Abstract
Background
Premature senescence can reduce the yield and quality of crops. WRKY transcription factors (TFs) play important roles during leaf senescence, but little is known about their ageing mechanisms in cotton.
Results
In this study, a group III WRKY TF, GhWRKY27, was isolated and characterized. The expression of GhWRKY27 was induced by leaf senescence and was higher in an early-ageing cotton variety than in a non-early-ageing cotton variety. Overexpression of GhWRKY27 in Arabidopsis promoted leaf senescence, as determined by reduced chlorophyll content and elevated expression of senescence-associated genes (SAGs). Yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays showed that GhWRKY27 interacted with an MYB TF, GhTT2. Putative target genes of GhWRKY27 were identified via chromatin immunoprecipitation followed by sequencing (ChIP-seq). Yeast one-hybrid (Y1H) assay and electrophoretic mobility shift assay (EMSA) revealed that GhWRKY27 binds directly to the promoters of cytochrome P450 94C1 (GhCYP94C1) and ripening-related protein 2 (GhRipen2–2). In addition, the expression patterns of GhTT2, GhCYP94C1 and GhRipen2–2 were identified during leaf senescence. Transient dual-luciferase reporter assay indicated that GhWRKY27 could activate the expression of GhCYP94C1 and GhRipen2–2.
Conclusions
Our work lays the foundation for further study of the functional roles of WRKY genes during leaf senescence in cotton. In addition, our data provide new insights into the senescence-associated mechanisms of WRKY genes in cotton.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1688-z) contains supplementary material, which is available to authorized users.
Keywords: ChIP-seq, Cotton, GhWRKY27, Leaf senescence, SAGs, Transcription factor
Background
Leaf senescence is the final stage of plant development and is influenced by both internal factors and environmental factors [1, 2]. When these factors are detected by plants, signal transduction occurs, causing changes in gene expression and physiological indices, eventually leading to plant senescence [3]. Although senescence can maximize the plant’s use of its own resources, early and abnormal senescence not only shortens the plant lifespan but also decreases crop yield and quality [4–6]. Therefore, it is essential to study the genes involved in mediating leaf senescence, which will be highly useful for crop genetic breeding.
Analysis of the senescence-associated transcriptome has shown that the expression of many genes can be strongly induced by leaf senescence. Among these genes, WRKY genes constitute the second largest family of transcription factors (TFs), which is smaller only than the family of NAC TFs, strongly demonstrating that WRKY TFs play important roles during leaf senescence [7, 8]. WRKY TFs are named for their conserved WRKY domains and can be classified into three main categories: groups I, II (IIa, IIb, IIc, IId and IIe) and III [9, 10]. These TFs can specifically bind to W-boxes in the promoters of target genes to regulate gene expression and participate in leaf senescence, pathogen defence, stress responses and plant growth and development [11–14]. W-boxes are also found in the promoter regions of many WRKY genes, suggesting the existence of a regulatory network among WRKY TFs and potential functional roles of W-boxes in leaf senescence [13, 15, 16]. Moreover, WRKY TFs can regulate leaf senescence by interacting with a variety of proteins via gibberellic acid (GA)-, jasmonic acid (JA)-, auxin-, and salicylic acid (SA)-mediated signalling pathways [17–19].
Recent functional studies have provided some strong and persuasive evidence that WRKY TFs participate in the senescence-associated regulatory network. Several WRKY genes, such as AtWRKY6 [20], AtWRKY18 [21], AtWRKY22 [22], AtWRKY45 [17], AtWRKY53 [23], AtWRKY54 [24], AtWRKY57 [18], AtWRKY70 [25] and AtWRKY75 [26], have been shown to be SAGs in Arabidopsis, and their corresponding regulatory mechanisms have been elucidated at the molecular level. For example, a recent report indicated that a positive regulator of ageing, AtWRKY75, is involved in a tripartite amplification loop in which mutual promotion of AtWRKY75, SA and reactive oxygen species (ROS) occurs via different mechanisms [26]. Overexpression of AtWRKY45 promotes leaf senescence, and AtWRKY45 can interact with the DELLA protein RGL1 to modulate leaf senescence via a GA-mediated signalling pathway [17]. The mechanisms by which WRKY TFs regulate ageing have also been identified in other species, such as Chinese flowering cabbage, litchi and rice [14, 27, 28]. BrWRKY65 has been shown to be upregulated during postharvest leaf senescence in the economically important leafy vegetable Chinese flowering cabbage and to directly bind to the downstream SAGs BrNYC1, BrSGR1 and BrDIN1 [14]. In litchi, LcNAC1 interacts with LcWRKY1 to form a complex that antagonistically regulates LcAOX1a expression in postharvest litchi fruit senescence [27]. OsWRKY42 represses the expression of OsMT1d by inducing ROS production, thus promoting leaf senescence in rice [28].
Cotton is an important economic crop as well as the dominant raw material of the textile industry. The cultivation of short-season cotton varieties is an effective way to solve the contradiction between grain and cotton. Early maturity is an important trait of short-season cotton varieties. However, some early-maturing cotton varieties are prone to pro-senescence [29]. To overcome this challenge, identification and functional characterization of genes involved in leaf senescence are important for cultivating early-maturing but non-early-ageing cotton varieties. Previously, we conducted a transcriptomic analysis to identify differentially expressed genes during leaf senescence in cotton [30]. It was found that leaf senescence induced a considerable proportion of genes encoding TFs, including WRKY TFs [30]. To date, very few WRKY TFs have been functionally characterized during leaf senescence, and little is known about their molecular mechanisms and the signalling pathways associated with leaf senescence in cotton. In the present study, we performed molecular and genetic assays to identify the functional roles of GhWRKY27 in age-triggered leaf senescence. Overexpression of GhWRKY27 in Arabidopsis can promote leaf senescence. In addition, GhTT2 can physically interact with GhWRKY27, and GhWRKY27 targets GhCYP94C1 and GhRipen2–2. GhWRKY27 activates the transcriptional expression of GhCYP94C1 and GhRipen2–2 in the regulatory pathway. Therefore, GhWRKY27 may be a potential ageing-related factor and participate in leaf senescence together with GhTT2, GhCYP94C1 and GhRipen2–2. Our studies provide new mechanistic insight into the roles of WRKY genes during leaf senescence in cotton.
Results
Isolation and sequence analysis of GhWRKY27
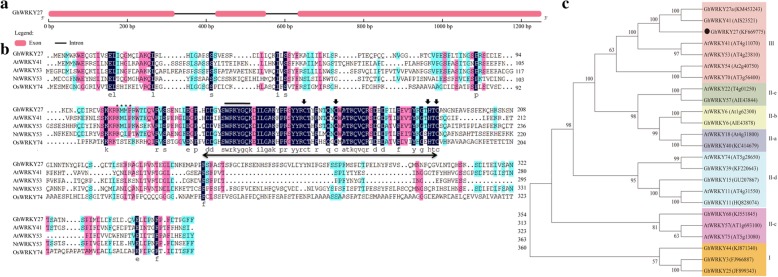
We previously showed that the GhWRKY27 (accession KF669775) gene is induced by leaf senescence and highly expressed in senescent leaves based on a transcriptome database [30, 31]. Thus, GhWRKY27 was selected for further functional characterization. The open reading frame (ORF) and genomic DNA fragments of GhWRKY27 were isolated from the cotton variety CCRI74. The ORF of GhWRKY27 is 1068 bp long and encodes 355 amino acid residues. The molecular weight (Mw) of the deduced protein is 40.06 kDa, and the isoelectric point (pI) is 5.48. The GhWRKY27 gene contains three exons and two introns (Fig. 1a). Multiple sequence alignment of the GhWRKY27 protein with its related proteins showed that the GhWRKY27 protein shares 41.30, 36.43, 49.59, and 28.61% identity with AtWRKY41 (AT4G11070), AtWRKY53 (AT4G23810), NtWRKY53 (XP_016508373) and OsWRKY74 (AAT84161), respectively. In addition, the putative GhWRKY27 protein contains a typical WRKY domain that includes the highly conserved amino acid sequence “WRKYGQK” and a C-X7-C-X23-H-X1-C zinc-finger motif (Fig. 1b), which is a typical feature of group III members according to Eulgem et al. [9]. A phylogenetic tree was constructed to assess the evolutionary relationships of the GhWRKY27 protein with other WRKY proteins from cotton and Arabidopsis. Phylogenetic analysis showed that GhWRKY27 clusters with proteins that are reported group III members (Fig. 1c).
Fig. 1.
Sequence characteristics and phylogenetic analyses of GhWRKY27. a Intron-exon structure of GhWRKY27. Exons are indicated by thick red lines, and introns are indicated by black lines. b Multiple sequence alignment analysis of the GhWRKY27 protein with its homologues in other species. The WRKY domain, composed of approximately 60 amino acids, is represented by a double-headed arrow. The highly conserved core amino acid sequence, WRKYGQK, in the WRKY domain is indicated with a horizontal line. The C and H residues in the zinc-finger motif are indicated by arrows. c Phylogenetic analysis of GhWRKY27 with other WRKY proteins from cotton and Arabidopsis. GhWRKY27 is indicated by a dot. Gh, Gossypium hirsutum; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Os, Oryza sativa
Expression patterns of GhWRKY27 during leaf senescence
To understand the potential role of GhWRKY27 during leaf senescence, the transcript levels of GhWRKY27 were determined at different stages of leaf senescence. Using the expression profile data published by Lin et al. [30], the expression levels of GhWRKY27 were found to be gradually upregulated during leaf senescence in 15-, 25-, 35-, 45-, 55-, and 65-day-old leaves of the CCRI36 variety (Fig. 2a). The transcript accumulation of GhWRKY27 was further detected in the early-ageing cotton variety CCRI74 and the non-early-ageing cotton variety PB-12 at different stages of leaf senescence. The expression level of GhWRKY27 gradually increased in both varieties and was significantly lower in PB-12 than in CCRI74 in 14- to 35-day-old leaves (Fig. 2b). In addition, the transcript level of GhWRKY27 was significantly higher in 35-day-old leaves than in 7-day-old leaves in CCRI74. However, there was no difference in expression at different stages of leaf senescence in PB-12 (Fig. 2b).
Fig. 2.
Expression patterns of GhWRKY27 during leaf senescence in cotton. a Expression profiles of the GhWRKY27 gene in 15-, 25-, 35-, 45-, 55-, and 65-day-old leaves of the CCRI36 cotton variety during leaf senescence using RNA-seq data [30]. The expression values were normalized using the transcript per million clean tags (TPM) algorithm. b Transcript levels of GhWRKY27 in the early-ageing cotton variety CCRI74 and non-early-ageing cotton variety PB-12. Values represent the means ± standard deviations (SDs) from three replicates. The significance of the data was determined using Student’s t-test. “+” represents a comparison with 7 days in the same variety (+P < 0.05). “**” and “*” represent a comparison between the same days of different varieties (**P < 0.01 and *P < 0.05)
Overexpression of GhWRKY27 promotes leaf senescence in transgenic Arabidopsis plants
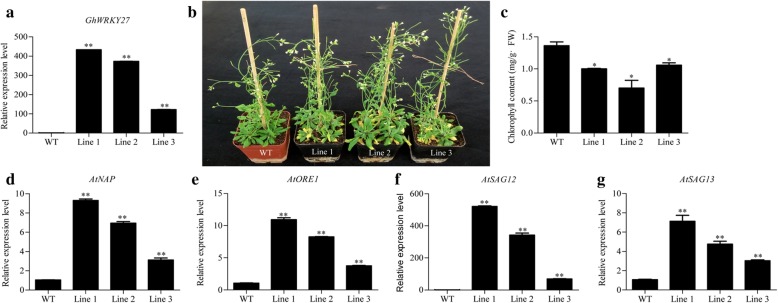
To further investigate the functional role of GhWRKY27 in leaf senescence, GhWRKY27 was transformed into Arabidopsis, and three independent transgenic lines were confirmed by quantitative real-time PCR (qRT-PCR) (Fig. 3a). The wild-type (WT) and transgenic plants were cultured on 1/2 MS medium. Two weeks later, the plants were transplanted to nutrient-rich soil in a greenhouse to observe the natural senescence phenotype. The transgenic lines exhibited a severe leaf senescence phenotype compared with that of WT plants (Fig. 3b). The chlorophyll content was significantly lower in transgenic plants than in WT plants (Fig. 3c). The transcript levels of four SAGs associated with positive regulation of leaf senescence (AtNAP, AtORE1, AtSAG12 and AtSAG13) were found to be significantly higher in the transgenic plants than in WT plants (Fig. 3d-g). In addition, the expression trends of these genes were consistent with those of GhWRKY27 in the three transgenic lines (Fig. 3d-g).
Fig. 3.
The GhWRKY27 gene accelerates age-triggered leaf senescence in transgenic Arabidopsis. a qRT-PCR analysis of GhWRKY27 transcript levels in WT and three transgenic lines. b Senescence phenotypes of WT plants and three transgenic lines. c Chlorophyll content of WT plants and three transgenic lines. d-g qRT-PCR analysis of the transcript levels of the SAGs AtNAP, AtORE1, AtSAG12 and AtSAG13 in Arabidopsis. AtUBQ10 was employed as a reference gene. Values represent the means ± SDs. The significance of the data was determined using Student’s t-test (**P < 0.01). The experiment was conducted with three repetitions
GhTT2 is an interacting partner of GhWRKY27 in yeast cells and Arabidopsis protoplasts
We constructed a yeast two-hybrid (Y2H) cDNA library using senescent cotton leaves and performed library screening using GhWRKY27 as bait to screen its interacting partners. Through library screening, approximately 164 positive colonies were isolated and sequenced. After discarding repeat sequences, 67 potential interacting proteins were identified. Functional annotation analysis showed that these candidate proteins were mainly involved in signal transduction, stress responses and regulation of growth and development (Additional file 1: Table S1).
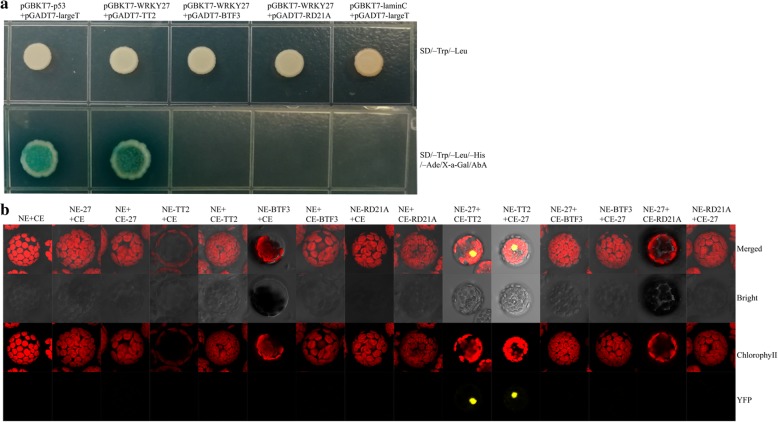
To confirm which proteins interact with GhWRKY27, three candidate genes, MYB domain protein gene TT2 (Gh_A07G0140, GhTT2), TF BTF3 (Gh_A08G2018, GhBTF3) and cysteine proteinase RD21a (Gh_D01G1044, GhRD21A), were selected and investigated using a Y2H system. The ORFs of the three genes were cloned into the pGADT7 vector. Each fusion construct and pGBKT7-GhWRKY27 plasmid was co-transformed into the yeast strain Y2HGold. Our results showed that the co-transformed yeast cells containing the positive control pGBKT7-p53 + pGADT7-largeT and the experimental group pGBKT7-GhWRKY27 + pGADT7-GhTT2 could grow well on SD/−Trp/−Leu/-His medium and turned blue on SD/−Trp/−Leu/−His/−Ade/X-a-Gal/AbA medium. However, no interaction was found in pGBKT7-GhWRKY27 + pGADT7-GhBTF3, pGBKT7-GhWRKY27 + pGADT7-GhRD21A and the negative control pGBKT7-laminC + pGADT7-largeT in yeast cells (Fig. 4a). Furthermore, the remaining genes in Additional file 1: Table S1 could not interact with GhWRKY27 (data not shown).
Fig. 4.
The interaction of GhWRKY27 with GhTT2, GhBTF3 and GhRD21A was determined in yeast cells and Arabidopsis mesophyll protoplasts. a Physical interaction of GhTT2, GhBTF3 and GhRD21A with GhWRKY27 in yeast cells. pGBKT7-p53 + pGADT7-largeT and pGBKT7-laminC + pGADT7-largeT were used as positive and negative controls, respectively. b The interaction of GhTT2, GhBTF3 and GhRD21A with GhWRKY27 in Arabidopsis mesophyll protoplasts. The yellow signal indicates YFP fluorescence, and the red signal indicates the autofluorescence of chlorophyll. The merged image is a combination of images showing the YFP and chlorophyll signals. No YFP signals were detected in the negative controls
To further confirm the interactions of GhWRKY27 with the three candidate proteins in plant cells, bimolecular fluorescence complementation (BiFC) assays were conducted in Arabidopsis protoplasts. The results showed that GhTT2 interacted with GhWRKY27 by emitting a yellow fluorescent signal in the nucleus, while GhBTF3 and GhRD21A did not exhibit any fluorescence (Fig. 4b). In addition, no yellow fluorescent signal was detectable when two empty vectors and one empty vector with a fusion construct were applied (Fig. 4b).
Screening for genomic binding sites of GhWRKY27 using a chromatin immunoprecipitation followed by sequencing (ChIP-seq) assay
ChIP-seq was used to identify the locations in the genome to which the GhWRKY27 protein can bind. The locations were characterized by significant numbers of mapped reads (peaks) [32]. The ChIP-seq results showed that the peaks came from 98 different target promoter regions, and the fold enrichment of the peaks varied from 15.59 (Gh_A10G0788) to 127.86 (Gh_A07G1258) (Additional file 2: Table S2). In addition, W-boxes were abundant in the sequencing data and were the most numerous among the cis-elements (Fig. 5). In the 1500-bp promoter region, 94 of the genes contained W-boxes, and several genes contained one or more W-boxes (Additional file 2: Table S2). The putative direct target genes included TFs, stress- and defence-related genes, growth- and development-related genes, photosynthesis genes, enzyme-encoding genes, ion transport-related genes and several unknown genes (Additional file 2: Table S2).
Fig. 5.

The cis-element accumulation in the ChIP-seq results
Yeast one-hybrid (Y1H) assay and electrophoretic mobility shift assay (EMSA) show that GhWRKY27 binds to the promoters of GhCYP94C1 and GhRipen2–2
To identify the downstream target genes participating in the regulatory network, 8 putative target genes were selected and validated by using the Y1H system. These genes encode hormone-related enzymes (GhIAA15A, Gh20ox2 and GhGH3.5), TFs (GhWRKY1), ripening-related proteins (GhRipen2–1 and GhRipen2–2), aspartic protease (GhASPG1) and cytochrome P450 (GhCYP94C1). Three tandem copies from the peak sequences, which represent the most likely binding sites in the promoter regions, were fused to the pHIS2 bait vector (Additional file 3: Table S3). GhWRKY27 was inserted into the pGADT7 prey vector. The prey construct was co-transformed into Y187 yeast cells with the bait carriers. The results showed that the transformation products harbouring the combination of GhWRKY27 with the specific promoter regions of GhCYP94C1 and GhRipen2–2 grew well on SD/−Trp/−Leu + 200 mM 3-AT selective medium, whereas the negative control and other candidate genes did not (Fig. 6a).
Fig. 6.
GhWRKY27 directly targets the promoters of GhCYP94C1 and GhRipen2–2. a Interaction between GhWRKY27 and the candidate target genes GhASPG1, GhWRKY1, GhRipen2–1, Gh20ox2, GhCYP94C1, GhGH3.5, GhIAA15A, and GhRipen2–2 in Y187 yeast cells. The binding ability of GhWRKY27 with the target promoter sequences was identified based on the growth status of transformed Y187 yeast cells on SD-TL and SD-TLH + 200 mM 3-AT medium. The combination of pGADT7-GhWRKY27 and pHIS2 was used as a negative control. b Wild and mutated promoter probe sequences of GhCYP94C1 were used in the EMSA assay. The W-box sequence 5′-TGACT-3′ was mutated to 5′-TGAAA-3′. c Wild and mutated promoter probe sequences of GhRipen2–2 were used in the EMSA assay. The W-box sequence 5′-TTGACT-3′ was mutated to 5′-TTGAAA-3′. d GhWRKY27 binds to the W-boxes in the promoter of GhCYP94C1 in the EMSA assay. e GhWRKY27 binds to the W-boxes in the promoter of GhRipen2–2 in the EMSA assay. A monoclonal antibody against GhWRKY27 (anti-GhWRKY27) was employed in the supershift assay. “–” indicates absence, while “+” indicates presence
The direct binding of the GhWRKY27 protein to the GhCYP94C1 and GhRipen2–2 promoters was further confirmed via EMSA. As shown in Fig. 6b and c, the wild and mutant probe sequences, which were the WRKY recognition regions containing the W-box, were present within the − 1345 bp to − 1295 bp upstream region of the ATG initiation codon of GhCYP94C1 and the − 506 bp to − 457 bp of GhRipen2–2. The EMSA results revealed that the GhWRKY27 protein was able to recognize and bind the labelled wild probes of GhCYP94C1 to cause a mobility shift (Fig. 6d; lanes 2, 6, 8, 9). In addition, in the presence of the labelled wild probe of GhCYP94C1, the unlabelled mutant probe did not affect the generation of the lagging band when overdosed at the same time (Fig. 6d; lane 4). However, the band was effectively abolished when the unlabelled wild probe of GhCYP94C1 was overdosed as a cold competitor (Fig. 6d; lane 5). In this case, most labelled wild probes were replaced by unlabelled wild probes. Therefore, the GhWRKY27 protein mainly bound to the competitors (unlabelled wild probes), leading to the disappearance of the band (Fig. 6d; lane 5). Furthermore, anti-GhWRKY27 was employed for supershift detection. When anti-GhWRKY27 was added, the antibody-protein-DNA complex formed a retarded band (Fig. 6d; lane 7). Similar results were found in GhRipen2–2 (Fig. 6e). Our findings clearly showed that the GhWRKY27 protein bound specifically to the W-box in the promoter regions of GhCYP94C1 and GhRipen2–2.
The expression patterns of GhTT2, GhCYP94C1 and GhRipen2–2 at different stages of leaf senescence
It has been demonstrated that the genes GhTT2, GhCYP94C1 and GhRipen2–2 interact with GhWRKY27. GhWRKY27 is a positive regulator of leaf senescence, which motivated us to study the roles of GhTT2, GhCYP94C1 and GhRipen2–2 during leaf senescence. The expression levels of GhTT2, GhCYP94C1 and GhRipen2–2 were examined at different stages of leaf senescence. The results showed that the transcript abundance of GhTT2 decreased gradually as the leaf aged and that the expression levels of GhTT2 were significantly lower in Stages 3–5 than in Stage 1 (Fig. 7a). The transcript levels of GhCYP94C1 showed no obvious differences among the different stages (Fig. 7b). However, the expression levels of the GhRipen2–2 gene gradually increased with the senescence of the leaves and were 10,000 times higher at Stage 5 than at Stage 1; this difference was highly significant (Fig. 7c).
Fig. 7.
Expression patterns of GhTT2, GhCYP94C1 and GhRipen2–2 at different stages of leaf senescence. a Transcript levels of GhTT2 during leaf senescence. b Transcript levels of GhCYP94C1 during leaf senescence. c Transcript levels of GhRipen2–2 during leaf senescence. Stage 1 to stage 5 indicate different stages of cotton leaves with different senescence areas, as described previously [35]. GhActin was used as an internal reference. Values represent the means ± SDs. The significance of the data was determined using Student’s t-test (**P < 0.01 and *P < 0.05). The experiment was conducted with three replications
GhWRKY27 increased transcriptional expression of GhCYP94C1 and GhRipen2–2
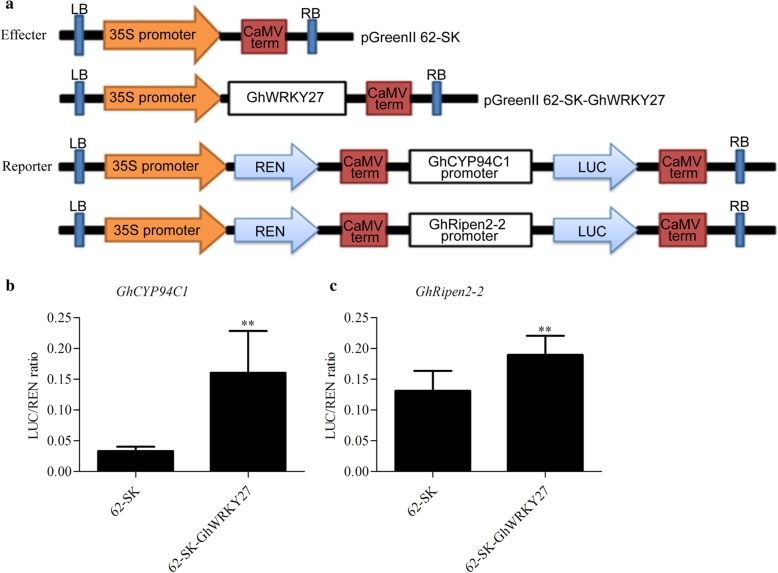
A transient dual-luciferase assay was performed in tobacco leaves to elucidate the functional role of GhWRKY27 in regulating the expression of GhCYP94C1 and GhRipen2–2 in vivo. The experiment was conducted using a double reporter plasmid, pGreenII0800-LUC, containing the REN luciferase driven by the 35S promoter and the LUC luciferase driven by the GhCYP94C1 or GhRipen2–2 promoter. In addition, the assay includes an effecter plasmid, pGreenII62-SK, expressing the GhWRKY27 TF. The constructs are shown in Fig. 8a. The ratio of LUC to REN was used to reflect the transcriptional activity. The results showed that compared with the control, GhWRKY27 activated the GhCYP94C1 and GhRipen2–2 promoters by significantly increasing the LUC/REN ratio (Fig. 8b, c), suggesting that GhWRKY27 activated the expression of GhCYP94C1 and GhRipen2–2.
Fig. 8.
GhWRKY27 activated the expression of GhCYP94C1 and GhRipen2–2 in the dual-luciferase reporter assay. a Sketch of the effecter and reporter constructs. The GhWRKY27 was cloned into the effecter vector pGreenII 62-SK. The promoters of GhCYP94C1 and GhRipen2–2 were cloned into the reporter vector pGreenII 0800-LUC. b GhWRKY27 activated the expression of GhCYP94C1. c GhWRKY27 activated the expression of GhRipen2–2. The data were indicated by the ratio of LUC to REN. The values represent the means ± SDs of six independent repeats. The significance of the data was determined using Student’s t-test (**P < 0.01)
Discussion
Many important agronomic traits, such as crop yield and quality, are affected by leaf senescence. However, the regulatory mechanism of leaf senescence in cotton remains poorly understood. In our previous work, RNA-seq was performed using leaves in different stages of senescence to comprehensively analyse gene expression in cotton [30]. Similar to the transcriptomic analysis of leaf senescence in Arabidopsis, our transcriptomic analysis revealed that the WRKY gene family is one of the largest TF families induced by leaf senescence [7, 30]. Subsequently, a genome-wide analysis of the WRKY gene family was performed in cotton, and the specific functional roles of the WRKY gene family during leaf senescence were explored [31]. Previous reports have shown that members of the group III WRKY subfamily participate in leaf senescence [31]. In Arabidopsis, several ageing-related WRKY genes have been studied, among which AtWRKY30, AtWRKY53, AtWRKY54 and AtWRKY70 belong to the group III subfamily [24]. Given the important roles of the group III subfamily in leaf senescence, the group III subfamily was further analysed in cotton, and GhWRKY27 was identified as a member of this subfamily [33]. In the present study, multiple sequence alignment and evolutionary analysis further demonstrated that GhWRKY27 belongs to the group III subfamily.
Leaf senescence is basically regulated by the developmental age and influenced by various internal and external factors, causing degradation of macromolecules and translocation of nutrients, eventually leading to cell death [2]. In the present study, GhWRKY27 was closely related to group III members. Previous reports showed that group III genes AtWRKY30/53/54/70 are senescence regulators in Arabidopsis [24]. For example, AtWRK70 negatively regulates developmental senescence and has been found to be involved in SA- and JA/ETH-mediated signalling pathways based on examining transgenic and mutant plants [25]. In addition, a positive regulator of leaf senescence, AtWRKY53, which shows the highest similarity with GhWRKY27, functions as a convergence node between senescence and H2O2-, SA- and JA-related stress response [23]. We found that the transcript levels of GhWRKY27 increased as leaf senescence progressed. Moreover, in the early-ageing cotton variety, the expression level of GhWRKY27 was significantly higher than that in the non-early-ageing cotton variety. Guo et al. showed that the expression of AtWRKY75 increases gradually with leaf ageing and that overexpression of AtWRKY75 in Arabidopsis plants accelerates leaf senescence [26]. In addition, GhNAC12 [34] and GhWRKY17 [35] have been found to be highly expressed in an early-ageing cotton variety, and leaf senescence is triggered in Arabidopsis plants transgenic for these genes. The high expression level of the GhWRKY27 gene in senescent leaves and in the early-ageing variety indicates the potential role of GhWRKY27 in leaf senescence. SAGs have been demonstrated to serve as indicators of leaf senescence [2, 17, 36]. SAGs such as AtNAP have been found to be upregulated during leaf senescence and to positively regulate leaf senescence in transgenic Arabidopsis [37]. AtORE1, an NAC TF in Arabidopsis, is involved in regulatory networks of age-dependent and salt-promoted senescence [38]. AtSAG12 and AtSAG13, which encode cysteine proteases, are upregulated during developmental senescence [17]. Four marker genes of leaf senescence, AtNAP, AtORE1, AtSAG12 and AtSAG13, are significantly upregulated in transgenic Arabidopsis, and overexpression of GhWRKY27 accelerates leaf senescence in transgenic Arabidopsis plants, which further demonstrates the positive role of GhWRKY27 during leaf senescence. In our previous work, the expression level of GhWRKY27 was significantly elevated after SA treatment [33], suggesting the important role of this gene in SA-related stress response.
Some WRKYs regulate leaf senescence by interacting with other proteins [24]. For example, JAZ4/8 and IAA29, which are repressors of the JA and indole-3-acetic acid (IAA) signalling pathways, respectively, competitively interact with AtWRKY57 to negatively regulate JA-induced leaf senescence [18]. The JA-inducible protein EPITHIOSPECIFYING SENESCENCE REGULATOR (ESR/ESP) and the HECT-domain E3 ubiquitin ligase (UPL5) have been determined to interact with the positive ageing regulator AtWRKY53 via distinct mechanisms [19, 39]. Here, we screened for putative interaction partners of GhWRKY27 using a Y2H system. GhTT2, an MYB family protein, was found to interact with GhWRKY27. Previous studies have demonstrated that TT2, which encodes an R2R3 MYB domain protein, is mainly involved in the regulation of proanthocyanidin metabolism [40, 41]. In Arabidopsis, AtTT2/AtMYB123 positively regulates anthocyanin metabolism and is also involved in flavonoid-related pathways [42]. In cotton, two TT2-type MYB genes, GhTT2-3A and GhTT2_A07, are associated with brown cotton fibres by influencing the accumulation of proanthocyanidin [43, 44]. However, MYB TFs such as AtMYBL [45] and AtMYB44 [46] have been shown to be involved in leaf senescence, and AtMYBL overexpression in Arabidopsis plants promotes leaf senescence [45]. Usually, CAB (chlorophyll a/b-binding protein) is used as a senescent marker gene, and the expression level of CAB gradually decreases with leaf senescence [47]. In our work, similar to the CAB gene, GhTT2 showed decreased expression during leaf senescence, indicating that GhTT2 might be involved in the early stage of leaf senescence. However, the relationship between GhTT2 and leaf senescence should be further clarified in transgenic materials.
W-box cis-elements in promoter regions have been reported to act as binding motifs for WRKY TFs [48]. The ChIP assay results showed that W-box elements accumulated in the sequencing data and that 94 of the 98 genes contained different numbers of W-boxes. The appearance of W-boxes in the promoter fragments indicated their potential functions as candidate target genes [13]. Some genes possessed one or more W-boxes in their promoter regions. Although a single W-box element in the promoter region is enough for participation in gene expression regulated by the WRKY gene, multiple W-box elements are frequently found to be clustered in promoter regions [9]. We discovered that the promoter regions of several genes contain no W-box elements. Previous studies have shown that AtWRKY6 and AtWRKY53 can directly target the promoter region of AtWRKY42, which lacks W-boxes [12, 13]. These findings indicate that the targets of WRKY TFs may be indirectly regulated or may be regulated via different elements in promoter regions [12, 13]. Therefore, it remains difficult to determine whether the GhWRKY27 gene directly targets candidate genes based solely on the presence or absence of W-boxes. The peak sequences of the potential target genes (GhASPG1, GhWRKY1, GhRipen2–1, Gh20ox2, GhCYP94C1, GhGH3.5, GhIAA15A and GhRipen2–2) were used as candidate sequences for Y1H verification. Whether the GhWRKY27 gene can bind to W-boxes in other regions of their promoters remains to be determined.
Our results showed that GhCYP94C1 and GhRipen2–2 are targets of GhWRKY27. GhCYP94C1 is a member of the cytochrome P450 family. Some studies have shown that cytochrome P450 genes such as CYP89A9 [49], CYP74C9 [50], CYP82E4 [51] and CYP76C2 [52] are involved in plant leaf senescence. For example, CYP89A9, a cytochrome P450 monooxygenase, plays an important role in the accumulation of nonfluorescent dioxobilin-type chlorophyll catabolites (NDCC), a major chlorophyll degradation product, during leaf senescence in Arabidopsis [49]. In our study, GhWRKY27 activated the expression of GhCPY91C1 in the dual-luciferase reporter system. However, we observed no differences in GhCYP94C1 expression at different stages of leaf senescence and the Ct values were greater than 33 at every stage. Tissue-specific expression analysis showed that GhCYP94C1 has very high expression levels in flower organs and very low levels in leaves (Additional file 4: Figure S1). The expression of GhCYP94C1 was further examined at different stages of flower bud development in different maturing varieties. The expression level of GhCYP94C1 is higher in early maturing varieties than in non-early maturing varieties and has the highest expression levels at the 3-leaf stage (flower bud differentiation stage) in Yanzao and Guoxin11 (Additional file 5: Figure S2). These results suggested that GhCYP94C1 might be associated with cotton maturity and flower bud differentiation. Early maturity of cotton is often accompanied by pro-senescence [29]. We speculate that GhCYP94C1 might affect cotton maturity via affecting reproductive growth, thereby affecting the senescence of cotton. However, the precise functions of GhCYP94C1 should be clarified by further studies.
However, no studies have yet addressed GhRipen2–2-related genes in cotton or their homologues in Arabidopsis, indicating that GhRipen2–2 might be a novel gene that requires further exploration in cotton. In addition, the expression of GhRipen2–2 was significantly induced during leaf senescence and activated by GhWRKY27, suggesting its potential roles in leaf senescence and ageing regulatory pathways.
Conclusions
In summary, a group III WRKY TF, GhWRKY27, was isolated and characterized. The gene expression profiles of cotton leaves from different senescent stages revealed that GhWRKY27 was induced by leaf senescence and predominantly expressed in the early-ageing cotton variety. Overexpression of GhWRKY27 accelerated leaf senescence in Arabidopsis, accompanied by the elevated accumulation of SAG transcripts. GhWRKY27 was shown to physically interact with GhTT2 and bind to the promoters of GhCYP94C1 and GhRipen2–2. GhTT2, GhCYP94C1 and GhRipen2–2 were differentially expressed at different stages of leaf senescence. In addition, GhWRKY27 activated the transcriptional expression of GhCYP94C1 and GhRipen2–2 during leaf senescence. Our results suggested that GhWRKY27 may positively regulate leaf senescence via interaction with or transcriptional regulation of other genes. These findings provide novel insight into the regulatory pathways of leaf senescence in cotton and a theoretical basis for genetic improvement of short-season cotton varieties to early-maturing but non-early-ageing cotton varieties.
Methods
Plant materials and growth conditions
One early-ageing cotton variety, CCRI74, and two non-early-ageing varieties, PB-12 and Liao4086, were used for leaf senescence analysis. In addition, two early-maturing varieties, CCRI50 and Yanzao, and two non-early-maturing varieties, Guoxin11 and Lu28, were used for maturing analysis. The cotton varieties were planted in the field of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences (Anyang, Henan, China). Different tissues, including root, stems, leaves, petals, pistils, stamens, fibre and ovules were harvested from CCRI74.
To detect gene expression during leaf senescence in cotton, the fourth leaves from the top of the CCRI74 and PB-12 plants were marked at the full-bloom stage. After one week, the leaves were picked weekly at five successive developmental stages (7, 14, 21, 28, and 35 days). In addition, five areas of CCRI74 true leaves with yellowing were harvested according to a previous method [35].
To examine gene expression at different stages of bud differentiation from different maturity varieties, the buds were collected from CCRI50, Yanzao, Guoxin11 and Lu28 at the cotyledon and 1-, 2-, 3-, 4- and 5-leaf stages. Every sample included three repetitions. All samples were quickly frozen in liquid nitrogen and stored at − 80 °C for subsequent experiments.
Gene clone and sequence analysis
The ORF and genomic DNA sequences of GhWRKY27 were cloned from the cDNA and DNA of CCRI74 leaves, respectively. The PCR fragments were then inserted into the pMD18-T vector (TAKARA) for sequencing. The intron-exon structure of the GhWRKY27 gene was generated using the online software GSDS2.0 (http://gsds.cbi.pku.edu.cn/). The conserved domains within the GhWRKY27 protein were analysed using the online software CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Mw and pI were predicted with ExPASy (http://web.expasy.org/compute_pi/). Multiple sequence alignment of the GhWRKY27 protein with its homologues from other species was performed using DNAMAN 6.0 software. A phylogenetic tree was constructed with the MEGA 7.0 program via the neighbour-joining method, with 1000 bootstrap replicates.
qRT-PCR
Total RNA was isolated using the RNAprep Pure Plant Kit (For Plants) (DP441; Tiangen, Beijing, China). Reverse transcription was conducted using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (RR047A; TaKaRa, Dalian, China). Transcript levels were measured via qRT-PCR with SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (RR420A; TaKaRa) in an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The G. hirsutum Actin (GhActin) and A. thaliana UBQ10 (AtUBQ10) genes were used as internal controls. The primers designed with Oligo 7 software are shown in Additional file 6: Table S4. Three biological replicates were performed. The qRT-PCR assays were conducted with three technical replicates.
Plasmid construction and plant transformation
The ORF of GhWRKY27 was amplified from the T vector containing GhWRKY27 and cloned into the pBI121 vector, driven by the cauliflower mosaic virus 35S promoter. The recombinant pBI121-GhWRKY27 plasmid was transformed into the Agrobacterium tumefaciens strain LBA4404. The LBA4404 strain harbouring the pBI121-GhWRKY27 plasmid was transformed into WT Arabidopsis thaliana (Colombia-0) using the floral dip method [53]. The transgenic seedlings were screened on 1/2 MS agar medium plates containing kanamycin (50 mg L− 1) in a chamber at 22 °C with a 16 h light/8 h dark cycle. The transgenic plants were further confirmed through PCR until the T3 generation. The ageing phenotype was observed in the T3 generation. The rosette leaves were collected for the expression analysis.
Determination of chlorophyll content
Approximately 0.1 g of rosette leaf sample was placed in a 15 ml tube to which 15 ml of extract (acetone: absolute ethanol = 1:1) was added; the tube was left for 24 h at room temperature in the dark. When the sample was completely white, the absorbance was measured at 645 nm and 663 nm. Chlorophyll content (mg/g· fresh weight) = (20.31D645 + 8.03D663)(mg L− 1)·V(mL)/(fresh weight (g)·1000).
Y2H library assay
Old CCRI74 leaves were used to construct a cDNA library to screen interacting proteins. The ORF of GhWRKY27 was amplified and inserted into the pGBKT7 vector containing the GAL4 DNA-binding domain. A Y2H library screening assay was performed following the manufacturer’s protocols (Clontech). The fusion construct pGBKT7-GhWRKY27 and the cDNA library plasmids were co-transformed into yeast strain Y2HGold competent cells (Clontech). The transformed cells were initially screened on lower-stringency SD/−Trp/−Leu/−His/−Ade agar plates with incubation at 30 °C for 3–5 days. Yeast colonies with diameters greater than 2 mm were patched onto the higher-stringency SD/−Trp/−Leu/−His/−Ade agar medium containing 20 μg mL− 1 X-α-Gal and 125 μg mL− 1 aureobasidin A (SD/−Trp/−Leu/−His/−Ade/X-a-Gal/AbA). Blue colonies were identified via PCR and sequencing. The sequences were aligned to the reference genome of Gossypium hirsutum [54] and annotated. The ORFs of GhTT2, GhBTF3 and GhRD21A, which encoded potential interacting proteins, were cloned into the pGADT7 vector. Each recombinant construct was co-transformed into the yeast strain Y2HGold with the pGBKT7-GhWRKY27 plasmid. The yeast transformation products were assayed on both SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade/X-a-Gal/AbA medium at 30 °C for 3–5 days. The pGBKT7-p53 and pGADT7-largeT plasmids were used as positive controls, and pGBKT7-laminC and pGADT7-largeT were used as negative controls.
BiFC
To detect protein interactions in vivo, the ORFs of the GhWRKY27, GhTT2, GhBTF3 and GhRD21A genes were individually cloned into the pSPYNE-35S and pSPYCE-35S vectors. Combinations of the plasmid constructs (pSPYNE + pSPYCE, pSPYNE-GhWRKY27 + pSPYCE, pSPYNE + pSPYCE-GhWRKY27, pSPYNE-GhTT2 + pSPYCE, pSPYNE + pSPYCE-GhTT2, pSPYNE-GhWRKY27 + pSPYCE-GhTT2, pSPYNE-GhTT2 + pSPYCE-GhWRKY27, pSPYNE-GhBTF3 + pSPYCE, pSPYNE + pSPYCE-GhBTF3, pSPYNE-GhWRKY27 + pSPYCE-GhBTF3, pSPYNE-GhBTF3 + pSPYCE-GhWRKY27, pSPYNE-GhRD21A + pSPYCE, pSPYNE + pSPYCE-GhRD21A, pSPYNE-GhWRKY27 + pSPYCE-GhRD21A and pSPYNE-GhRD21A + pSPYCE-GhWRKY27) were co-transformed into Arabidopsis protoplasts. The BiFC assay was performed according to previously described protocols [55, 56]. The fluorescent signal was detected using an Olympus FV 1000 confocal microscope.
Production of the recombinant GhWRKY27 protein and anti-GhWRKY27 preparation
The ORF of GhWRKY27 was cloned into the pET-28a(+) prokaryotic expression vector to generate the pET-28a(+)-GhWRKY27 fusion plasmid. The plasmid that encodes the 6 × His-tagged fusion protein was transformed into the Escherichia coli strain BL21 (DE3). To obtain the fusion protein, BL21 (DE3) cells harbouring the pET-28a(+)-GhWRKY27 construct were cultured in LB liquid medium and induced by 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 28 °C. The induced cells were then sonicated until the solution became clear. The supernatant was collected via centrifugation, and the recombinant proteins were purified using a His-tag Protein Purification Kit (Beyotime). The purified proteins were employed to immunize mice for the preparation of an anti-GhWRKY27 monoclonal antibody (Loyel, Zhengzhou).
ChIP-seq
ChIP-seq was used to identify GhWRKY27 protein binding sites at the genome-wide scale in vivo. The ChIP assay was conducted according to protocols described previously [57]. Approximately 5-g senescent cotyledon samples of Liao4086 were collected and treated with 1% formaldehyde to crosslink and fix the DNA-protein complexes in the cells. The cells were then lysed, and chromatin was broken via ultrasound into DNA fragments with sizes of approximately 100–500 bp. The anti-GhWRKY27 antibody was employed to immunoprecipitate the DNA-GhWRKY27 protein complexes. Beads were added, and the bead-bound complexes were collected. The complexes were subsequently eluted and reverse cross-linked, and the DNA fragments were purified for construction of the sequencing library. The ends of the purified DNA fragments were repaired and attached to adapter sequences using a Paired-End DNA Sample Prep Kit (Illumina). The DNA fragments were amplified via PCR, and a fragment size of 100–500 bp was selected. The qualified library was used for HiSeq 2500 sequencing (Novogene, Beijing). The adapter sequence and low-quality data were removed from the raw sequencing data. The obtained clean reads were aligned to the reference genome of Gossypium hirsutum [54] using Short Oligonucleotide Analysis Package (SOAP2) [58]. Model-based Analysis for ChIP-Seq (MACS) was employed to identify peak calls at the whole-genome scale [59]. Anti-lgG was used as a control.
Y1H assay
Based on the ChIP-seq results, 8 potential downstream target genes encoding proteins such as aspartic protease in guard cell 1 (Gh_A07G1258, GhASPG1), WRKY TF 1 (Gh_A11G1391, GhWRKY1), ripening-related protein 2 (Gh_A12G0955, GhRipen2–1), gibberellin 20 oxidase 2 (Gh_D05G2274, Gh20ox2), cytochrome P450 94C1 (Gh_D08G0085, GhCYP94C1), IAA-amido synthetase GH3.5 (Gh_D11G1006, GhGH3.5), auxin-induced protein 15A (Gh_D12G0291, GhIAA15A) and ripening-related protein 2 (Gh_D12G1102, GhRipen2–2) were verified using a Y1H system. Three copies of specific promoter fragments were cloned into the pHIS2 vector. The fragments came from the highly enriched promoter regions observed through ChIP-seq analysis. The fragment sequences are listed in Additional file 3: Table S3. The ORF of GhWRKY27 was amplified and cloned into the pGADT7 vector. Each pHIS2 bait vector containing a target gene was co-transformed into the competent yeast strain Y187 with the pGADT7-GhWRKY27 prey vector. The transformants were selected on SD/−Trp/−Leu medium, and the co-transformed cells were further identified on SD/−Trp/−Leu/−His medium containing 200 mM 3-amino-1,2,4-triazole (3-AT) (SD/−Trp/−Leu/−His + 200 mM 3-AT). The combination of pGADT7-GhWRKY27 and pHIS2 was employed as a negative control plasmid.
EMSA
For EMSA, the specific promoter fragments of GhCYP94C1 and GhRipen2–2 containing the W-box and mutated W-box were synthesized as biotin end-labelled and unlabelled oligonucleotides. The unlabelled W-box oligonucleotide served as a competitor. The anti-GhWRKY27 antibody was employed for supershift identification. The assay was performed using the LightShift® Chemiluminescent EMSA Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
Transient reporter expression was assayed in tobacco leaves using a dual-luciferase reporter system [60]. The GhCYP94C1 and GhRipen2–2 promoters were amplified and inserted into the pGreenII0800-LUC vector as the reporter plasmid. The ORF of GhWRKY27 was amplified and inserted into the pGreenII 62-SK vector as the effecter plasmids. The fusion construct plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (pSoup-p19). The GV3101 (pSoup-p19) cells containing the recombinant plasmids were incubated in LB liquid medium containing 50 mg L− 1 kanamycin, gentamycin and rifampin until the OD600 value reached 0.5–0.6. Subsequently, the culture was adjusted to an OD600 value of 0.2 with infiltration buffer (10 mM MgCl2, 10 mM MES and 100 μM acetosyringone). The culture suspensions were left for 2 h at room temperature. The constructed effecter and reporter suspensions were mixed in a 1:1 ratio and co-infiltrated into tobacco leaves. After 2 days of infiltration, LUC and REN luciferase activity was detected using a dual-luciferase® reporter assay system (Promega, USA) on a Glomax 20/20 Luminometer (Promega, USA) according to the manufacturer’s instructions. At least six independent replicates were performed.
Additional files
Table S1. Partial result from library screening by the Y2H assay (DOCX 23 kb)
Table S2. Detailed information regarding the target genes of GhWRKY27 identified by ChIP-seq (DOCX 84 kb)
Table S3. Nucleic acid sequences of the potential target genes of GhWRKY27 used for the Y1H system (DOCX 24 kb)
Fig. S1. The expression levels of GhCYP94C1 in different tissues (TIFF 2064 kb)
Fig. S2. The expression levels of GhCYP94C1 at different stages of flower bud differentiation from different maturing varieties. (TIFF 124 kb)
Table S4. Primers used in this study (DOCX 40 kb)
Acknowledgements
We thank all the authors for their contributions to the article. We also appreciate the reviewers and editors for the patience to the work.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2016YED0101006). The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional file.
Abbreviations
- ChIP-seq
chromatin immunoprecipitation followed by sequencing
- EMSA
Electrophoretic mobility shift assay
- JA
Jasmonic acid
- ORF
Open reading frame
- qRT-PCR
quantitative real-time PCR
- SA
Salicylic acid
- SAGs
Senescence-associated genes
- TFs
Transcription factors
- WT
Wild-type
Authors’ contributions
SY designed the research programme. HLW, LD, YG and HTW analysed the data. LL, CW and LM revised the language. LG performed the experiment and wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lijiao Gu, Email: gulijiao1990@126.com.
Lingling Dou, Email: xjyldll@126.com.
Yaning Guo, Email: guoyaning01308060@163.com.
Hantao Wang, Email: w.wanghantao@163.com.
Libei Li, Email: lilibei0412@gmail.com.
Congcong Wang, Email: wangcongc1992@163.com.
Liang Ma, Email: maliang3417@163.com.
Hengling Wei, Email: henglingwei@163.com.
Shuxun Yu, Email: Ysx195311@163.com.
References
- 1.Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant journal : for cell and molecular biology. 2005;42(4):567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 2.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 3.Gan S. Mitotic and postmitotic senescence in plants. Sci Aging Knowledge Environ. 2003;2003(38):RE7. doi: 10.1126/sageke.2003.38.re7. [DOI] [PubMed] [Google Scholar]
- 4.Wu XY, Kuai BK, Jia JZ, Jing HC. Regulation of leaf senescence and crop genetic improvement. J Integr Plant Biol. 2012;54(12):936–952. doi: 10.1111/jipb.12005. [DOI] [PubMed] [Google Scholar]
- 5.Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. Autophagy, plant senescence, and nutrient recycling. J Exp Bot. 2014;65(14):3799–3811. doi: 10.1093/jxb/eru039. [DOI] [PubMed] [Google Scholar]
- 6.Guo Y, Gan SS. Translational researches on leaf senescence for enhancing plant productivity and quality. J Exp Bot. 2014;65(14):3901–3913. doi: 10.1093/jxb/eru248. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004;27(5):521–549. doi: 10.1111/j.1365-3040.2003.01158.x. [DOI] [Google Scholar]
- 8.Bakshi M, Oelmuller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9(2):e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 10.Rinerson CI, Rabara RC, Tripathi P, Shen QJ, Rushton PJ. The evolution of WRKY transcription factors. BMC Plant Biol. 2015;15:66. doi: 10.1186/s12870-015-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto S, Nakano T, Suzuki K, Shinshi H. Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Bba-Gene Struct Expr. 2004;1679(3):279–287. doi: 10.1016/j.bbaexp.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16(9):1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55(6):853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 14.Fan ZQ, Tan XL, Shan W, Kuang JF, Lu WJ, Chen JY. BrWRKY65, a WRKY Transcription Factor, Is Involved in Regulating Three Leaf Senescence-Associated Genes in Chinese Flowering Cabbage. Int J Mol Sci. 2017;18(6). [DOI] [PMC free article] [PubMed]
- 15.Dong JX, Chen CH, Chen ZX. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51(1):21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Xu W, Hu X, Liu H, Lin Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci Rep. 2016;6:20881. doi: 10.1038/srep20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LG, Xiang SY, Chen YL, Li DB, Yu DQ. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant. 2017;10(9):1174–1189. doi: 10.1016/j.molp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Jiang YJ, Liang G, Yang SZ, Yu DQ. Arabidopsis WRKY57 functions as a node of convergence for Jasmonic acid- and auxin-mediated signaling in Jasmonic acid-induced leaf senescence. Plant Cell. 2014;26(1):230–245. doi: 10.1105/tpc.113.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao Y, Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 2007;19(3):819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001;28(2):123–133. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 21.Potschin M, Schlienger S, Bieker S, Zentgraf U. Senescence networking: WRKY18 is an upstream regulator, a downstream target gene, and a protein interaction partner of WRKY53. J Plant Growth Regul. 2014;33(1):106–118. doi: 10.1007/s00344-013-9380-2. [DOI] [Google Scholar]
- 22.Zhou X, Jiang YJ, Yu DQ. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecules and cells. 2011;31(4):303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zentgraf U, Laun T, Miao Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol. 2010;89(2–3):133–137. doi: 10.1016/j.ejcb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 24.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63(7):2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulker B, Mukhtar MS, Somssich IE. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. 2007;226(1):125–137. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- 26.Guo PR, Li ZH, Huang PX, Li BS, Fang S, Chu JF, Guo HW. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell. 2017;29(11):2854–2870. doi: 10.1105/tpc.17.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang GX, Yan HL, Wu FW, Zhang DD, Zeng W, Qu HX, Chen F, Tan L, Duan XW, Jiang YM. Litchi fruit LcNAC1 is a target of LcMYC2 and regulator of fruit senescence through its interaction with LcWRKY1. Plant Cell Physiol. 2017;58(6):1075–1089. doi: 10.1093/pcp/pcx054. [DOI] [PubMed] [Google Scholar]
- 28.Han M, Kim CY, Lee J, Lee SK, Jeon JS. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Molecules and cells. 2014;37(7):532–539. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu SX, Song MZ, Fan SL, Wang W, Yuan RH. Biochemical genetics of short-season cotton cultivars that express early maturity without senescence. J Integr Plant Biol. 2005;47(3):334–342. doi: 10.1111/j.1744-7909.2005.00029.x. [DOI] [Google Scholar]
- 30.Lin M, Pang CY, Fan SL, Song MZ, Wei HL, Yu SX. Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol. 2015:15, 43. [DOI] [PMC free article] [PubMed]
- 31.Dou L, Zhang X, Pang C, Song M, Wei H, Fan S, Yu S. Genome-wide analysis of the WRKY gene family in cotton. Molecular genetics and genomics : MGG. 2014;289(6):1103–1121. doi: 10.1007/s00438-014-0872-y. [DOI] [PubMed] [Google Scholar]
- 32.Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, Madrigal P, Taslim C, Zhang J. Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput Biol. 2013;9(11):e1003326. doi: 10.1371/journal.pcbi.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou LL, Guo YN, Evans O, Pang CY, Wei HL, Song MZ, Fan SL, Yu SX. Identification and expression analysis of group III WRKY transcription factors in cotton. J Integr Agr. 2016;15(11):2469–2480. doi: 10.1016/S2095-3119(15)61306-5. [DOI] [Google Scholar]
- 34.Zhao FL, Ma JH, Li LB, Fan SL, Guo YN, Song MZ, Wei HL, Pang CY, Yu SX. GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L) Gene. 2016;576(1):268–274. doi: 10.1016/j.gene.2015.10.042. [DOI] [PubMed] [Google Scholar]
- 35.Gu L, Li L, Wei H, Wang H, Su J, Guo Y, Yu S. Identification of the group IIa WRKY subfamily and the functional analysis of GhWRKY17 in upland cotton (Gossypium hirsutum L.) PloS one. 2018;13(1):e0191681. doi: 10.1371/journal.pone.0191681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. Large-scale identification of leaf senescence-associated genes. Plant J. 2003;36(5):629–642. doi: 10.1046/j.1365-313X.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 37.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant journal : for cell and molecular biology. 2006;46(4):601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 38.Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62(2):250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- 39.Miao Y, Zentgraf U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. The Plant journal : for cell and molecular biology. 2010;63(2):179–188. doi: 10.1111/j.1365-313X.2010.04233.x. [DOI] [PubMed] [Google Scholar]
- 40.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13(9):2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gesell A, Yoshida K, Tran LT, Constabel CP. Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta. 2014;240(3):497–511. doi: 10.1007/s00425-014-2098-y. [DOI] [PubMed] [Google Scholar]
- 42.Heppel SC, Jaffe FW, Takos AM, Schellmann S, Rausch T, Walker AR, Bogs J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol Biol. 2013;82(4–5):457–471. doi: 10.1007/s11103-013-0074-8. [DOI] [PubMed] [Google Scholar]
- 43.Yan Q, Wang Y, Li Q, Zhang Z, Ding H, Zhang Y, Liu H, Luo M, Liu D, Song W, et al. Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol J. 2018. [DOI] [PMC free article] [PubMed]
- 44.Hinchliffe DJ, Condon BD, Thyssen G, Naoumkina M, Madison CA, Reynolds M, Delhom CD, Fang DD, Li P, McCarty J. The GhTT2_A07 gene is linked to the brown colour and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibres. J Exp Bot. 2016;67(18):5461–5471. doi: 10.1093/jxb/erw312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, Kim CS. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011;52(1):138–148. doi: 10.1093/pcp/pcq180. [DOI] [PubMed] [Google Scholar]
- 46.Jaradat MR, Feurtado JA, Huang DQ, Lu YQ, Cutler AJ. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013;13. [DOI] [PMC free article] [PubMed]
- 47.Gombert J, Etienne P, Ourry A, Le Dily F. The expression patterns of SAG12/cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J Exp Bot. 2006;57(9):1949–1956. doi: 10.1093/jxb/erj142. [DOI] [PubMed] [Google Scholar]
- 48.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol. 2008;68(1–2):81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christ B, Sussenbacher I, Moser S, Bichsel N, Egert A, Muller T, Krautler B, Hortensteiner S. Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis. Plant Cell. 2013;25(5):1868–1880. doi: 10.1105/tpc.113.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Ishida H, Reisen D, Hanson MR. Upregulation of a tonoplast-localized cytochrome P450 during petal senescence in Petunia inflata. BMC Plant Biol. 2006;6:8. doi: 10.1186/1471-2229-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarti M, Bowen SW, Coleman NP, Meekins KM, Dewey RE, Siminszky B. CYP82E4-mediated nicotine to nornicotine conversion in tobacco is regulated by a senescence-specific signaling pathway. Plant Mol Biol. 2008;66(4):415–427. doi: 10.1007/s11103-007-9280-6. [DOI] [PubMed] [Google Scholar]
- 52.Godiard L, Sauviac L, Dalbin N, Liaubet L, Callard D, Czernic P, Marco Y. CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett. 1998;438(3):245–249. doi: 10.1016/S0014-5793(98)01309-X. [DOI] [PubMed] [Google Scholar]
- 53.Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1(2):641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 54.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 55.Schutze K, Harter K, Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol. 2009;479:189–202. doi: 10.1007/978-1-59745-289-2_12. [DOI] [PubMed] [Google Scholar]
- 56.Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40(3):428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 57.Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2(3):213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 58.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu T. Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol. 2014;1150:81–95. doi: 10.1007/978-1-4939-0512-6_4. [DOI] [PubMed] [Google Scholar]
- 60.Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Partial result from library screening by the Y2H assay (DOCX 23 kb)
Table S2. Detailed information regarding the target genes of GhWRKY27 identified by ChIP-seq (DOCX 84 kb)
Table S3. Nucleic acid sequences of the potential target genes of GhWRKY27 used for the Y1H system (DOCX 24 kb)
Fig. S1. The expression levels of GhCYP94C1 in different tissues (TIFF 2064 kb)
Fig. S2. The expression levels of GhCYP94C1 at different stages of flower bud differentiation from different maturing varieties. (TIFF 124 kb)
Table S4. Primers used in this study (DOCX 40 kb)
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional file.