcina acetivorans. J. Am. Soc. Mass Spectrom 20, 1743–1750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellie JF et al. Robust analysis of the yeast proteome under 50 kDa by molecular-mass-based fractionation and top-down mass spectrometry. Anal. Chem 84, 209–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng F et al. Processing complex mixtures of intact proteins for direct analysis by mass spectrometry. Anal. Chem 74, 2923–2929 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Meng F et al. Molecular-level description of proteins from Saccharomyces cerevisiae using quadrupole FT hybrid mass spectrometry for top down proteomics. Anal. Chem 76, 2852–2858 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ntai I et al. Applying label-free quantitation to top down proteomics. Anal. Chem 86, 4961–4968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran JC & Doucette AA Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal. Chem 80, 1568–1573 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Tran JC & Doucette AA Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Anal. Chem 81, 6201–6209 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Hardman M & Makarov AA Interfacing the orbitrap mass analyzer to an electrospray ion source. Anal. Chem 75, 1699–1705 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Hu Q et al. The Orbitrap: a new mass spectrometer. J. Mass Spectrom 40, 430–443 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Michalski A et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111.011015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalski A et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics 11, O111.013698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen JV et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senko MW et al. Novel parallelized quadrupole/linear ion trap/Orbitrap tribrid mass spectrometer improving proteome coverage and peptide identification rates. Anal. Chem 85, 11710–11714 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ahlf DR et al. Evaluation of the compact high-field orbitrap for top-down proteomics of human cells. J. Proteome Res 11, 4308–4314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai W et al. MASH Suite Pro: a comprehensive software tool for top-down proteomics. Mol. Cell. Proteomics 15, 703–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guner H et al. MASH Suite: a user-friendly and versatile software interface for high-resolution mass spectrometry data interpretation and visualization. J. Am. Soc. Mass Spectrom 25, 464–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kou Q, Xun L & Liu X TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 32, 3495–3497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leduc RD & Kelleher NL Using ProSight PTM and related tools for targeted protein identification and characterization with high mass accuracy tandem MS data. Curr. Protoc. Bioinformatics Chapter 13: Unit 13.6 (2007). [DOI] [PubMed] [Google Scholar]
- 31.LeDuc RD et al. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 32, W340–W345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X et al. Protein identification using top-down. Mol. Cell. Proteomics 11, M111.008524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor GK et al. Web and database software for identification of intact proteins using “top down” mass spectrometry. Anal. Chem 75, 4081–4086 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Zamdborg L et al. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 35, W701–W706 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellie JF et al. Quantitative measurement of intact alpha-synuclein proteoforms from post-mortem control and Parkinson’s disease brain tissue by intact protein mass spectrometry. Sci. Rep 4, 5797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laouirem S et al. Progression from cirrhosis to cancer is associated with early ubiquitin post-translational modifications: identification of new biomarkers of cirrhosis at risk of malignancy. J. Pathol 234, 452–463 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Martelli C et al. Integrated proteomic platforms for the comparative characterization of medulloblastoma and pilocytic astrocytoma pediatric brain tumors: a preliminary study. Mol. Biosyst 11, 1668–1683 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Zhang J et al. Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J. Proteome Res 10, 4054–4065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desiderio C et al. Cerebrospinal fluid top-down proteomics evidenced the potential biomarker role of LVV- and VV-hemorphin-7 in posterior cranial fossa pediatric brain tumors. Proteomics 12, 2158–2166 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Cabras T et al. Significant modifications of the salivary proteome potentially associated with complications of Down syndrome revealed by top-down proteomics. Mol. Cell. Proteomics 12, 1844–1852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iavarone F et al. Characterization of salivary proteins of schizophrenic and bipolar disorder patients bytop-down proteomics. J. Proteomics 103, 15–22 (2014). [DOI] [PubMed] [Google Scholar]
- 42.De Petris L et al. Tumor expression of S100A6 correlates with survival of patients with stage I non-small-celllung cancer. Lung Cancer 63, 410–417 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Florell SR et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol. Mod. Pathol 14, 116–128 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Savaryn JP et al. Comparative top down proteomics of peripheral blood mononuclear cells from kidney transplant recipients with normal kidney biopsies or acute rejection. Proteomics 16, 2048–2058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toby TK et al. Proteoforms in peripheral blood mononuclear cells as novel rejection biomarkers in liver transplant recipients. Am. J. Transplant 17, 2458–2467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeHart CJ et al. Bioinformatics analysis of top-down mass spectrometry data with ProSight Lite. Methods Mol. Biol 1558, 381–394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fellers RT et al. ProSight Lite: graphical software to analyze top-down mass spectrometry data. Proteomics 15, 1235–1238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeDuc RD et al. The C-score: a Bayesian framework to sharply improve proteoform scoring in high-throughput top down proteomics. J. Proteome Res 13, 3231–3240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rader JS et al. A unified sample preparation protocol for proteomic and genomic profiling of cervical swabs to identify biomarkers for cervical cancer screening. Proteomics Clin. Appl 2, 1658–1669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson LC et al. Identification and characterization of human proteoforms by top-down LC-21 Tesla FT-ICR mass spectrometry. J. Proteome Res 16, 1087–1096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ntai I, Toby TK, LeDuc RD & Kelleher NL A method for label-free, differential top-down proteomics. Methods Mol. Biol 1410, 121–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denisov E, Damoc E, Lange O & Makarov A Orbitrap mass spectrometry with resolving powers above1,000,000. Int. J. Mass Spectrom 325, 80–85 (2012). [Google Scholar]
- 53.Olsen JV et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Durbin KR et al. Quantitation and identification of thousands of human proteoforms below 30 kDa. J. Proteome Res 15, 976–982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afgan E et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giardine B et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X et al. Deconvolution and database search of complex tandem mass spectra of intact proteins: acombinatorial approach. Mol. Cell. Proteomics 9, 2772–2782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carvalho PC et al. YADA: a tool for taking the most out of high-resolution spectra. Bioinformatics 25, (2734–2736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng F et al. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nat. Biotechnol. 19, 952–957 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Toby TK et al. Proteoforms in peripheral blood mononuclear cells as novel rejection biomarkers in liver transplant recipients. Am. J. Transplant 17, 2458–2467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kessner D et al. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higdon R, Haynes W & Kolker E Meta-analysis for protein identification: a case study on yeast data. OMICS 14, 309–314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fornelli L et al. Analysis of intact monoclonal antibody IgG1 by electron transfer dissociation Orbitrap FTMS. Mol. Cell. Proteomics 11, 1758–1767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fornelli L et al. Advancing top-down analysis of the human proteome using a benchtop quadrupole Orbitrap mass spectrometer. J. Proteome Res 16, 609–618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durbin KR et al. Autopilot: an online data acquisition control system for the enhanced high-throughput characterization of intact proteins. Anal. Chem 86, 1485–1492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richards AL et al. One-hour proteome analysis in yeast. Nat. Protoc 10, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fornelli L et al. Top-down analysis of immunoglobulin G isotypes 1 and 2 with electron transfer dissociationon a high-field Orbitrap mass spectrometer. J. Proteomics 159, 67–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shevchenko A, Wilm M, Vorm O & Mann M Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Wessel D & Flugge UI A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]