Abstract
Vast repertoires of unique antigen receptors are created in developing lymphocytes. The antigen receptor loci contain many variable (V), diversity (D), and joining (J) gene segments that are arrayed across very large genomic expanses and are joined to form variable-region exons. This process creates the potential for an organism to respond to large numbers of different pathogens. Here, we consider the underlying molecular mechanisms that favor some V genes for recombination prior to selection of the final antigen receptor repertoire. We discuss chromatin structures that form in antigen receptor loci to permit spatial proximity among the V, D, and J gene segments and how these relate to the generation of antigen receptor diversity.
Keywords: antibody genes, B cells, V(D)J recombination
Introduction
In vertebrates, the adaptive immune response is capable of recognizing pathogens using antigen-specific receptors expressed on B and T lymphocytes. The B-cell receptor (BCR) is composed of two identical immunoglobulin (Ig) heavy chains (IgH) and two identical light chains (Igκ or Igλ). There are two lineages of T cells that are distinguished by the type of T-cell receptor (TCR) expressed. TCRαβ is encoded by the Tcra and Tcrb loci, whereas TCRγδ is encoded by the Tcrg and Tcrd loci. Antigen receptors are composed of variable (V) and constant (C) regions. The organization of the Igh and Igκ loci are schematically depicted ( Figure 1A and Figure 2A). Igh variable-region exons are produced by the joining of one each of the many variable (V), diversity (D), and joining (J) gene segments, whereas Igκ and Igλ are created by joining one each of the V and J gene segments, all by V(D)J recombination during lymphocyte development ( Figure 1B). V(D)J recombination is a stepwise process during which D H-to-J H recombination occurs first followed by V H-to-D HJ H rearrangement. This process depends on the lymphocyte-specific V(D)J recombinase, RAG1/2, which recognizes recombination signal sequences (RSSs) that flank all V, D, and J gene segments 3. During V(D)J recombination, two RSSs adjacent to V, D, or J gene segments partner such that cleavage and rejoining occur. RAG1 contains endonuclease activity and targets the RSS, and RAG2 is recruited to the epigenetically modified histone 3 when it is trimethylated on lysine 4 3. Within each antigen receptor locus, the RAG recombinase concentrates in the recombination center (RC) that is focused to the J segment–containing domain. Double-strand DNA breaks are generated at RSSs by RAG1/2, and the V, D, and J exons are joined together through non-homologous end joining 4.
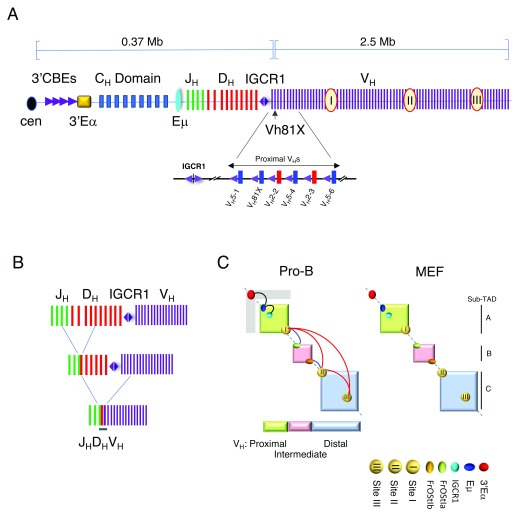
Figure 1. Overview of the Igh locus.
The Igh locus spans 2.9 Mb and contains about 100 V H gene segments. ( A) (Upper panel) Schematic diagram of the Igh locus showing the V Hs, Ds, J Hs, and C H exons and regulatory elements (not to scale). The V H7183 and V HQ52 families—blue and red bars, respectively (lower panel)—are located at the D HJ H-proximal end of the locus. Each D HJ H-proximal V H gene segment is paired with a recombination signal sequence (not shown) and a CTCF-binding element (CBE) (purple triangles). The CBE associated with the V H5-1 segment is non-functional (gray triangle). CBE orientation is indicated by the direction of the triangle. V H gene segment names indicate their position along the locus. V H81X (V H5-2) is the original name of the second gene segment relative to intergenic control region 1 (IGCR1) and is used because it is well known by this nomenclature. The intermediate V H segments include the V HS107 family along with nine smaller V H families. At the 5′ end of the locus, the interspersed distal V H segments are composed of the V HJ558 and V H3609 families. Regulatory elements include intronic Eμ and 3′Eα super-enhancers and IGCR1, which is composed of two divergent CBEs. A cluster of at least nine CBEs is located at the 3′ boundary of the Igh locus and is adjacent to 3′Eα. The 3′CBEs and 3′Eα are referred to as the 3′ regulatory region (3′RR). Sites I, II, and III (red circles) engage in exceptionally long-range looping interactions and may mediate locus compaction. Sub-topologically associating domain (Sub-TADs) A, B, and C are indicated. ( B) Diagram of the stepwise process of V(D)J recombination. D-J rearrangement precedes V-DJ recombination. ( C) A schematic of the Igh TAD in pro-B cells that is subdivided into three sub-TADs A, B, and C. Looping interactions between Eμ:3′Eα and Eμ:IGCR1 (black arcs), Sites I and II, Sites II and III, Sites I to III (red arcs), Site I-FrOStIa, and Site II-FrOStIb (blue arcs) were detected and are not described here in detail 1.
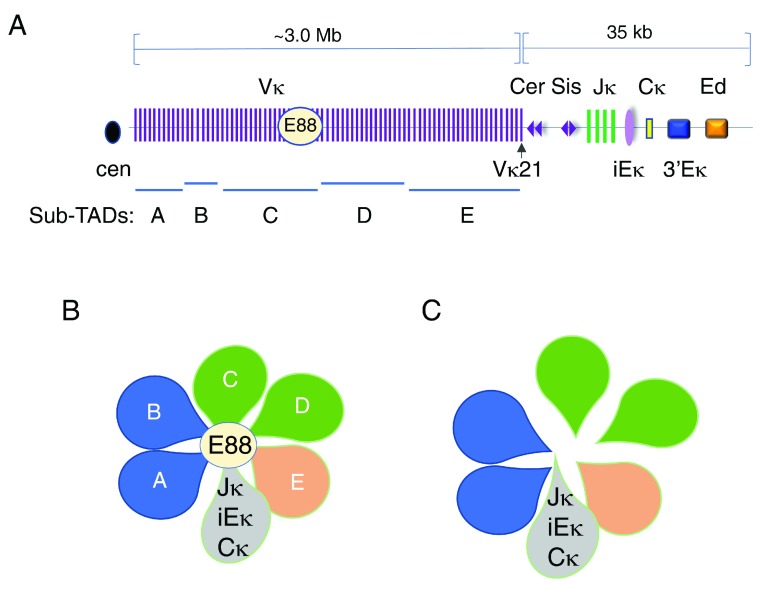
Figure 2. Three-dimensional conformation of the Igκ locus.
The Igκ locus spans 3.2-Mb topologically associating domain (TAD) and contains about 120 functional Vκ gene segments. ( A) Schematic diagram of the Igκ locus showing the Vs, Js, and C exons and regulatory elements (not to scale). Regulatory elements include intronic Eκ (iEκ), 3′Eκ, Ed, and E88 elements. Contracting element for recombination (Cer) and silencer in the intervening sequence (Sis) are located between the V and J domains and are composed of CTCF-binding elements (CBEs) (purple triangles). The orientation of each CBE is indicated. The Igκ locus is subdivided into five sub-TADs (A–E) as indicated. ( B) Sub-TAD structure of the Igκ locus as determined by Hi-C 2. Each loop represents a sub-TAD that is labeled A–E. The regulatory region containing the Jκ genes, the three distal enhancers, and the constant region are in gray. ( C) Deletion of E88 results in untethering of sub-TADs C and D from the regulatory region.
Antigen receptor gene rearrangement is tightly regulated during lymphocyte development; in turn, lymphocyte development is strictly dependent on V(D)J recombination 5, 6. The composition and complexity of antigen receptor repertoires depend on the number of V, D, and J gene segments and the degree to which those segments are available for rearrangement. However, V gene usage in the pre-selected Igh repertoire is only quasi-random since it has been shown that V genes rearrange at very different intrinsic frequencies 7– 14. No one factor, or combination of factors, could fully account for unequal V gene usage in studies considering V germline transcript levels, transcription factor (TF) binding, RSS quality, and the distribution of a variety of epigenetic marks 7– 9, 11, 15. Hence, the mechanisms underlying V gene rearrangement frequencies remain to be determined.
Antigen receptor loci are quite large spanning 0.67 Mb- 3.0 Mb and containing up to 100 functional V genes. Chromatin conformational changes in antigen receptor loci are important determinants for long-distance V(D)J recombination events 6. Developmental stage-specific contraction of Ig and TCR loci promotes proximity of J-distal V genes with (D-)J segments and generally is thought to facilitate recombination but this has not been formally proven 16– 19. Two currently unresolved questions in the formation of the antigen receptor repertoires are (1) what is the molecular basis for locus contraction that is hypothesized to support V->DJ recombination over exceptionally long genomic distances and (2) what underlies the unequal rearrangement potential of individual V genes? Here, we focus on the murine Igh and Igκ loci to address these questions. The molecular principles resulting from these studies may be generally applicable to all antigen receptor loci.
Locus contraction is a feature of antigen receptor loci
Developmental activation of the Igh locus is a stepwise process that features acquisition of epigenetic modifications, DNase I hypersensitive sites, and the onset of sense and anti-sense transcription 16, 20– 23. Additionally, the Igh locus undergoes large-scale locus contraction during development that is detected by using three-dimensional (3D) DNA fluorescence in situ hybridization (FISH) methods 24, 25.
The early observations by Kosak revealed two fundamental findings regarding the disposition of the Igh locus in the nucleus 24. First, the Igh locus is located at the nuclear periphery in non-B cells and relocates to the nuclear center at the pro-B cell stage 24 through a process that requires active dislocation from the nuclear lamina 26. Second, the Igh locus is in an extended conformation in non-B cells and lymphoid progenitors, whereas both Igh alleles are contracted in pro-B cells, a developmental stage coincident with V(D)J recombination 24, 25. These pioneering studies have led to the recognition that all of the large antigen receptor loci undergo developmentally regulated conformational changes before rearrangement at that locus 24, 25, 27– 30. The contracted Igh locus in pro-B cells undergoes decontraction at the pre-B cell stage of development to prevent a second round of V H-D HJ H rearrangement on the second Igh allele, presumably aiding allelic exclusion 28. Degrees of locus compaction have been inferred from the relationship of inter-probe nuclear distances derived from 3D DNA FISH versus genomic distances and these measurements have limited resolution (100–1000 nm). Consequently, it has been difficult to identify DNA elements that mediate locus contraction.
Igh locus contraction depends on the TFs Pax5, Ikaros, and YY1 25, 31, 32. Loss of Igh locus compaction is correlated with preferential usage of the most D H-proximal V H genes 25, 31, 33, indicating that spatial access to the more distally positioned V H gene segments has been lost. Although depletion of any of these TFs reduces distal V H rearrangement, chromatin accessibility remains unchanged 25, 31, 33. Low-level transcription over V H genes and intergenic regions occurs as the locus is preparing to undergo rearrangement 34– 37. The highest level of non-coding RNA (ncRNA) in the Igh locus is found at elements called Pax5-activated intergenic repeats (PAIRs) and these ncRNAs are dependent upon the presence of Pax5 and YY1 37, 38. Although the function of TFs in locus contraction remains speculative, PAIR elements have been suggested to induce long-range chromatin looping by relocating to transcription factories where they associate with the 3′ proximal Eμ-J H-D H domain 37. However, the molecular mechanism that mediates locus contraction remains unclear.
The Igh locus is conformationally distinct in pro-B cells
Eukaryotic chromosomes are organized into higher-order spatial configurations of multiple-length scales as determined by using high-resolution chromosome conformation capture (3C)-based approaches and microscopy-based methods, including 3D DNA FISH and live cell imaging 39– 47. For example, insulators and enhancers often engage in physical interactions with their target promoters 48– 51, indicating that regulatory elements can control distant gene expression through direct long-range molecular contact. However, not all long-range chromatin interactions are directed toward regulating gene expression. For example, intra-chromosomal interactions are required to regulate V(D)J recombination and Ig class-switch recombination (CSR) 6, 16, 17, 52. In CSR, the constant (C H)-region exons encoding IgM are substituted with a downstream C H gene such that IgM is no longer produced and instead IgG, IgE, or IgA is made in conjunction with the original recombined variable-region exons. CSR is dependent on 3D chromatin architecture mediated by long-range intra-chromosomal interactions between distantly located transcriptional elements 53– 56. During V(D)J recombination, antigen receptor genes undergo ordered rearrangement with D H-to-J H joining preceding V H-to-D HJ H recombination 4. To produce a fully representative Ig repertoire, it is essential that the distal V H genes achieve spatial proximity with the RC and D HJ H domain. Murre and colleagues have shown that Igh locus topology is best described as a series of three large chromatin loops joined by linkers in pre-pro-B cells but that these loops have intermingled and provide equal access of the D H-distal and -proximal V H gene segments with rearranged 3′ D HJ H in pro-B cells 57. The time interval for D HJ H to gain proximity with a V H gene segment is on the order of minutes, and spatial confinement of topological domains largely regulates first-passage times for chromatin interactions in vivo 58. Although it is clear that Igh locus conformation is structured, the DNA elements that anchor chromatin looping in support of V(D)J recombination remain largely undefined.
The Igh and Igκ loci are configured as topologically associating domains
Topologically associating domains (TADs) are megabase sized and represent regions of high-frequency self-interacting chromatin contacts as defined in 3C-based studies 59, 60. The organization of interphase chromatin is largely conserved between cell types, especially with regard to TAD boundaries 44, 50, 61. Strikingly, the Igh locus is contained within a 2.9-Mb TAD in pro-B cells 1.
The murine Igh TAD is partitioned into two highly structured sub-TADs A and C—corresponding to the D H-proximal and D H-distal V H gene families, respectively—and flank a less structured sub-TAD B that includes the intermediate V H gene segments 1 ( Figure 1C). Sub-TADs are zones within a TAD in which chromatin contacts are more frequent than with sites outside the sub-domain, and contacts can be tissue-specific and can contribute to the overall architectural structure of the TAD 49, 62, 63.
V genes can be subdivided into V families based on sequence relatedness and this reflects gene duplication and divergence of primordial V genes. The correspondence of sub-TAD structure with the murine V H gene family distribution profile is striking ( Figure 1C). In the murine Igh locus, V H families tend to be clustered, but in most other antigen receptor loci and in other species, the members of individual V H families generally are interspersed.
The Igκ locus is also contained within a 3.5-Mb TAD which is subdivided into five sub-domains 2 ( Figure 2A, B). However, because Vκ gene families are interspersed across the locus, there is no correspondence between Vκ families and sub-TAD structure. One unique feature of the Igκ locus is that about one third of Vκ genes are present in the reverse orientation such that they rearrange to Jκ genes by inversion as opposed to the predominant deletional rearrangement found at other antigen receptor loci. However, there is no correlation between sub-TAD structure and the inversional or deletional orientation of Vκ genes. The conservation of Igκ TAD and sub-TAD structure has not been examined in different cell types.
Igh TAD conformation is sculpted by developmentally specific chromatin looping
TADs can be thought of as scaffolds for constitutive architectural interactions. Nevertheless, interactions within TADs may vary significantly between cell types or developmental stages and for private enhancer–promoter contacts 44, 50, 64– 68. Igh sub-TADs A, B, and C become juxtaposed in pro-B cells via megabase-scale chromatin looping but these contacts are absent in non-B cells 1. The loop anchors located in sub-TAD A and C are termed sites I, II, and III ( Figure 1C). Our FISH studies indicated that sites I, II, and III participate in three-way physical contacts in about 32% of pro-B cells and in less than 5% of non-B cells and may functionally create proximity between the distal V H domain with the RC/D H/J H region to facilitate efficient access of all V H gene segments for recombination 1.
The structure of sub-TAD A is worthy of additional consideration as it contains the Eμ and 3′Eα enhancers, the RC located in the J H-D H domain, intergenic control region 1 (IGCR1) (an insulator which will be discussed in detail in sections below), and the proximal V H genes ( Figure 1A). Sub-TAD A becomes modified in pro-B as compared with non-B cells. In non-B cells, Igh sub-TAD A encompasses the proximal V H genes spanning from site I to Eμ ( Figure 1C). In pro-B cells, sub-TAD A becomes subsumed within a larger topological fold that extends from site I to the 3′Eα enhancer ( Figure 1C). The higher-order chromatin structure in pro-B cells may have significant implications for D H-proximal V H gene usage during V(D)J recombination.
Several earlier observations have shown that D H-proximal V H genes are regulated differently from the rest of the V H genes. Although distal V H gene recombination is reduced in Pax5-, YY1-, and Ezh2-deficient pro-B cells, D H-proximal V H genes recombine normally 25, 31– 33. Thus, the localization of the D H-proximal V H genes within the same conformational sub-TAD as the RC/D H/J H region distinguishes them from distal V H genes that lie within sub-TADs B and C.
Pax5 organizes sub-TAD C that spans the distal V HJ558 gene family. Site III within sub-TAD C fails to associate with sites I and II in Pax5-deficient pro-B cells, thus providing a possible explanation for reduced V HJ558 rearrangements in Pax5-deficient pro-B cells 1. Notably, 14 PAIR elements that were proposed to mediate locus compaction via Pax5 are all situated within sub-TAD C, and PAIR motifs 10 and 11 overlap with site III 1, 38. PAIR elements are bound by the TFs Pax5, E2A, and CTCF (CCCTC-binding factor) in pro-B cells 38. It is not known whether transcriptional activity at PAIR elements regulates chromatin looping. Our studies provide a potential molecular definition of locus contraction by identifying loop anchor sites that are key mediators of this process.
CTCF mediates insulator function at TAD boundaries
TAD boundaries are frequently enriched for CTCF binding and CTCF-binding elements (CBEs) 45, 59, 60, 62, 63. CTCF is a ubiquitously expressed zinc-finger protein that binds DNA, functions as an insulator in vertebrates 69, and plays a key role in chromatin looping 45, 63, 70, 71. There is an observed inward or convergent orientation of CBEs flanking TADs 45, 70, 72, 73. Insulators were originally defined as genomic elements that act as a barrier to position effects caused by the spreading of chromatin marks and they block enhancer activity 74, 75. Although loci situated within TADs are relatively insulated from loci outside the domain, these same elements readily interact with other loci within the same domain. CRISPR/Cas9-mediated rearrangements of TAD boundaries and regulatory elements facilitate or prevent looping interactions with distal regulatory elements 76– 78. Acute depletion of CTCF leads to loss of loop domains and impaired regulation of nearby genes through loss of enhancer insulation 79.
High-resolution in situ Hi-C studies demonstrated that mammalian genomes are partitioned into contact domains 45. Contact domains with end points that anchor a loop are referred to as loop domains 45, 70. TADs are most frequently loop domains but not all loop domains are TADs. In the context of V(D)J recombination, RAG recombinase activity was shown to be confined to loop domains that are defined by convergent CTCF-bound elements. RAG primarily initiates double-stranded breaks (DSBs) at RSSs within the antigen receptor loci. However, RAG can also initiate low-frequency DSBs at off-target sites that have sequence similarity to RSSs and cause chromosomal rearrangements and translocations 52, 80– 82. Notably, when RAG was experimentally directed to chromosomal domains outside of antigen receptor loci, off-target DSBs were confined within loop domains and deletion of convergent CBEs extended the range of RAG activity 83.
CTCF partners with cohesin to mediate chromatin looping
CTCF-based long-range looping interactions are dependent on co-binding with cohesin 84, 85. The cohesin complex is thought to form a ring around two CTCF proteins bound to DNA 85, 86. Different combinations of architectural proteins may mediate context-specific genomic organization 63, 87. Promoter–enhancer interactions are disrupted in embryonic stem cells 88 and in thymocytes 89 when cohesin is depleted. There is a rich CTCF-cohesin landscape in the Igh locus. One hundred thirty-two sites are bound by CTCF and cohesin and the majority of these are located at a distance of 1 to 32 kb from V H gene segments in the Igh locus 90, 91. Strikingly, all of the rearranging D H-proximal V H genes are closely paired with CBEs that are located within 68 base pairs (bp) of the RSS ( Figure 1A) 90. However, CBEs in the non-rearranging D H-proximal V H genes are located more than 1 kb from the RSS in the two most D H-proximal V H gene families. As described below, close proximity to the adjacent CBE has functional consequences for these V H genes 7, 91, 92. In addition, a cluster of nine CBEs marks the 3′ boundary of the Igh TAD 92, and two CBEs located within IGCR1 mark the boundary between the D HJ H domain and the D H-proximal V H genes ( Figure 1B) 20, 93, 94. Similarly, the Tcrb and Tcrd loci have CBEs located between the V and J gene segments 30, 95, 96. In the Igκ locus, CBEs, termed contracting element for recombination (Cer) and silencer in the intervening sequence (Sis), are located between the V and J gene segments, and many CBEs are found throughout the Vκ domain ( Figure 2A) 91, 97, 98.
In the Igh and TCRβ loci, all bound CTCF sites in the V exon domains upstream of the D-J-C-regions are oriented toward them, and the CBEs in D-J-C regions of those loci are oriented toward the V exons. In contrast, the other two large antigen receptor loci (TCRα/δ and Igκ) have more complex patterns with the bound CTCF sites in the V gene portion of each locus found in both orientations 91. A role for the CTCF-cohesin complex in Igh locus looping has been suggested by shRNA knockdown studies in pro-B cells demonstrating that the Igh locus is less contracted after CTCF is knocked down 99.
A convergence of loop extrusion and directional RAG tracking?
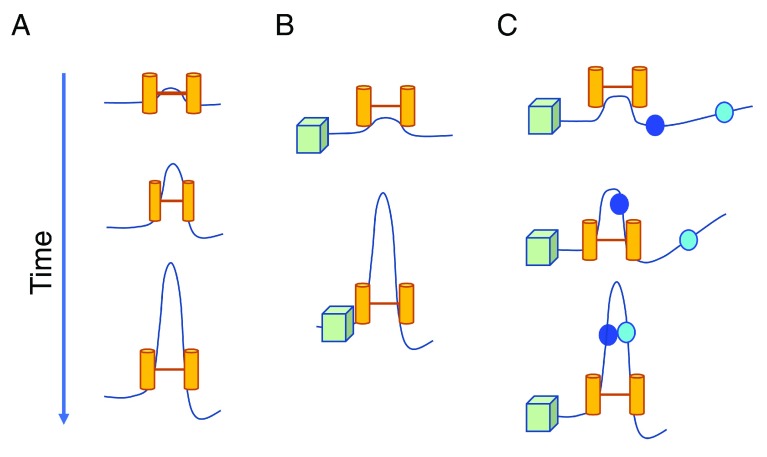
TADs and loop domains have been implicated in regulating gene expression in mammalian cells 40, 86, with convergent CBEs in a large subset of cases 72, 73, 100, 101. It has been proposed that TADs can be formed by the loop extrusion activity of cohesin ( Figure 3A) 100, 102. When cohesin is bound to chromatin, it forms a progressively larger loop until it encounters an obstacle formed by another cohesin or boundary protein including CTCF ( Figure 3B). The association of CTCF with widely separated convergent CBEs may involve cohesin that is halted upon arriving at convergent CTCF-bound loop anchors 40, 100, 102, 103. It has been proposed that loop extrusion may also facilitate close-range contacts between regulatory elements, including promoters and enhancers, by bringing them into molecular contact 40 ( Figure 3C). Promoter–enhancer interactions may preferentially occur within chromatin domains that are insulated by extrusion blocking factors.
Figure 3. Loop extrusion as a topologically associating domain generating machine.
( A) The chromatin fiber extrudes over time through an extruding factor (possibly cohesin; yellow cylinders). ( B) A boundary element (possibly CTCF, green cube) can block loop extrusion when the CTCF-binding element is in the proper orientation. It has been proposed that CTCF can block extrusion by one of the cohesin extruding motors while the second motor will be unobstructed and continue to extrude the loop 102. ( C) Regulatory elements may come into close molecular contact by the process of loop extrusion. These interactions will occur only within a topologically associating domain and in the presence of extrusion blocking elements.
In a situation strikingly analogous to convergent CBE-mediated loop formation, RAG-dependent recombination involves interactions between distant convergent RSSs with the exception of inverted RSSs in some antigen receptor loci. The Alt group has shown that RAG off-target activity within CTCF loop domains spanning 2 Mb depends on orientation-specific RSSs 83. It was inferred from DNA sequencing data that RAG can travel directionally from a physiological or ectopically introduced RC within a convergent CBE-based loop domain of megabase size 83. Long-range directional exploration by RAG can be blocked by an encounter with cohesin-bound convergent CBE pairs and possibly by other impediments that create chromatin sub-domains within TADs 83, 104. Alt and colleagues proposed that RAG complexes bind one RSS and then track along the chromatin fiber in a linear fashion to the next convergent RSS 82, 104. Several different topological machine models have been postulated to explain directional cis-guided long-range looping interactions 40. It remains unclear whether RAG tracking occurs via loop extrusion or by a mechanistically different activity.
IGCR1 is an insulator that partitions the D HJ H domain from V H genes
CTCF has been implicated as a mediator of transcriptional insulation through its ability to participate in chromatin looping 71. The striking number and organization of CBEs across antigen receptor loci have led to the proposal for a role of CBE in V(D)J recombination 93, 99. The Igh sub-TAD A contains several important looping contacts, including Eμ-IGCR looping interactions in pro-B cells ( Figure 1C) 93, 105. IGCR1 contains a pair of divergent CBEs that demarcate the boundary of the RC/D HJ H domain and function as an insulator that prevents D H-to-V H joining prior to D HJ H rearrangements 93, 94. However, the relationship of CTCF-anchored chromatin looping for V H:IGCR1 and antigen receptor rearrangement frequency remains unclear.
V H CBEs are convergently oriented with respect to the upstream IGCR1 CBE, and 3′ CBEs are convergently oriented relative to the downstream IGCR1 CBE 93 ( Figure 1A). Although CBE-dependent Eμ:IGCR1 looping is prominent in pro-B cells, it is striking that Eμ:V H81X contacts are largely undetectable in the wild-type context, indicating that the RC located between Eμ and IGCR1 is sequestered away from all V H genes 106, 107. The functional D H-proximal V H genes are very closely paired with CBEs 90. For example, V H81X is the first functional proximal V H gene located about 100 kb from IGCR1 and it is immediately adjacent to a CBE ( Figure 1A). Two new studies have shown that when the IGCR1 CBEs are deleted, Eμ-V H81X contacts are newly observed, indicating that IGCR1 CBEs prevent looping interactions between the Eμ-RC/D HJ H domain and the proximal V H genes 106, 107. Strikingly, IGCR1:V H81X interactions are dependent on the V H81X CBE, as shown by deletion of the V H81X-flanking CBE.
Interestingly, the Igκ locus contains the Cer/Sis CBEs in the V-J intervening region ( Figure 2A). Deletion or inversion of Cer leads to preferential usage of Jκ-proximal Vκ genes 97, 108, highlighting the importance of convergent CTCF-mediated long-range interactions that facilitate spatial proximity of the distal Vκ with the J segments. Cer/Sis and IGCR1 are similarly located between the V genes and the (D)J genes, and both are involved in mediating chromatin looping. Cer also functions as a transcriptional insulator 108.
Proximal V H gene rearrangement frequencies are determined by CTCF looping
To begin, one might expect that the V H5-1 gene segment would be highly used in V H-to-D HJ H rearrangements since it is most proximal to the RC/D HJ H domain ( Figure 1A). However, despite being paired with a highly conserved RSS, V H5-1 is not used in V(D)J recombination. In contrast, V H81X (V H5-2), the next V H gene segment along the genome, is the most frequently used in V(D)J recombination. The question of why V H81X and not V H5-1 is used is long-standing. Two groups have explored the relationship between CTCF-mediated chromatin looping and proximal V H gene usage during V(D)J recombination 106, 107.
CBEs adjacent to the functional D H-proximal V H genes are found within 68 bp downstream of the RSSs 90. Mutagenesis analyses have revealed that proximal V H CBEs dramatically influence the frequency of V(D)J rearrangement of that V H gene 106, 107. Mutation of the CBE associated with V H81X (V H5-2) ( Figure 1A) greatly reduced both looping with IGCR1 and its rearrangement frequency and boosted the rearrangement frequency of the next most upstream V H gene, V H2-2 106. Genomic editing of the non-functional V H5-1 CBE into a functional motif turns this non-rearranging V H gene into the most frequently rearranging gene 106 ( Figure 1A). Thus, as discussed below, CBE quality and chromatin looping between IGCR1 and the D H-proximal VH gene segments are significant factors determining V H gene usage in V(D)J recombination.
The antigen receptor loci have a much higher density of CTCF sites than the genome overall, making CTCF/cohesin a candidate for forming multiple long-range loops within these loci 90, 91. Although it is clear that TAD boundaries are usually formed between convergent CBEs 45, 72, relatively little is known regarding the CBE orientation dependence in anchoring chromosome loops within the V domains of Ig loci. All of the bound CBEs in the V H domain are oriented toward the 3′ regulatory region (3′RR) and a single CBE within IGCR1 ( Figure 1A). If CTCF-mediated looping occurs only between convergent CBE, one would predict that the orientation of motifs adjacent to proximal V H genes will be critically required for looping and V(D)J rearrangement. However, when the V H81X CBE was inverted, usage of V H81X in V(D)J rearrangement was only modestly decreased 106, indicating that the orientation specificity inside the V H sub-TADs is not strictly required.
Together, these studies demonstrate that the proximal V H gene CBE’s quality determines looping efficiency with IGCR1 and determines that V H gene’s recombination efficiency. It is noteworthy that most V H and all Vκ genes do not have any CTCF sites in close proximity, in contrast with the location of CBEs for the proximal V H genes 90, 91. Thus, for the majority of V genes, CBE-mediated looping with IGCR1 may have a less straightforward impact on V H gene rearrangement frequency.
Vκ rearrangement frequency is determined by enhancer E88
In addition to long-range loops mediated by CTCF, other long-range loops can be enhancer-mediated. The Igκ locus is encompassed within a TAD that is subdivided into at least five sub-TADs A–E based on Hi-C studies ( Figure 2A) 2. We identified a novel enhancer element, E88, which is located close to the boundary separating sub-TADs C and D and which becomes active at the pro-B cell stage prior to V-J rearrangement ( Figure 2A) 2. E88 is the major site of interaction with iEκ as detected by 4C analyses in pro-B cells. In pre-B cells, the stage at which V-J recombination occurs, E88 continues to interact strongly with iEκ and also contacts many more sites throughout the locus ( Figure 2B). Strikingly, deletion of E88 results in significant changes in long-range looping interactions and in reduction in rearrangement levels of adjacent Vκ genes ( Figure 2C). Its deletion also results in a modest but consistent reduction of rearrangement of almost all Vκ genes in a 1.5 Mb region surrounding E88 that corresponds to sub-TADs C and D ( Figure 2C). Most Vκ genes that are upstream and downstream of sub-TADs C and D—located in sub-TADs A and B and sub-TADs E, respectively—were modestly increased in relative rearrangement frequency 2. Thus, our studies revealed the novel concept that Vκ rearrangement is regulated in a domain-specific manner and suggest that sub-TAD structure has functional ramifications.
Future questions
Chromatin conformation is now recognized as an important feature regulating gene expression and recombination. Although locus contraction has been a recognized feature of antigen receptor loci for more than 15 years, its underlying molecular mechanism remains largely undefined. Recent studies have provided new insights regarding the convergence of chromatin conformation, TAD and sub-TAD structure, and CTCF-cohesin-mediated looping with V(D)J recombination. These studies revealed that the conformational organization of the Igh, Igκ, and TCRα/δ loci has significant implications for locus contraction and likely influences skewed V gene usage that together affects the composition of the pre-selected repertoires. Going forward, studies focused on the relationship of CTCF- and promoter-enhancer-mediated chromatin looping with locus contraction are likely to provide new insights. Studies designed to clarify the relationship of CTCF-dependent looping and D H-distal V gene rearrangement will be important. It is likely that new enhancers, similar to Igκ E88, will be characterized. The influence of individual enhancers on the frequency of individual V gene usage during initial repertoire formation will be important. The emergence of extremely high-resolution DNA FISH is likely to provide additional insights into locus conformation. Finally, studies that determine the extent to which the pre-selected repertoire determines the shape of the peripheral repertoire will yield new insights.
Abbreviations
3C, chromosome conformation capture; 3D, three-dimensional; bp, base pairs; C, constant; CBE, CTCF-binding element; Cer, contracting element for recombination; CSR, class-switch recombination; CTCF, CCCTC-binding factor; D, diversity; DSB, double-stranded break; FISH, fluorescence in situ hybridization; Ig, immunoglobulin; IGCR1, intergenic control region 1; IgH, immunoglobulin heavy chain; J, joining; ncRNA, non-coding RNA; PAIR, Pax5-activated intergenic repeat; RC, recombination center; RSS, recombination signal sequence; Sis, silencer in the intervening sequence; TAD, topologically associating domain; TCR, T-cell receptor; TF, transcription factor; V, variable
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Yehudit Bergman, Department of Developmental Biology and Cancer Research, Hebrew University of Jerusalem, Jerusalem, Israel
Harry W Schroeder, Department of Genetics, University of Alabama at Birmingham, Birmingham, Alabama, USA
Craig H Bassing, Department of Pathology and Laboratory Medicine, Children's Hospital of Philadelphia, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, 19104, USA
Funding Statement
This work was supported by NIH RO1AI121286 and R21AI137865 to ALK and R56AI19092, R21137867, and R01AI082918 to AJF.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved]
References
- 1. Montefiori L, Wuerffel R, Roqueiro D, et al. : Extremely Long-Range Chromatin Loops Link Topological Domains to Facilitate a Diverse Antibody Repertoire. Cell Rep. 2016;14(4):896–906. 10.1016/j.celrep.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barajas-Mora EM, Kleiman E, Xu J, et al. : A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol Cell. 2019;73(1):48–60.e45. 10.1016/j.molcel.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teng G, Schatz DG: Regulation and Evolution of the RAG Recombinase. Adv Immunol. 2015;128:1–39. 10.1016/bs.ai.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 4. Alt FW, Zhang Y, Meng FL, et al. : Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152(3):417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krangel MS: Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21(2):133–139. 10.1016/j.coi.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bossen C, Mansson R, Murre C: Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. 10.1146/annurev-immunol-020711-075003 [DOI] [PubMed] [Google Scholar]
- 7. Choi NM, Loguercio S, Verma-Gaur J, et al. : Deep sequencing of the murine IgH repertoire reveals complex regulation of nonrandom V gene rearrangement frequencies. J Immunol. 2013;191(5):2393–2402. 10.4049/jimmunol.1301279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolland DJ, Koohy H, Wood AL, et al. : Two Mutually Exclusive Local Chromatin States Drive Efficient V(D)J Recombination. Cell Rep. 2016;15(11):2475–2487. 10.1016/j.celrep.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Kleiman E, Loguercio S, Feeney AJ: Epigenetic Enhancer Marks and Transcription Factor Binding Influence Vκ Gene Rearrangement in Pre-B Cells and Pro-B Cells. Front Immunol. 2018;9:2074. 10.3389/fimmu.2018.02074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aoki-Ota M, Torkamani A, Ota T, et al. : Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J Immunol. 2012;188(5):2305–2315. 10.4049/jimmunol.1103484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matheson LS, Bolland DJ, Chovanec P, et al. : Local Chromatin Features Including PU.1 and IKAROS Binding and H3K4 Methylation Shape the Repertoire of Immunoglobulin Kappa Genes Chosen for V(D)J Recombination. Front Immunol. 2017;8:1550. 10.3389/fimmu.2017.01550 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Williams GS, Martinez A, Montalbano A, et al. : Unequal V H gene rearrangement frequency within the large V H7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J Immunol. 2001;167(1):257–263. 10.4049/jimmunol.167.1.257 [DOI] [PubMed] [Google Scholar]
- 13. Feeney AJ, Goebel P, Espinoza CR: Many levels of control of V gene rearrangement frequency. Immunol Rev. 2004;200:44–56. 10.1111/j.0105-2896.2004.00163.x [DOI] [PubMed] [Google Scholar]
- 14. Feeney AJ, Lugo G, Escuro G: Human cord blood kappa repertoire. J Immunol. 1997;158(8):3761–3768. [PubMed] [Google Scholar]
- 15. Degner-Leisso SC, Feeney AJ: Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Semin Immunol. 2010;22(6):346–352. 10.1016/j.smim.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumari G, Sen R: Chromatin Interactions in the Control of Immunoglobulin Heavy Chain Gene Assembly. Adv Immunol. 2015;128:41–92. 10.1016/bs.ai.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 17. Ebert A, Hill L, Busslinger M: Spatial Regulation of V-(D)J Recombination at Antigen Receptor Loci. Adv Immunol. 2015;128:93–121. 10.1016/bs.ai.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 18. Carico Z, Krangel MS: Chromatin Dynamics and the Development of the TCRα and TCRδ Repertoires. Adv Immunol. 2015;128:307–361. 10.1016/bs.ai.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 19. Majumder K, Bassing CH, Oltz EM: Regulation of Tcrb Gene Assembly by Genetic, Epigenetic, and Topological Mechanisms. Adv Immunol. 2015;128:273–306. 10.1016/bs.ai.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 20. Featherstone K, Wood AL, Bowen AJ, et al. : The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem. 2010;285(13):9327–9338. 10.1074/jbc.M109.098251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garrett FE, Emelyanov AV, Sepulveda MA, et al. : Chromatin architecture near a potential 3' end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25(4):1511–1525. 10.1128/MCB.25.4.1511-1525.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maes J, Chappaz S, Cavelier P, et al. : Activation of V(D)J recombination at the IgH chain JH locus occurs within a 6-kilobase chromatin domain and is associated with nucleosomal remodeling. J Immunol. 2006;176(9):5409–5417. 10.4049/jimmunol.176.9.5409 [DOI] [PubMed] [Google Scholar]
- 23. Chowdhury D, Sen R: Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 2001;20(22):6394–6403. 10.1093/emboj/20.22.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kosak ST, Skok JA, Medina KL, et al. : Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Fuxa M, Skok J, Souabni A, et al. : Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18(4):411–422. 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Zullo JM, Demarco IA, Piqué-Regi R, et al. : DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149(7):1474–1487. 10.1016/j.cell.2012.04.035 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Sayegh CE, Jhunjhunwala S, Riblet R, et al. : Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19(3):322–327. 10.1101/gad.1254305 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Roldán E, Fuxa M, Chong W, et al. : Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6(1):31–41. 10.1038/ni1150 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Shih HY, Krangel MS: Distinct contracted conformations of the Tcra/Tcrd locus during Tcra and Tcrd recombination. J Exp Med. 2010;207(9):1835–1841. 10.1084/jem.20100772 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Chen L, Carico Z, Shih HY, et al. : A discrete chromatin loop in the mouse Tcra- Tcrd locus shapes the TCRδ and TCRα repertoires. Nat Immunol. 2015;16(10):1085–1093. 10.1038/ni.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Liu H, Schmidt-Supprian M, Shi Y, et al. : Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21(10):1179–1189. 10.1101/gad.1529307 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Reynaud D, Demarco IA, Reddy KL, et al. : Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9(8):927–936. 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Hesslein DG, Pflugh DL, Chowdhury D, et al. : Pax5 is required for recombination of transcribed, acetylated, 5' IgH V gene segments. Genes Dev. 2003;17(1):37–42. 10.1101/gad.1031403 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Yancopoulos GD, DePinho RA, Zimmerman KA, et al. : Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5(12):3259–3266. 10.1002/j.1460-2075.1986.tb04637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolland DJ, Wood AL, Johnston CM, et al. : Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5(6):630–637. 10.1038/ni1068 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Bolland DJ, Wood AL, Afshar R, et al. : Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer E mu. Mol Cell Biol. 2007;27(15):5523–5533. 10.1128/MCB.02407-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verma-Gaur J, Torkamani A, Schaffer L, et al. : Noncoding transcription within the Igh distal V H region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A. 2012;109(42):17004–17009. 10.1073/pnas.1208398109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ebert A, McManus S, Tagoh H, et al. : The distal V H gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34(2):175–187. 10.1016/j.immuni.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 39. Gibcus JH, Dekker J: The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dekker J, Mirny L: The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164(6):1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Bickmore WA, van Steensel B: Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 42. Fraser J, Williamson I, Bickmore WA, et al. : An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol Mol Biol Rev. 2015;79(3):347–372. 10.1128/MMBR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsieh TH, Weiner A, Lajoie B, et al. : Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 2015;162(1):108–119. 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Lieberman-Aiden E, van Berkum NL, Williams L, et al. : Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Rao SS, Huntley MH, Durand NC, et al. : A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Shachar S, Voss TC, Pegoraro G, et al. : Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping. Cell. 2015;162(4):911–923. 10.1016/j.cell.2015.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Tang Z, Luo OJ, Li X, et al. : CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163(7):1611–1627. 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carter D, Chakalova L, Osborne CS, et al. : Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32(4):623–626. 10.1038/ng1051 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Li G, Ruan X, Auerbach RK, et al. : Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Sanyal A, Lajoie BR, Jain G, et al. : The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Tolhuis B, Palstra RJ, Splinter E, et al. : Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–1465. 10.1016/S1097-2765(02)00781-5 [DOI] [PubMed] [Google Scholar]
- 52. Kenter AL, Feldman S, Wuerffel R, et al. : Three-dimensional architecture of the IgH locus facilitates class switch recombination. Ann N Y Acad Sci. 2012;1267:86–94. 10.1111/j.1749-6632.2012.06604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar S, Wuerffel R, Achour I, et al. : Flexible ordering of antibody class switch and V(D)J joining during B-cell ontogeny. Genes Dev. 2013;27(22):2439–2444. 10.1101/gad.227165.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feldman S, Achour I, Wuerffel R, et al. : Constraints contributed by chromatin looping limit recombination targeting during Ig class switch recombination. J Immunol. 2015;194(5):2380–2389. 10.4049/jimmunol.1401170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wuerffel R, Wang L, Grigera F, et al. : S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27(5):711–722. 10.1016/j.immuni.2007.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Sellars M, Reina-San-Martin B, Kastner P, et al. : Ikaros controls isotype selection during immunoglobulin class switch recombination. J Exp Med. 2009;206(5):1073–1087. 10.1084/jem.20082311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jhunjhunwala S, van Zelm MC, Peak MM, et al. : The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133(2):265–279. 10.1016/j.cell.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Lucas JS, Zhang Y, Dudko OK, et al. : 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158(2):339–352. 10.1016/j.cell.2014.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Nora EP, Lajoie BR, Schulz EG, et al. : Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Dixon JR, Selvaraj S, Yue F, et al. : Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Peric-Hupkes D, Meuleman W, Pagie L, et al. : Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol cell. 2010;38(4):603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Hanssen LLP, Kassouf MT, Oudelaar AM, et al. : Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol. 2017;19(8):952–961. 10.1038/ncb3573 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Phillips-Cremins JE, Sauria ME, Sanyal A, et al. : Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295. 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Mifsud B, Tavares-Cadete F, Young AN, et al. : Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598–606. 10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Hughes JR, Roberts N, McGowan S, et al. : Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46(2):205–212. 10.1038/ng.2871 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Jin F, Li Y, Dixon JR, et al. : A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Phanstiel DH, Van Bortle K, Spacek D, et al. : Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol Cell. 2017;67(6):1037–1048.e6. 10.1016/j.molcel.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Siersbaek R, Madsen JGS, Javierre BM, et al. : Dynamic Rewiring of Promoter-Anchored Chromatin Loops during Adipocyte Differentiation. Mol Cell. 2017;66(3):420–435.e5. 10.1016/j.molcel.2017.04.010 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Bell AC, West AG, Felsenfeld G: The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–396. 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- 70. Vietri Rudan M, Barrington C, Henderson S, et al. : Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10(8):1297–1309. 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ong CT, Corces VG: CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–246. 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. de Wit E, Vos ES, Holwerda SJ, et al. : CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60(4):676–684. 10.1016/j.molcel.2015.09.023 [DOI] [PubMed] [Google Scholar]
- 73. Guo Y, Xu Q, Canzio D, et al. : CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162(4):900–910. 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Phillips JE, Corces VG: CTCF: master weaver of the genome. Cell. 2009;137(7):1194–1211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Giles KE, Gowher H, Ghirlando R, et al. : Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harb Symp Quant Biol. 2010;75:79–85. 10.1101/sqb.2010.75.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andrey G, Montavon T, Mascrez B, et al. : A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340(6137):1234167. 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Dowen JM, Fan ZP, Hnisz D, et al. : Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159(2):374–387. 10.1016/j.cell.2014.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lupiáñez DG, Kraft K, Heinrich V, et al. : Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Nora EP, Goloborodko A, Valton AL, et al. : Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169(5):930–944.e922. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Küppers R, Dalla-Favera R: Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20(40):5580–5594. 10.1038/sj.onc.1204640 [DOI] [PubMed] [Google Scholar]
- 81. Nussenzweig A, Nussenzweig MC: Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141(1):27–38. 10.1016/j.cell.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsai AG, Lu H, Raghavan SC, et al. : Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135(6):1130–1142. 10.1016/j.cell.2008.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Hu J, Zhang Y, Zhao L, et al. : Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163(4):947–959. 10.1016/j.cell.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Rao SSP, Huang SC, Glenn St Hilaire B, et al. : Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171(2):305–320.e324. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Vietri Rudan M, Hadjur S: Genetic Tailors: CTCF and Cohesin Shape the Genome During Evolution. Trends Genet. 2015;31(11):651–660. 10.1016/j.tig.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 86. Merkenschlager M, Nora EP: CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Van Bortle K, Nichols MH, Li L, et al. : Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15(6):R82. 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kagey MH, Newman JJ, Bilodeau S, et al. : Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. Seitan VC, Hao B, Tachibana-Konwalski K, et al. : A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476(7361):467–471. 10.1038/nature10312 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 90. Degner SC, Wong TP, Jankevicius G, et al. : Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182(1):44–48. 10.4049/jimmunol.182.1.44 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 91. Loguercio S, Barajas-Mora EM, Shih HY, et al. : Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci. Front Immunol. 2018;9:425. 10.3389/fimmu.2018.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Volpi SA, Verma-Gaur J, Hassan R, et al. : Germline deletion of Igh 3' regulatory region elements hs 5, 6, 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol. 2012;188(6):2556–2566. 10.4049/jimmunol.1102763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Guo C, Yoon HS, Franklin A, et al. : CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477(7365):424–430. 10.1038/nature10495 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 94. Lin SG, Guo C, Su A, et al. : CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc Natl Acad Sci U S A. 2015;112(6):1815–1820. 10.1073/pnas.1424936112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Majumder K, Koues OI, Chan EA, et al. : Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med. 2015;212(1):107–120. 10.1084/jem.20141479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lin SG, Ba Z, Du Z, et al. : Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc Natl Acad Sci U S A. 2016;113(28):7846–7851. 10.1073/pnas.1608649113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Xiang Y, Park SK, Garrard WT: Vκ gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vκ-Jκ intervening region. J Immunol. 2013;190(4):1819–1826. 10.4049/jimmunol.1203127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xiang Y, Zhou X, Hewitt SL, et al. : A multifunctional element in the mouse Igκ locus that specifies repertoire and Ig loci subnuclear location. J Immunol. 2011;186(9):5356–5366. 10.4049/jimmunol.1003794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Degner SC, Verma-Gaur J, Wong TP, et al. : CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108(23):9566–9571. 10.1073/pnas.1019391108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sanborn AL, Rao SS, Huang SC, et al. : Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112(47):E6456–6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ruiz-Velasco M, Kumar M, Lai MC, et al. : CTCF-Mediated Chromatin Loops between Promoter and Gene Body Regulate Alternative Splicing across Individuals. Cell Syst. 2017;5(6):628–637.e6. 10.1016/j.cels.2017.10.018 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 102. Fudenberg G, Imakaev M, Lu C, et al. : Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15(9):2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 103. Nichols MH, Corces VG: A CTCF Code for 3D Genome Architecture. Cell. 2015;162(4):703–705. 10.1016/j.cell.2015.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhao L, Frock RL, Du Z, et al. : Orientation-specific RAG activity in chromosomal loop domains contributes to Tcrd V(D)J recombination during T cell development. J Exp Med. 2016;213(9):1921–1936. 10.1084/jem.20160670 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Guo C, Gerasimova T, Hao H, et al. : Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147(2):332–343. 10.1016/j.cell.2011.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Jain S, Ba Z, Zhang Y, et al. : CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell. 2018;174(1):102–116.e14. 10.1016/j.cell.2018.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 107. Qiu X, Kumari G, Gerasimova T, et al. : Sequential Enhancer Sequestration Dysregulates Recombination Center Formation at the Igh Locus. Mol Cell. 2018;70(1):21–33.e26. 10.1016/j.molcel.2018.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Kleiman E, Xu J, Feeney AJ: Cutting Edge: Proper Orientation of CTCF Sites in Cer Is Required for Normal Jκ-Distal and Jκ-Proximal Vκ Gene Usage. J Immunol. 2018;201(6):1633–1638. 10.4049/jimmunol.1800785 [DOI] [PMC free article] [PubMed] [Google Scholar]