Abstract
Importance
Based on a small retrospective study, rechallenge with cetuximab-based therapy for patients with KRAS wild-type metastatic colorectal cancer (mCRC) who were previously treated with the same anti–epidermal growth factor receptor–based regimen might be efficacious. Recent data suggest the role of liquid biopsy as a tool to track molecular events in circulating tumor DNA (ctDNA).
Objective
To prospectively assess the activity of cetuximab plus irinotecan as third-line treatment for patients with RAS and BRAF wild-type mCRC who were initially sensitive to and then resistant to first-line irinotecan- and cetuximab-based therapy.
Design, Setting, and Participants
Multicenter phase 2 single-arm trial conducted from January 7, 2015, to June 19, 2017. Liquid biopsies for analysis of ctDNA were collected at baseline. Main eligibility criteria included RAS and BRAF wild-type status on tissue samples; prior first-line irinotecan- and cetuximab-based regimen with at least partial response, progression-free survival of at least 6 months with first-line therapy, and progression within 4 weeks after last dose of cetuximab; and prior second-line oxaliplatin- and bevacizumab-based treatment.
Interventions
Biweekly cetuximab, 500 mg/m2, plus irinotecan, 180 mg/m2.
Main Outcomes and Measures
Overall response rate according to the Response Evaluation Criteria in Solid Tumors, version 1.1. Secondary end points included progression-free survival and overall survival and, as an exploratory analysis, RAS mutations in ctDNA.
Results
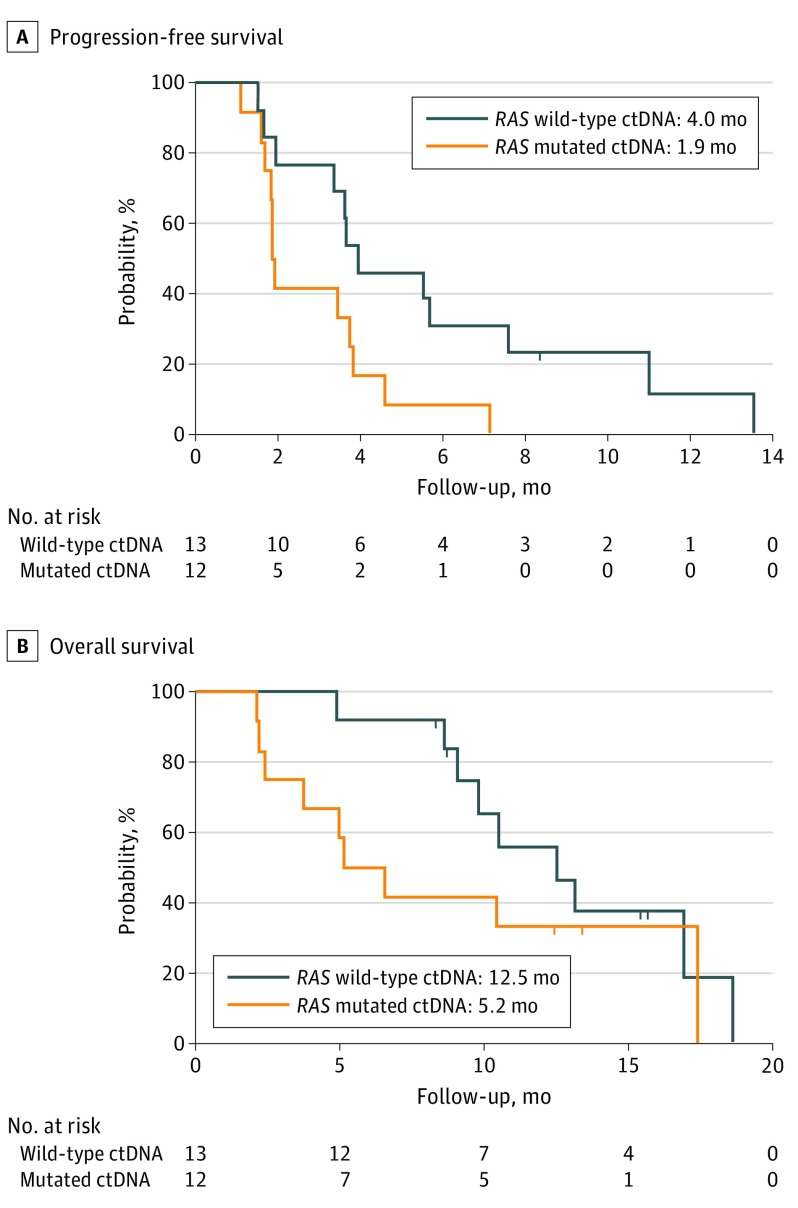
Twenty-eight patients (9 women and 19 men; median age, 69 years [range, 45-79 years]) were enrolled. Six partial responses (4 confirmed) and 9 disease stabilizations were reported (response rate, 21%; 95% CI, 10%-40%; disease control rate, 54%; 95% CI, 36%-70%). Primary end point was met because lower limit of 95% CI of response rate was higher than 5%. RAS mutations were found in ctDNA collected at rechallenge baseline in 12 of 25 evaluable patients (48%). No RAS mutations were detected in samples from patients who achieved confirmed partial response. Patients with RAS wild-type ctDNA had significantly longer progression-free survival than those with RAS mutated ctDNA (median progression-free survival, 4.0 vs 1.9 months; hazard ratio, 0.44; 95% CI, 0.18-0.98; P = .03).
Conclusions and Relevance
This is the first prospective demonstration that a rechallenge strategy with cetuximab and irinotecan may be active in patients with RAS and BRAF wild-type mCRC with acquired resistance to first-line irinotecan- and cetuximab-based therapy. The evaluation of RAS mutational status on ctDNA might be helpful in selecting candidate patients.
Trial Registration
ClinicalTrials.gov Identifier: NCT02296203
This multicenter, phase 2, single-arm trial assesses the activity of cetuximab plus irinotecan as third-line treatment for patients with RAS and BRAF wild-type metastatic colorectal cancer who were initially sensitive to and then resistant to first-line irinotecan- and cetuximab-based therapy.
Key Points
Question
Is third-line cetuximab plus irinotecan an active option for patients with RAS and BRAF wild-type metastatic colorectal cancer who have acquired resistance to first-line irinotecan- and cetuximab-based therapy?
Findings
In this phase 2 single-arm clinical trial, rechallenge with cetuximab plus irinotecan was active in 21% of patients with RAS and BRAF wild-type metastatic colorectal cancer. Preplanned circulating tumor DNA profiling revealed that only patients with RAS and BRAF wild-type circulating tumor DNA at the time of rechallenge could derive benefit.
Meaning
These findings lay the foundation for further evaluating the efficacy of anti–epidermal growth factor receptor rechallenge in larger studies including only patients with no mechanisms of acquired resistance detectable in circulating tumor DNA.
Introduction
The combination of an anti–epidermal growth factor receptor (anti-EGFR) monoclonal antibody (cetuximab or panitumumab) with a chemotherapy doublet is a first-line treatment option for patients with RAS (KRAS: OMIM, 190070; and NRAS: OMIM, 164790) and BRAF (OMIM, 164757) wild-type metastatic colorectal cancer (mCRC).1,2,3 A retrospective study highlighted the potential efficacy of reintroducing cetuximab for patients with acquired resistance to a previous treatment with chemotherapy plus cetuximab, followed by at least 1 intervening line of therapy.4 Although the study was limited by its retrospective nature, the finding is currently supported by an intriguing biological rationale. The emergence of RAS mutations in tumors that were initially RAS wild-type is a well-recognized mechanism of acquired resistance to anti-EGFR monoclonal antibodies.5,6,7,8 It is currently unclear whether this event might be due to the late acquisition of these mutations by cellular subclones or to the progressive selection of initially undetectable mutated subclones. According to the latter hypothesis, an anti-EGFR–based therapy would be able to substantially decrease the bulk of sensitive (wild-type) cells, thus making the resistant (mutant) clones progressively predominant until the clinical evidence of disease progression. During a subsequent treatment that was not anti-EGFR based, sensitive clones would be at least partially restored, thus laying the foundation for the potential and reported activity of anti-EGFR rechallenge.5
More recently, a growing amount of molecular evidence highlighted the intratumoral heterogeneity of colorectal cancer and the dynamism of clonal evolution under the pressure exerted by treatments. In particular, preliminary proof of concept results pointed out the biological relevance of circulating tumor DNA (ctDNA) as an extremely sensitive tool to document the complexity of the tumor and to potentially drive strategies of therapy adaptation.5,9,10,11,12 The emergence of RAS mutations at the time of disease progression to first-line chemotherapy plus anti-EGFR monoclonal antibodies may be followed by a dwindling of the fractional abundance of acquired RAS mutations—even to undetectable levels—after withdrawal of EGFR blockade.5
The CRICKET (Cetuximab Rechallenge in Irinotecan-Pretreated mCRC, KRAS, NRAS and BRAF Wild-Type Treated in 1st line With Anti-EGFR Therapy) trial was designed to prospectively evaluate the activity of a rechallenge strategy with irinotecan and cetuximab as third-line treatment for patients experiencing an initial response and then progression with a first-line irinotecan- and cetuximab-containing therapy, and receiving second-line chemotherapy plus bevacizumab. The prospective collection of liquid biopsy samples from enrolled patients was planned to investigate whether the analysis of potential mechanisms of acquired resistance to anti-EGFR monoclonal antibodies in ctDNA could help determine the benefit of this strategy.
Methods
Patient Population
The CRICKET trial (trial protocol in Supplement 1) was a prospective, open-label, multicenter, single-arm phase 2 trial that recruited patients with mCRC from 9 Italian oncology units from January 7, 2015, to June 19, 2017. The main inclusion criteria were histologically confirmed colorectal adenocarcinoma; RAS and BRAF wild-type status of primary colorectal cancer and/or related metastasis; older than 18 years of age; an Eastern Cooperative Oncology Group performance status of 0 to 2; measurable metastatic disease according to the Response Evaluation Criteria in Solid Tumors, version 1.113; first-line irinotecan-based cetuximab-containing therapy (FOLFIRI [fluorouracil and leucovorin combined with irinotecan] or FOLFOXIRI [fluorouracil, leucovorin, oxaliplatin, and irinotecan] plus cetuximab) producing at least a partial response; first-line progression-free survival (PFS) of 6 months or more; documentation of progression to first-line therapy within 4 weeks after the last administration of cetuximab; time between the end of first-line therapy and the start of third-line therapy of 4 months or more; and progression to a second-line oxaliplatin-based bevacizumab-containing therapy (FOLFOXIRI, FOLFOX [leucovorin, fluorouracil, and oxaliplatin], or XELOX [capecitabine and oxaliplatin] plus bevacizumab). The trial was conducted in accordance with the Declaration of Helsinki14 and adhered to the international Good Clinical Practice guidelines. The protocol was approved by local ethics committees of participating sites (Centre of Pisa and Pontedera: Comitato Etico Area Vasta Nord Ovest and Sezione Autonoma Del Comitato Etico Regionale Per La Sperimentazione Clinica, Centre of Roma Campus Biomedico: Comitato Etico dell’Università Campus Biomedico di Roma, Centre of Parma: Comitato Etico per Parma, Centre of Padova: Comitato Etico per la Sperimentazione Clinica dell’ Istituto Oncologico Veneto, Centre of Roma–Isola Tiberina: Comitato Etico Lazio 1 c/o Farmacia dell’Ospedale San Camillo, Centre of Udine: Comitato Etico Regionale Unico, Centre of Roma-Gemelli: Comitato Etico Università Cattolica del Sacro Cuore, and Centre of Rimini: Comitato Etico di Area Vasta Romagna). All patients provided written informed consent to study procedures.
Study Treatment and Procedures
Patients were treated with intravenous cetuximab, 500 mg/m2, and intravenous irinotecan 180, mg/m2, repeated biweekly until disease progression, patient’s refusal, unacceptable toxic effects, or withdrawal of consent. The response was evaluated according to the Response Evaluation Criteria in Solid Tumors, version 1.1. A computed tomographic scan was recommended every 8 weeks.13 Investigator-reported measurements were subsequently centrally reviewed. Adverse events were recorded and graded according to the National Cancer Institute’s Common Terminology Criteria for Adverse Events guidelines, version 4.0.15 Patients’ registration and data collection were centralized by the Gruppo Oncologico del Nord Ovest.
Molecular Analyses
Liquid biopsies were collected at the rechallenge baseline. Circulating tumor DNA was analyzed with droplet digital polymerase chain reaction for specific RAS and BRAF mutations and then by means of ultra-deep next-generation sequencing with Ion Torrent S5 XL (Thermo Fisher Scientific).
Six milliliters of blood were collected in EDTA tubes and centrifuged at 4°C for 10 minutes at 3000 rpm within 2 hours after blood collection. Plasma samples were stored at −80°C until analysis. Circulating tumor DNA was extracted using a QIAmp Circulating nucleic acid Kit (Qiagen) from 2 to 3 mL of plasma following the manufacturer’s protocol, and the ctDNA was eluted in 50 μL of elution buffer. The analysis was performed with a droplet digital polymerase chain reaction KRAS and NRAS Screening Multiplex Kit, and the results were confirmed using single mutation assays (BioRad). BRAF analysis for the V600E mutation was conducted using the droplet digital polymerase chain reaction BRAF V600E Mutation Assay (BioRad). The droplet reader (BioRad) was used for the fluorescence signal quantification, the QuantaSoft (BioRad) software was used to measure the number of positive vs negative droplets for both fluorophores (5(6)-carboxyfluorescein and 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein [FAM/HEX]), and their ratio was fitted to a Poisson distribution to determine the copy number per milliliter of the target molecule in the input reaction. A fluorescence intensity threshold of 3000 was set as a cutoff point, and all droplets above this threshold were scored as positive for RAS and BRAF mutations.
The next-generation sequencing was performed using the Ion AmpliSeq Cancer Hotspot Panel (Thermo Fisher); this panel is designed to amplify 207 amplicons covering approximately 2800 COSMIC (Catalogue Of Somatic Mutations In Cancer) mutations from 50 oncogenes and tumor suppressor genes commonly mutated in human cancers, including RAS and BRAF mutations (eTable 1 in Supplement 2). The libraries were prepared by IonAmpliSeq Library kit 2.0 (Thermo Fisher).16 Emulsion polymerase chain reaction and chip loading were performed on the IonChef System (Thermo Fisher), according to the manufacturer’s instructions. Sequencing was performed on the ION S5 XL System (Thermo Fisher) using Ion 540 Chips and Ion 540 Kit-Chef according to the manufacturer’s instructions (MAN0010846). Data were processed by using Torrent Suite (Thermo Fisher); the variant calling from sequencing data was generated by using the Variant Caller plugin. The resulting variants were annotated using the Ensemble Variant Effect Predictor pipeline, Ion Reporter analysis software, the COSMIC database,17 the dbSNP database,18 and the ClinVar database of the National Center for Biotechnology Information.19 The filtered variants were examined using the Integrative Genomic Viewer IGV tool (Broad Institute) to check their quality level and confirm the variant’s presence on both the positive and the negative strand.20 For all plasma ctDNA samples, the coverage depth of most amplicons was over 5000×.
Statistical Analysis
The primary end point of the study was the overall response rate, defined as the proportion of patients achieving complete response or partial response. Patients whose disease was not reassessed and those who were unavailable for follow-up or who were dead before disease reassessment were considered not to be responders for the purpose of the primary end-point analysis. According to the Fleming single-stage design, selecting P = .05 (overall response rate in the null hypothesis) based on results with second-line irinotecan-based therapies21,22 and selecting P = .20 (overall response rate in the alternative hypothesis) as a potential target of interest for future studies, with α (1-sided) errors of .05 and β errors of .20, we found that a total of 27 patients were required. The null hypothesis would have been rejected if at least 4 patients had achieved a response.23,24 Secondary end points included PFS, overall survival (OS) calculated from the start of treatment, the toxicity profile, and the results of translational analyses. Progression-free survival and OS were summarized using the Kaplan-Meier method; hazard ratios and corresponding 95% CIs were estimated with the Cox proportional hazards regression model. Patients not experiencing disease progression or death were censored at the date of the last follow-up visit. The median period of follow-up was calculated for the entire study cohort according to the reverse Kaplan-Meier method. Statistical analyses were performed by using MedCalc Statistical Software, version 14.8.1 (MedCalc Software) and GraphPad Prism, version 6.00 for Windows (GraphPad Software).
Results
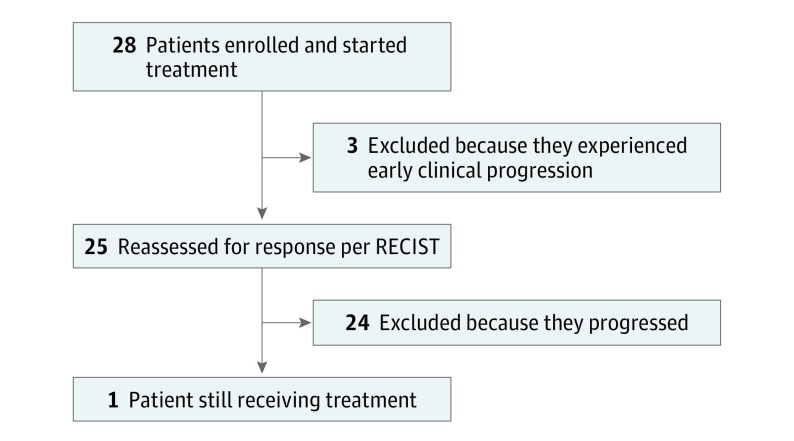
From January 7, 2015, to June 19, 2017, 28 patients were enrolled in 9 Italian oncology units (Figure 1). The patients’ main characteristics are reported in eTable 2 in Supplement 2: the median age was 69 years (range, 45-79 years), 18 patients (64%) had an Eastern Cooperative Oncology Group performance status of 0, metastases’ presentation was synchronous in 20 cases (71%), 25 patients (89%) had undergone resection of the primary tumor, and 21 patients (75%) had multiple sites of metastases, while metastases were still limited to the liver in 5 patients (18%). The median time from the diagnosis of metastatic disease to study entry was 24.4 months (95% CI, 20.2-31.7 months).
Figure 1. Study CONSORT Diagram.
RECIST indicates Response Evaluation Criteria in Solid Tumors.
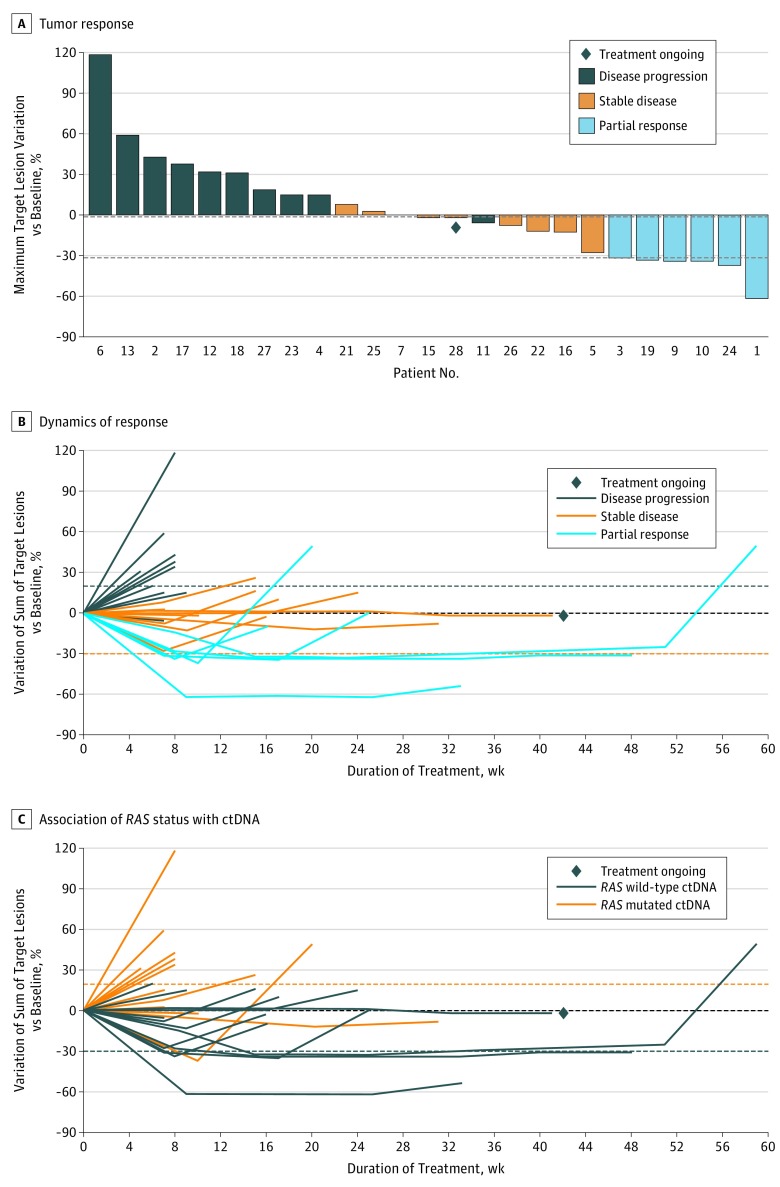
At the time of data cutoff on March 1, 2018, the median follow-up was 15.4 months (interquartile range, 4.35-13.25 months). Six patients (21%) achieved a partial response that was confirmed in 4 patients at the subsequent computed tomographic scan assessment (Table). The overall response rate was 21% (95% CI, 10%-40%). Nine patients (32%) reported stable disease, for a disease control rate of 54% (95% CI, 36%-70%). Thirteen of 25 patients (52%) assessed for a radiologic response had tumor shrinkage (Figure 2A). Three patients (11%) had clinically detectable disease progression before undergoing the first computed tomographic scan reassessment. The median duration of disease control was 9.9 weeks (95% CI, 8.1-23.1 weeks) (Figure 2B). The median PFS was 3.4 months (95% CI, 1.9-3.8 months), and the median OS was 9.8 months (95% CI, 5.2-13.10 months) (eFigure in Supplement 2).
Table. Data on Responses of Patients.
| Type of Response | No. (%) (N = 28) |
|---|---|
| Complete response | 0 |
| Partial response | 6 (21) |
| Confirmed | 4 (14) |
| Unconfirmed | 2 (7) |
| Stable disease | 9 (32) |
| Progressive disease | 10 (36) |
| Not evaluable | 3 (11) |
| Objective response ratea (95% CI) | 6 (21) (10-40) |
| Disease control rateb (95% CI) | 15 (54) (36-70) |
Complete response or partial response.
Complete response, partial response, or stable disease.
Figure 2. Radiographic Response.
A, Tumor response in 25 evaluable patients. The bars show the best percentage change in the target lesions from baseline. Three patients progressed before the first disease assessment. The dashed horizontal line at –30 indicates the threshold value to define partial response. B, Dynamics of response according to best response in 25 evaluable patients. The individual lines represent the percentage variation of the sum of target lesions at different time points. C, Association of RAS status with circulating tumor DNA (ctDNA) in 25 evaluable patients. The longitudinal assessment of the sum of target lesions is shown according to RAS mutational status of ctDNA at rechallenge baseline.
eTable 3 in Supplement 2 shows treatment-related grade 3 or higher adverse events. The most common adverse events were diarrhea (5 [18%]), acneiform skin rash (4 [14%]), neutropenia (4 [14%]), and hand-foot syndrome (2 [7%]). No grade 5 adverse events occurred. No patient interrupted treatment because of adverse events, and treatment withdrawal was not requested by any patient.
A total of 202 cycles (median, 4.5 cycles per patient) were administered. Treatment was delayed for any reason in 29 cycles (14.4%) or because of an adverse event in 11 cycles (5.4%) and was administered with reduced dosage in 35 cycles (17.3%). The mean (SD) relative dose intensities were 87.3% (20.8%) for irinotecan and 94.8% (13.4%) for cetuximab.
As described in eTable 4 in Supplement 2, RAS mutations were found in liquid biopsy samples collected at the rechallenge baseline in 12 (48%) of 25 patients reassessed by computed tomographic scan (6 KRAS G12D, 5 KRAS G12V with 1 also harboring a Q61H mutation, and 1 NRAS Q61L). No BRAF or PIK3CA (OMIM 171834) mutations were found.
No RAS mutations were detected in ctDNA from patients who achieved a confirmed partial response compared with 12 (57%) of 21 patients who did not achieve a partial response (P = .10 determined by the Fisher exact test). Patients with RAS wild-type ctDNA (n = 13) had significantly longer PFS than did those with RAS mutated ctDNA (n = 12), with a median PFS of 4.0 vs 1.9 months (hazard ratio, 0.44; 95% CI, 0.18-0.98; P = .03) (Figure 3A), while no significant differences were reported in terms of OS (median OS, 12.5 vs 5.2 months; hazard ratio, 0.58; 95% CI, 0.22-1.52; P = .24) (Figure 3B). The percentage variation of the sum of target lesions at different time points according to ctDNA RAS status at rechallenge baseline is shown in Figure 2C.
Figure 3. Kaplan-Meier Estimates of Progression-Free Survival and Overall Survival According to RAS and BRAF Circulating Tumor DNA (ctDNA) Status.
A, Hazard ratio, 0.44 (95% CI, 0.18-0.98; P = .03). B, Hazard ratio, 0.58 (95% CI, 0.22-1.52; P = .24).
Discussion
The therapeutic landscape of later lines of treatment of mCRC has recently become more complex owing to the availability of 2 drugs with demonstrated efficacy in prolonging survival of chemorefractory patients when compared with placebo: the multitarget tyrosine kinase inhibitor regorafenib25,26 and the novel fluoropyrimidine trifluridine-tipiracil, also known as TAS-102.27,28 The magnitude of OS benefit provided by both these agents is limited because half of treated patients are unable to derive any advantage in terms of PFS. Moreover, no molecular or clinical factors associated with benefit from these drugs have been identified so far, making their cost-benefit ratio quite narrow. Alternatively, data from uncontrolled studies support other therapeutic strategies for small molecular subgroups, such as dual-targeted ERBB2/HER2 (OMIM 164870) therapy for HER2-positive tumors,29 immune checkpoints for microsatellite instable tumors,30,31 and tyrosine kinase inhibition for gene fusion–positive tumors.32,33
Here we provide the first prospective demonstration, to our knowledge, of the potential usefulness of another treatment option for a molecularly and clinically defined subgroup of patients with mCRC: rechallenge with cetuximab and irinotecan for patients with RAS and BRAF wild-type mCRC who experienced an initial benefit and then became resistant to a first-line cetuximab-containing regimen and received second-line oxaliplatin-based chemotherapy plus bevacizumab. Although acknowledging the intrinsic limitations of a single-arm phase 2 study, our results provide a clear signal of activity for the anti-EGFR rechallenge in the third-line setting, in patients with strict clinical and molecular criteria for defining acquired resistance to first-line treatment based on anti-EGFR monoclonal antibodies. Given the increasing amount of evidence of the role of anti-EGFR–based maintenance after first-line induction regimens, the clinical scenario of anti-EGFR rechallenge might be frequently faced in future clinical practice.
The analysis of ctDNA revealed that approximately 50% of these patients still had detectable RAS mutations at the time of rechallenge, and it highlights the actual reliability of liquid biopsy as a tool to inform therapeutic decisions. None of the patients with RAS mutations in ctDNA at the start of rechallenge achieved response, thus making the choice of rechallenge inappropriate for them. Only 1 patient with a small fractional abundance of RAS mutation in ctDNA experienced a transient response to rechallenge. A potential explanation is that, even if RAS mutation was still detectable at the time of study entry, a significant drop in its frequency from the time of disease progression at first-line treatment had occurred, thus suggesting a dwindling of the mutant clones and a contemporary increasing prevalence of wild-type cells at the tumor level during second-line therapy. Unfortunately, the lack of longitudinal paired samples during previous lines of therapy does not allow drawing any conclusion about this hypothesis.
Lack of RAS mutations in ctDNA is associated in our small series with a probability of 31% of achieving response (13 patients had no RAS mutations in ctDNA; 4 of these patients had a response [eTable 4 in Supplement 2]). Although we did not identify any BRAF or PIK3CA mutation in analyzed samples, other mechanisms of acquired resistance to anti-EGFR monoclonal antibodies may occur or co-occur with RAS mutations and have been identified in liquid biopsy samples from patients with disease progression.5,6,9,11,34,35,36
However, despite intriguing preliminary proof-of-concept evidence about the potential role of liquid biopsy in driving therapeutic choices, the translation of these findings from the bench to the bedside will definitely need robust confirmation from biomarker-driven clinical trials. A further step forward on this route will be marked by the currently ongoing CHRONOS (Phase II Trial of Rechallenge With Panitumumab Driven by RAS Clonal-Mediated Dynamic of Resistance) study (NCT03227926), in which patients eligible for anti-EGFR rechallenge are eligible only if a decrease of at least 50% in the fractional abundance of RAS mutations in ctDNA is evident at the time of rechallenge when compared with the time of progression to the first-line anti-EGFR–containing therapy.
Limitations
This study has some limitations. Owing to its phase 2 single-arm design, the CRICKET trial is a proof-of-concept study, able to provide signals of activity and to generate preliminary evidence, supported by a sound translational background, to be further confirmed in a larger clinical trial. The registration of patients at the time of rechallenge (ie, after the second evidence of disease progression) prevents us from deriving any conclusion about how many patients with RAS and BRAF wild-type tumors who are receiving chemotherapy plus anti-EGFR monoclonal antibodies as initial treatment may be candidates for this third-line approach.
Conclusions
Based on results of the CRICKET trial, whereas trifluridine-tipiracil or regorafenib do still represent third-line options with the highest levels of evidence for patients with chemorefractory mCRC, anti-EGFR rechallenge could be a tailored strategy for selected patients. Although markers of EGFR dependency are still lacking, patients with RAS and BRAF wild-type left-sided tumors, possibly not showing other molecular mechanisms of intrinsic resistance to EGFR inhibition,37 who derived clinically meaningful benefit from first-line anti-EGFR–containing therapy, and with undetectable markers of acquired resistance to anti-EGFR monoclonal antibodies in tissue and/or liquid biopsy samples at the time of retreatment, might be the optimal candidates for rechallenge.
Trial Protocol
eTable 1. List of Oncogenes and Tumour Suppressor Genes analyzed in ctDNA
eTable 2. Baseline Patients’ and Disease Characteristics
eTable 3. Most Common Grade 3/4 Adverse Events
eTable 4. RAS Status on ctDNA Collected at Rechallenge Baseline and Best Response According to RECIST 1.1
eFigure. Kaplan-Meier Estimates of Progression-Free Survival and Overall Survival in the Study Population
References
- 1.National Comprehensive Cancer Network NCCN clinical practice guidelines in oncology—colon cancer; version 2. https://www.nccn.org/professionals/physician_gls/default.aspx#site. Accessed March 28, 2018.
- 2.Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386-1422. doi: 10.1093/annonc/mdw235 [DOI] [PubMed] [Google Scholar]
- 3.Salvatore L, Aprile G, Arnoldi E, et al. Management of metastatic colorectal cancer patients: guidelines of the Italian Medical Oncology Association (AIOM). ESMO Open. 2017;2(1):e000147. doi: 10.1136/esmoopen-2016-000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santini D, Vincenzi B, Addeo R, et al. Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol. 2012;23(9):2313-2318. doi: 10.1093/annonc/mdr623 [DOI] [PubMed] [Google Scholar]
- 5.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795-801. doi: 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:13665. doi: 10.1038/ncomms13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532-536. doi: 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21(9):2157-2166. doi: 10.1158/1078-0432.CCR-14-2821 [DOI] [PubMed] [Google Scholar]
- 9.Montagut C, Argilés G, Ciardiello F, et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses: a phase 2 randomized clinical trial. JAMA Oncol. 2018;4(4):e175245. doi: 10.1001/jamaoncol.2017.5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537-540. doi: 10.1038/nature11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietrantonio F, Vernieri C, Siravegna G, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23(10):2414-2422. doi: 10.1158/1078-0432.CCR-16-1863 [DOI] [PubMed] [Google Scholar]
- 12.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8(6):1095-1111. doi: 10.1016/j.molonc.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228-247. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 14.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 15.National Cancer Institute Common terminology criteria for adverse events, version 4.0. Bethesda, MD: May 2009.
- 16.Meazza C, Belfiore A, Busico A, et al. AKT1 and BRAF mutations in pediatric aggressive fibromatosis. Cancer Med. 2016;5(6):1204-1213. doi: 10.1002/cam4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777-D783. doi: 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.dbSNP: short genetic variations. Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine. https://www.ncbi.nlm.nih.gov/SNP/. Accessed June 15, 2018.
- 19.ClinVar https://www.ncbi.nlm.nih.gov/clinvar/. Accessed June 15, 2018.
- 20.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178-192. doi: 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tournigand C, André T, Achille E, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22(2):229-237. doi: 10.1200/JCO.2004.05.113 [DOI] [PubMed] [Google Scholar]
- 22.Sobrero AF, Maurel J, Fehrenbacher L, et al. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(14):2311-2319. doi: 10.1200/JCO.2007.13.1193 [DOI] [PubMed] [Google Scholar]
- 23.Fleming TR. One-sample multiple testing procedure for phase II clinical trials. Biometrics. 1982;38(1):143-151. doi: 10.2307/2530297 [DOI] [PubMed] [Google Scholar]
- 24.A’Hern RP. Sample size tables for exact single-stage phase II designs. Stat Med. 2001;20(6):859-866. doi: 10.1002/sim.721 [DOI] [PubMed] [Google Scholar]
- 25.Li J, Qin S, Xu R, et al. ; CONCUR Investigators . Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2015;16(6):619-629. doi: 10.1016/S1470-2045(15)70156-7 [DOI] [PubMed] [Google Scholar]
- 26.Grothey A, Van Cutsem E, Sobrero A, et al. ; CORRECT Study Group . Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381(9863):303-312. doi: 10.1016/S0140-6736(12)61900-X [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Kim TW, Shen L, et al. Results of a randomized, double-blind, placebo-controlled, phase III trial of trifluridine/tipiracil (TAS-102) monotherapy in Asian patients with previously treated metastatic colorectal cancer: the TERRA study. J Clin Oncol. 2018;36(4):350-358. doi: 10.1200/JCO.2017.74.3245 [DOI] [PubMed] [Google Scholar]
- 28.Mayer RJ, Van Cutsem E, Falcone A, et al. ; RECOURSE Study Group . Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372(20):1909-1919. doi: 10.1056/NEJMoa1414325 [DOI] [PubMed] [Google Scholar]
- 29.Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738-746. doi: 10.1016/S1470-2045(16)00150-9 [DOI] [PubMed] [Google Scholar]
- 30.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773-779. doi: 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 31.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182-1191. doi: 10.1016/S1470-2045(17)30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartore-Bianchi A, Ardini E, Bosotti R, et al. Sensitivity to entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. J Natl Cancer Inst. 2015;108(1):djv306. doi: 10.1093/jnci/djv306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drilon A, Siena S, Ou SI, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017;7(4):400-409. doi: 10.1158/2159-8290.CD-16-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 2016;6(2):147-153. doi: 10.1158/2159-8290.CD-15-1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J-M, Wang Y, Wang Y-L, et al. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23(16):4602-4616. doi: 10.1158/1078-0432.CCR-16-2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oddo D, Siravegna G, Gloghini A, et al. Emergence of MET hyper-amplification at progression to MET and BRAF inhibition in colorectal cancer. Br J Cancer. 2017;117(3):347-352. doi: 10.1038/bjc.2017.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cremolini C, Morano F, Moretto R, et al. Negative hyper-selection of metastatic colorectal cancer patients for anti-EGFR monoclonal antibodies: the PRESSING case-control study. Ann Oncol. 2017;28(12):3009-3014. doi: 10.1093/annonc/mdx546 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial Protocol
eTable 1. List of Oncogenes and Tumour Suppressor Genes analyzed in ctDNA
eTable 2. Baseline Patients’ and Disease Characteristics
eTable 3. Most Common Grade 3/4 Adverse Events
eTable 4. RAS Status on ctDNA Collected at Rechallenge Baseline and Best Response According to RECIST 1.1
eFigure. Kaplan-Meier Estimates of Progression-Free Survival and Overall Survival in the Study Population