Abstract
The threat of antibiotic resistance has attracted strong interest during the last two decades, thus stimulating stewardship programs and research on alternative antimicrobial therapies. Conversely, much less attention has been given to the directly related problem of resistance toward antiseptics and biocides. While bacterial resistances toward triclosan or quaternary ammonium compounds have been considered in this context, the bis-biguanide chlorhexidine (CHX) has been put into focus only very recently when its use was associated with emergence of stable resistance to the last-resort antibiotic colistin. The antimicrobial effect of CHX is based on damaging the bacterial cytoplasmic membrane and subsequent leakage of cytoplasmic material. Consequently, mechanisms conferring resistance toward CHX include multidrug efflux pumps and cell membrane changes. For instance, in staphylococci it has been shown that plasmid-borne qac (“quaternary ammonium compound”) genes encode Qac efflux proteins that recognize cationic antiseptics as substrates. In Pseudomonas stutzeri, changes in the outer membrane protein and lipopolysaccharide profiles have been implicated in CHX resistance. However, little is known about the risk of resistance toward CHX in oral bacteria and potential mechanisms conferring this resistance or even cross-resistances toward antibiotics. Interestingly, there is also little awareness about the risk of CHX resistance in the dental community even though CHX has been widely used in dental practice as the gold-standard antiseptic for more than 40 years and is also included in a wide range of oral care consumer products. This review provides an overview of general resistance mechanisms toward CHX and the evidence for CHX resistance in oral bacteria. Furthermore, this work aims to raise awareness among the dental community about the risk of resistance toward CHX and accompanying cross-resistance to antibiotics. We propose new research directions related to the effects of CHX on bacteria in oral biofilms.
Keywords: antiseptic, chlorhexidine, CHX, resistance, adaptation, tolerance, persistence, efflux pump
Introduction
The 2016 Review on Antimicrobial Resistance has predicted an alarming scenario that the number of annual deaths attributable to antimicrobial resistance will increase globally from the current 700,000 to 10 million in the year 2050 (O’Neill, 2016). In this light, resistance toward antibiotics has attracted strong interest in the scientific as well as the medical community during the last few decades, and provided a major stimulus for establishing stewardship programs (Allerberger et al., 2009) and in the search for alternative antimicrobial measures such as antimicrobial peptides, natural compounds, cold atmospheric plasma, or light-based approaches (Karygianni et al., 2015; Czaplewski et al., 2016; Wainwright et al., 2017; Cieplik et al., 2018a). Furthermore, the oral cavity has recently been highlighted as a potential reservoir of antibiotic resistance genes that can be transferred via horizontal gene transfer among the bacteria present in oral biofilms (Roberts and Mullany, 2010; Al-Ahmad et al., 2014; Jiang et al., 2018).
Conversely, much less attention has been paid to the directly related problem of resistance toward the different classes of antiseptics and biocides (Forman et al., 2016; Kampf, 2016; Venter et al., 2017). This is somewhat surprising as bacterial resistance toward commonly used antiseptics such as benzalkonium chloride (BAC; a quaternary ammonium compound, QAC), triclosan (TCS; a polychloro phenoxy phenol) and chlorhexidine (CHX), as well as induction of cross-resistances between these agents and a range of clinically important antibiotics, have been known for many years (Yamamoto et al., 1988; McDonnell and Russell, 1999; Stickler, 2002; Russell, 2004; Yazdankhah et al., 2006).
While TCS has already been banned from its use in household wash products by the FDA in 2016 due to its risk of triggering cross-resistances (McNamara and Levy, 2016) and resistance toward QACs like BAC has also been highlighted in numerous reports (e.g., Buffet-Bataillon et al., 2012; Jaglic and Cervinkova, 2012), CHX has come into the spotlight only recently (Kampf, 2016; Venter et al., 2017; Wand et al., 2017). For example, it was reported that exposure to CHX may be associated with emergence of stable resistance to the last-resort antibiotic colistin, which may be associated with mutations in the two-component regulator phoPQ (Wand et al., 2017). The lack of focus on CHX resistance is somewhat surprising since it was found as early as 1980 that the extensive and liberal use of CHX in the management of patients with long-term bladder catheters resulted in urinary tract infections from CHX-resistant Gram-negative bacteria (i.e., strains of Proteus mirabilis, Providencia stuartii, and Pseudomonas aeruginosa), which were also found to be multidrug-resistant (Stickler and Thomas, 1980; Stickler, 2002). More recently, a high prevalence of reduced susceptibility toward CHX (69%) was found in microorganisms that caused central line-associated bloodstream infections in a large academic medical center with significantly higher proportion in patients who received daily CHX bathing (86% vs. 64%; p = 0.028) (Suwantarat et al., 2014).
Interestingly, the use of CHX in the field of dentistry for oral biofilm control has not been considered so far in this context as a possible source for the development of resistance against CHX itself or cross-resistances against other antiseptics or antibiotics (Sreenivasan and Gaffar, 2002). This is in spite of the widespread use of CHX by dental clinicians as gold-standard antiseptic (Jones, 1997), e.g., for plaque control and managing gingivitis (Van der Weijden et al., 2015) or for treatment of periodontitis (Teughels et al., 2009). In addition, CHX is included in a wide range of oral care consumer products (Sanz et al., 2013; Cieplik et al., 2018b).
Therefore, the objectives of this work are to review the evidence for resistance in oral bacteria toward CHX and to summarize our understanding of potential resistance mechanisms toward CHX. This review further aims to raise awareness among the dental community about the potential risk of resistance toward CHX as well as concomitant cross-resistances toward antibiotics and to propose future research directions related to CHX resistance in oral bacteria.
Chemistry and Historical Aspects
Chlorhexidine is a symmetric bis-biguanide molecule comprising two chloroguanide chains that are connected by a central hexamethylene chain and carries two positive charges at physiological pH (see Figure 1A for structural formula). It acts as a strong base and is practically insoluble in water but reacts with acids to form salts that exhibit varying water solubility characteristics (Davies et al., 1954; Denton, 2001; Lim and Kam, 2008). Today, mostly the digluconate or diacetate salts are used, whereby the water solubility of CHX digluconate is considerably higher than that of CHX diacetate (50% w/v vs. 2% w/v) (Kampf, 2018b). For the sake of clarity, in the present paper the acronym CHX will be used for all of its salts.
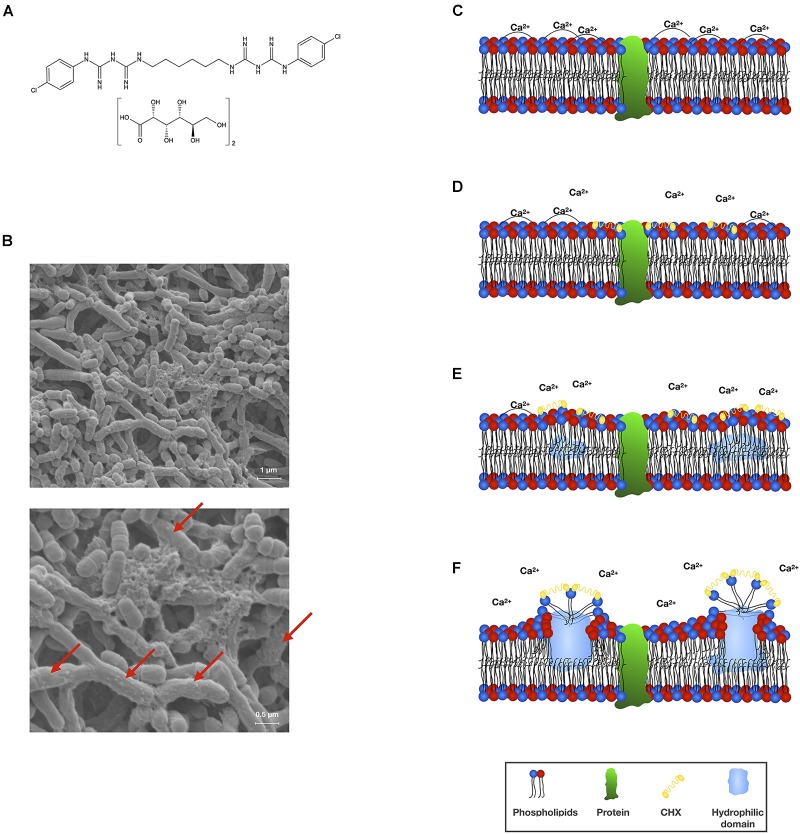
FIGURE 1.
(A) Chemical structural formula of CHX digluconate. (B) Scanning electron microscopic (SEM) visualization of an in vitro polymicrobial biofilm comprising Actinomyces naeslundii, Actinomyces odontolyticus, and S. mutans (methodology as described in Cieplik et al., 2018b,c) following treatment with CHX (0.2%; 10 min). Vesicle-like structures on the surfaces of bacterial cells indicate membrane damage (indicated by red arrows). SEM images are reprinted from Cieplik et al. (2018b). (C–F) Scheme depicting the mode of action of CHX toward bacterial cytoplasmic membranes. The bacterial cytoplasmic membrane carries a net negative charge and is composed of a phospholipid bilayer with embedded proteins. The phospholipid bilayer is stabilized by divalent cations such as Ca2+ and forms a hydrophobic environment, which is essential to moderate the functionality of the embedded proteins (C). CHX (as a cationic agent) binds to the negatively charged bacterial cell surface and initially interacts with the cytoplasmic membrane. Thereby, CHX bridges between pairs of phospholipid headgroups and displaces the associated divalent cations (D). Progressive decrease in fluidity of the outer phospholipid layer with creation of hydrophilic domains within the bilayer affecting the osmoregulation and metabolic activity of the cytoplasmic membrane and its associated enzymes (E,F). This scheme was adopted and modified from Gilbert and Moore (2005).
Chlorhexidine was first synthesized in Great Britain in the early 1950s within the scope of a broad screening exercise for active agents against malaria by Imperial Chemical Industries (Manchester, United Kingdom) (Lim and Kam, 2008; Kampf, 2016). It was described by Davies et al. (1954) under the name “Hibitane®” as “a new antibacterial agent of high potency” using its diacetate and dihydrochloride salts to ensure water solubility. Since then CHX has extensively been used in various medical fields (e.g., urology, gynecology, ophthalmology, otolaryngology) as well as for disinfection of surgical environments before it was introduced to dentistry in the late 1960s (Gjermo, 1974).
Antibacterial Mechanism of Action
In general, it is often difficult to distinguish the primary mechanism of action of a given antiseptic or biocide from secondary effects that are merely a consequence of the action (Maris, 1995). CHX is usually described to act as a bacteriostatic agent at low concentrations and to be bactericidal at higher concentrations (Jones, 1997). Its antibacterial mechanism of action is described as damage of bacterial membranes and subsequent leakage of cytoplasmic components (see Figure 1B for scanning electron microscopic images of CHX-mediated damage to bacterial cell surfaces) (McDonnell and Russell, 1999; Gilbert and Moore, 2005). First-line targets of CHX (at lower concentrations) are cytoplasmic membrane integrity as well as function of membrane-bound enzymes (see Figure 1C–F for a scheme describing the detailed interaction of CHX with bacterial cytoplasmic membranes), while secondary effects (at higher concentrations) are cytoplasmic leakage and, ultimately, coagulation and precipitation of intracellular constituents such as proteins and nucleic acids (Denyer and Stewart, 1998; McDonnell and Russell, 1999; Gilbert and Moore, 2005). As cytoplasmic membranes are the main action sites of CHX, the outer membrane in Gram-negative bacteria may act as a permeability barrier for CHX and limit its antibacterial efficacy. For instance, cationic CHX molecules may be “locked up” in the outer membrane due to interactions with negatively charged lipopolysaccharide-moieties and thus may not be able to even reach the cytoplasmic membrane (Cheung et al., 2012). Furthermore, outer membrane vesicles released by Gram-negative bacteria like Porphyromonas gingivalis may bind CHX, thus allowing protection of bacteria (Grenier et al., 1995). As there is not much data on the actual role or influence of the outer membrane in Gram-negative bacteria with regard to the action of CHX, this point merits closer attention.
Studies of the effects of CHX at sub-inhibitory, bacteriostatic, concentrations are very scarce. Analogous to QACs or antibiotics at sub-inhibitory concentrations, the effects of CHX at such concentrations may be far more complicated including multiple processes such as loss of osmoregulation around the cytoplasmic membrane, disturbance of processes related to transport and respiratory activity, dissipation of proton motive force and oxidative stress, which triggers SOS responses and in turn induces error-prone DNA replication leading to mutations and gene transfers (Andersson and Hughes, 2014; Tezel and Pavlostathis, 2015).
Resistance Mechanisms
Intrinsic resistance is defined as natural property of an organism, while acquired resistance is resulting from genetic changes and arising either by mutation or by the acquisition of the genetic material, e.g., via plasmids (Russell, 1995). Furthermore, one has to clearly distinguish between phenotypic adaptation, which is reversible when the exposure to the agent ends, and acquired resistance, which is genetically defined and therefore stable (Meyer and Cookson, 2010). Resistance must also be differentiated from tolerance, whereby resistance means the inherited ability of microorganisms to survive high concentrations of an antimicrobial drug (quantified by the minimum inhibitory concentration, MIC), while tolerance describes the ability to withstand a transient exposure to high concentrations of an antimicrobial (which otherwise would be lethal) without a change in MIC mostly due to the deceleration of metabolic processes (Brauner et al., 2016). In contrast, persistence describes the presence of a sub-population that is able to survive a treatment with concentrations higher than the MIC of a given antimicrobial despite the population being clonal (Brauner et al., 2016). These terms have often been used synonymously in the literature (especially in older studies), which hampers differentiation.
Furthermore, the definition of “resistance” toward antiseptics is not entirely clear (Chapman, 2003; Sheldon, 2005; Maillard, 2007). There are internationally recognized and standardized methods for susceptibility testing of antibiotics with defined breakpoints by means of which given isolates can be classified as resistant, intermediate or susceptible. This practically means that given isolates whose MIC values exceed the breakpoint concentrations are deemed resistant to a given antibiotic (Chapman, 2003). In contrast, similar frameworks are missing for susceptibility testing of biocides like CHX and there is a lack of well-defined MIC cut-off values indicating resistance (Vijayakumar and Sandle, 2018). While in vitro methods originally developed for systemic antibiotics (e.g., MICs) are still used for antiseptics, their results must be interpreted with caution (Sheldon, 2005). In this context, measurable MIC increases by a given factor (often 4- to 16-fold) have been considered relevant and may be seen as a parallel definition of “resistance” (Chapman, 2003), while on the other hand “in-use” concentrations of these biocides may be much higher than the measured MICs (Maillard, 2007; Vijayakumar and Sandle, 2018). Despite these limitations, investigation of MICs can still be valuable for antiseptics for studying potential resistance mechanisms in vitro (Maillard, 2007).
Intrinsic resistance to CHX is known from bacterial spores and mycobacteria and is due to their outer cell layers which form an impermeable barrier to the ingress of CHX molecules (Horner et al., 2012). Acquired genetically defined mechanisms conferring resistance toward CHX include multidrug efflux pumps and cell membrane changes (Jaglic and Cervinkova, 2012; Kampf, 2018b). The latter have been suspected to be responsible for CHX resistance in Pseudomonas stutzeri, where changes in the outer membrane protein and lipopolysaccharide profiles were found in CHX-resistant strains as compared to CHX-sensitive ones (Tattawasart et al., 2000b). Accordingly, electron-microscopic investigations confirmed that treatment with CHX led to considerably greater morphological changes in CHX-sensitive cells, while CHX-resistant cells showed no structural damage. Furthermore, it was confirmed by energy-dispersive spectroscopy that there was less uptake of CHX in resistant cells (Tattawasart et al., 2000a). The CHX-resistant strains also showed decreased susceptibility toward different classes of antibiotics which was suggested to be due to a non-specific decrease in cell permeability limiting uptake of chemically unrelated molecules into the resistant cells (Tattawasart et al., 1999).
Multidrug efflux pumps are membrane proteins that contain multiple transmembrane domains forming channels to remove toxic substances from the cytoplasm and the cytoplasmic membrane (see Figure 2 for schematic depiction of two exemplary efflux systems conferring CHX resistance in Gram-positive or Gram-negative bacteria, respectively) (Wassenaar et al., 2015). For instance, the plasmid-borne qac (standing for “quaternary ammonium compound”) gene family (e.g., qacA, qacB, qacC; qacA, and qacB usually recorded as qacA/B and qacC synonymously termed smr) is well known in Gram-positive bacteria (mainly in staphylococci). These qac genes encode for Qac efflux proteins that belong to the “Major Facilitator Superfamily” (MFS, i.e., QacA/B) or to the “Small Multidrug Resistance” (SMR) family (i.e., Smr) and have cationic biocides like CHX as substrates (Littlejohn et al., 1991; Poole, 2007; Buffet-Bataillon et al., 2012; Jaglic and Cervinkova, 2012; Liu et al., 2015; Kampf, 2016). Accordingly, introduction of a CHX-based surface antiseptic protocol in an intensive care unit with methicillin-resistant Staphylococcus aureus (MRSA) prevalence of about 20% was found to lead to an immediate and sustained reduction in transmission of susceptible MRSA strains while strains carrying qacA/B genes were not affected after introduction of this protocol (Batra et al., 2010). In contrast, a recent study found no correlation in S. aureus isolates between susceptibility toward CHX and carriage of qacA/B genes (Hardy et al., 2018). Interestingly, when comparing two pairs of isolates with fourfold differences in susceptibility to CHX (as shown by minimum bactericidal concentrations, MBCs) but very close relation according to phylogeny data, single-nucleotide polymorphisms within chromosomal efflux systems encoded by norA or norB were identified indicating that norA/norB may functionally complement qacA/qacB in these strains (Hardy et al., 2018).
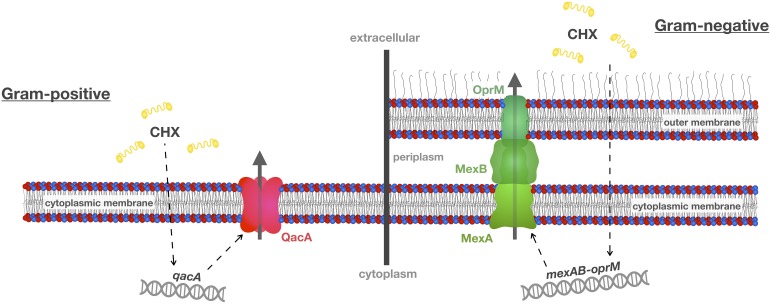
FIGURE 2.
Efflux mechanisms conferring resistance toward CHX. A common resistance mechanism toward antibacterial agents such as CHX is up-regulation of multidrug efflux pumps. This scheme shows two well-known efflux systems, i.e., qacA in Gram-positive S. aureus and mexAB-oprM in Gram-negative P. aeruginosa. These efflux pumps may also be present in oral bacteria and recognize not only CHX as their substrate but also other antiseptics and antibiotics, and, thus, may contribute to cross-resistances between CHX and antibiotics. This scheme was adopted and modified from Venter et al. (2017).
In Gram-negative bacteria, efflux proteins from the SMR family (e.g., QacE, QacEΔ1, QacF, QacG) and from the “Resistance-Nodulation-Division” (RND) superfamily (like the Mex efflux systems in P. aeruginosa) have been described (Poole, 2007; Liu et al., 2015). For instance, the activity of the MexCD-OprJ multidrug efflux system was found to be upregulated in P. aeruginosa upon exposure to sub-inhibitory concentrations of CHX or BAC (Morita et al., 2014). Expression of this efflux gene was found in various mutants, suggesting that MexCD-OprJ is a determinant of CHX resistance (Fraud et al., 2008). Furthermore, the four RND superfamily efflux pumps MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM are also well recognized to confer resistance to fluoroquinolones (Morita et al., 2014). Likewise, the “Proteobacterial Antimicrobial Compound Efflux” (PACE) family is wide-spread in Gram-negative bacteria conferring resistance to a wide range of biocides (Hassan et al., 2018) whose first member, the active CHX efflux protein AceI, was identified by investigating the transcriptomic response in Acinetobacter baumannii toward CHX (Hassan et al., 2013). In the same study, it was also found that expression of the RND efflux protein AdeAB was even more up-regulated upon CHX exposure (Hassan et al., 2013).
The close association frequently found between antibiotic and antiseptic resistance may be explained by the fact that genetic determinants of resistance are commonly linked to each other; accordingly, the qac genes are frequently located on plasmids with various other resistance genes (Jaglic and Cervinkova, 2012). For example, a clinical strain of S. aureus with high-level resistance to vancomycin isolated in June 2002 harbored a 57.9 kb multi-resistance conjugative plasmid, pLW1043, comprising elements that encode for resistance toward vancomycin (vanA), trimethoprim (dfrA), β-lactams (blaZ), aminoglycosides (aacA-aphD) as well as antiseptics (qacC) (Weigel et al., 2003). Likewise, the plasmid pSAJ1 from a methicillin- and gentamicin-resistant strain of S. aureus isolated in the 1980s conferred resistance toward CHX (most likely due to qacA) along with resistance toward kanamycin, gentamicin, tobramycin, amikacin, BAC, acriflavine, and ethidium bromide (Yamamoto et al., 1988).
Recently, it was found that adaptation of clinical Klebsiella pneumoniae isolates to CHX in vitro can lead not only to stable resistance toward CHX but also cross-resistance toward the last line antibiotic colistin (Wand et al., 2017). Whole genome sequencing revealed mutations in phoPQ and/or smvR, whereby the smvR mutation in turn may lead to upregulation of smvA which encodes for expression of SmvA, an efflux pump belonging to the MFS, which was suggested to play a major role in CHX resistance of K. pneumoniae strains. The cross-resistance toward colistin upon adaptation to CHX was likely due to the mutation in PhoPQ affecting the regulatory targets pmrD and pmrK. The operon pmrK functions to alter lipopolysaccharides by replacement of phosphate groups by 4-amino-4-deoxy-L-arabinose, which in turn reduces the net negative charge of lipid A and results in a reduction of its binding affinity to colistin (Wand et al., 2017).
Evidence for Resistance Toward CHX in Oral Bacteria?
Chlorhexidine has been extensively used in dental practice since 1970, when Löe and Schiøtt (1970) described total inhibition of plaque formation and gingivitis development in patients applying a 0.2% CHX mouth rinse twice daily despite stopping all other oral hygiene measures (e.g., tooth brushing) (see Gjermo, 1974). Already in 1972 two studies were published reporting clinical isolates of Streptococcus sanguinis that showed slightly reduced susceptibility toward CHX after long-term use of CHX-containing mouth rinses, which was, however, considered to be “relatively inconspicuous” (Emilson et al., 1972; Schiøtt and Löe, 1972). Isolates of S. sanguinis with reduced susceptibility to CHX were also found in a later clinical study upon daily tooth brushing with 0.5% CHX-containing gel, whereby the authors concluded that it was unclear whether this finding was a result of an adaptation toward CHX or due to a selection for less sensitive mutants within the original oral microbiota (Emilson and Fornell, 1976).
Westergren and Emilson (1980) described adaptation to CHX in three CHX-sensitive strains of S. sanguinis when grown in vitro as continuous cultures in a fermenter containing medium with increasing CHX concentrations. This resistance persisted after continuous growth in CHX-free medium and, interestingly, extracted DNA from the resistant mutants transformed competent sensitive S. sanguinis strains to increased CHX resistance. Therefore, they concluded that appearance of less susceptible isolates in the oral cavity after long-term use of CHX may be explained by genetic changes and not solely by selection for naturally occurring less susceptible strains (Westergren and Emilson, 1980). Likewise, when screening 315 isolates from subgingival plaque for their susceptibility to a mouth rinse containing 0.2% CHX, evidence for what the authors called “relative resistance” in different strains of Streptococcus mitis, S. sanguinis, and Capnocytophaga spp., was found (Wade and Addy, 1989). Maynard et al. (1993) reported significantly higher MICs for CHX in bacteria recovered from patients that had been using a 1% CHX toothpaste for 6 months as compared to those using a toothpaste without CHX. However, this finding was not thought to be of clinical significance (Maynard et al., 1993).
Passaging 24 periodontitis-associated bacteria (i.e., 10 oral streptococci, 8 P. gingivalis strains, 4 Aggregatibacter actinomycetemcomitans strains, and 2 enterobacteria) on agar plates containing sub-inhibitory concentrations of CHX resulted in a transitory moderate increase in the tolerance to CHX in five of the tested isolates (i.e., 1 oral streptococcus strain, 3 P. gingivalis strains, and 1 A. actinomycetemcomitans strain) after 25 passages, which vanished again after 50 passages in spite of the agar still containing sub-inhibitory concentrations of CHX (Eick et al., 2011). Kulik et al. (2015) reported two- to fourfold increased MICs in two out of five P. gingivalis strains after culturing for 20–30 passages in sub-inhibitory concentrations of CHX, whereas no increase in MIC was observed for two Streptococcus mutans and two Streptococcus sobrinus strains.
Recently, Kitagawa et al. (2016) found that repeated exposure of Enterococcus faecalis to CHX by serial passaging (10 cycles of exposure to CHX for MIC testing followed by re-growth in CHX-free medium) resulted in continuous increase in its MIC. The adapted cells showed increased surface hydrophobicity. Furthermore, SDS-PAGE of the adapted mutant strain revealed a novel protein of about 19-kDa (Kitagawa et al., 2016) that had also been found in vancomycin-resistant enterococci (Cho et al., 2008). The authors searched the proteome of E. faecalis 62 in the NCBI database for a candidate protein and reported that the most relevant protein in the 19-kDa range with regard to resistance was a 19.93-kDa MFS efflux pump protein (Kitagawa et al., 2016). This protein is unusually small for an MFS transporter and it is not clear whether it directly exports vancomycin. However, it is noteworthy that the candidate protein is a partial match to a larger 45-kDa MFS protein of E. faecalis V583, encoded by ef2068 (Yan et al., 2015; Kitagawa et al., 2016).
Wang et al. (2017a) evaluated changes in MIC in eight common oral bacterial species over 10 passages of CHX-challenge and re-growth in CHX-free medium and reported adaptation in Streptococcus gordonii, E. faecalis, Fusobacterium nucleatum, and P. gingivalis. Further analysis of the adapted S. gordonii strain showed a delayed growth with prolonged log-phase and decelerated growth rate as compared to its parental strain indicating reduced metabolic activity that may be responsible for the reduced susceptibility toward CHX (Wang et al., 2017a).
In another recent study, bacteria were isolated from dental plaque from five healthy individuals to screen for strains resistant toward CHX (Saleem et al., 2016). CHX-resistant isolates also displayed variable resistance to a range of antibiotics including ampicillin, kanamycin, gentamicin and tetracycline. Exposure of the most resistant isolate, a strain identified as Chryseobacterium indologenes, to CHX (16 μg/mL) resulted in a 19-fold up-regulation of the gene CIN01S_RS05745 encoding expression of the HlyD-like periplasmic adaptor protein of a tripartite efflux pump. However, it was not investigated whether the other components of this efflux pump were upregulated as well (Saleem et al., 2016). The authors highlighted the requirement for increased vigilance for the presence of multidrug resistant bacteria within dental plaque and raised awareness of the potential risk of long-term use of oral care products containing antimicrobial agents such as CHX for the control of oral biofilms.
Interestingly, treatment with CHX has also been shown to induce the formation of antifungal-tolerant persister cells in Candida albicans biofilms in vitro (LaFleur et al., 2006). In another study, 150 isolates of C. albicans and Candida glabrata obtained from cancer patients, who were at high risk for the development of oral candidiasis and who had been treated with topical CHX once a day, were investigated in terms of the development of persisters (LaFleur et al., 2010). The authors found that persister cells are clinically relevant and that antimicrobial therapy with CHX and long-term carriage of Candida select for high-persister strains in vivo. There are also reports of the induction of S. mutans persisters by CHX in single-species biofilms (Wang et al., 2017b).
The increased clinical use of CHX makes it important to be alert to the possibility of the emergence of new clones with reduced susceptibility (Horner et al., 2012). It seems reasonable that microorganisms will be exposed to sub-inhibitory concentrations of antiseptics in a clinical environment (Block and Furman, 2002). Such sub-inhibitory concentrations can be reached in the oral cavity upon treatment with CHX, as the antimicrobial effect of CHX seems to be diminished in the presence of organic substance such as saliva or serum due to inactivation by salivary or serum proteins (Portenier et al., 2006; Abouassi et al., 2014). Also inside oral biofilms, treatment with CHX results in a concentration gradient from the biofilm surface toward its lower strata which may lead to biofilm layers with sub-inhibitory concentrations of CHX (Thurnheer et al., 2003). Accordingly, the limited antimicrobial effects of CHX in the inner layers of oral biofilms have been shown using confocal laser scanning microscopy in combination with LIVE/DEAD staining (see Figure 3) (Zaura-Arite et al., 2001; Karygianni et al., 2014; Al-Ahmad et al., 2016). Furthermore, starvation may play a major role in these biofilms, which may increase adaptation of the bacteria within the biofilm. It was shown that S. mutans exhibited significantly increased MBCs toward CHX under starvation conditions in vitro than in nutrient-rich medium (Tong et al., 2011).
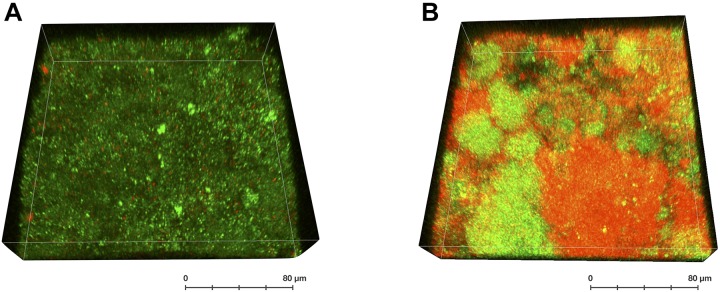
FIGURE 3.
3D-reconstruction of a LIVE/DEAD-stained confocal image stack of a supragingival oral biofilm after treatment with CHX. The biofilm was formed in situ on a bovine enamel slab for 72 h (methodology as described in Al-Ahmad et al., 2015) and was either left untreated (A) or was treated with 0.2% CHX for 5 min (B). Bacteria with intact (green; considered “live”) or compromised bacterial membranes (red; considered “dead”) are depicted indicating “pockets of viable cells” within the biofilm.
Conclusion and Future Directions
The question, whether there already is reason for concern with regard to enhanced tolerance or even resistance toward CHX in oral bacteria, cannot be answered that easily and must be treated in a more differentiated way: On the one hand, CHX is still very effective in its various fields of clinical application, e.g., intensive care, hand soaps and as an oral antiseptic. On the other hand, there unquestionably are reports about emergence of isolates exhibiting enhanced tolerance toward CHX. True resistance toward CHX will have horrendous consequences for infection control. For example, Copin et al. (2019) recently reported the spread of a unique USA300 clone of community-acquired MRSA in an Orthodox Jewish Community in Brooklyn. Genetic analyses showed that a metabolic change (mutation in pyrA) and acquisition of a clone-specific prophage (Φ11) probably have primed the clonal variant for success by promoting colonization and abscess formation. However, it was also found that emergence of a dominant clone coincided with acquisition and evolution of a plasmid (pBSRC1) with genes conferring resistance to CHX (qacA/B) and mupirocin (mupA), strongly suggesting that resistance toward antimicrobials used for decolonization therapy like CHX and mupirocin were key elements in the spread of this clone (Copin et al., 2019). Moreover, low level exposure to CHX (as potentially occurring in deeper layers of oral biofilms) may result in development of cross-resistances toward antibiotics as it has already been described in biocide-sensitive strains from organic foods (Kampf, 2018a). As there currently is little awareness about the potential risks accompanying the widespread use of CHX in dentistry, this research subject should be particularly highlighted as a hot topic in dental research. From the authors’ point of view, there are two key questions relating to CHX resistance that urgently need to be addressed:
-
simple •
Does the widespread use of CHX lead to an enrichment of resistant strains in oral biofilms and does it further encourage the development of cross-resistances in oral biofilms?
-
simple •
What are the molecular mechanisms conferring CHX resistance in oral bacteria?
The emergence of resistant bacteria as a result of the widespread use of CHX in dental practice has not been studied systematically so far. Culturing biofilms in oral in situ biofilm models and treating them with CHX sequentially would give insights into ecological changes of the microbiota of the oral biofilm after long-term treatment with this antimicrobial agent. In this light, it will be important to analyze the impact of CHX by microbiome analyses by high-throughput sequencing methods as well as by culture techniques that can distinguish between viable and non-viable cells. In addition, subsequent testing of antibiotic resistance in representative oral isolates may facilitate the study of potential effects of CHX on cross-resistance toward antibiotics and emergence of multidrug resistant bacteria. What remains unclear is the clinical relevance of persister cells in oral biofilms which have frequently been treated with CHX. The research on persister development in oral bacteria under the pressure of CHX treatment is still in its early stage.
Furthermore, novel techniques like transposon sequencing (Tn-seq) are now available that can be used to identify key genetic mechanisms of CHX resistance in oral bacteria by determining gene disruptions differentially represented in mutant populations on a genome-wide scale (van Opijnen et al., 2009; Burby et al., 2017). For this purpose, resistant strains must either be isolated from patient samples or induced in vitro by multiple passaging of bacteria under CHX-treatment (see Figure 4), whereby the latter has the advantage that the mutant strain exhibiting phenotypic adaptation to CHX can be compared to the wildtype strain. Finally, it is important to consider the respective fitness cost of a given mutation related to CHX resistance as the magnitude of this cost is the main biological parameter determining the rate of development of resistance (Andersson and Hughes, 2010; Cai et al., 2017).
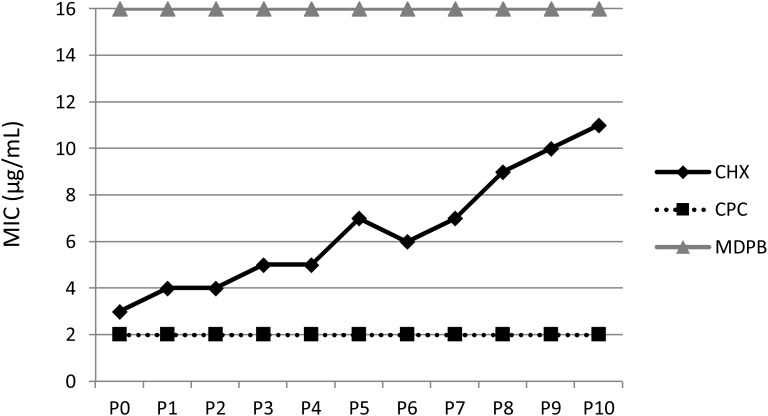
FIGURE 4.
Multiple passaging of bacteria under CHX-treatment in vitro. MICs of CHX, cetylpyridinium chloride (CPC) and methacryloyloxydodecylpyridinium bromide (MDPB) repeatedly performed from passages 0–10 (P0–P10) against E. faecalis. This figure is reprinted from Kitagawa et al. (2016) with kind permission from the publisher.
Data Availability
All datasets generated for this study are included in the manuscript.
Author Contributions
FC and AA-A took the lead in writing the manuscript. NJ, WB, TM, and EH provided the critical feedback and reviewed the manuscript for high intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Marie Follo and Andreas Vollmer for their help with preparing Figure 3.
Footnotes
Funding. FC thanks for funding from the University Medical Center Regensburg within the ReForM B program and from the International Association for Dental Research (IADR) as recipient of the IADR STAR Network Academy Fellowship 2018. This review was further supported in part by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; Grant AL 1179/2-1 to AA-A).
References
- Abouassi T., Hannig C., Mahncke K., Karygianni L., Wolkewitz M., Hellwig E., et al. (2014). Does human saliva decrease the antimicrobial activity of chlorhexidine against oral bacteria? BMC Res. Notes 7:711. 10.1186/1756-0500-7-711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ahmad A., Ameen H., Pelz K., Karygianni L., Wittmer A., Anderson A. C., et al. (2014). Antibiotic resistance and capacity for biofilm formation of different bacteria isolated from endodontic infections associated with root-filled teeth. J. Endod. 40 223–230. 10.1016/j.joen.2013.07.023 [DOI] [PubMed] [Google Scholar]
- Al-Ahmad A., Bucher M., Anderson A. C., Tennert C., Hellwig E., Wittmer A., et al. (2015). Antimicrobial photoinactivation using visible light plus water-filtered infrared-A (VIS + wIRA) alters in situ oral biofilms. PLoS One 10:e0132107. 10.1371/journal.pone.0132107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ahmad A., Walankiewicz A., Hellwig E., Follo M., Tennert C., Wittmer A., et al. (2016). Photoinactivation using visible light plus water-filtered infrared-A (vis+wIRA) and chlorine e6 (Ce6) eradicates planktonic periodontal pathogens and subgingival biofilms. Front. Microbiol. 7:1900. 10.3389/fmicb.2016.01900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerberger F., Gareis R., Jindrák V., Struelens M. J. (2009). Antibiotic stewardship implementation in the EU: the way forward. Expert Rev. Anti Infect. Ther. 7 1175–1183. 10.1586/eri.09.96 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8 260–271. 10.1038/nrmicro2319 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12 465–478. 10.1038/nrmicro3270 [DOI] [PubMed] [Google Scholar]
- Batra R., Cooper B. S., Whiteley C., Patel A. K., Wyncoll D., Edgeworth J. D. (2010). Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 50 210–217. 10.1086/648717 [DOI] [PubMed] [Google Scholar]
- Block C., Furman M. (2002). Association between intensity of chlorhexidine use and micro-organisms of reduced susceptibility in a hospital environment. J. Hosp. Infect. 51 201–206. 10.1053/jhin.2002.1246 [DOI] [PubMed] [Google Scholar]
- Brauner A., Fridman O., Gefen O., Balaban N. Q. (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14 320–330. 10.1038/nrmicro.2016.34 [DOI] [PubMed] [Google Scholar]
- Buffet-Bataillon S., Tattevin P., Bonnaure-Mallet M., Jolivet-Gougeon A. (2012). Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds–a critical review. Int. J. Antimicrob. Agents 39381–389. 10.1016/j.ijantimicag.2012.01.011 [DOI] [PubMed] [Google Scholar]
- Burby P. E., Nye T. M., Schroeder J. W., Simmons L. A. (2017). Implementation and data analysis of Tn-seq, whole-genome resequencing, and single-molecule real-time sequencing for bacterial genetics. J. Bacteriol. 199:e00560-16. 10.1128/JB.00560-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Liao Y., Brandt B. W., Wei X., Liu H., Crielaard W., et al. (2017). The fitness cost of fluoride resistance for different streptococcus mutans strains in biofilms. Front. Microbiol. 8:260. 10.3389/fmicb.2017.01630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. S. (2003). Biocide resistance mechanisms. Int. Biodeterior. Biodegradation 51 133–138. 10.1016/s0964-8305(02)00097-5 [DOI] [Google Scholar]
- Cheung H.-Y., Wong M. M.-K., Cheung S.-H., Liang L. Y., Lam Y.-W., Chiu S.-K. (2012). Differential actions of chlorhexidine on the cell wall of Bacillus subtilis and Escherichia coli. PLoS One 7:e36659. 10.1371/journal.pone.0036659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. S., Lee H. S., Kim J. M., Lee M. H., Yoo H. S., Park Y. H., et al. (2008). Immunogenic proteins in the cell envelope and cytoplasm of vancomycin-resistant enterococci. J. Immunoassay Immunochem. 29 319–331. 10.1080/15321810802329252 [DOI] [PubMed] [Google Scholar]
- Cieplik F., Deng D., Crielaard W., Buchalla W., Hellwig E., Al-Ahmad A., et al. (2018a). Antimicrobial photodynamic therapy - what we know and what we don’t. Crit. Rev. Microbiol. 44 571–589. 10.1080/1040841X.2018.1467876 [DOI] [PubMed] [Google Scholar]
- Cieplik F., Kara E., Muehler D., Enax J., Hiller K.-A., Maisch T., et al. (2018b). Antimicrobial efficacy of alternative compounds for use in oral care toward biofilms from caries-associated bacteria in vitro. Microbiologyopen 10.1002/mbo3.695 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplik F., Wimmer F., Muehler D., Thurnheer T., Belibasakis G. N., Hiller K.-A., et al. (2018c). Phenalen-1-one-mediated antimicrobial photodynamic therapy and chlorhexidine applied to a novel caries biofilm model. Caries Res. 52 447–453. 10.1159/000487815 [DOI] [PubMed] [Google Scholar]
- Copin R., Sause W. E., Fulmer Y., Balasubramanian D., Dyzenhaus S., Ahmed J. M., et al. (2019). Sequential evolution of virulence and resistance during clonal spread of community-acquired methicillin-resistant Staphylococcus aureus. PNAS 116 1745–1754. 10.1073/pnas.1814265116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplewski L., Bax R., Clokie M., Dawson M., Fairhead H., Fischetti V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16 239–251. 10.1016/S1473-3099(15)00466-1 [DOI] [PubMed] [Google Scholar]
- Davies G. E., Francis J., Martin A. R., Rose F. L., Swain G. (1954). 1:6-Di-4′-chlorophenyldiguanidohexane (hibitane); laboratory investigation of a new antibacterial agent of high potency. Br. J. Pharmacol. 9 192–196. 10.1111/j.1476-5381.1954.tb00840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton G. W. (2001). “Chlorhexidine,” in Disinfection, Sterilization, and Preservation, ed. Block S. S. (Philadelphia: Lippincott Williams & Wilkins; ), 321–336. [Google Scholar]
- Denyer S. P., Stewart G. (1998). Mechanisms of action of disinfectants. Int. Biodeterior. Biodegradation 41 261–268. 10.1016/S0964-8305(98)00023-7 [DOI] [Google Scholar]
- Eick S., Goltz S., Nietzsche S., Jentsch H., Pfister W. (2011). Efficacy of chlorhexidine digluconate-containing formulations and other mouthrinses against periodontopathogenic microorganisms. Quintessence Int. 42 687–700. [PubMed] [Google Scholar]
- Emilson C. G., Ericson T., Heyden G., Lilia J. (1972). Effect of chlorhexidine on human oral streptococci. J. Periodont. Res. 7 189–191. 10.1111/j.1600-0765.1972.tb00644.x [DOI] [PubMed] [Google Scholar]
- Emilson C. G., Fornell J. (1976). Effect of toothbrushing with chlorhexidine gel on salivary microflora, oral hygiene, and caries. Scand. J. Dent. Res. 84 308–319. 10.1111/j.1600-0722.1976.tb00495.x [DOI] [PubMed] [Google Scholar]
- Forman M. E., Fletcher M. H., Jennings M. C., Duggan S. M., Minbiole K. P. C., Wuest W. M. (2016). Structure-resistance relationships: interrogating antiseptic resistance in bacteria with multicationic quaternary ammonium dyes. ChemMedChem 11 958–962. 10.1002/cmdc.201600095 [DOI] [PubMed] [Google Scholar]
- Fraud S., Campigotto A. J., Chen Z., Poole K. (2008). MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob. Agents Chemother. 52 4478–4482. 10.1128/AAC.01072-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P., Moore L. E. (2005). Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99 703–715. 10.1111/j.1365-2672.2005.02664.x [DOI] [PubMed] [Google Scholar]
- Gjermo P. (1974). Chlorhexidine in dental practice. J. Clin. Periodontol. 1143–152. 10.1111/j.1600-051X.1974.tb01250.x [DOI] [PubMed] [Google Scholar]
- Grenier D., Bertrand J., Mayrand D. (1995). Porphyromonas gingivalis outer membrane vesicles promote bacterial resistance to chlorhexidine. Oral Microbiol. Immunol. 10 319–320. 10.1111/j.1399-302X.1995.tb00161.x [DOI] [PubMed] [Google Scholar]
- Hardy K., Sunnucks K., Gil H., Shabir S., Trampari E., Hawkey P., et al. (2018). Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. mBio 9:533. 10.1128/mBio.00894-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K. A., Jackson S. M., Penesyan A., Patching S. G., Tetu S. G., Eijkelkamp B. A., et al. (2013). Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. PNAS 110 20254–20259. 10.1073/pnas.1317052110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K. A., Liu Q., Elbourne L. D. H., Ahmad I., Sharples D., Naidu V., et al. (2018). Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169 450–454. 10.1016/j.resmic.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner C., Mawer D., Wilcox M. (2012). Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J. Antimicrob. Chemother. 67 2547–2559. 10.1093/jac/dks284 [DOI] [PubMed] [Google Scholar]
- Jaglic Z., Cervinkova D. (2012). Genetic basis of resistance to quaternary ammonium compounds – the qac genes and their role: a review. Veterinární Medicína 57 275–281. 10.17221/6013-vetmed [DOI] [Google Scholar]
- Jiang S., Zeng J., Zhou X., Li Y. (2018). Drug resistance and gene transfer mechanisms in respiratory/oral bacteria. J. Dent. Res. 97 1092–1099. 10.1177/0022034518782659 [DOI] [PubMed] [Google Scholar]
- Jones C. G. (1997). Chlorhexidine: is it still the gold standard? Periodontol. 2000 15 55–62. 10.1111/j.1600-0757.1997.tb00105.x [DOI] [PubMed] [Google Scholar]
- Kampf G. (2016). Acquired resistance to chlorhexidine - is it time to establish an “antiseptic stewardship” initiative? J. Hosp. Infect. 94 213–227. 10.1016/j.jhin.2016.08.018 [DOI] [PubMed] [Google Scholar]
- Kampf G. (2018a). Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 7:110. 10.3390/antibiotics7040110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G. (2018b). “Chlorhexidine digluconate,” in Antiseptic Stewardship Biocide Resistance and Clinical Implications ed. Kampf G. (Cham: Springer; ), 429–534. 10.1007/978-3-319-98785-9_13 [DOI] [Google Scholar]
- Karygianni L., Al-Ahmad A., Argyropoulou A., Hellwig E., Anderson A. C., Skaltsounis A. L. (2015). Natural antimicrobials and oral microorganisms: a systematic review on herbal interventions for the eradication of multispecies oral biofilms. Front. Microbiol. 6:1529. 10.3389/fmicb.2015.01529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karygianni L., Ruf S., Follo M., Hellwig E., Bucher M., Anderson A. C., et al. (2014). Novel broad-spectrum antimicrobial photoinactivation of in situ oral biofilms by visible light plus water-filtered infrared A. Appl. Environ. Microbiol. 80 7324–7336. 10.1128/AEM.02490-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H., Izutani N., Kitagawa R., Maezono H., Yamaguchi M., Imazato S. (2016). Evolution of resistance to cationic biocides in Streptococcus mutans and Enterococcus faecalis. J. Dent. 47 18–22. 10.1016/j.jdent.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Kulik E. M., Waltimo T., Weiger R., Schweizer I., Lenkeit K., Filipuzzi-Jenny E., et al. (2015). Development of resistance of mutans streptococci and Porphyromonas gingivalis to chlorhexidine digluconate and amine fluoride/stannous fluoride-containing mouthrinses, in vitro. Clin. Oral Investig. 19 1547–1553. 10.1007/s00784-014-1379-y [DOI] [PubMed] [Google Scholar]
- LaFleur M. D., Kumamoto C. A., Lewis K. (2006). Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50 3839–3846. 10.1128/AAC.00684-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur M. D., Qi Q., Lewis K. (2010). Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob. Agents Chemother. 54 39–44. 10.1128/AAC.00860-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.-S., Kam P. C. A. (2008). Chlorhexidine–pharmacology and clinical applications. Anaesth. Intensive Care 36 502–512. 10.1177/0310057X0803600404 [DOI] [PubMed] [Google Scholar]
- Littlejohn T. G., DiBerardino D., Messerotti L. J., Spiers S. J., Skurray R. A. (1991). Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene 101 59–66. 10.1016/0378-1119(91)90224-Y [DOI] [PubMed] [Google Scholar]
- Liu Q., Zhao H., Han L., Shu W., Wu Q., Ni Y. (2015). Frequency of biocide-resistant genes and susceptibility to chlorhexidine in high-level mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MuH MRSA). Diagn. Microbiol. Infect. Dis. 82 278–283. 10.1016/j.diagmicrobio.2015.03.023 [DOI] [PubMed] [Google Scholar]
- Löe H., Schiøtt C. R. (1970). The effect of mouthrinses and topical application of chlorhexidine on the development of dental plaque and gingivitis in man. J. Periodont. Res. 5 79–83. 10.1111/j.1600-0765.1970.tb00696.x [DOI] [PubMed] [Google Scholar]
- Maillard J. Y. (2007). Bacterial resistance to biocides in the healthcare environment: should it be of genuine concern? J. Hosp. Infect. 65(Suppl. 2), 60–72. 10.1016/S0195-6701(07)60018-8 [DOI] [PubMed] [Google Scholar]
- Maris P. (1995). Modes of action of disinfectants. Rev. Sci. Tech. Off. Int. Epiz. 14 47–55. 10.20506/rst.14.1.829 [DOI] [PubMed] [Google Scholar]
- Maynard J. H., Jenkins S. M., Moran J., Addy M., Newcombe R. G., Wade W. G. (1993). A 6-month home usage trial of a 1% chlorhexidine toothpaste (II). Effects on the oral microflora. J. Clin. Periodontol. 20 207–211. 10.1111/j.1600-051X.1993.tb00345.x [DOI] [PubMed] [Google Scholar]
- McDonnell G., Russell A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12 147–179. 10.1128/CMR.12.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P. J., Levy S. B. (2016). Triclosan: an instructive tale. Antimicrob. Agents Chemother. 60 7015–7016. 10.1128/AAC.02105-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Cookson B. (2010). Does microbial resistance or adaptation to biocides create a hazard in infection prevention and control? J. Hosp. Infect. 76 200–205. 10.1016/j.jhin.2010.05.020 [DOI] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. 10.3389/fmicb.2013.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. (ed.) (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available at: google.co.in/books/about/Tackling_Drug_resistant_Infections_Globa.html?id=aa6lAQAACAAJ&redir_google.co.in/books/about/esc=y. [Google Scholar]
- Poole K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39 162–176. 10.1080/07853890701195262 [DOI] [PubMed] [Google Scholar]
- Portenier I., Waltimo T., Ørstavik D., Haapasalo M. (2006). Killing of Enterococcus faecalis by MTAD and chlorhexidine digluconate with or without cetrimide in the presence or absence of dentine powder or BSA. J. Endod. 32 138–141. 10.1016/j.joen.2005.10.027 [DOI] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P. (2010). Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev. Anti Infect. Ther. 8 1441–1450. 10.1586/eri.10.106 [DOI] [PubMed] [Google Scholar]
- Russell A. D. (1995). Mechanisms of bacterial resistance to biocides. Int. Biodeterior. Biodegradation 36 247–265. 10.1016/0964-8305(95)00056-9 [DOI] [Google Scholar]
- Russell A. D. (2004). Bacterial adaptation and resistance to antiseptics, disinfectants and preservatives is not a new phenomenon. J. Hosp. Infect. 57 97–104. 10.1016/j.jhin.2004.01.004 [DOI] [PubMed] [Google Scholar]
- Saleem H. G. M., Seers C. A., Sabri A. N., Reynolds E. C. (2016). Dental plaque bacteria with reduced susceptibility to chlorhexidine are multidrug resistant. BMC Microbiol. 16:661. 10.1186/s12866-016-0833-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz M., Serrano J., Iniesta M., Santa Cruz I., Herrera D. (2013). Antiplaque and antigingivitis toothpastes. Monogr. Oral Sci. 23 27–44. 10.1159/000350465 [DOI] [PubMed] [Google Scholar]
- Schiøtt C. R., Löe H. (1972). The sensitivity of oral streptococci to chlorhexidine. J. Periodont. Res. 7 192–194. 10.1111/j.1600-0765.1972.tb00645.x [DOI] [PubMed] [Google Scholar]
- Sheldon A. T. (2005). Antiseptic “resistance”: real or perceived threat? Clin. Infect. Dis. 40 1650–1656. 10.1086/430063 [DOI] [PubMed] [Google Scholar]
- Sreenivasan P., Gaffar A. (2002). Antiplaque biocides and bacterial resistance: a review. J. Clin. Periodontol. 29 965–974. 10.1034/j.1600-051X.2002.291101.x [DOI] [PubMed] [Google Scholar]
- Stickler D. J. (2002). Susceptibility of antibiotic-resistant Gram-negative bacteria to biocides: a perspective from the study of catheter biofilms. J. Appl. Microbiol. 92 163S–170S. 10.1046/j.1365-2672.92.5s1.6.x [DOI] [PubMed] [Google Scholar]
- Stickler D. J., Thomas B. (1980). Antiseptic and antibiotic resistance in Gram-negative bacteria causing urinary tract infection. J. Clin. Pathol. 33 288–296. 10.1136/jcp.33.3.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwantarat N., Carroll K. C., Tekle T., Ross T., Maragakis L. L., Cosgrove S. E., et al. (2014). High prevalence of reduced chlorhexidine susceptibility in organisms causing central line–associated bloodstream infections. Infect. Control Hosp. Epidemiol. 35 1183–1186. 10.1086/677628 [DOI] [PubMed] [Google Scholar]
- Tattawasart U., Hann A. C., Maillard J. Y., Furr J. R., Russell A. D. (2000a). Cytological changes in chlorhexidine-resistant isolates of Pseudomonas stutzeri. J. Antimicrob. Chemother. 45 145–152. 10.1093/jac/45.2.145 [DOI] [PubMed] [Google Scholar]
- Tattawasart U., Maillard J. Y., Furr J. R., Russell A. D. (2000b). Outer membrane changes in Pseudomonas stutzeri resistant to chlorhexidine diacetate and cetylpyridinium chloride. Int. J. Antimicrob. Agents 16 233–238. [DOI] [PubMed] [Google Scholar]
- Tattawasart U., Maillard J. Y., Furr J. R., Russell A. D. (1999). Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J. Hosp. Infect. 42 219–229. 10.1053/jhin.1999.0591 [DOI] [PubMed] [Google Scholar]
- Teughels W., Dekeyser C., Van Essche M., Quirynen M. (2009). One-stage, full-mouth disinfection: fiction or reality? Periodontol. 2000 50 39–51. 10.1111/j.1600-0757.2008.00292.x [DOI] [PubMed] [Google Scholar]
- Tezel U., Pavlostathis S. G. (2015). Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 33 296–304. 10.1016/j.copbio.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Thurnheer T., Gmür R., Shapiro S., Guggenheim B. (2003). Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl. Environ. Microbiol. 69 1702–1709. 10.1128/AEM.69.3.1702-1709.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Tao R., Jiang W., Li J., Zhou L., Tian Y., et al. (2011). In vitro study of the properties of Streptococcus mutans in starvation conditions. Arch. Oral Biol. 56 1306–1311. 10.1016/j.archoralbio.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Van der Weijden F. A., Van der Sluijs E., Ciancio S. G., Slot D. E. (2015). Can chemical mouthwash agents achieve plaque/gingivitis control? Dent. Clin. North Am. 59 799–829. 10.1016/j.cden.2015.06.002 [DOI] [PubMed] [Google Scholar]
- van Opijnen T., Bodi K. L., Camilli A. (2009). Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6 767–772. 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter H., Henningsen M. L., Begg S. L. (2017). Antimicrobial resistance in healthcare, agriculture and the environment: the biochemistry behind the headlines. Essays Biochem. 61 1–10. 10.1042/EBC20160053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar R., Sandle T. (2018). A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes—special notes on pharmaceutical environmental isolates. J. Appl. Microbiol. 27:139. 10.1111/jam.14118 [DOI] [PubMed] [Google Scholar]
- Wade W. G., Addy M. (1989). In vitro activity of a chlorhexidine–containing mouthwash against subgingival bacteria. J. Periodontol. 60 521–525. 10.1902/jop.1989.60.9.521 [DOI] [PubMed] [Google Scholar]
- Wainwright M., Maisch T., Nonell S., Plaetzer K., Almeida A., Tegos G. P., et al. (2017). Photoantimicrobials—are we afraid of the light? Lancet Infect. Dis. 17 e49–e55. 10.1016/S1473-3099(16)30268-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand M. E., Bock L. J., Bonney L. C., Sutton J. M. (2017). Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 61:e01162-16. 10.1128/AAC.01162-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang H., Ren B., Li H., Weir M. D., Zhou X., et al. (2017a). Do quaternary ammonium monomers induce drug resistance in cariogenic, endodontic and periodontal bacterial species? Dent. Mater. 33 1127–1138. 10.1016/j.dental.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Wang S., Zhou C., Ren B., Li X., Weir M. D., Masri R. M., et al. (2017b). Formation of persisters in Streptococcus mutans biofilms induced by antibacterial dental monomer. J. Mater. Sci.: Mater. Med. 28:178. 10.1007/s10856-017-5981-9 [DOI] [PubMed] [Google Scholar]
- Wassenaar T. M., Ussery D., Nielsen L. N., Ingmer H. (2015). Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. 5 44–61. 10.1556/EUJMI-D-14-00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., Flannagan S. E., et al. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302 1569–1571. 10.1126/science.1090956 [DOI] [PubMed] [Google Scholar]
- Westergren G., Emilson C.-G. (1980). In vitro development of chlorhexidine resistance in Streptococcus sanguis and its transmissibility by genetic transformation. Scand. J. Dent. Res. 88 236–243. 10.1111/j.1600-0722.1980.tb01220.x [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tamura Y., Yokota T. (1988). Antiseptic and antibiotic resistance plasmid in Staphylococcus aureus that possesses ability to confer chlorhexidine and acrinol resistance. Antimicrob. Agents Chemother. 32932–935. 10.1128/AAC.32.6.932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Budin-Verneuil A., Verneuil N., Gilmore M. S., Artigaud S., Auffray Y., et al. (2015). Transcriptomic response of Enterococcus faecalis V583 to low hydrogen peroxide levels. Curr. Microbiol. 70 156–168. 10.1007/s00284-014-0691-8 [DOI] [PubMed] [Google Scholar]
- Yazdankhah S. P., Scheie A. A., Høiby E. A., Lunestad B.-T., Heir E., Fotland T.Ø.et al. (2006). Triclosan and antimicrobial resistance in bacteria: an overview. Microb. Drug Resist. 12 83–90. 10.1089/mdr.2006.12.83 [DOI] [PubMed] [Google Scholar]
- Zaura-Arite E., van Marle J., ten Cate J. M. (2001). Confocal microscopy study of undisturbed and chlorhexidine-treated dental biofilm. J. Dent. Res. 80 1436–1440. 10.1177/00220345010800051001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the manuscript.