This study assesses the ability of retinal specialists to distinguish real from synthetic retinal fundus images, assess gradability, and diagnose age-related macular degeneration (AMD) using real images from the Age-Related Eye Disease Study and synthetic images generated therefrom.
Key Points
Question
Can deep learning be used to synthesize fundus images of age-related macular degeneration (AMD) that appear realistic to retinal specialists?
Findings
In this study of fundus images from 4613 study participants from the Age-Related Eye Disease Study and 133 821 fundus images, the ability of 2 retinal specialists to distinguish real from synthetic fundus images of varying stages of AMD was close to chance, and their diagnostic accuracy similar for real and synthetic images. Machines trained with only synthetic images showed performance nearing that resulting from training on real images.
Meaning
These findings suggest that deep-learning–synthesized fundus images of AMD are realistic and could be used for education of humans across various levels of expertise and for machine training.
Abstract
Importance
Deep learning (DL) used for discriminative tasks in ophthalmology, such as diagnosing diabetic retinopathy or age-related macular degeneration (AMD), requires large image data sets graded by human experts to train deep convolutional neural networks (DCNNs). In contrast, generative DL techniques could synthesize large new data sets of artificial retina images with different stages of AMD. Such images could enhance existing data sets of common and rare ophthalmic diseases without concern for personally identifying information to assist medical education of students, residents, and retinal specialists, as well as for training new DL diagnostic models for which extensive data sets from large clinical trials of expertly graded images may not exist.
Objective
To develop DL techniques for synthesizing high-resolution realistic fundus images serving as proxy data sets for use by retinal specialists and DL machines.
Design, Setting, and Participants
Generative adversarial networks were trained on 133 821 color fundus images from 4613 study participants from the Age-Related Eye Disease Study (AREDS), generating synthetic fundus images with and without AMD. We compared retinal specialists’ ability to diagnose AMD on both real and synthetic images, asking them to assess image gradability and testing their ability to discern real from synthetic images. The performance of AMD diagnostic DCNNs (referable vs not referable AMD) trained on either all-real vs all-synthetic data sets was compared.
Main Outcomes and Measures
Accuracy of 2 retinal specialists (T.Y.A.L. and K.D.P.) for diagnosing and distinguishing AMD on real vs synthetic images and diagnostic performance (area under the curve) of DL algorithms trained on synthetic vs real images.
Results
The diagnostic accuracy of 2 retinal specialists on real vs synthetic images was similar. The accuracy of diagnosis as referable vs nonreferable AMD compared with certified human graders for retinal specialist 1 was 84.54% (error margin, 4.06%) on real images vs 84.12% (error margin, 4.16%) on synthetic images and for retinal specialist 2 was 89.47% (error margin, 3.45%) on real images vs 89.19% (error margin, 3.54%) on synthetic images. Retinal specialists could not distinguish real from synthetic images, with an accuracy of 59.50% (error margin, 3.93%) for retinal specialist 1 and 53.67% (error margin, 3.99%) for retinal specialist 2. The DCNNs trained on real data showed an area under the curve of 0.9706 (error margin, 0.0029), and those trained on synthetic data showed an area under the curve of 0.9235 (error margin, 0.0045).
Conclusions and Relevance
Deep learning–synthesized images appeared to be realistic to retinal specialists, and DCNNs achieved diagnostic performance on synthetic data close to that for real images, suggesting that DL generative techniques hold promise for training humans and machines.
Introduction
Progress has been made applying deep learning (DL) for discriminative retinal image analysis tasks, including automated classification of fundus images for referable diabetic retinopathy,1,2,3,4,5 retinopathy of prematurity,6 age-related macular degeneration (AMD),7,8,9,10 and granular (9-step) AMD severity classification.11,12 Deep learning has achieved performance exceeding traditional machine learning13,14 and close to that of retinal specialists.
Nevertheless, deep convolutional neural networks (DCNNs) require large, diverse, and well-balanced image training data sets with labels defining structures. Although increased sharing of data sets among researchers15 may address this problem, sharing for this purpose, as well as sharing retinal images for educational purposes, may be impeded by institutional review board (ethics committee) concerns, laws regulating personally identifiable information, or proprietary data restrictions. Alternatively, these needs might be addressed using generative DL methods, such as generative adversarial networks16 (GANs), to synthesize new fundus retinal images from a training data set of real images. This study investigated the ability of high-resolution GANs17 to synthesize realistic fundus images that can serve as proxy data sets for use by retinal specialists and DL machines for education and training.
Methods
This study used AMD18,19 fundus images from the National Institutes of Health Age-Related Eye Disease Study (AREDS) data set7,9,20,21,22 that were collected from 4613 participants from whom written informed consent was obtained. Use of the AREDS deidentified data set of 133 821 fundus photographs (see eTable 1 in the Supplement) was performed after Johns Hopkins University Institutional Review Board approval. One stereo pair was taken for each eye, if available, at each visit. At some visits, only 1 eye was imaged and available for analysis. Variable numbers of visits were available for each participant; some participants were followed up for 12 years, whereas others had shorter follow-up times. Both stereo images were used, but stereo pairs were not used for both training and testing. In addition, patient partitioning was used (ie, patients in training, validation, or testing were distinct, as described in the eTable in the Supplement). Criterion standard gradings for this data set were available from quantitative gradings by trained experts and certified graders at a fundus photograph reading center.20,22 Before use, all images underwent preprocessing, as described in the eAppendix in the Supplement.
Classification Structure
This study focused on a 2-class AMD referral challenge derived from the original 4-step AREDS enrollment scale9: nonreferable AMD (examples are shown in eFigure 1 in the Supplement) vs referable AMD (Figure). Referable AMD was defined as intermediate or advanced AMD and nonreferable AMD as no or early AMD9 (details are given in the eAppendix in the Supplement). This 4-class scale has been used clinically to discriminate between individuals with referable AMD who might be referred to physicians to monitor for progression of the intermediate to the advanced stage and to consider use of dietary supplements (such as that used in AREDS) to reduce risk of progression from the intermediate to the advanced stage or to consider use of anti–vascular endothelial growth factor for the choroidal neovascular form of AMD. Nonreferable AMD included cases for which referral for monitoring and supplements typically would not be indicated.
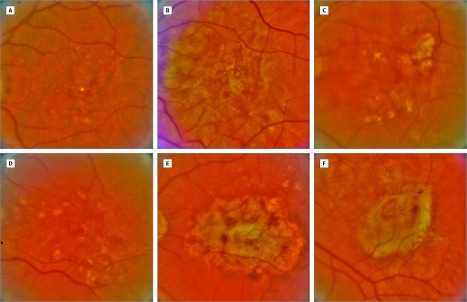
Figure. Synthetically Created and Real Fundus Images of Eyes With Referable Age-Related Macular Degeneration.
Examples of 3 synthetically created and 3 real fundus images with referable age-related macular degeneration. The answer to which are real and which are synthetic is given in the Additional Information at the end of the Article Information section. A continuum of similar synthetic images is given in Video 1.
Image Synthesis Method
All development was done in TensorFlow (Google). For image synthesis with DL, progressively grown generative adversarial networks (ProGANs)17 were used. Generative adversarial networks are adversarially trained to perform antagonistic tasks, including a discriminative network (D) to discriminate between real vs synthetic images and a generative network (G) to generate synthetic images ideally realistic enough to fool network D. Network D takes a low-dimensional (so-called latent) vector as input and maps using upconvolution to a synthetic image. ProGANs address early limitations of GAN architectures that had issues generating high-resolution images. ProGANs achieve high-resolution synthetization (in this study, 512 × 512 pixels) by progressively growing networks G and D; they start with simple networks that perform very low-resolution (2 × 2–pixel images) generative and discriminative tasks. Subsequently, these are grown by alternating training and adding new network layers. With each additional layer, the resolution is doubled (eg, 4 × 4, 8 × 8) allowing the generation of increasingly higher-resolution images. When the GAN has been trained sufficiently, an input latent vector to the generative network should give rise to a bona fide synthetic image. However, in some cases, synthetization may have problems, including mode collapse (see below).
For this investigation, 2 separate synthetic data sets were generated, 1 for referable AMD and 1 for nonreferable AMD, by using 2 separate ProGANs: referable GAN and nonreferable GAN, respectively. The referable GAN was exclusively trained on referable AMD images and only synthesized referable AMD cases. Similarly, the nonreferable GAN was trained only on nonreferable AMD and synthesized only nonreferable AMD images. With use of this process, synthetic images were generated along with their class label (nonreferable or referable AMD). Once trained, GANs can generate a quasi-unlimited number of synthetic images; in this study, approximately 120 519 synthetic images were used for our human retinal specialist and machine-based training and testing experiments (exact numbers are given in the eTable in the Supplement).
Evaluation Methodology
To assess whether synthetic images are fit for use by humans and machines, appear realistic to retinal specialists, do not introduce novel artifacts, and can be used for DL training, evaluations using a dual-pronged approach involving retinal specialists and testing the performance of these synthetic images to train DL algorithms were conducted. The human experiments used 2 retinal specialists who had more than 5 (retinal specialist 1; T.Y.A.L.) and more than 10 (retinal specialist 2; K.D.P.) years of clinical experience.
For experiments focusing on retinal specialists, both retinal specialists performed manual annotation of 600 images, which included approximately equal numbers of real and synthetic as well as referable and nonreferable AMD images. Distributions for each type of image were not revealed to minimize bias of the retinal specialists’ answers. The first 2 experiments or tasks requested of the specialists included the following.
Retinal Specialist Experiments
Human Diagnostic Ability
In experiment 1, diagnostic ability and performance of retinal specialists in classifying AMD compared with certified human graders from AREDS was tested on real vs synthetic images to assess image realism and the possible incidence of synthetization artifacts on typical human diagnosis. The following question was asked: “For each image, what is the diagnosis: referable or nonreferable AMD?”
Gradability of Real and Synthetic Images
In experiment 2, for each image, the following question was asked: “Is the image quality sufficient for grading?” The goal was to determine whether there was a quality issue with synthetic images or whether a sufficient number of artifacts in synthetic images rendered them nongradable as assessed by the retinal specialists.
When asked to perform experiments 1 and 2, the retinal specialists were not told that the annotation data set of 600 images included any synthetic images to avoid confirmation bias in their answers. After this annotation was completed, the retinal specialists were told that the annotation data set was composed of a mixture of real and synthetic images. The retinal specialists were then asked to perform the following experiments and annotations.
Discrimination Between Real and Synthetic Images by Retinal Specialists
Experiment 3 examined discrimination between real and synthetic images by retinal specialists. The following question was asked for this task: “Is the image synthetic?”
Deep Learning Experiments
Evaluation of Synthetic Images Used for DL Classification
In experiment 4, DCNNs were used to perform a diagnostic task of nonreferable vs referable AMD classification by comparing 2 DL models, 1 trained exclusively with real images (DCNN-R) and 1 trained only with synthetic images (DCNN-S). The 2 DL models used ResNet,23 specifically, a pretrained ResNet-50 fine-tuned with retinal images (either all real for DCNN-R or all synthetic for DCNN-S). Stochastic gradient descent was used with Nesterov momentum of 0.9 and categorical cross-entropy loss function. Dynamic learning rate scheduling was used by multiplying the learning rate by 0.5 when the training loss did not improve for 10 epochs. If the validation data set loss did not improve for 20 epochs, training was stopped, and model weights (saved on every epoch) with the best validation data set loss were saved as the final model. A base learning rate of 0.001 and a batch size of 32 were used using hyperparameter selection on the validation data set. Data augmentation was used for training improvement and to capture geometric variations not included in the original data set. This included horizontal flipping, modest blurring, sharpening, and changes to saturation, brightness, contrast, and color balance.
For the 2 DCNNs, real and synthetic images were partitioned as previously described9 into training, validation, and testing data sets as follows: DCNN-R used training and validation images directly taken from the AREDS data set, and DCNN-S used training and validation images generated by the GANs (GAN-NR and GAN-R). Then, both DCNN-R and DCNN-S were tested on the same testing data set composed of real images from AREDS. This testing data set did not include any image used to train the GANs to preserve strict train and test data separation. The DCNNs’ classification performance was then compared for DCNN-R and DCNN-S.
Additional Analysis of Expressiveness and Redundancy
Experiment 5 was performed to ascertain whether ProGAN was sufficiently expressive to generate a wide swath of interesting, distinct, and realistic retinal structures and AMD conditions. A possible concern mentioned above was mode collapse, which would result in the inability to synthesize images sufficiently distinct from each other. Another concern was generating images too similar to the ProGAN training images. One way to check for this is to navigate inside the latent (input) space and inspect the resulting generated (output) images. A continuum of synthetic images was synthesized by generating all images obtained by following rectilinear trajectories between several vectors’ positions in latent space, demonstrating, for visual inspection, how the latent space sees and encodes variations in retinal structure and AMD lesions. A total of 12 navigation trajectories shown in 2 videos were generated, 1 each for referable AMD (Video 1) and nonreferable AMD (Video 2). We also used a nearest-neighbor procedure applied to DL features,17 linking synthetic images to their closest real counterparts, checking to determine whether images generated were sufficiently distinct from real training images (eFigures 2 and 3 in the Supplement).
Video 1. Continuum Of Synthetically Generated Images With Referable Age-Related Macular Degeneration (AMD).
The video shows a continuum of synthetic images generated with AMD by traversing the latent space, demonstrating some of the variations in synthetic images that can be generated by the methods, including smoothly varying the retinal vasculature, the macular positions within the image, and the size and placement of lesions associated with AMD. The video shows breadth of the synthetization possible showing hundreds of variations of synthetically generated images with AMD and demonstrating the realism of these images.
Video 2. Continuum Of Synthetically Generated Images Without Referable Age-Related Macular Degeneration (AMD).
The video shows a continuum of synthetic images generated without AMD by traversing the latent space, demonstrating some of the variations in synthetic images that can be generated by the methods, including smoothly varying the retinal vasculature and the macular positions within the image.
Statistical Analysis
The statistical analyses for experiments 1 and 3 were done using metrics including accuracy, sensitivity, specificity, negative predictive value, positive predictive value, and κ score. Experiment 4 used accuracy and area under the curve (AUC). Evaluation for experiments 1 and 4 was done by comparing retinal specialist and DL algorithm performance with AREDS fundus photograph graders’ reference criterion. Experiment 5 used visual inspection. The error margin, when added to the value, provides the ± of the 95% CI.
Results
Human Evaluation Results by Retina Specialists
Human Diagnostic Ability
For experiment 1, the diagnostic performance metrics of the retinal specialists on real vs synthetic images (Table 1, Figure, and eFigure 1 in the Supplement) revealed that retinal specialist 1 had an accuracy that was similar on real vs synthetic images (84.54% [error margin, 4.06%] vs 84.12% [error margin, 4.16%]); this accuracy was similar to that for retinal specialist 2 (89.47% [error margin, 3.45%] vs 89.19% [error margin, 3.54%]).
Table 1. Retinal Specialist Determination of Diagnosis as Referable vs Nonreferable Age-Related Macular Degeneration Compared With Certified Human Graders (Experiment 1).
| Annotator, Data | Accuracy, % (Error Margin) | Sensitivity, % (Error Margin) | Specificity, % (Error Margin) | PPV, % (Error Margin) | NPV, % (Error Margin) | κ |
|---|---|---|---|---|---|---|
| Retinal specialist 1 | ||||||
| All | 84.33 (2.91) | 91.75 (3.10) | 76.77 (4.80) | 80.12 (4.20) | 90.12 (3.68) | 0.6862 |
| Real | 84.54 (4.06) | 93.96 (3.83) | 75.48 (6.77) | 78.65 (6.02) | 92.86 (4.50) | 0.6918 |
| Synthetic | 84.12 (4.16) | 89.61 (4.82) | 78.17 (6.79) | 81.66 (5.84) | 87.40 (5.77) | 0.6806 |
| Retinal specialist 2 | ||||||
| All | 89.33 (2.47) | 88.45 (3.60) | 90.24 (3.38) | 90.24 (3.38) | 88.45 (3.60) | 0.7867 |
| Real | 89.47 (3.45) | 89.93 (4.83) | 89.03 (4.92) | 88.74 (5.04) | 90.2 (4.71) | 0.7894 |
| Synthetic | 89.19 (3.54) | 87.01 (5.31) | 91.55 (4.57) | 91.78 (4.46) | 86.67 (5.44) | 0.7839 |
Abbreviations: NPV, negative predictive value; PPV, positive predictive value.
Gradability of Real and Synthetic Images
Results of experiment 2 showed that both retinal specialists rated synthetic images as having approximately the same fraction of gradable images as real images (Table 2); for example, retinal specialist 1 rated 98.03% (error margin, 1.56%) of real images to be gradable vs 98.31% (error margin, 1.47%) of synthetic images.
Table 2. Determination of Image Gradability and Quality Sufficiency for Diagnosis (Experiment 2).
| Annotator, Data | Sufficient Quality, % (Error Margin)a |
|---|---|
| Retinal specialist 1 | |
| All | 98.17 (1.07) |
| Real | 98.03 (1.56) |
| Synthetic | 98.31 (1.47) |
| Retinal specialist 2 | |
| All | 91.17 (2.27) |
| Real | 88.82 (3.54) |
| Synthetic | 93.58 (2.79) |
For each image, the following question was asked: “Is the image quality sufficient for grading?” Values indicate the percentage of images with sufficient quality among all images in each category.
Discrimination Between Real and Synthetic Images by Retinal Specialists
For experiment 3, Table 3 shows that the retinal specialists’ ability to discriminate real from synthetic images was limited, with an accuracy of 59.50% (error margin, 3.93%) for retinal specialist 1 and 53.67% (error margin, 3.99%) for retinal specialist 2.
Table 3. Determination of Real vs Synthetic Fundus Image.
| Annotator | Accuracy, % (Error Margin) | Sensitivity, % (Error Margin) | Specificity, % (Error Margin) | PPV, % (Error Margin) | NPV, % (Error Margin) | κ |
|---|---|---|---|---|---|---|
| Retinal specialist 1 | 59.50 (3.93) | 23.31 (4.82) | 94.74 (2.51) | 81.18 (8.31) | 55.92 (4.29) | 0.1822 |
| Retinal specialist 2 | 53.67 (3.99) | 18.92 (4.46) | 87.5 (3.72) | 59.57 (9.92) | 52.57 (4.35) | 0.0648 |
Abbreviations: NPV, negative predictive value; PPV, positive predictive value.
Overall, results from the retinal specialists showed that they had few differences in the capability to annotate either real vs synthetic images and an approximately equal preponderance of gradability on synthetic and real images. These findings suggest that the retinal specialists did not see additional artifacts in synthetic images and that they had limited ability to discern real from synthetic images.
Evaluation of Synthetic Images Used for DL Classification
For experiment 4, the AUC for DCNN-R trained on real images was 0.9706 (error margin, 0.0029), and the accuracy was 91.12% (error margin, 0.48%). The AUC of the DCNN-S trained exclusively on synthetic images was 0.9235 (error margin, 0.0045), and the accuracy was 82.92% (error margin, 0.64%). These findings suggest moderate performance decrease using the synthetic training data set, but the performance was still considered to be good.
Additional Analysis of Expressiveness and Redundancy
For experiment 5, generating a continuum of synthetic images by navigating the latent space and producing fly-through video sequences of synthetic images and visually inspecting results (Video 1 and Video 2) suggested that ProGANs were able to generate a continuum of novel synthetic images with interesting and realistic variations in retinal and lesion structure, including changes in extent, shape, and location of AMD lesions. In addition, the nearest-neighbor analysis (eFigures 2 and 3 in the Supplement) demonstrated that ProGAN synthetic images were distinct from their closest real nearest neighbors.
Discussion
Work in DL used in medical imaging has principally involved discriminative tasks, such as discriminating fundus images along a 9-step detailed AMD severity scale.12 More recently, novel applications of generative tasks (ie, generative networks and adversarial training)24,25,26,27,28 have been used for aims other than pure synthesis, including (1) a multistep method using vasculature segmentation, vessel generation, and image synthesis used for generating healthy-only fundus images and tested on vessel detection24; (2) for optical coherence tomography anomaly detection of lesion27 performed in latent space and applied at pixel level; (3) in radiology,25 applied for denoising low-dose computed tomography25; (4) for magnetic resonance imaging to computed tomography translation26 to help generate computed tomographic images from preoperative magnetic resonance imaging, which could help guide interventions; (5) for deformable soft-tissue cross-modality medical image registration28; and (6) in dermatology, for melanoma lesion synthesis.29 These applications suggest possible applications of generative methods for retinal image registration, fundus to optical coherence tomography translation, or fundus or optical coherence tomography image denoising.30
This study focused on using and evaluating ProGANs dedicated to generating high-resolution fundus images with and without referable AMD. The findings of experiments 1 through 5 suggest that synthetic fundus images may serve as proxy, replacement, or augmentation of real image data sets for use by humans and machines. Retinal specialists were equally accurate in grading real and synthetic images, highlighting the realism and quality of the synthetization. These findings suggest the potential use of such synthetic images by retinal specialists for tasks such as education or training. In experiment 2, more than 90% of the synthetic images were noted to be of sufficient quality to be adequately graded. Moreover, differences in the gradability between real and synthetic images were approximately within the 95% CI error margin, suggesting that quality degradation did not result from image generation. In experiment 3, when retinal specialists were made aware of image synthesis and asked to grade whether an image was real or synthetic, their accuracy was close to 50%, indicating that the retinal specialists were no better than random chance at correctly identifying synthetic fundus images.
In experiment 4, the diagnostic performance of the DCNNs showed small reductions in AUC and accuracy after the images used for training of the DLS were changed from only real fundus images to only synthetic images. Reduction in performance was expected. However, it was relatively modest; this result was encouraging in that the DL system still achieved respectable results with performance not far from human performance based on earlier studies9 even though machine training was based completely on synthetic data.
In experiment 5, visual inspection of movies showing generated images and nearest neighbor analysis suggested that a variety of realistic and distinct synthetic images could be generated. In addition, it provided evidence that synthetization when training from images that include small or few medium-sized drusen, corresponding to no AMD or the early stage of AMD, did not lead to the generation of a synthetic image with merged or confluent, larger drusen, consistent with the intermediate stage of AMD. The method did not synthesize images classifiable as referable from images previously classified as nonreferable. It also confirmed that training with an image with small numbers (eg, 15) of medium-sized drusen, corresponding to early-stage AMD and therefore nonreferable AMD, did not result in synthetic images that had more than 20 medium-sized drusen, which would have then led to the synthetic image being considered as showing intermediate (ie, referable) AMD.
Uses of these techniques could include training, education, testing, or augmenting data sets without having to find real images. The GAN-generated synthetic images could be used to create data sets for data augmentation and exchange between research groups, which is currently hampered by personally identifiable information, institutional review board requirements, or other proprietary restrictions. Personally identifiable information for real retinal images may become an irremediable problem considering that retinal vasculature is uniquely identifiable.31 The problem may grow in importance as databases used for biometrics with personally identifiable information are stored with increasing frequency in parallel with recent increases in electronic privacy laws, such as the recently enacted European Union.32 Such laws could possibly preclude the use of such images regardless of the original individual study participant consent or local institutional review board permission.
The techniques in this investigation may address conditions less common than AMD or diabetic retinopathy, wherein a few expertly graded images of rare diseases could be used to synthesize other images for training. This future work, however, would require more progress in GANs to address training with reduced data sets. Another possible use includes research in methods to defeat possible nefarious uses of synthetically developed retinal images, which could be applied to provide false prevalence of disease in a clinical setting or “ghost” study participants.
Limitations
This study considered only a 2-class (referable or nonreferable) AMD classification. Although in theory the method could be extended to more granular classifications11,12 (eg, AREDS’ 9-step detailed AMD severity scale), other pathologic processes (eg, diabetic retinopathy1,2,3 or retinopathy of prematurity8), or other retinal modalities (eg, optical coherence tomography27,30), testing will be required to determine how amenable such extensions are to this approach. In addition, images synthesized in our study were 512 × 512 and corresponded to an effective fundus resolution of 724 × 724 (512*sqrt[2], because we preprocessed images by taking the inscribed square), which is adequate to evaluate most lesions but is a lower resolution than was used in the original AREDS (approximately equal or greater than 2K × 2K); the resolution used in AREDS would be preferable to look at very small lesions. Although there was no intrinsic theoretical limitation of ProGANs to generate higher resolutions (eg, 1024 × 1024 or above), a longer training time would be required (weeks to a month), which was impractical for this study. Future work will involve evaluations at higher resolutions (2K × 2K or above) using similar experimental design that will be useful for evaluating very small lesions.
Conclusions
Experiments suggest that realistic synthetic fundus images of retinal diseases can be generated using GANs and used by clinicians in place of real images (eg, for training and data exchange); these images also hold promise for augmenting DL models while preserving anonymity and obviating constraints owing to institutional review board or other use restrictions. Although unscrupulous uses of the technology could be envisioned, this work might help motivate research into mitigation methods. The proposed generative approach also might alleviate issues of data imbalance and rarity of positive cases, which affects current data sets, such as geographic atrophy cases in AREDS20 or very severe nonproliferative diabetic retinopathy in the Eye Picture Archive Communication System (EyePACS), and might help improve generalizability of DL diagnostic models.
eFigure 1. Parts A through C (top row) and D through F (bottom row)
eTable. Showing Exact Number of Images Used for Training/Validation/Testing for the Various GANs and DCNNs
eFigure 2. The Figure Shows the Nearest Neighbors’ Analysis for Real AREDS Images for Non-Referable Cases
eFigure 3. Showing the Nearest Neighbors’ Analysis for Real AREDS Images for Referable Cases
eAppendix.
References
- 1.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402-2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 2.Quellec G, Charrière K, Boudi Y, Cochener B, Lamard M. Deep image mining for diabetic retinopathy screening. Med Image Anal. 2017;39:178-193. doi: 10.1016/j.media.2017.04.012 [DOI] [PubMed] [Google Scholar]
- 3.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124(7):962-969. doi: 10.1016/j.ophtha.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 4.Ting DSW, Cheung CY, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318(22):2211-2223. doi: 10.1001/jama.2017.18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342-1350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 6.Brown JM, Campbell JP, Beers A, et al. ; Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium . Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136(7):803-810. doi: 10.1001/jamaophthalmol.2018.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM. Comparing humans and deep learning performance for grading AMD: A study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med. 2017;82:80-86. doi: 10.1016/j.compbiomed.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burlina P, Freund DE, Joshi N, Wolfson Y, Bressler NM. Detection of age-related macular degeneration via deep learning. In: 2016 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2016 - Proceedings. Piscataway, NJ: IEEE; 2016:184-188. [Google Scholar]
- 9.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135(11):1170-1176. doi: 10.1001/jamaophthalmol.2017.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burlina P, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Utility of deep learning methods for referability classification of age-related macular degeneration. JAMA Ophthalmol. 2018;136(11):1305-1307. doi: 10.1001/jamaophthalmol.2018.3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grassmann F, Mengelkamp J, Brandl C, et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology. 2018;125(9):1410-1420. doi: 10.1016/j.ophtha.2018.02.037 [DOI] [PubMed] [Google Scholar]
- 12.Burlina PM, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Use of deep learning for detailed severity characterization and estimation of 5-year risk among patients with age-related macular degeneration [published online September 14, 2018]. JAMA Ophthalmol. doi: 10.1001/jamaophthalmol.2018.4118 [DOI] [PMC free article] [PubMed]
- 13.Kankanahalli S, Burlina PM, Wolfson Y, Freund DE, Bressler NM. Automated classification of severity of age-related macular degeneration from fundus photographs. Invest Ophthalmol Vis Sci. 2013;54(3):1789-1796. doi: 10.1167/iovs.12-10928 [DOI] [PubMed] [Google Scholar]
- 14.Burlina P, Freund DE, Dupas B, Bressler N. Automatic screening of age-related macular degeneration and retinal abnormalities. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:3962-3966. [DOI] [PubMed] [Google Scholar]
- 15.Ting DSW, Liu Y, Burlina P, Xu X, Bressler NM, Wong TY. AI for medical imaging goes deep. Nat Med. 2018;24(5):539-540. doi: 10.1038/s41591-018-0029-3 [DOI] [PubMed] [Google Scholar]
- 16.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;2672-2680. [Google Scholar]
- 17.Karras T, Aila T, Laine S, Lehtinen J. Progressive growing of GANs for improved quality, stability, and variation. Preprint. Published online October 27, 2017. arXiv 1710:10196.
- 18.Velez-Montoya R, Oliver SC, Olson JL, Fine SL, Quiroz-Mercado H, Mandava N. Current knowledge and trends in age-related macular degeneration: genetics, epidemiology, and prevention. Retina. 2014;34(3):423-441. doi: 10.1097/IAE.0000000000000036 [DOI] [PubMed] [Google Scholar]
- 19.Bressler NM. Age-related macular degeneration is the leading cause of blindness.... JAMA. 2004;291(15):1900-1901. doi: 10.1001/jama.291.15.1900 [DOI] [PubMed] [Google Scholar]
- 20.Age-Related Eye Disease Study Research Group. The age-related eye disease study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the age-related eye disease study report number 6. Am J Ophthalmol. 2001;132(5):668-681. [DOI] [PubMed] [Google Scholar]
- 21.Bressler NM, Bressler SB, Congdon NG, et al. ; Age-Related Eye Disease Study Research Group . Potential public health impact of Age-Related Eye Disease Study results: AREDS report no. 11. Arch Ophthalmol. 2003;121(11):1621-1624. doi: 10.1001/archopht.121.11.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Age-Related Eye Disease Study Research Group The Age-Related Eye Disease Study (AREDS): design implications. AREDS report no. 1. Control Clin Trials. 1999;20(6):573-600. doi: 10.1016/S0197-2456(99)00031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. CVPR. 2016:771-778. [Google Scholar]
- 24.Costa P, Galdran A, Meyer MI, et al. End to end adversarial retinal image synthesis. IEEE Trans Med Imaging. 2018;37(3):781-791. doi: 10.1109/TMI.2017.2759102 [DOI] [PubMed] [Google Scholar]
- 25.Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging. 2018;31(5):655-669. doi: 10.1007/s10278-018-0056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie D, Trullo R, Lian J, et al. Medical Image Synthesis with Deep Convolutional Adversarial Networks. IEEE Transactions on Biomedical Engineering, 2018: https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=8310638. Accessed November 19, 2018. [DOI] [PMC free article] [PubMed]
- 27.Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In: Niethammer M, Styner M, Aylward S, et al, eds. International Conference on Information Processing in Medical Imaging; 25th International Conference, IPMI 2017, Boone, NC, USA, June 25-30, 2017, Proceedings New York, NY: Springer International Publishing; 2017:146-157. [Google Scholar]
- 28.Mahapatra D, Bhavna A, Suman S, Rahil G Deformable medical image registration using generative adversarial networks. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) Piscataway, NJ: IEEE; 2018:1449-1453. [Google Scholar]
- 29.Baur C, Albarqouni S, Navab N MelanoGANs: high resolution skin lesion synthesis with GANs. Preprint. Published online April 12, 2018. arXiv. 1804.04338.
- 30.Pekala M, Joshi N, Freund DE, Bressler NM, Cabrera Debuc D, Burlina P Deep learning based retinal OCT segmentation. Preprint. Published online January 29, 2018. arXiv. 1801.
- 31.Jain AK, Arun R, Salil P. An introduction to biometric recognition. IEEE Trans Circ Syst Video Tech. 2004;14(1):4-20. doi: 10.1109/TCSVT.2003.818349 [DOI] [Google Scholar]
- 32.Bowles N. After Cambridge Analytica, privacy experts get to say ‘I told you so.’ New York Times April 12, 2018. https://www.nytimes.com/2018/04/12/technology/privacy-researchers-facebook.html. Accessed July 24, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Parts A through C (top row) and D through F (bottom row)
eTable. Showing Exact Number of Images Used for Training/Validation/Testing for the Various GANs and DCNNs
eFigure 2. The Figure Shows the Nearest Neighbors’ Analysis for Real AREDS Images for Non-Referable Cases
eFigure 3. Showing the Nearest Neighbors’ Analysis for Real AREDS Images for Referable Cases
eAppendix.