Abstract
Background
CD14 polymorphisms are associated with an increased risk of cardiovascular events. So far, many studies have been conducted, whereas the results were not always consistent.
Materials and methods
Twenty-six articles involving thirty-seven datasets were recruited to evaluate the association between rs2569190 (9413 patients and 7337 controls), C-159T (4813 patients and 2852 controls) polymorphisms and cardiovascular diseases in a meta-analysis. The random or fixed effect models were used to evaluate the pooled odds ratios (ORs) and their corresponding 95% confidence intervals.
Results
The strongest association was observed between rs2569190 and CVD in overall population (T vs. C, OR = 1.169, 95% CI: 1.087–1.257, p = 2.44 × 10− 5). Analysis after stratification by ethnicity indicated that rs2569190 was related to CVD in East Asian population (T vs. C, OR = 1.370, 95% CI; 1.226–1.531, p = 2.86 × 10− 8) and a potential relationship in European (T vs. C, OR = 1.100, 95% CI: 1.019–1.189, p = 0.015). In the stratification of endpoints, the associations were found in CHD subgroup (T vs. C, OR = 1.357, 95% CI: 1.157–1.592, p = 2.47 × 10− 7) and in AMI subgroup (T vs. C, OR = 1.152, 95% CI: 1.036–1.281, p = 0.009). However, we did not find any association between C-159T polymorphism with cardiovascular disease under any model.
Conclusions
The SNP rs2569190 significantly contribute to susceptibility and development of cardiovascular disease, particularly in the East Asian population and in the subtype CHD group, in addition, a potential association was observed in the AMI group, T allele acts as a risk factor for cardiovascular disease.
Electronic supplementary material
The online version of this article (10.1186/s12944-019-1018-3) contains supplementary material, which is available to authorized users.
Keywords: CD14, Cardiovascular disease, Polymorphism, Meta-analysis
Introduction
Cardiovascular disease (CVD) is a major public health problem owing to associate increased risk of human mortality [1]. By far the most common cause of acute coronary syndrome (ACS) is atherosclerosis and coronary artery stenosis, these lesions are the pathological foundation of CAD [2]. According to the number of coronary artery stenoses and the diverse clinical manifestation, CVD was defined to various clinical phenotypes (like coronary heart disease (CHD), acute myocardial infarction (AMI), myocardial infarction (MI) and so on [3]). Atherosclerosis is a process of progressive thickening and hardening of the walls of medium-sized and large arteries as a result of fat deposits on their inner lining [4]. Atherosclerosis is a pathological condition that underlies several important averse vascular events including coronary artery disease (CAD), stroke, and peripheral arterial disease, responsible for most of the cardiovascular morbidity and mortality [5]. In the 1990s, it was first time to demonstrate a strong association between inflammation and atherosclerosis, and suggested that atherosclerosis was one of chronic inflammatory diseases [5]. Recent studies have suggested that inflammation was an important factor in the initiation and development of atherosclerosis [2, 6, 7], the inflammatory reaction which in the coronary artery atherosclerosis plaque, leads to the intima damage, plaque rupture and acute cardiac ischemia [7–9]. These points revealed that infection may enhance the inflammatory processes present in atherosclerosis and CAD.
The cluster of differentiation antigen 14 (CD14) is a lipopolysaccharide (LPS) receptor located on the surface of monocytes and macrophages and it is a multiple function inflammation cytokine which is mainly produced by mature mononuclear macrophage [10]. CD14 is known as a surface marker, being glycosylated phosphatidylinositol anchored in the cell membrane (mCD14) [11]. In addition, CD14 could specially combine with LPS, transfer the activation signal to the downstream pathway through the TLR4 and bone marrow differentiation protein-2 [12, 13]. By this way, the monocytes-macrophage system was launched and many pro-inflammation cytokines as TNF-α, IL-1, IL-6 and so on were released [14]. These cytokines have multi-functions in the process of mediating initial immune response and inflammation reaction which causing the endothelia damage, disturbance of immunologic function and vascular smooth muscle cells proliferation [2]. Thus, CD14 was considered to be a key role in the process of atherosclerosis and complications.
It is generally accepted that genetic predisposition is a major risk factoring for atherosclerosis leading to CAD. In 1999, since Hubacek et al. [15] first reported that CD14 gene single nucleotide polymorphism (SNP) rs2569190, related to the translation start site at upstream of promoter region, was apparently relevant to atherosclerosis in the Czech population, the allele T of rs2569190 was also reported being a risk factoring for MI in European population. Then, a bunch of studies were carried out to verify the causal relationship between rs2569190 and CAD in diverse ethnics. Differently, the negative results that the polymorphism was not associated with CHD and MI were observed in European participants [16]. In addition, the variant of rs2569190 whether effected CAD on Han Chinese has been a research hotpot [17, 18]. Meanwhile, a new promoter polymorphism C-159T in the gene of LPS receptor was also found to be associated with CAD by Unkelbach et al. [19]. However, the inverse finding that there was not interaction between C-159T and CAD was observed by two groups [20, 21]. Besides, the allele T of C-159T was considered to be a risk factor for CAD in a Chinese population [22], then the same conclusion was obtained in Chinese Yanbian population [23]. On the contrary, the T-to-C exchange in C-159T was considered to be risk allele in European populations, especially in older patients with a low atherosclerotic risk profile [19].
Although many studies have been conducted so far to investigate the relationship between CD14 gene polymorphisms and CVD, the results were inconsistent. Population stratification might lead to inconsistent results, especially when allele frequency and incidence rate of the disease vary across ethnic groups. Meanwhile, inclusion of data that didn’t satisfy the requirement of meta-analysis would produce a spurious association. Therefore, in order to reduce the limitations of single study and to overcome the possible random errors, a large-scale meta-analysis involving multifarious ethnics was performed by us.
Materials and methods
Identification of eligible studies
To analyze the association between CD14 gene (SNP rs2569190, C-159T) and CVD, all published literature before December 2017 that researched the relationship between these polymorphisms and CVD risk were concluded. The electronic databases were used including PubMed databases (National Center for Biotechnology, National Library of Medicine), CNKI (China National Knowledge Infrastructure), and Web of Science were retrieved by using the keywords “CD14”, “C-260T”, “C-159T”, “rs2569190”, “polymorphism” connect to “CVD”, “coronary artery disease”, “coronary heart disease”, “myocardial infarction”, or “atherosclerosis” without language restrictions. Finally, we extracted data from the published articles, not included meetings or any conference abstracts. All of the included studies used either case-control or nested case-control design. Appropriate diagnosis criteria (e.g., arteriography confirmed; changes of electrocardiographic and clinical symptoms according to the WHO criteria; a documented history of coronary intervention) and proper genotyping methods were used in most of the studies.
Selection criteria and data extraction
Such major criteria must be followed for included studies (1) Original papers containing complete data, (2) Case–control or cohort studies that assessed the association of CD14 gene -159C/T and rs2569190 polymorphisms with CVD, and (3) Sufficient data to calculate the odds ratio (OR) or P value, (4) Relevant cardiovascular outcomes were angiographically confirmed (generally by WHO criteria) and coronary stenosis was diagnosed as at least one coronary artery stenosis (no less than 50% by a coronary angiography [24, 25]), (5) The genotype distribution in the control group for each individual study should follow Hardy-Weinberg equilibrium (HWE).
The primary reasons for excluded studies: (1) Case-only studies, review or meta-analysis articles, (2) Deviate from the major selection criteria, (3) Overlapping or that supplied inadequate data, (4) Repeated publications or the same authors employed similar data in different papers, the data was only used once.
The study data was extracted based on standard protocol. For studies in which data could not be separated according to type of CVD from published data, cases were classified in the more inclusive category of coronary stenosis for the purposes of subsidiary analyses. Disagreement was settled by a consensus between all authors. Where essential information was not presented in articles, every effort was made to contact the authors. All procedures conformed to the guidelines for meta-analysis of observational studies in epidemiology. The following information were extracted independently by individual in our study: first author, year of publication, ethnicity, study design, types of CVD endpoints, HWE status among control, sample size of case and control, number of genotype and allele frequency.
Statistical analysis
We calculated the allele frequency for each study in allele counting method, the Hardy–Weinberg equilibrium (HWE) was tested by using the Chi square test [26]. We employed pooled ORs and 95% confidence intervals (CIs) to evaluate the strength of association between polymorphisms and cardiovascular disease for every eligible study [27]. The methodology of Cochran’s Q-statistic was used to evaluate the heterogeneity, which similar to the previous study in our lab [28]. If the P value in heterogeneity test was higher than 0.1, the fixed effect model was used. Moreover, the random effect model was used [29–31]. We used the following formula to quantified the effect of heterogeneity :I2 = 100 % × (Q − dƒ )/Q. The proportion of between-study variability attributable to heterogeneity was indicated by I2 value, and I2 values of 25, 50 and 75% were considered to be of low, moderate and high heterogeneity, respectively. If study groups revealed no heterogeneity, the similar results were produced in fixed and random effects models and, otherwise the random effects model usually produced wider CIs than the fixed effects model. In this meta-analysis, P value of less than 0.05 was considered a statistically significant.
In order to get exacting search results, we evaluated possible publication bias by Egger’s linear regression text [32]. If P value < 0.05 the statistical publication bias was considered [33]. Moreover the Begg’s test also used a funnel plot to evaluate the publication bias [32]. For sensitivity analysis, we removed one study orderly from the total and tested residual studies [34]. Statistical analysis was carried out using the software program STATA12.0 (Stata Corporation, College Station, Texas).
Results
Studies included in the meta-analysis
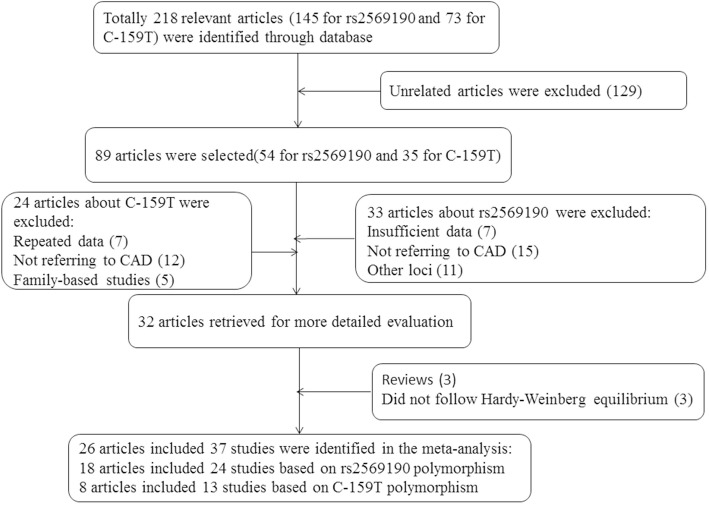
In this meta-analysis, totally 218 relevant articles (145 for rs2569190 and 73 for C-159T) were searched. After reading titles and abstracts, we excluded irrelevant studies, 89 articles (54 for rs2569190 and 35 for C-159T) for further reading. Then, we excluded 57 articles (33 for rs2569190 and 24 for C-159T), because of no data, insufficient data, repeated date, family-based studies and not referring to cardiovascular disease (CVD). Thus, 32 articles (23 for rs2569190 and 9 for C-159T) met the study inclusion criteria. Lastly, after excluding three groups of data in which the control populations deviated from HWE [17, 35, 36] and three reviews [37–39] about rs2569190 and C-159T. After filtering, 26 eligible studies involved 37 data sets were finally included [15, 16, 18–23, 40–55], in which two articles contained dual data (which articles included two type data about rs2569190 and C-159T) [19, 40]. Eventually, 26 studies providing 14,226 cases and 10,189 controls (rs2569190: 9413 patients and 7337 controls; C-159T: 4813 patients and 2852 controls) were pooled to evaluate the relationship between SNPs of CD14 and CVD in the meta-analysis (Table 1). The flowchart of selecting article process is presented in Fig. 1.
Table 1.
The basic information of every studies included in this meta-analysis
| Polymorphisms | Study | Year | Ethnicity | design | Endpoint | P(HWE) | Sample size | Genotypes | Allele frequencies (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |||||||||||||
| CC | CT | TT | CC | CT | TT | C | T | C | T | |||||||||
| rs2569190 (C-260T) | Wei et al. [40] | 2006 | East Asian | CC | CHD | 0.22 | 246 | 258 | 58 | 112 | 76 | 83 | 118 | 57 | 46.3 | 53.7 | 55.0 | 45.0 |
| Zhang et al. [18] | 2006 | East Asian | CC | CHD | 0.05 | 193 | 225 | 20 | 95 | 78 | 39 | 92 | 94 | 35.0 | 65.0 | 37.8 | 62.2 | |
| Li et al. [41] | 2007 | East Asian | CC | CHD | 0.14 | 193 | 225 | 29 | 95 | 69 | 47 | 124 | 54 | 40.0 | 60.0 | 48.0 | 52.0 | |
| Xie et al. [42] | 2008 | East Asian | CC | CHD | 0.24 | 241 | 149 | 49 | 127 | 65 | 56 | 65 | 28 | 46.7 | 53.3 | 59.4 | 40.6 | |
| Liu et al. [55] | 2010 | East Asian | CC | AMI | 0.19 | 12O | 130 | 23 | 56 | 41 | 43 | 57 | 30 | 42.5 | 57.5 | 55.0 | 45.0 | |
| Shimada et al. [43] | 2000 | East Asian | CC | MI | 0.83 | 128 | 83 | 27 | 49 | 52 | 21 | 43 | 19 | 40.0 | 60.0 | 51.0 | 49.0 | |
| Hohda et al. [44] | 2003 | East Asian | CC | MI | 0.2 | 502 | 527 | 97 | 242 | 163 | 115 | 278 | 134 | 43.4 | 56.6 | 48.2 | 51.8 | |
| Nauck et al. [16] | 2002 | European | NCC | AMI | 0.43 | 2559 | 697 | 675 | 1262 | 622 | 188 | 358 | 151 | 51.0 | 49.0 | 53.0 | 47.0 | |
| Nauck et al. [16] | 2002 | European | NCC | MI | 0.43 | 1599 | 697 | 444 | 758 | 397 | 188 | 358 | 151 | 51.0 | 49.0 | 53.0 | 47.0 | |
| Unkelbach et al. [19] | 1999 | European | CC | MI | 0.99 | 1053 | 1175 | 292 | 520 | 241 | 339 | 584 | 252 | 52.0 | 48.0 | 54.0 | 46.0 | |
| Koenig et al. [45] | 2002 | European | CC | AMI | 0.62 | 312 | 476 | 75 | 164 | 73 | 126 | 243 | 107 | 50.3 | 49.7 | 52.0 | 48.0 | |
| Longobardo et al. [46] | 2003 | European | CC | AMI | 0.38 | 215 | 215 | 44 | 101 | 70 | 55 | 101 | 59 | 44.0 | 56.0 | 49.0 | 51.0 | |
| Morange et al. [47] | 2005 | European | CC | MI | 0.38 | 194 | 197 | 42 | 98 | 54 | 39 | 104 | 54 | 47.0 | 53.0 | 46.0 | 54.0 | |
| Giacconi et al. [48] | 2007 | European | CC | CHD | 0.14 | 146 | 148 | 39 | 69 | 38 | 48 | 80 | 20 | 50.4 | 49.6 | 59.0 | 41.0 | |
| Hubacek et al. [15] | 1999 | European | CC | MI | 0.11 | 178 | 135 | 52 | 77 | 49 | 61 | 53 | 21 | 50.8 | 49.2 | 64.8 | 35.2 | |
| Lorenzova et al. [49] | 2007 | European | CC | AMI | 0.32 | 230 | 562 | 63 | 116 | 51 | 166 | 268 | 128 | 53.0 | 47.0 | 53.0 | 47.0 | |
| Arroyo-Espliguro [50] | 2005 | European | CC | AMI | 0.35 | 194 | 94 | 45 | 85 | 64 | 31 | 42 | 21 | 45.1 | 54.9 | 55.3 | 44.7 | |
| Arroyo-Espliguro [50] | 2005 | European | CC | CHD | 0.35 | 140 | 94 | 34 | 78 | 28 | 31 | 42 | 21 | 52.1 | 47.9 | 55.3 | 44.7 | |
| Morange et al. [47] | 2005 | European | CC | MI | 0.41 | 54 | 70 | 12 | 28 | 14 | 24 | 31 | 15 | 48.0 | 52.0 | 56.0 | 44.0 | |
| Morange et al. [47] | 2005 | European | CC | MI | 0.19 | 99 | 121 | 20 | 57 | 22 | 29 | 53 | 39 | 49.0 | 51.0 | 46.0 | 54.0 | |
| Morange et al. [51] | 2004 | European | NCC | MI | 0.09 | 128 | 253 | 43 | 59 | 26 | 69 | 113 | 71 | 57.0 | 43.0 | 50.0 | 50.0 | |
| Morange et al. [51] | 2004 | European | NCC | CHD | 0.75 | 123 | 243 | 31 | 58 | 34 | 61 | 124 | 58 | 49.0 | 51.0 | 51.0 | 49.0 | |
| Morange et al. [47] | 2005 | European | CC | MI | 0.2 | 179 | 176 | 60 | 94 | 25 | 65 | 77 | 34 | 60.0 | 40.0 | 59.0 | 41.0 | |
| Zee et al. [52] | 2001 | America | NCC | MI | 0.99 | 387 | 387 | 98 | 215 | 74 | 108 | 193 | 86 | 53.0 | 47.0 | 53.0 | 47.0 | |
| C-159T | Jin et al. [23] | 2016 | East Asian | CC | EH-LVH | 0.99 | 116 | 108 | 27 | 58 | 31 | 25 | 54 | 29 | 48.3 | 51.7 | 48.3 | 51.7 |
| Jin et al. [23] | 2016 | East Asian | CC | EH-NLVH | 0.99 | 107 | 108 | 24 | 46 | 37 | 25 | 54 | 29 | 43.8 | 56.2 | 48.3 | 51.7 | |
| Jin et al. [23] | 2016 | East Asian | CC | EH-LVH | 1 | 103 | 108 | 23 | 52 | 28 | 26 | 54 | 28 | 47.6 | 52.4 | 49.1 | 50.9 | |
| Jin et al. [23] | 2016 | East Asian | CC | EH-NLVH | 1 | 100 | 108 | 23 | 44 | 33 | 26 | 54 | 28 | 45.0 | 55.0 | 49.1 | 50.9 | |
| Wei et al. [40] | 2006 | East Asian | CC | CHD | 0.09 | 246 | 258 | 47 | 128 | 71 | 39 | 139 | 80 | 45.1 | 54.9 | 42.1 | 57.9 | |
| Li et al. [22] | 2005 | East Asian | CC | CHD | 0.2 | 162 | 196 | 24 | 75 | 63 | 54 | 89 | 53 | 38.0 | 62.0 | 59.2 | 40.8 | |
| Haberbosch et al. [21] | 2009 | European | CC | AMI | 0.46 | 54 | 252 | 16 | 24 | 14 | 71 | 131 | 50 | 51.9 | 48.1 | 54.2 | 45.8 | |
| Haberbosch et al. [21] | 2009 | European | CC | MI | 0.46 | 146 | 252 | 50 | 69 | 27 | 71 | 131 | 50 | 57.9 | 42.1 | 54.2 | 45.8 | |
| Koch et al. [20] | 2002 | European | CC | AMI | 0.43 | 998 | 340 | 273 | 498 | 227 | 88 | 177 | 75 | 52.0 | 48.0 | 52.0 | 48.0 | |
| Koch et al. [20] | 2002 | European | CC | MI | 0.43 | 793 | 340 | 232 | 390 | 171 | 88 | 177 | 75 | 54.0 | 46.0 | 52.0 | 48.0 | |
| Unkelbach et al. [19] | 1999 | European | CC | AMI | 0.36 | 1727 | 501 | 491 | 864 | 372 | 140 | 240 | 121 | 53.0 | 47.0 | 52.0 | 48.0 | |
| Damiano et al. [53] | 2012 | European | CC | CHD | 0.07 | 51 | 49 | 7 | 23 | 21 | 13 | 18 | 18 | 36.0 | 64.0 | 44.0 | 56.0 | |
| Banerjee et al. [54] | 2009 | Indian | CC | AMI | 0.14 | 210 | 232 | 45 | 116 | 49 | 38 | 126 | 68 | 49.0 | 51.0 | 44.0 | 56.0 | |
CHD coronary heart disease, AMI acute myocardial infarction, MI myocardial infarction, EH –LVH/ EH –LVH essential hypertension with left/not left ventricular hypertrophy, CC case–control, NCC nested case–control, Endpoint diseases of cases, HWE Hardy-Weinberg Equilibrium
Fig. 1.
The process of the articles selected in this meta-analysis
Meta-analysis results
SNP rs2569190 and cardiovascular disease risk
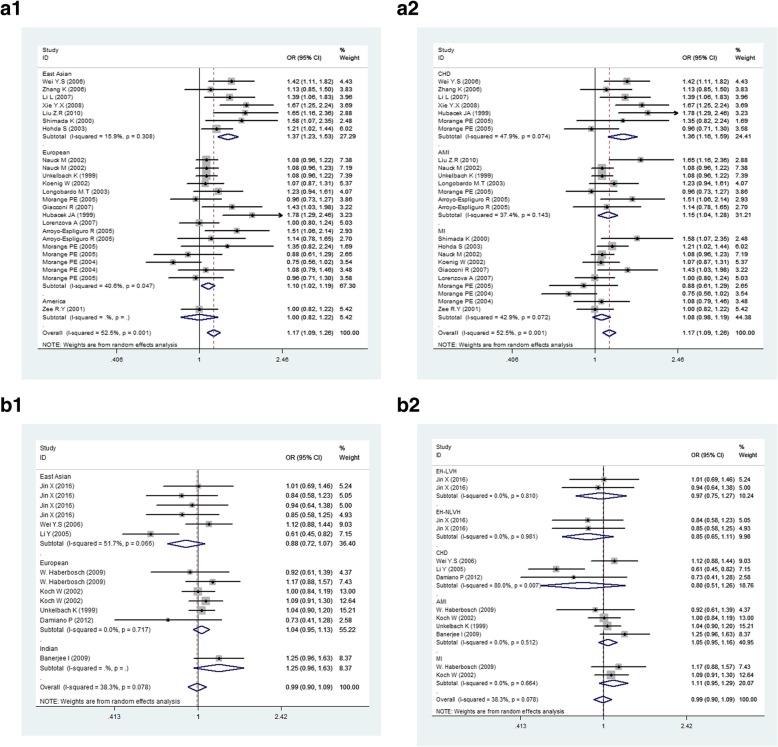
The test of heterogeneity indicated that there was potential heterogeneity in the overall population (p = 0.001, I2 = 52.5%), but after stratified by ethnicity, the heterogeneity was resolved in any subgroup (p = 0.308, I2 = 15.9%). An outstanding association was found in the overall population, under the random effect model (T vs. C, OR = 1.169, 95% CI: 1.087–1.257, P = 2.44 × 10− 5, Table 2, Fig. 2a1). Accordingly, dominant and recessive models were also tested to estimate the relationship between rs2569190 and CVD risk, the significant associations were observed in overall population (TT + CT vs. CC, OR = 1.233, 95% CI: 1.110–1.370, p = 3.79 × 10− 5, Table 2, Additional file 1: Figure S1a; TT vs. CC + CT, OR = 1.195, 95% CI: 1.062–1.345, p = 0.003, Table 2, Additional file 1: Figure S1b). In the subgroup’s analysis by ethnicity, we obtained the most significant relationship between rs2569190 with CVD, particularly in East Asian population under allele model (T vs. C, OR = 1.370, 95% CI: 1.226–1.531, p = 2.86 × 10− 8, Table 2, Fig. 2a1), dominant model (TT + TC vs. CC, OR = 1.574, 95% CI: 1.278–1.938, p = 9.11 × 10− 7, Table 2, Additional file 1: Figure S1a) and recessive model (TT vs. TC + CC, OR = 1.492, 95% CI: 1.236–1.801, p = 3.05 × 10− 5, Table 2, Additional file 1: Figure S1b). Besides, the results revealed that rs2569190 was associated with CVD in European under allele and dominant models (T vs. C, OR = 1.100, 95% CI: 1.019–1.189, p = 0.015, Table 2, Additional file 1: Figure S1a; TT + TC vs. CC, OR = 1.113, 95% CI: 1.006–1.232, p = 0.039, Table 2, Additional file 1: Figure S1b). Meanwhile, no relationship under any model was found in American population (Table 2, Fig. 2a1).
Table 2.
Meta-analysis of the association between rs2569190 (C-260T) polymorphism and CVD risk
| Sub-group analysis | No of data sets | No of cases/controls | Allele model (T VS.C) | Dominant model (CC VS.TT + CT) | Recessive model (TT VS.CC + CT) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | POR | PH | OR (95% CI) | POR | PH | OR (95% CI) | POR | PH | ||
| Overall | 24 | 9413 | 7337 | 1.169(1.087–1.257) | 2.44 × 10−5* | 0.001 | 1.233(1.110–1.370) | 3.79 × 10−5* | 0.02 | 1.195(1.062–1.345) | 0.003* | 0.002 |
| Ethnicity | ||||||||||||
| East Asian | 7 | 1503 | 1597 | 1.370(1.226–1.531) | 2.86 × 10−8* | 0.308 | 1.574(1.278–1.938) | 9.11 × 10−7* | 0.217 | 1.492(1.236–1.801) | 3.05 × 10−5* | 0.22 |
| European | 16 | 7403 | 5353 | 1.100(1.019–1.189) | 0.015 | 0.047 | 1.113(1.006–1.232) | 0.039 | 0.237 | 1.112(0.975–1.269) | 0.113 | 0.04 |
| America | 1 | 387 | 387 | 1.000(0.819–1.221) | 1 | – | 1.142(0.830–1.571) | 0.416 | – | 0.827(0.584–1.173) | 0.287 | – |
| Endpoint | ||||||||||||
| CHD | 7 | 1019 | 1005 | 1.357(1.157–1.592) | 2.47 × 10−7* | 0.074 | 1.669(1.388–2.007) | 2.48 × 10−6* | 0.433 | 1.342(1.011–1.781) | 0.042 | 0.035 |
| AMI | 7 | 762 | 1244 | 1.152(1.036–1.281) | 0.009 | 0.143 | 1.186(0.997–1.412) | 0.054 | 0.131 | 1.162(1.030–1.310) | 0.015 | 0.483 |
| MI | 10 | 4641 | 3915 | 1.077(0.976–1.188) | 0.139 | 0.072 | 1.061(0.949–1.186) | 0.297 | 0.799 | 1.118(0.907–1.378) | 0.297 | 0.002 |
OR odd ratio, 95%CI 95% confidence interval, PORP value for the test of association, PHP value for heterogeneity analysis
∗Significant P-value
Fig. 2.
Forest plot for the meta-analysis of the association between CD14 gene polymorphisms and CVD. a1 rs2569190 and CVD (T VS.C), stratification by ethnicity. a2 rs2569190 and CVD (T VS.C), stratification by endpoint. b1 C-159T and CVD (T VS.C), stratification by ethnicity. b2 C-159T and CVD (T VS.C), stratification by endpoint
Similarly, we carried out subgroup analysis by endpoints to estimate the relationship between rs2569190 and the specific isoforms of CVD, a very significant association was identified between rs2569190 and CHD under allelic model (T vs. C, OR = 1.357, 95% CI: 1.157–1.592, p = 2.47 × 10− 7, Table 2, Fig. 2a2), dominant model (TT + TC vs. CC, OR = 1.669, 95% CI: 1.388–2.007, p = 2.48 × 10− 6, Table 2, Additional file 1: Figure S2a) and a potential association in recessive model (TT vs. CC + CT, OR = 1.342, 95% CI: 1.011–1.781, p = 0.042, Table 2, Additional file 1: Figure S2b). Besides, the association between rs2569190 and AMI was also observed under allelic model (T vs. C, OR = 1.152, 95% CI: 1.036–1.281, p = 0.009, Table 2, Fig. 2a2), and recessive model (TT vs. CC + CT, OR = 1.162, 95% CI: 1.030–1.310, p = 0.015, Table 2, Additional file 1: Figure S2b).
C (− 159) T and cardiovascular disease risk
The association between C-159T and CVD was conducted in 13 independent studies with 7665 participants in this meta-analysis and all of datasets followed the inclusion criteria. The test of heterogeneity in the overall population was not significant (p = 0.324, I2 = 12.1%), suggesting that the fixed effect model could be used.
We didn’t observe association in overall population under allele model (T vs. C, OR = 1.009, 95% CI: 0.941–1.082, p = 0.854, Table 3, Fig. 2b1). We also tested the dominant and recessive genetic models in overall population, but no associations were found in the two models (Table 3, Additional file 1: Figure S1a, d). In the subgroup analysis by ethnicity, no potential association was found between C-159T and the risk of CVD in any populations under all genetic models (Table 3, Fig. 2b1, Additional file 1: Figure S1c, d). Furthermore, we also performed subgroup analysis by endpoints in three models, respectively, which indicated that no statistically significant association was discovered in these isoforms of CVD (Table 3, Fig. 2b2, Additional file 1: Figure S2c, d). In essence, the results did not reveal any allele-specific or genotype-specific relation of the C-159T with CVD (Table 3).
Table 3.
Meta-analysis of the association between C-159T polymorphism and CVD risk
| Sub-group analysis | No of data sets | No of cases/controls | Allele model (T VS.C) | Dominant model (CC VS.TT + CT) | Recessive model (TT VS.CC + CT) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (b95% CI) | POR | PH | OR (95% CI) | POR | PH | OR (95% CI) | POR | PH | ||
| Overall | 13 | 4813 | 2852 | 1.009(0.941–1.082) | 0.854 | 0.078 | 1.049(0.936–1.176) | 0.41 | 0.164 | 1.009(0.900–1.132) | 0.878 | 0.324 |
| Ethnicity | ||||||||||||
| East Asian | 6 | 834 | 886 | 0.881(0.723–1.075) | 0.212 | 0.066 | 1.120(0.888–1.413) | 0.34 | 0.109 | 1.198(0.974–1.474) | 0.087 | 0.327 |
| European | 6 | 3769 | 1734 | 1.039(0.953–1.132) | 0.387 | 0.717 | 0.923(0.806–1.058) | 0.249 | 0.488 | 0.962(0.832–1.113) | 0.602 | 0.751 |
| Indian | 1 | 210 | 232 | 1.249(0.958–1.628) | 0.101 | – | 0.718(0.445–1.160) | 0.176 | – | 0.734(0.479–1.125) | 0.156 | – |
| Endpoint | ||||||||||||
| EH-LVH | 2 | 219 | 216 | 0.974(0.746–1.271) | 0.845 | 0.81 | 1.045(0.669–1.631) | 0.847 | 0.817 | 1.028(0.672–1.573) | 0.898 | 0.87 |
| EH-NLVH | 2 | 207 | 216 | 0.846(0.646–1.109) | 0.227 | 0.981 | 1.051(0.669–1.653) | 0.828 | 0.967 | 1.424(0.938–2.162) | 0.097 | 0.957 |
| CHD | 3 | 459 | 503 | 0.804(0.514–1.257) | 0.338 | 0.007 | 1.292(0.953–1.785) | 0.121 | 0.007 | 1.186(0.904–1.556) | 0.217 | 0.1 |
| AMI | 4 | 2989 | 1325 | 1.048(0.950–1.156) | 0.351 | 0.512 | 0.926(0.790–1.085) | 0.342 | 0.729 | 0.917(0.778–1.080) | 0.3 | 0.314 |
| MI | 2 | 939 | 592 | 1.109(0.952–1.293) | 0.184 | 0.664 | 0.817(0.642–1.038) | 0.098 | 0.669 | 0.957(0.735–1.246) | 0.743 | 0.851 |
OR odd ratio, 95%CI 95% confidence interval, PORP value for the test of association, PHP value for heterogeneity analysis
Allele frequency of the rs2569190 and comparing to the 1000 genome phase 3 population
We displayed the alleles frequencies of different ethnicities in our meta-analysis and 1000 genomes alleles frequencies of rs2569190 in Table 4. In view of the sample size and population, the allelic frequencies of rs2569190 in this meta-analysis were consistent with the allelic frequencies in the 1000 Genome Project EAS (East Asian ancestry), EUR (European ancestry), respectively, however, there was distinction between the allele frequencies in AMR (Admixed American) and 1000 Genomes Project. Although such inconsistent allele frequencies in AMR were observed in the comparison to 1000 Genomes Project, the approximate allele frequency was obtained in overall population to 1000 Genomes Project. Because of the deficient data, we fail to make a comparison for C-159T to1000 Genomes Project (Table 4).
Table 4.
The allele frequency comparison between the meta-analysis and 1000 Genomes Project
| Polymorphism | Populations | Meta-analysis (alleles frequencies) | 1000 genomes (alleles frequencies) | ||||
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| C | T | C | T | C | T | ||
| rs2569190 (C-260T) | East Asian | 0.43 | 0.57 | 0.5 | 0.5 | 0.43(EAS) | 0.57(EAS) |
| European | 0.51 | 0.49 | 0.53 | 0.47 | 0.51(EUR) | 0.49(EUR) | |
| America | 0.53 | 0.47 | 0.53 | 0.47 | 0.47(AMR) | 0.53(AMR) | |
| All | 0.51 | 0.49 | 0.52 | 0.48 | 0.53 | 0.47 | |
| C-159T | East Asian | 0.44 | 0.56 | 0.47 | 0.53 | NA | NA |
| European | 0.53 | 0.47 | 0.52 | 0.48 | NA | NA | |
| Indian | 0.49 | 0.51 | 0.44 | 0.46 | NA | NA | |
| All | 0.49 | 0.51 | 0.5 | 0.5 | NA | NA | |
NA Not Available, EAS East Asian ancestry, EUR European ancestry, AMR Admixed American, All overall individuals from Phase 3 of the 1000 Genomes Project
Publication bias and sensitivity analysis
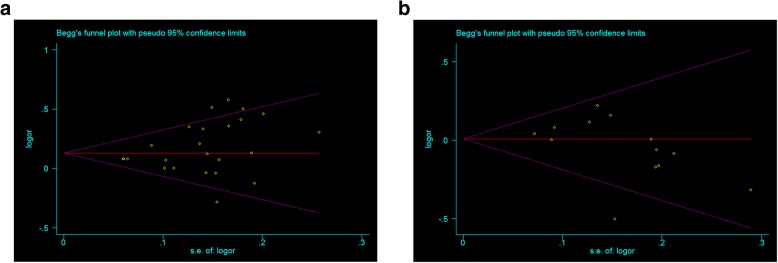
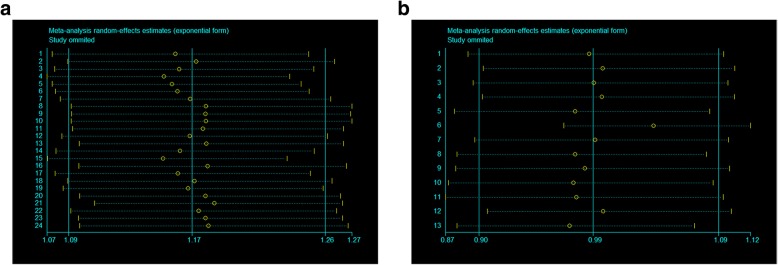
Begg’s funnel plot and Egger’s test were performed to estimate publication bias (Fig. 3a-b). There were no evidence of publication bias for SNP rs2569190 (p = 0.118) and C-159T (p = 0.077) under allele genetic model (Additional file 1: Table S1; Fig. 3a-b; Additional file 1: Figure S2 a-d). In addition, no significant difference in the Egger’s test neither, suggesting no obvious bias of publication in the present meta-analysis. We also conducted sensitivity analysis to assess the influence of individual studies on the pooled ORs. We found the pooled OR was not substantially altered, when a single study involved in the meta-analysis was deleted each time (Fig. 4a-b; Additional file 1: Figure S3 a-d).
Fig. 3.
Begg’s funnel plot of publication bias in the meta-analysis of the association of CD14 polymorphisms with CVD risk. a rs2569190 and CVD (T vs. C). b C-159T and CVD (T vs. C)
Fig. 4.
Sensitivity analysis to assess the stability of the meta-analysis. a rs2569190 and CVD (T vs. C). b C-159T and CVD (T vs. C)
Discussion
In the present study, we conducted a meta-analysis to evaluate the association between rs2569190 and C-159T with the susceptibility of cardiovascular disease (CVD). It was verified that the T allele of rs2569190 apparently increased the risk of CVD, particularly in East Asian population. In stratified analysis by endpoints, rs2569190 was significantly associated with CHD, whereas, no any allele-specific or genotype-specific relation of the C-159T with cardiovascular disease was observed in diverse ethnicities.
The SNP rs2569190, locating in the promoter region of the CD14 gene was considered modulating the capacity to stimulate inflammation through the regulation of CD14 gene expression and plasma soluble CD14 (sCD14) levels [56]. A potential functional role of rs2569190 on CD14 has been suggested as it alters a Sp1 transcription factor binding site and modulates the activity of promoter, the T allele was associated with higher transcription. This SNP was identified to be associated with multiple inflammatory diseases [57, 58]. In previous studies, rs2569190 was indicated to be associated with inflammatory bowel disease [59] and eczema [60]; afterward, studies showed the relevance between rs2569190 and diverse vascular events, such as atherosclerosis [61]. In 1999, rs2569190 was first shown to influence the interaction between CD14 receptor and human coronary artery atherosclerosis and complications [5].
Recently, in the genome-wide association studies (GWAS) of Alex P et al. [62], a robust result was discovered that rs2569190 was significantly associated with CAD in older European-American adults and a potential association in black (European Americans P = 6.15e-08, Blacks P = 0.04), meanwhile the same findings were also obtained by other investigators in another European population [50]. To check this association in different ethnicities, we carried out a comprehensive ethnicity-specific meta-analysis involving 9413 cases and 7337 controls with 24 separate comparisons and used subgroup analysis as well as the random effect model to deal with the heterogeneity. We found that the association between rs2569190 and CAD achieved significant level in overall population and reached genome-wide significance in East Asian population (T vs. C, OR = 1.370, 95% CI: 1.226–1.531, P = 2.86 × 10− 8) as well as a certain relevance in the European, but no association in America population. Similarly, in the previous meta-analysis with 28 case–control articles testified a significant association between rs2569190 with CHD in East Asian, but they acquired a negative result in European populations under any genetic models [37], which was associated result in our study. After carefully read, we found that the data in their study were not convinced. Some studies about C-159T were included by researchers [20, 21, 54], and three studies with data deviated from HWE [17, 35, 36]. Papers of this kind were not supposed to pass the inclusion criteria of meta-analysis may deviate from the true results. Therefore, we excluded these studies to carry out a correct meta-analysis.
In our meta-analysis with rs2569190, 16 studies for the European were included. After analyzed, the marginal significance was discovered in European subgroup (P = 0.015), which was negative result in another meta-analysis study [37]. It was easy for us to see that the frequencies of the T allele in our European population approximate to that in the overall population. Meanwhile, the T allele of control was discordant with C allele in European and C allele frequency was distinctly larger than T. Besides, we could notice that the alleles frequencies are concordant in the comparison of European subgroup to another meta-analysis study [37]. In addition, we made a comparison for rs2569190 to1000 Genomes Project and found the allele frequencies of rs2569190 in this meta-analysis were consistent with the allele frequencies in the 1000 Genome Project EUR (European ancestry), EAS (East Asian ancestry), respectively, however, there was distinction between the allele frequencies in AMR (Admixed American) and 1000 Genomes Project due to the weak number of studies about American population. More studies with statistical enough power for American race are needed for deeply evaluation.
Of course, so far some research had reported that there was no association was found between rs2569190 and CVD in European population [16, 46], which was contradictory with our findings. It’s normal that such distinct consequences were obtained in separated studies. CVD is considered to be a common multifactor syndrome due to its complicated pathogenesis [63]. A part of previous studies verified that the rs2569190 was associated with CVD in European population [50] and the diametrical results were also carried out in other participants [46]. It was validated that the BMI and smoking will significantly contribute to susceptibility and development of CVD [64]. In addition, the gender difference was the key role in CVD morbidity [65]. However, the basic data which lack of BMI level in participants might lead to inconsistent results. These phenomena and discrepancy needed further investigation on the basis of large sample size.
C-159T base at − 159 lies 49 bp adjacent to an experimentally detected binding site for transcription factor Sp1 at − 110 and 1 bp adjacent to a putative Ap2 site at − 158, it was considered to be an important CD14-activating mediator of inflammatory responses that may result in atherosclerosis, coronary heart disease (CAD), thrombus formation and myocardial infarction (MI) [54]. Previous studies had confirmed that C-159T polymorphism was associated with CAD and indicated that C-159T expression was obviously higher in CAD cases than in controls [19, 23], but there were distinct conflictions among different researchers in the different ethnics [53].
In 2015, Li et al. [38] performed a meta-analysis included 2798 cases and 1669 controls from 7 articles. The results indicated that C-159T could increase the risk of CAD under allelic model (p = 0.05), recessive (p = 0.01) in the whole population with marginal significance, no significant association was found between them under dominant (p = 0.10). A significant association was detected in Chinese population (p < 0.05), while there was no significant association in the European subgroup (p > 0.05). To precisely explore the association between C-159T and CAD, we analyzed the data that were consistent with HWE. Ultimately, we carried out a comprehensive meta-analysis with 7665 participants. In contrast, we obtained the ineffective relationship between C-159T with any phenotypes of CVD in all subgroups from our study. Then, we compared to previous meta-analysis, and we found that they only included 5 articles about Chinese and 2 studies about European, such deficient data sets may lead to imprecise results. Simultaneously, we realized that the marginal significance under the allelic model and recessive model will be leaded by limited data sizes for C-159T, whereas our study included 13 researches to perform a more comprehensive study which will be more persuasive. In addition, the significant heterogeneity was easily found under all models in their study, of which the heterogeneity reached threshold (I2 = 83%) in the most relative recessive model. So many limitations were discovered, which did not emerge in our study, will produce contradictory conclusion.
Although no significant association between C-159T and CAD was found in our study, the positive results were discovered by previous studies [22]. We found that the participants which were included by the positive study, were stratified in the gender and the males were much higher than females [19]. Besides, the age was inconsistent in diverse studies, the youngest cohort is 47.9 ± 5.8 and the oldest is 65.30 ± 10.53 in cases of CAD [21, 36]. It’s likely that such differences may, at less in part, attributed to the conclusion difference.
In addition, CD14 gene, located in 5q23–31, spans 3.9 kb, which encodes the glycoprotein with 375 amino acids [66]. The two identified SNPs, rs2569190, in upstream of the CD14 promoter at base pair − 260 from the major transcription start site and C-159T, in the 5′ flanking region of the CD14 gene at position − 159 [67–69]. Many previous studies had suggested that the two polymorphisms will increase sCD14 levels in homozygous carriers of T allele [19, 50] and the base cytosine (C) is replaced by thymine (T) in CD14 gene polymorphisms rs2569190 and C-159T has been reported being associated with a higher risk of CAD [22]. Thus, the relationship that C-159T whether dependent on rs2569190 to alter the activity of CD14 gene promoter affect CD14 gene expression and lead to atherosclerosis, increase the risk of CAD, was not to be researched.
Although we revealed some new discoveries in this study, there were still several limitations should be taken into consideration. In our study, the overall sample size is large, but the size of each study is relatively small, the smallest sample is 54 cases and 70 controls [47], and we need numerous data to validate the relationship between rs2569190 and CVD for further study in American and other populations. For C-159T, the included data in current meta-analysis for ethnicity more from population with European and East Asian origin, and the findings are applicable to only these populations, more studies are required in Indian and other populations. Secondly, we had to indicate that significant between-study heterogeneity that was detected. We used the random effect model to deal with the heterogeneity, but this might induce an imprecise statistic because fixed effect model and random effect model address different research questions. Additionally, we are unable to analyze the actual impact of immanent factors on cardiovascular disease because of the incomplete data. Age is a powerful predictor of cardiovascular adverse events. It is vague why older patients continue to have poor outcomes after ACS despite improved access to contemporary treatment [70]. Meanwhile, the cardiovascular morbidity will also vary due to gender differences [71]. In this study, we did not investigate the contribution of age and gender to the onset of cardiovascular disease due to the insufficient data. Furthermore, the mechanism of CVD is considered to be comprehensive, including gene-gene and gene-environment interactions. To sum up, more studies with enough statistical power are needed for deeply evaluation.
Conclusions
We conducted a meta-analysis to evaluate the effects of CD14 polymorphisms (rs2569190 and C-159T) on the risk of cardiovascular disease. This meta-analysis indicated that SNP rs2569190 significantly contribute to susceptibility and development of CVD, particularly in the East Asian population and in the subtype CHD group, in addithon, a potential association was observed in the AMI group, T allele acts as a risk factor for cardiovascular disease. However, we failed to acquire the positive association between rs2569190 and other subtypes of CVD. Meanwhile, the associations between C-159T polymorphism with CVD were not observed under any model. Further efforts should be put on investigating the association between the functional mutations within CD14 gene and CVD, and the interactions in potential gene-gene and gene-environment should be comprehensively analyzed.
Additional file
Table S1. The information of publication bias in all Polymorphisms. Figure S1. Forest plot for the meta-analysis of the association between CD14 gene polymorphisms and CVD under the Dominant and Recessive models. Figure S2. Begg’s funnel plot of publication bias in the meta-analysis of the association of CD14 polymorphisms with CVD risk. Figure S3. Sensitivity analysis to assess the stability of the meta-analysis. (PDF 576 kb)
Acknowledgments
The funding agencies had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors wish to thank all colleagues and friends who helped us in writing this article. The kind and helpful advice from Dr. Peikuan Cong is gratefully acknowledged. We thank the peer reviewers for their thorough and helpful review of this manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81871831), the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars of China (LR17H070001).
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its Additional information files].
Abbreviations
- 95% CI
95% confidence interval
- AMI
Acute myocardial infarction
- AMR
Admixed American
- CC
Case–control
- CHD
Coronary heart disease
- CVD
Cardiovascular disease
- EAS
East Asian ancestry
- EH –LVH/EH –LVH
Essential hypertension with left/not left ventricular hypertrophy
- EUR
European ancestry
- HWE
Hardy-Weinberg Equilibrium
- MI
Myocardial infarction
- NA
Not available
- NCC
Nested case–control
- OR
Odd ratio
Authors’ contributions
HFZ and JJX conceived of the study, participated in the design, and drafted the manuscript. KQL, ZMY and XWZ carried out the study searches, XJX, PPZ and WYB collected the data. MCQ, PKC and XWZ performed the statistical analyses. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mokdad AH. Burden of cardiovascular diseases in the eastern Mediterranean region, 1990–2015: findings from the global burden of disease 2015 study. Int J Public Health. 2018;63:1–13. doi: 10.1007/s00038-017-1012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;353:429–430. doi: 10.1056/NEJM200507283530425. [DOI] [PubMed] [Google Scholar]
- 3.Luo Y, Xie XM, Liu HX. Related inflammation markers among different types of coronary heart disease. J Central South Univ Med Sci. 2004;29:227–229. [PubMed] [Google Scholar]
- 4.Baroldi G. Different types of myocardial necrosis in coronary heart disease: a pathophysiologic review of their functional significance. Am Heart J. 1975;89:742. doi: 10.1016/0002-8703(75)90189-1. [DOI] [PubMed] [Google Scholar]
- 5.Ross R. Atherosclerosis is an inflammatory disease. Am Heart J. 1999;138:S419–S420. doi: 10.1016/S0002-8703(99)70266-8. [DOI] [PubMed] [Google Scholar]
- 6.Bernardo E, Angiolillo DJ, Ramirez C, Cavallari U, Trabetti E, Sabate M, et al. Influence of the CD14 C260T promoter polymorphism on C-reactive protein levels in patients with coronary artery disease. Am J Cardiol. 2006;98:1182–1184. doi: 10.1016/j.amjcard.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Becker AE, de Boer OJ, Ac VDW. The role of inflammation and infection in coronary artery disease. Annu Rev Med. 2001;52:289–297. doi: 10.1146/annurev.med.52.1.289. [DOI] [PubMed] [Google Scholar]
- 8.Avanzas P, Arroyoespliguero R, Cosínsales J, Aldama G, Pizzi C, Quiles J, et al. Markers of inflammation and multiple complex stenoses (pancoronary plaque vulnerability) in patients with non-ST segment elevation acute coronary syndromes. Heart. 2004;90:847. doi: 10.1136/hrt.2003.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenhagen P, Tuzcu EM, Ellis SG. Plaque vulnerability, plaque rupture, and acute coronary syndromes: (multi)-focal manifestation of a systemic disease process. Circulation. 2002;106:760–762. doi: 10.1161/01.CIR.0000025708.36290.05. [DOI] [PubMed] [Google Scholar]
- 10.Mendel I, Feige E, Yacov N, Salem Y, Levi I, Propheta-Meiran O, et al. VB-201, an oxidized phospholipid small molecule, inhibits CD14- and toll-like receptor-2-dependent innate cell activation and constrains atherosclerosis. Clin Exp Immunol. 2014;175:126. doi: 10.1111/cei.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol (Baltimore, Md. : 1950) 1988;141:547–552. [PubMed] [Google Scholar]
- 12.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 13.Estruch M, Bancells C, Beloki L, Sanchez-Quesada JL, Ordóñez-Llanos J, Benitez S. CD14 and TLR4 mediate cytokine release promoted by electronegative LDL in monocytes[J] Atherosclerosis. 2013;229:356–362. doi: 10.1016/j.atherosclerosis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Fernándezreal JM, Broch M, Richart C, Vendrell J, Lópezbermejo A, Ricart W. CD14 monocyte receptor, involved in the inflammatory Cascade, and insulin sensitivity. J Clin Endocrinol Metabol. 2003;88:1780–1784. doi: 10.1210/jc.2002-020173. [DOI] [PubMed] [Google Scholar]
- 15.Hubacek JA, Pit’ha J, Škodová Z, Stanĕk V, Poledne R. C (− 260)→ T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation. 1999;99:3218–3220. doi: 10.1161/01.CIR.99.25.3218. [DOI] [PubMed] [Google Scholar]
- 16.Nauck M, Winkelmann BR, Hoffmann MM, Böhm BO, Wieland H, März W. C(−260)T polymorphism in the promoter of the CD14 gene is not associated with coronary artery disease and myocardial infarction in the Ludwigshafen risk and cardiovascular health (LURIC) study. Am J Cardiol. 2002;90:1249–1252. doi: 10.1016/S0002-9149(02)02845-X. [DOI] [PubMed] [Google Scholar]
- 17.PAN M, W-d GU, CHEN F. C (−260) T molecular variation in the promoter of the CD14 monocyte receptor gene is associated with coronary heart disease in Chinese. Chin J Misdiagn. 2003;10:010. [Google Scholar]
- 18.Zhang K, Lei LI, Qiu F, Guang-Yu GU, Luo XY, Chen JH, et al. Association of C(−260)T polymorphism in CD14 gene promoter with coronary heart disease in Han population of Jiangsu region. Chin J Clin Lab Sci. 2006;24:413–415. [Google Scholar]
- 19.Unkelbach K, Gardemann A, Kostrzewa M, Philipp M, Tillmanns H, Haberbosch W. A new promoter polymorphism in the gene of lipopolysaccharide receptor CD14 is associated with expired myocardial infarction in patients with low atherosclerotic risk profile. Arterioscler Thromb Vasc Biol. 1999;19:932–938. doi: 10.1161/01.ATV.19.4.932. [DOI] [PubMed] [Google Scholar]
- 20.Koch W, Kastrati A, Mehilli J, Von BN, Schömig A. CD14 gene -159C/T polymorphism is not associated with coronary artery disease and myocardial infarction. Am Heart J. 2002;143:971–976. doi: 10.1067/mhj.2002.122512. [DOI] [PubMed] [Google Scholar]
- 21.Haberbosch W, Unkelbach K, Schuster D, Gardemann A, Tillmanns H, Hölschermann H. CD14 promoter polymorphism (− 159C-->t) is not associated with myocardial infarction or coronary artery disease in patients with assumed high genetic risk. Thorac Cardiovasc Surg. 2009;57:386–390. doi: 10.1055/s-0029-1185876. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Xiong XQ, Zhang PA, Ming KH. Association of C-159T polymorphism in promoter region of CD14 and coronary heart disease. Chin J Med Genet. 2005;22:687. [PubMed] [Google Scholar]
- 23.Xing Jin J-lC, Zheng S-h, Jin G-m. Correlation between CD14 promoter gene-159 (C-T) polymorphism and essential hypertension with left ventricular hypertrophy in China Yanbian area. China J Mod Med. 2016;17:05. [Google Scholar]
- 24.Castelli WP, Garrison RJ, Wilson PW, Abbott RD, Kalousdian S, Kannel WB. Incidence of coronary heart disease and lipoprotein cholesterol levels: the Framingham study. Jama. 1986;256:2835–2838. doi: 10.1001/jama.1986.03380200073024. [DOI] [PubMed] [Google Scholar]
- 25.Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51:606. doi: 10.1016/S0002-9149(83)80105-2. [DOI] [PubMed] [Google Scholar]
- 26.Thakkinstian A, Mcelduff P, D'Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 27.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 28.Hu L-Y, Cheng Z, Zhang B, Yin Q, Zhu X-W, Zhao P-P, et al. Associations between PTPN22 and TLR9 polymorphisms and systemic lupus erythematosus: a comprehensive meta-analysis. Arch Dermatol Res. 2017;309:461–477. doi: 10.1007/s00403-017-1745-0. [DOI] [PubMed] [Google Scholar]
- 29.Thompson SG, Sharp SJ. Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med. 1999;18:2693–2708. doi: 10.1002/(SICI)1097-0258(19991030)18:20<2693::AID-SIM235>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 30.Thompson SG, Prevost TC, Sharp SJ. Investigating underlying risk as a source of heterogeneity in meta-analysis. Stat Med. 1997;16:2741–2758. doi: 10.1002/(SICI)1097-0258(19971215)16:23<2741::AID-SIM703>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Houwelingen HCV, Senn SJ. Letter to the editor: investigating underlying risk as a source of heterogeneity in meta-analysis. Stat Med. 1999;18:110–115. doi: 10.1002/(SICI)1097-0258(19990115)18:1<110::AID-SIM14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 32.Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ: Br Med J. 1998;316:469. doi: 10.1136/bmj.316.7129.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterne JAC. Systematic reviews in health care: investigating and dealing with publication and other biases in meta-analysis. Br Med J. 2001;323:101–105. doi: 10.1136/bmj.323.7304.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Copas J, Shi JQ. Meta-analysis, funnel plots and sensitivity analysis. Biostatistics. 2000;1:247–262. doi: 10.1093/biostatistics/1.3.247. [DOI] [PubMed] [Google Scholar]
- 35.Sediri Y, Hammami S, Kallel A, Mourali MS, Feki M, Elasmi M, et al. C(−260)T polymorphism in the promoter of CD 14 gene is not associated with myocardial infarction in the Tunisian population. Exp Mol Pathol. 2011;90:276–279. doi: 10.1016/j.yexmp.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Hu R, LX C, He MA. Polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for coronary heart disease in a Wuhan population. Chin J Geriatr Heart Brain Vessel Dis. 2006;3:017. [Google Scholar]
- 37.Pu H, Yin J, Wu Y, Zhang D, Wang Y, Zhou R, et al. The association between CD14 gene C-260T polymorphism and coronary heart disease risk: a meta-analysis. Mol Biol Rep. 2013;40:4001–4008. doi: 10.1007/s11033-012-2478-y. [DOI] [PubMed] [Google Scholar]
- 38.Li YY, Wang XM, Zhou CW, Xu J, Qian Y. CD14 gene-159C/T polymorphism and coronary artery disease: a meta-analysis involving 4467 subjects. Int J Clin Exp Med. 2015;8:12149–12160. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang HF, Zhong BL, Zhu WL, Xie SL, Qiu LX, Zhu LG, et al. CD14 C-260T gene polymorphism and ischemic heart disease susceptibility: a HuGE review and meta-analysis. Genet Med. 2009;11:403–408. doi: 10.1097/GIM.0b013e3181a16cb0. [DOI] [PubMed] [Google Scholar]
- 40.Wei YS, Liu YG, Tang RG. Association between CD14 gene promo-ter region polymorphisms and coronary heart disease. J Fourth Mil Med Univ. 2006;27:630–633. [Google Scholar]
- 41.Li L, Zhang K, Qiu F, Gu GY, Luo XY, Chen JH, et al. Association of C(−159)T polymorphism in the promoter of CD14 gene with coronary heart disease in Han population of Jiangsu province. J Clin Rehab Tissue Eng Res. 2007;11:8749–8752. [Google Scholar]
- 42.Y-x XIE, Ai-jiaer B, Ma Y-T. Distribution of-260 C/T single nucleotide polymorphisms (SNP) in the promoter region of CD14 gene and its significance for coronary heart disease and plasma lipids levels between the Han and the Uyghur people in Xinjiang. Chin J Cardiovasc Rev. 2008;6:003. [Google Scholar]
- 43.Shimada K, Watanabe Y, Mokuno H, Iwama Y, Daida H, Yamaguchi H. Common polymorphism in the promoter of the CD14 monocyte receptor gene is associated with acute myocardial infarction in Japanese men. Am J Cardiol. 2000;86:682–684. doi: 10.1016/S0002-9149(00)01054-7. [DOI] [PubMed] [Google Scholar]
- 44.Hohda S, Kimura A, Sasaoka T, Hayashi T, Ueda K, Yasunami M, et al. Association study of CD14 polymorphism with myocardial infarction in a Japanese population. Jpn Heart J. 2003;44:613–622. doi: 10.1536/jhj.44.613. [DOI] [PubMed] [Google Scholar]
- 45.Koenig W, Khuseyinova N, Hoffmann MM, März W, Fröhlich M, Hoffmeister A, et al. CD14 C(−260)-->T polymorphism, plasma levels of the soluble endotoxin receptor CD14, their association with chronic infections and risk of stable coronary artery disease. J Am Coll Cardiol. 2002;40:34–42. doi: 10.1016/S0735-1097(02)01937-X. [DOI] [PubMed] [Google Scholar]
- 46.Longobardo M, Cefalu A, Pezzino F, Noto D, Emmanuele G, Barbagallo C, et al. The C (−260)> T gene polymorphism in the promoter of the CD14 monocyte receptor gene is not associated with acute myocardial infarction. Clin Exp Med. 2003;3:161–165. doi: 10.1007/s10238-003-0020-1. [DOI] [PubMed] [Google Scholar]
- 47.Morange PE, Saut N, Alessi MC, Frere C, Hawe E, Yudkin JS, et al. Interaction between the C-260T polymorphism of the CD14 gene and the plasma IL-6 concentration on the risk of myocardial infarction: the HIFMECH study. Atherosclerosis. 2005;179:317–323. doi: 10.1016/j.atherosclerosis.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Giacconi R, Caruso C, Lio D, Muti E, Cipriano C, Costarelli L, et al. CD14 C (−260)T polymorphism, atherosclerosis, elderly: role of cytokines and metallothioneins. Int J Cardiol. 2007;120:45–51. doi: 10.1016/j.ijcard.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 49.Lorenzová A, Stanek V, Gebauerová M, Bohuslavová R, Stávek P, Hubácek JA, et al. High-sensitivity C-reactive protein concentration in patients with myocardial infarction-environmental factors, and polymorphisms in interleukin-10 and CD14 genes. Clin Chem Lab Med. 2007;45:855–861. doi: 10.1515/CCLM.2007.157. [DOI] [PubMed] [Google Scholar]
- 50.Arroyo-Espliguero R, Vazquez-Rey E, El-Sharnouby K, Kalidas K, Gimeno-Blanes JR, Cole D, et al. C(−260)→T polymorphism in the promoter of CD14 receptor gene is associated with the risk of acute coronary events in patients with angina pectoris. J Am Coll Cardiol. 2003;41:275. doi: 10.1016/S0735-1097(03)82290-8. [DOI] [Google Scholar]
- 51.Morange PE, Tiret L, Saut N, Luc G, Arveiler D, Ferrieres J, et al. TLR4/Asp299Gly, CD14/C-260T, plasma levels of the soluble receptor CD14 and the risk of coronary heart disease: the PRIME study. Eur J Hum Genet. 2004;12:1041–1049. doi: 10.1038/sj.ejhg.5201277. [DOI] [PubMed] [Google Scholar]
- 52.Zee RYL, Lindpaintner K, Struk B, Hennekens CH, Ridker PM. A prospective evaluation of the CD14 C(−260) T gene polymorphism and the risk of myocardial infarction. Atherosclerosis. 2001;154:699–702. doi: 10.1016/S0021-9150(00)00698-5. [DOI] [PubMed] [Google Scholar]
- 53.Pasqualini D, Bergandi L, Palumbo L, Borraccino A, Dambra V, Alovisi M, et al. Association among oral health, apical periodontitis, CD14 polymorphisms, and coronary heart disease in middle-aged adults. J Endod. 2012;38:1570–1577. doi: 10.1016/j.joen.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee I, Pandey U, Hasan OM, Parihar R, Tripathi V, Ganesh S. Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thromb Thrombolysis. 2009;27:88–94. doi: 10.1007/s11239-007-0184-8. [DOI] [PubMed] [Google Scholar]
- 55.Z-R LIU, Y-G LIU, Y-S WEI. Serum CD14 levels and the genotypes of CD14 gene in patients with acute myocardial infarction. Chin J Gerontol. 2010;6:009. [Google Scholar]
- 56.Griga T, Klein W, Epplen JT, Hebler U, Stachon A, May B. CD14 expression on monocytes and soluble CD14 plasma levels in correlation to the promotor polymorphism of the endotoxin receptor CD14 gene in patients with inactive Crohn's disease[J] Hepato-gastroenterology. 2004;52:808–811. [PubMed] [Google Scholar]
- 57.Mattila K, Valtonen V, Nieminen MS, Asikainen S. Role of infection as a risk factor for atherosclerosis, myocardial infarction, and stroke. Clin Infect Dis. 1998;26:719–734. doi: 10.1086/514570. [DOI] [PubMed] [Google Scholar]
- 58.Woods A, Brull DJ, Humphries SE, Montgomery HE. Genetics of inflammation and risk of coronary artery disease: the central role of interleukin-6. Eur Heart J. 2000;21:1574. doi: 10.1053/euhj.1999.2207. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, Hu J, Fan R, Zhou J, Zhong J. Association between CD14 gene C-260T polymorphism and inflammatory bowel disease: a meta-analysis. PLoS One. 2012;7:e45144. doi: 10.1371/journal.pone.0045144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litonjua AA, Belanger K, Celedón JC, Milton DK, Bracken MB, Kraft P, et al. Polymorphisms in the 5’ region of the CD14 gene are associated with eczema in young children. J Allergy Clin Immunol. 2005;115:1056–1062. doi: 10.1016/j.jaci.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Giacconi R, Caruso C, Lio D, Muti E, Cipriano C, Costarelli L, et al. CD14 C (−260) T polymorphism, atherosclerosis, elderly: role of cytokines and metallothioneins. Int J Cardiol. 2007;120:45–51. doi: 10.1016/j.ijcard.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 62.Reiner AP, Lange EM, Jenny NS, Chaves PHM, Ellis J, Li J, et al. Soluble CD14: genome-wide association analysis and relationship to cardiovascular risk and mortality in the older adults. Arterioscler Thromb Vasc Biol. 2013;33:158. doi: 10.1161/ATVBAHA.112.300421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwong RY, Schussheim AE, Rekhraj S, Aletras AH, Geller N, Davis J, et al. Detecting acute coronary syndrome in the emergency department with cardiac magnetic resonance imaging. Circulation. 2003;43:674–675. doi: 10.1161/01.cir.0000047527.11221.29. [DOI] [PubMed] [Google Scholar]
- 64.Wienke A, Herskind AM, Christensen K, Skytthe A, Yashin AI. The heritability of CHD mortality in danish twins after controlling for smoking and BMI. Twin Res Hum Genet. 2005;8:53. doi: 10.1375/twin.8.1.53. [DOI] [PubMed] [Google Scholar]
- 65.Brandon LJ, Mullis RM, Jonnalagadda SS, Hughes MH. Relationships and CHD risks of BMI, lipoproteins, lipids, and blood pressure in African-American men and women. Prev Med. 2005;40:349–354. doi: 10.1016/j.ypmed.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Rollins J, Paigen B, Wang X. Genetic and genomic insights into the molecular basis of atherosclerosis. Cell Metab. 2007;6:164. doi: 10.1016/j.cmet.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watkins H, Farrall M. Genetic susceptibility to coronary artery disease: from promise to progress. Nat Rev Genet. 2006;7:163–173. doi: 10.1038/nrg1805. [DOI] [PubMed] [Google Scholar]
- 68.Levan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, Martinez FD, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 69.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A polymorphism* in the 5 ′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with Total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 70.Granger CB, Goldberg RJ, Dabbous O, Pieper KS, Eagle KA, Cannon CP, et al. Predictors of hospital mortality in the global registry of acute coronary events. Arch Intern Med. 2004;13:13. doi: 10.1001/archinte.163.19.2345. [DOI] [PubMed] [Google Scholar]
- 71.Beveridge LA, Khan F, Struthers AD, Armitage J, Barchetta I, Bressendorff I, et al. Effect of vitamin D supplementation on markers of vascular function: a systematic review and individual participant meta-analysis. J Am Heart Assoc. 2018;7:e008273. doi: 10.1161/JAHA.117.008273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The information of publication bias in all Polymorphisms. Figure S1. Forest plot for the meta-analysis of the association between CD14 gene polymorphisms and CVD under the Dominant and Recessive models. Figure S2. Begg’s funnel plot of publication bias in the meta-analysis of the association of CD14 polymorphisms with CVD risk. Figure S3. Sensitivity analysis to assess the stability of the meta-analysis. (PDF 576 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its Additional information files].