Abstract
Background
Leukemia stem cell (LSC)-enriched genes have been shown to be highly prognostic in acute myeloid leukemia (AML). However, the prognostic value of tumor suppressor genes (TSGs) that are repressed early in LSC remains largely unknown.
Methods
We compared the public available expression/methylation profiling data of LSCs with that of hematopoietic stem cells (HSCs), in order to identify potential tumor suppressor genes in LSC. The prognostic relevance of PCDH17 was analyzed on a cohort of 173 AML patients from The Cancer Genome Atlas (TCGA), and further validated in three independent cohorts (n = 339).
Results
We identified protocadherin17 (PCDH17) and demonstrated that it was significantly down-regulated and hypermethylated in LSCs compared with HSCs. Our analyses of primary AML patient samples also confirmed these deregulations. Clinically, low PCDH17 expression was associated with female sex (P = 0.01), higher WBC (P < 0.0001), higher percentages of blasts in bone marrow (BM) and peripheral blood (PB) (P = 0.04 and < 0.001, respectively), presence of FLT3-internal tandem duplications (P = 0.002), mutated NPM1 (P = 0.02), and wild-type TP53 (P = 0.005). Moreover, low PCDH17 expression predicted worse overall survival (OS) in four independent cohorts as well as in the molecularly defined subgroups of AML patients. In multivariable analyses, low PCDH17 expression retained independent prognostic value for OS. Biologically, PCDH17 expression-associated gene signatures were characterized by deregulations of EMT- and Wnt pathway-related genes.
Conclusions
PCDH17 gene was silenced by DNA methylation in AML. Low PCDH17 expression is associated with distinct clinical and biological features and improves risk stratification in patients with AML.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1851-1) contains supplementary material, which is available to authorized users.
Keywords: Protocadherin17, Acute myeloid leukemia, DNA methylation, Gene expression, Prognosis
Background
Acute myeloid leukemia (AML) is a group of hematological malignancies marked by heterogeneity in its biological features and clinical outcomes. The biological heterogeneity mainly consists of cytogenetic [1], genetic [2, 3], epigenetic [4], and transcriptional diversities [5, 6]. These aberrations together contribute to the clinical heterogeneity of AML, which makes it difficult for risk stratification and targeted therapy of the disease. To date, karyotypic analysis remains as the mainstay of risk classification in AML, yet 40 to 50% of patients lack clonal chromosome aberrations [1]; molecular markers, especially recurrent somatic mutations, have shown their ability to subdivide this large group of patients [7]. As described by the latest WHO classification of AML [8], mutations in NPM1 (without concomitant FLT3-ITD) and biallelic mutations of CEBPA in cytogenetically normal patients are associated with a favorable prognosis, whereas RUNX1 mutations have been implicated in worse overall survival in AML patients. Many other mutations, however, are not included in current clinical practice, primarily due to inconsistent prognostic findings and a lack of validation in large cohorts. Moreover, there is considerable prognostic variability even in the genetically defined subclasses. Thus, novel biomarkers, like gene expression alterations, are needed to improve outcome prediction in the context of established prognostic markers in AML.
There is now ample evidence that AML is initiated and maintained by leukemia stem cells (LSCs) [9]; this subset of cells is deemed to mediate drug resistance and disease relapse of AML. Importantly, recent studies have generated LSC-specific gene expression signatures that are highly prognostic in multiple independent cohorts of AML patients [10–13], demonstrating the potential utility of testing LSC genes expression in clinical practice. Previously, it was found that haploinsufficiency of Dnmt1, a DNA methyltransferase, effectively impaired LSC function and lead to prolonged survival of MLL-AF9 induced leukemic models [14]. This leads us to speculate that some survival-related tumor suppressor genes (TSGs) may be repressed by DNA methylation in LSCs. Indeed, repression of stem cell-related TSGs has been shown to correlate with poor outcome and cancer progression in solid tumors [15]. As promoter methylation is a major mechanism in silencing TSGs expression, it would be of interest to identify aberrantly hypermethylated and repressed TSGs in LSCs, especially those genes that have profound biological and clinical implications.
Protocadherin17 (PCDH17), a member of the cadherin superfamily, is predominantly expressed in the nervous system and has its major roles in axon extension, synapse formation, and dendrite arborization [16]. The gene locates on chromosome 13q21, a region for which homozygous deletion often occurred in cancers [17]; this suggests PCDH17 as a potential tumor suppressor beyond its role in neural development. In line with this, PCDH17 gene is found transcriptionally silenced in various human cancers, due to mechanisms including deletion, mutation, and more often, promoter methylation [18, 19]. In esophageal squamous cell carcinoma (ESCC), PCDH17 was preferentially silenced in poorly differentiated tumors and re-expression of PCDH17 reduced proliferation and migration of ESCC cells [20]. Frequent epigenetic inactivation of PCDH17 was also reported in gastric and colorectal cancers, in which PCDH17 exerts its tumor suppressive activity by inducing apoptosis and autophagy [21]. Importantly, PCDH17 received as much attention for its diagnostic and prognostic implications—as in urologic neoplasms, where PCDH17 methylation was reported to be a highly sensitive and specific diagnostic marker [22]. Despite progress in understanding its tumor-suppressive functions and clinical relevance in solid tumors, the role of PCDH17 in hematological malignancies remains largely unknown. Indeed, a recent finding in pediatric acute lymphoblastic leukemia has shown that PCDH17 methylation was significantly associated with disease relapse and worse outcome [23], thereby suggesting a potential involvement of PCDH17 in leukemogenesis.
In this study, we identified six potential tumor suppressors that are specifically repressed and hypermethylated in LSCs, including PCDH17. The clinical impact of aberrant PCDH17 expression was further explored in the context of known molecular prognosticators in AML. Furthermore, to gain insights into molecular features of PCDH17-deregulated AML, we generated gene- and microRNA-expression signatures associated with changes in PCDH17 expression levels, using RNA and microRNA sequencing data derived from the Cancer Genome Atlas (TCGA) AML project [3].
Methods
Patients and samples
To analyze the prognostic impact of PCDH17 in AML patients, four independent cohorts with survival information were included in this study: (1) a discovery cohort including 200 de novo AML patients derived from the TCGA AML study [3]; (2) two validation cohorts of 242 cytogenetically normal AML patients described by Metzeler et al. [24]; (3) an independent (Chinese) cohort consisted of 184 AML patients enrolled from 2005 to 2016 and treated in the Affiliated People’s Hospital of Jiangsu University. Detailed patient characteristics and treatment of the three published cohorts were described in Additional file 1: Additional Methods. For the Chinese cohort, all patients received induction and consolidation chemotherapy as described previously [25]. Of the 184 patients studied, PCDH17 expression data were obtained by real-time quantitative PCR (RT-qPCR) for 126 patients and 97 of them have survival data (OS only), while PCDH17 methylation levels were detected by targeted bisulfite sequencing for 107 patients. To test the association between PCDH17 expression and chemotherapy, we collected 20 paired BM samples from 10 patients both at diagnosis and during complete remission. Mutation and cytogenetic analyses were performed as previously described [25]. For comparison with normal controls, bone marrow samples from 66 healthy donors were collected. This study was approved by the Institutional Ethics Committee of the Affiliated People’s Hospital of Jiangsu University.
Public datasets
We used eight public available AML datasets from TCGA and Gene Expression Omnibus in this study (see Additional file 1: Table S1 for details). Of these data sets, two of them consists of expression data for bulk primary AML samples (GSE12417 and the TCGA dataset), and four datasets contain both healthy and AML BM samples sorted by fluorescence-activated cell sorting (FACS) (GSE24006, GSE30029, GSE63270, and GSE63409). Profiling data of the HSC and LSC subpopulations extracted from GSE63270 to GSE63409 were used for discovering TSGs that are down-regulated and hypermethylated in LSCs. For GSE24006 and GSE30029—both of which were available for expression data from CD34+ and CD34− samples—only data of the CD34+ populations were utilized, as the majority of LSCs were found to reside in this compartment [11]. We also used a gene-expression dataset of normal hematopoietic cells (GSE42519) to assess the expression pattern of PCDH17 at various stages of normal hematopoiesis. In addition, two manually-aggregated datasets from BloodSpot (http://servers.binf.ku.dk/bloodspot/) were used, that is, HemaExplorer, which contains samples of normal hematopoietic cell lineages from four studies (GSE17054, GSE19599, GSE11864, and E-MEXP-1242), and BloodPool, a large dataset containing bulk AML samples and normal hematopoietic cell samples assembled from 6 independent studies (GSE13159, GSE15434, GSE61804, GSE14468, TCGA, and GSE42519). For each microarray dataset, the probe set with the highest average expression was selected to represent PCDH17 expression level. See Additional file 1: Additional Methods for detailed information and analyses of these data sets.
Real-time quantitative reverse transcriptase-PCR (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA as described previously [26]. Real-time quantitative PCR (RT-qPCR) was performed using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA) on a 7500 Thermo cycler (Applied Biosystems, CA). Reaction conditions and PCR cycling were conducted as previously described [26], adjusting only for optimized primer annealing temperatures (60 °C). PCR primers were designed using Primer Premier 6 (Premier Biosoft, Palo Alto, CA, USA), and the primer sequences were listed in Additional file 1: Table S2. The relative quantification was calculated using the ΔΔCT method and normalized to the ABL1 housekeeping gene.
Targeted bisulfite sequencing
Genomic DNA was extracted from bone marrow cells with Puregene Blood Core Kit B (QIAGEN Sciences, MD, USA). The DNA Concentration and purity were assessed using Nanodrop 2000 (Thermo Fisher Scientific, MA, USA), then DNA was subjected to bisulfite modification by EZ DNA Methylation-Gold Kit (ZYMO, CA, USA), following the manufacturer’s instructions. Primers were designed using primer3 (http://primer3.ut.ee/) based on the bisulfite converted DNA. To prepare for high-throughput DNA sequencing, the target sequence was subjected to multiplexed PCR amplification followed by index PCR, the PCR products were further purified to generate sequencing library. Paired-end sequencing (2 × 150 bp) was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA). After quality assessment and filtering, paired reads generated from the Illumina MiSeq platform were merged using FLASH (Fast length adjustment of short reads) [27], the merged reads were then aligned (mapped) to the human GRCh37/hg19 genome assembly (in silico bisulfite-converted human reference genome [GRCh37]) using blast + [28]. CpG methylation values were calculated as methylated reads counts/total read counts of every CG point. Principal component analysis (PCA) was conducted using the “FactoMineR” package in R, to determine similarities of methylation levels between the two groups; two principal components (PC1 and PC2, representing the largest two variances) were plotted using “fviz_pca_ind” function from “factoextra” R package.
Cell culture, 5-aza-dC treatment and real-time methylation-specific polymerase chain reaction (RQ-MSP)
Human leukemia cell line HL60 (American Type Culture Collection, Manassas, VA, USA) was cultured in RPMI 1640 medium plus 10% fetal calf serum (ExCell Bio, Shanghai, China). Cells were incubated in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were placed at a density of 5 × 105 cells/ml in 5 mL medium 24 h before treatment. Graded doses (0 μM, 2 μM, and 4 μM) of 5-aza-2′-deoxycytidine (5-aza-dC) (Sigma-Aldrich, Steinheim, USA) were then added to the medium. After 3 days of treatment, the cells were harvested. Extractions of RNA and DNA were performed as described above. To assess the changes in methylation level, real-time methylation-specific polymerase chain reaction (RQ-MSP) was employed using primers listed in Additional file 1: Table S2. PCR conditions were as described previously [26], except that the annealing temperature for methylation primers was 59 °C.
Statistical analysis and bioinformatics
Overall survival (OS) was defined as the time from the date of diagnosis to death due to any cause. Disease-free survival (DFS) was defined as the period after primary treatment during which there are no signs and symptoms of the disease. For the datasets analyzed, patients were stratified as high PCDH17 expression and low PCDH17 expression, according to the median expression value. Survival probabilities of the two groups were estimated by the Kaplan–Meier method and compared by log-rank test. Those variables with log-rank p-values less than or equal to 0.20 were further included in a multivariate Cox regression model. All survival analyses were performed using the “survival” R package, with the survival curves drawn by R package “survminer”.
To test for the significance of the differences across groups, Chi square tests or Fisher’s exact test was used for categorical data. For continuous data, a nonparametric test such as the Wilcoxon rank-sum test was used. Spearman correlation analysis was performed to determine the association between two continuous variables. All basic statistical analyses were performed using the base functions in R version 3.4 (https://www.r-project.org). Differential gene expression analysis for RNA/microRNA sequencing data was calculated using the raw read counts with the R/Bioconductor package “edgeR”, while package “limma” was used for gene-expression and DNA-methylation microarray data, all analyses were controlled for the false discovery rate (FDR) by the Benjamini–Hochberg procedure. Gene set enrichment analysis (GSEA) was performed on the TCGA dataset using GSEA v3.0 software (http://www.broad.mit.edu/gsea). All plots and graphs were generated using the R package “ggplot2” unless otherwise stated. For details of bioinformatics see Additional file 1: Additional Methods.
Results
PCDH17 is significantly down-regulated in AML and dynamic expressed during normal hematopoiesis
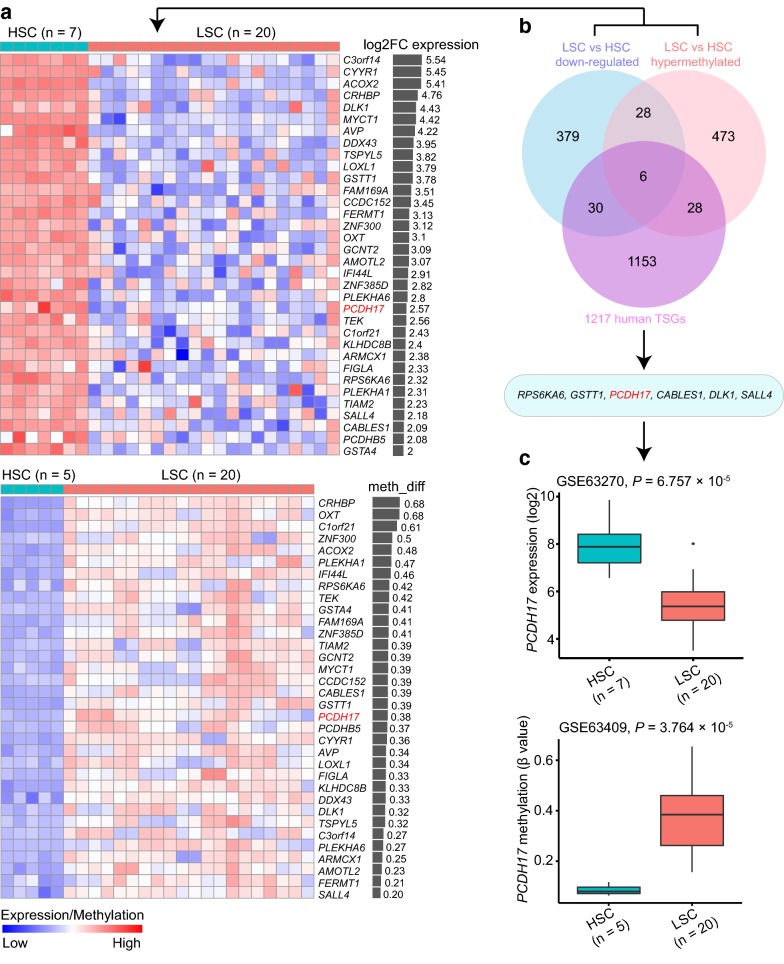
To identify candidate tumor suppressor genes in LSCs, the dataset GSE63270 (gene-expression data, n = 104) and GSE63409 (DNA-methylation data, n = 74) were utilized [12]. The two datasets were profiled from FACS sorted BM samples from 15 AML patients and 5 healthy controls. For our purpose, we only extracted data concerning the LSC fractions (n = 20 for both datasets) and the HSC subpopulations (GSE63270, n = 7; GSE63409, n = 5), in which the LSC phenotype was determined by their surface immunophenotype and leukemic engraftment ability [12]. Differential expression/methylation analyses were then performed between these two populations. Our analyses yielded 443 significantly down-regulated genes/probes (FDR < 0.05, log2 FC > 2, Additional file 2) in LSCs versus HSCs. For DNA methylation data, 863 hypermethylated CpG sites located in the promoter regions (FDR < 0.05, methylation difference > 20%, Additional file 3) were identified, corresponding to 535 unique genes. By intersecting the two gene lists, we identified that 34 genes were simultaneously down-regulated and hypermethylated in LSCs compared to HSCs (Fig. 1a). The 34 genes were further checked in the tumor suppressor gene database including 1217 TSGs [29]. Six genes (RPS6KA6, GSTT1, PCDH17, CABLES1, DLK1, and SALL4) were at last screened (The overlap of three gene sets was shown as a Venn diagram in Fig. 1b). Among them, PCDH17 has been reported as a negative regulator of the Wnt/β-catenin pathway [30], and since our group has already established the prognostic significance of several Wnt antagonists in AML [31–33], subsequent studies focused on the clinical impact of PCDH17 gene.
Fig. 1.
Identification of potential tumor suppressor genes that are repressed early in LSC. a Heatmaps showing down-regulated genes (upper panel) and hypermethylated genes (lower panel) in LSCs versus HSCs, from GEO: GSE63270 and GEO: GSE63409, respectively. Log2 fold changes of gene expression (log2 FC expression, upper panel) and β-value differences in DNA methylation (meth_diff, lower panel) are displayed as bar graphs on the right. The heatmaps only represent the 34 genes that are simultaneously down-regulated and hypermethylated in LSCs versus HSCs. For LSC, n = 20 represents 20 LSC fractions from 10 patients. b Venn diagram showing the overlap of three gene sets including: 443 down-regulated genes (FDR < 0.05, log2 FC > 2) in LSCs versus HSCs, 535 hypermethylated genes (FDR < 0.05, methylation difference > 20%) in LSCs versus HSCs, and 1217 TSGs from the tumor suppressor gene database. The overlapping region (6) represents the finally screened tumor suppressor genes. c Boxplots showing gene expression (upper panel) and DNA methylation (lower panel) changes of PCDH17 in LSCs versus HSCs, from GEO: GSE63270 and GEO: GSE63409, respectively. The P values calculated with a Wilcoxon rank-sum test are shown
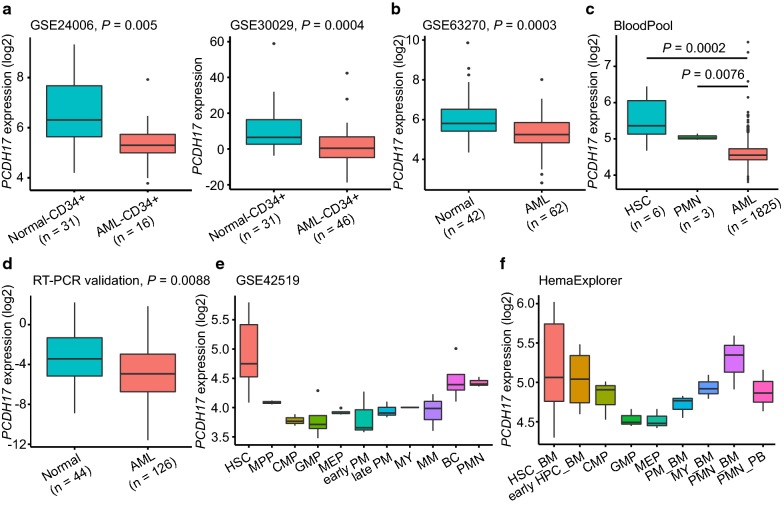
The expression and DNA methylation changes of PCDH17 were displayed as boxplots regarding the two discovery datasets (Fig. 1c). Further comparing PCDH17 expression between CD34+ AML and CD34+ normal samples in two independent datasets (GSE24006 and GSE30029), we also detected a significant reduction of PCDH17 expression in CD34+ AML cells (GSE24006, n = 16; GSE30029, n = 46), as compared with CD34+ normal cells (n = 31 for both datasets) (Fig. 2a). A similar result was observed when the total AML—(n = 62) and normal—(n = 42) mononuclear cell (MNC) fractions of the GSE63270 dataset were analyzed (Fig. 2b). Next, using the BloodPool dataset from BloodSpot, we show that PCDH17 expression is significantly lower in patients with AML (n = 1825) in comparison to HSC (n = 6) and polymorphonuclear cell (PMN) (n = 3) from healthy donors (Fig. 2c). This repression was further verified through RT-qPCR on 126 bulk primary AML and 44 normal BM samples (Fig. 2d). Together, these results demonstrate that transcriptional silencing of PCDH17 is a common feature in AML patients.
Fig. 2.
PCDH17 expression in AML and normal hematopoiesis. a, b Box plots illustrating the validation of PCDH17 expression changes in AML in three independent datasets. a Datasets using CD34 + cells from normal controls and AML patients, as indicated. b Dataset using whole bone marrow cells from normal controls and AML patients, as indicated. c Box plots showing that PCDH17 expression is significantly lower in patients with AML in comparison to HSC and PMN from healthy donors, using a large aggregated data set (BloodPool) from the BloodSpot website (http://servers.binf.ku.dk/bloodspot/). d Box plot comparing PCDH17 expression levels in normal controls and AML as assessed by RT-qPCR. Comparisons were made by the Wilcoxon rank-sum test. e Box plot showing the expression pattern of PCDH17 during differentiation of the major hematopoietic lineages using published dataset (GSE42519). f Box plot showing the expression pattern of PCDH17 during differentiation of the major hematopoietic lineages using a merged data set (HemaExplorer) from the BloodSpot website (http://servers.binf.ku.dk/bloodspot/). HSC hematopoietic stem cell, MPP multipotent progenitor, CMP common myeloid progenitor, GMP granulocyte-monocyte progenitor, MEP megakaryocyte-erythrocyte progenitor, PM promyelocyte, MY myelocyte, MM metamyelocytes, BC band cell, PMN polymorphonuclear cells, HPC hematopoietic progenitor cells, BM bone marrow, PB peripheral blood
To extend these findings and determine the expression pattern of PCDH17 during normal hematopoiesis, the level of PCDH17 transcripts was analyzed using a published data (GSE42519), in which gene expression profiling was performed with samples from normal hematopoiesis [34]. We found PCDH17 transcripts was highest in BM HSCs, with a sharp reduction in committed progenitors (common myeloid progenitor, granulo-monocyte progenitor, megakaryocyte-erythroid progenitor), the expression remained low and then gradually increased during myeloid maturation (Fig. 2e). The dynamic expression pattern of PCDH17 during normal hematopoiesis was further confirmed in a merged data set (HemaExplorer) from BloodSpot (Fig. 2f). This dynamic regulation of PCDH17 transcripts suggests a potential role of PCDH17 in HSC biology.
Repression of PCDH17 is mediated by promoter methylation in AML
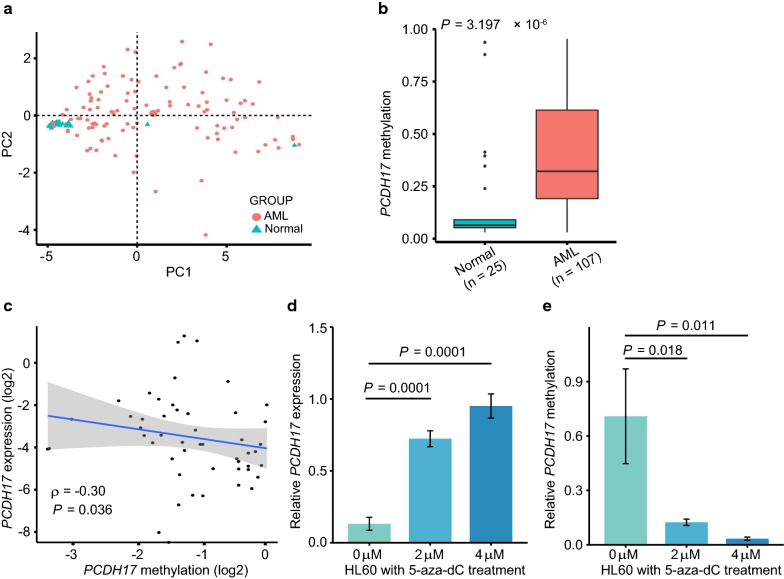
To assess the methylation pattern of PCDH17 in primary AML samples and to see whether its repression is mediated by methylation, we used targeted bisulfite sequencing to measure the methylation levels of 20 CpG sites in the first exon of PCDH17 (chr13:58,206,837-58207013). Detailed methylation data of this sequenced region was provided in Additional file 4. PCA analysis of our methylation data showed a clear discrimination between AML and normal BM-derived MNCs (Fig. 3a). The β values of the 20 CpG sites were then averaged as the DNA methylation level of PCDH17 gene, which was significantly higher in AML patients (n = 107; median, 0.3219; range, 0.0303-0.9542; P < 0.0001) compared with the healthy controls (n = 25; median, 0.0649; range, 0.0302-0.9372) (Fig. 3b).
Fig. 3.
Methylation-mediated silencing of PCDH17 expression in AML. a Principal component analysis (PCA) of PCDH17 methylation levels determined by targeted bisulfite sequencing. Methylation data of the measured 20 CpG sites across 132 samples (107 AML and 25 normal BM- derived MNCs) is provided in Additional file 4. b Box plot showing average PCDH17 methylation differences in normal controls and AML patients, calculated using the Wilcoxon rank-sum test. c Scatterplot showing the correlation between PCDH17 methylation and PCDH17 expression in AML patients. Spearman’s correlation coefficient (ρ) and P value are indicated. d, e PCDH17 expression (d) and methylation (e) in HL60 cell line before and after 5-aza-dC treatment. The data are shown as mean ± S.D. of triplicate analyses. Statistical significances were determined using Student’s t-tests
Furthermore, as shown in Fig. 3c, PCDH17 methylation levels were negatively correlated with mRNA levels (ρ = − 0.30, P = 0.036). To determine whether aberrant PCDH17 methylation could be reverted in vitro, we treated HL60 cells with demethylation reagent 5-aza-dC. Our results showed that 5-aza-dC treatment significantly up-regulated PCDH17 expression (Fig. 3d); this was supported by the MSP analyses, which also showed a change in the methylation status of the PCDH17 promoter (Fig. 3e). In summary, these results suggested PCDH17 expression is inactivated, at least in part, by its promoter methylation in AML.
PCDH17 expression is associated with clinical characteristics and molecular markers
For the discovery cohort, the association of the clinical and genetic characteristics with PCDH17 expression was summarized in Table 1. At diagnosis, patients with low PCDH17 expression were more likely to be females (P = 0.01), with higher white blood cell (WBC) counts (P < 0.0001), higher percentage of blasts in BM and PB (P = 0.04 and < 0.001, respectively). There were no other significant differences in presenting clinical characteristics between these two groups including age (P = 0.41) and karyotype classification (P = 0.35).
Table 1.
Patient characteristics with respect to PCDH17 expression
| Patient characteristics | AML (TCGA dataset) | ||
|---|---|---|---|
| High PCDH17 (n = 86) | Low PCDH17 (n = 87) | P | |
| Age, years | 0.41 | ||
| Median | 59 | 57 | |
| Range | 18–88 | 21–82 | |
| Sex (male/female) | 54/32 | 38/49 | 0.01 |
| WBC count, × 109/L | < 0.0001 | ||
| Median | 9.3 | 34 | |
| Range | 0.4–116.2 | 0.8–297.4 | |
| BM blasts, % | 0.04 | ||
| Median | 68.5 | 75 | |
| Range | 30–100 | 32–100 | |
| PB blasts, % | < 0.001 | ||
| Median | 18 | 49 | |
| Range | 0–97 | 0–98 | |
| Karyotype | 0.343 | ||
| Favorable | 16 | 16 | |
| Intermediate | 46 | 55 | |
| Adverse | 23 | 14 | |
| Unknown | 1 | 2 | |
| FLT3-ITD | 0.002 | ||
| Present | 6 | 21 | |
| Absent | 80 | 66 | |
| NPM1 | 0.02 | ||
| Mutated | 17 | 31 | |
| Wild-type | 69 | 56 | |
| CEBPA | 1 | ||
| Single mutated | 4 | 4 | |
| Double mutated | 2 | 3 | |
| Wild-type | 80 | 80 | |
| IDH1 | 0.28 | ||
| Mutated | 10 | 6 | |
| Wild-type | 76 | 81 | |
| IDH2 | 0.82 | ||
| Mutated | 8 | 9 | |
| Wild-type | 78 | 78 | |
| RUNX1 | 0.769 | ||
| Mutated | 8 | 7 | |
| Wild-type | 78 | 80 | |
| DNMT3A | 0.755 | ||
| Mutated | 20 | 22 | |
| Wild-type | 66 | 65 | |
| TP53 | 0.005 | ||
| Mutated | 12 | 2 | |
| Wild-type | 74 | 85 | |
| ERG expressiona | 0.08 | ||
| High | 37 | 49 | |
| Low | 49 | 38 | |
| BAALC expressiona | 0.494 | ||
| High | 45 | 41 | |
| Low | 41 | 46 | |
| MN1 expressiona | 0.704 | ||
| High | 44 | 42 | |
| Low | 42 | 45 | |
| WT1 expressiona | 0.323 | ||
| High | 40 | 47 | |
| Low | 46 | 40 | |
AML acute myeloid leukemia, TCGA The Cancer Genome Atlas, WBC white blood cells, BM bone marrow, PB peripheral blood, ITD internal tandem duplication
aThe median expression value was used as a cut point
With respect to known prognostic markers, patients with low PCDH17 expression more often had FLT3-ITD (P = 0.002) and NPM1 mutations (P = 0.02) and less often had TP53 mutations (P = 0.005) than patients with high PCDH17 expression. These associations were confirmed by comparing PCDH17 expression levels as stratified by mutation status of the three genes (Additional file 1: Figure S1a–c). No significant differences were observed between low- and high- PCDH17 expression patients regarding other molecular alternations, although low expressers had a trend toward more often having high expression of ERG (P = 0.08).
Reduced PCDH17 expression predicts inferior survival in four independent cohorts
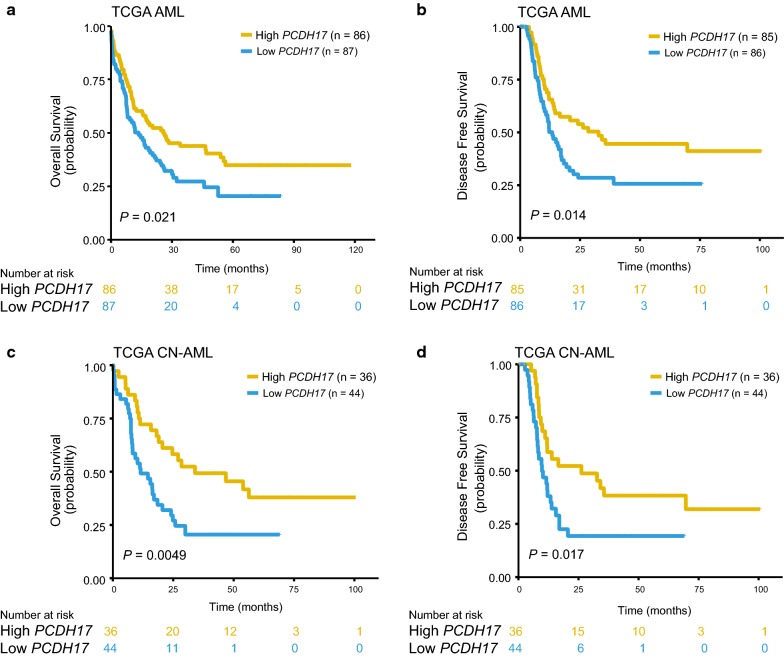
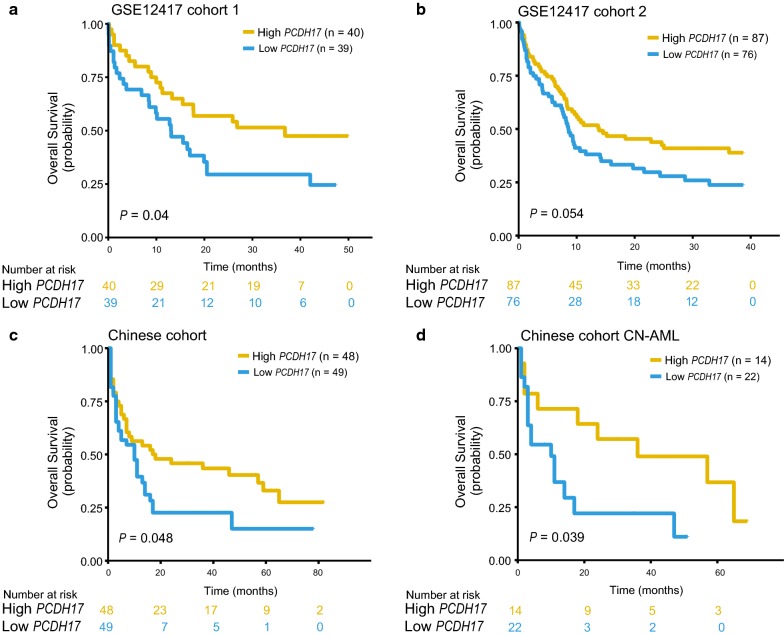
To test the prognostic relevance of deregulated PCDH17 expression, we investigated the correlation between PCDH17 expression and clinical outcomes in four independent AML cohorts, all cohorts were dichotomized according to the median expression value of PCDH17. In the TCGA AML cohort (n = 173), patients with low PCDH17 expression had significantly reduced OS (P = 0.021, Fig. 4a) and DFS (P = 0.014, Fig. 4b). This survival difference was also found for the subset of cytogenetically normal (CN)-AML patients (n = 80) (OS, P = 0.0049, Fig. 4c; DFS, P = 0.017, Fig. 4d). The prognostic value for OS was further validated in two CN-AML cohorts from GSE12417 (cohort1, n = 79, P = 0.04, Fig. 5a; cohort 2, n = 163, P = 0.054, Fig. 5b). When applied to the RT-qPCR data in our cohort, low PCDH17 expression remained an adverse indicator for OS both in the whole cohort (n = 97) (P = 0.048, Fig. 5c) and in the CN-AML subsets (n = 36) (P = 0.039, Fig. 5d).
Fig. 4.
Prognostic significance of PCDH17 in the discovery (TCGA) cohort. a, b The association of PCDH17 expression levels with OS (a) and DFS (b) for the whole TCGA cohort. c, d The association of PCDH17 expression levels with OS (c) and DFS (d) in the TCGA CN-AML patients. Patients were separated into high and low PCDH17 group based on the median expression levels of PCDH17
Fig. 5.
Validation of the prognostic value of PCDH17 in three independent AML patient cohorts. a, b Kaplan–Meier analysis of overall survival (OS) in two CN-AML cohorts: GSE12417 cohort 1 (n = 79) (a) and GSE12417 cohort 2 (n = 163) (b). c, d Kaplan–Meier estimate for OS of the Chinese cohort in the whole (c) as well as in the CN-AML (d) patients. Patients were separated into high and low PCDH17 group based on the median expression levels of PCDH17
Prognostic value of PCDH17 expression in the context of other risk factors in AML
To assess whether the prognostic impact of PCDH17 expression is independent from knowing clinical factors, especially recurrent mutations found in AML, a univariate analysis was first conducted for the TCGA cohort (Detailed results were shown in Additional file 1: Table S3), variables considered significant (P ≤ 0.2) were further included in the multivariate Cox proportional hazard model. As shown in Table 2, low PCDH17 expression maintained significant independent prognostic value with respect to OS (HR = 2.04; 95% CI, 1.37-3.04; P < 0.001), after adjustment for age group (P < 0.0001), cytogenetic risk group (P = 0.001), TP53 mutation status (P = 0.004), DNMT3A mutation status (P = 0.03), and RUNX1 mutation status (P = 0.08). The independent predictive value of PCDH17 was also true for DFS (HR = 1.89; 95% CI 1.19–2.98; P = 0.007). In CN-AML patients, PCDH17 down-regulation was still an independent poor prognostic factor for OS (HR = 2.02; 95% CI 1.13–3.62; P = 0.02) and a borderline poor prognostic factor for DFS (HR = 1.79; 95% CI 0.93–3.44; P = 0.08). Importantly, the independent prognostic value of PCDH17 for OS was also validated in the Chinese cohort (Table 3).
Table 2.
Multivariate analysis of PCDH17 expression for overall survival and disease-free survival in the TCGA cohort
| Variables | Overall survival | Disease-free survival | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |
| Full cohort | (n = 173) | (n = 171) | ||
| PCDH17 a | 2.04 (1.37–3.04) | < 0.001 | 1.89 (1.19–2.98) | 0.007 |
| Ageb | 2.90 (1.94–4.34) | < 0.0001 | 1.52 (0.96–2.43) | 0.08 |
| WBC countc | – | – | 2.14 (1.36–3.39) | 0.001 |
| Cytogenetic riskd | 1.84 (1.28–2.65) | 0.001 | 1.39 (0.95–2.04) | 0.09 |
| TP53 e | 2.76 (1.37–5.55) | 0.004 | 2.64 (0.93–7.47) | 0.07 |
| DNMT3A e | 1.61 (1.05–2.48) | 0.03 | 1.41 (0.85–2.33) | 0.18 |
| RUNX1 e | 1.75 (0.94–3.26) | 0.08 | – | – |
| CN-AML | (n = 80) | (n = 80) | ||
|---|---|---|---|---|
| PCDH17 a | 2.02 (1.13–3.62) | 0.02 | 1.79 (0.93–3.44) | 0.08 |
| Ageb | 2.38 (1.35–4.19) | 0.003 | 2.19 (1.10–4.36) | 0.03 |
| WBC countc | 1.42 (0.81–2.48) | 0.22 | 1.65 (0.89–3.04) | 0.11 |
| FLT3-ITDf | – | – | 1.34 (0.60–2.99) | 0.47 |
| TP53 e | – | – | 4.09 (0.46–36.02) | 0.20 |
| DNMT3A e | 1.99 (1.13–3.52) | 0.02 | 2.15 (1.15–4.03) | 0.02 |
| IDH1 e | 0.61 (0.21–1.78) | 0.36 | 0.68 (0.22–2.08) | 0.49 |
Hazard Ratio > 1 or Hazard Ratio < 1 indicate a higher or lower risk. Only variables with a univariable P ≤ 0.20 were included in the multivariable models. See Additional file 1: Table S3 for a full list of variables evaluated in univariable analysis
TCGA The Cancer Genome Atlas, CI confidence interval, WBC white blood cells, CN-AML cytogenetically normal AML, ITD internal tandem duplication
aLow vs high expression
b > 60 vs ≤ 60 years
c ≥ 30 vs < 30 × 109/L
dAdverse vs intermediate vs favorable
eMutated vs wild type
fPresent vs absent
Table 3.
Univariate and multivariate analysis of PCDH17 expression for overall survival in the Chinese cohort
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | P | Hazard Ratio (95% CI) | P | |
| Full cohort | (n = 97) | (n = 97) | ||
| PCDH17 a | 1.65 (1–2.72) | 0.05 | 2.01 (1.19–2.98) | 0.01 |
| Ageb | 3.07 (1.87–5.04) | < 0.0001 | 2.10 (1.18–3.61) | 0.02 |
| WBC countc | 3.14 (1.92–5.14) | < 0.0001 | 2.06 (1.16–3.64) | 0.01 |
| Cytogenetic riskd | 2.04 (1.49–2.78) | < 0.0001 | 1.60 (1.05–2.43) | 0.03 |
| FLT3-ITDe | 0.80 (0.36–1.77) | 0.58 | – | – |
| NPM1 f | 0.97 (0.39–2.43) | 0.95 | – | – |
| CEBPA f | 1.18 (0.53–2.62) | 0.69 | – | – |
| DNMT3A f | 1.66 (0.71–3.89) | 0.24 | – | – |
| IDH2 f | 6.13 (0.8–47) | 0.08 | 5.35 (0.63–45.16) | 0.12 |
| NRAS f | 1.18 (0.37–3.81) | 0.78 | – | – |
| KRAS f | 8.80 (2.53–30.55) | < 0.001 | 5.16 (1.13–23.59) | 0.03 |
| U2AF1 f | 14.43 (1.81–115.4) | 0.01 | 4.99 (0.37–67.94) | 0.23 |
| SRSF2 f | 3.28 (0.78–13.85) | 0.11 | 0.95 (0.12–7.46) | 0.96 |
| SETBP1 f | 0.82 (0.11–5.96) | 0.85 | – | – |
| KIT f | 0.64 (0.16–2.64) | 0.54 | – | – |
| CN-AML | (n = 36) | (n = 36) | ||
|---|---|---|---|---|
| PCDH17 a | 2.58 (1.02–6.52) | 0.04 | 2.95 (1.04–8.32) | 0.04 |
| Ageb | 1.39 (0.61–3.14) | 0.43 | – | – |
| WBC countc | 3.05 (1.34–6.93) | 0.008 | 3.26 (1.35–7.87) | 0.009 |
| FLT3-ITDe | 0.48 (0.14–1.63) | 0.24 | – | – |
| NPM1 f | 0.50 (0.15–1.69) | 0.27 | – | – |
| CEBPA f | 1.19 (0.40–3.50) | 0.76 | – | – |
| DNMT3A f | 1.25 (0.42–3.68) | 0.69 | – | – |
| IDH2 f | 6.37 (0.76–53.48) | 0.09 | 13.30 (1.23–143.32) | 0.03 |
| NRAS f | 3.55 (0.45–27.94) | 0.23 | – | – |
| KRAS f | 6.46 (1.37–30.46) | 0.02 | 9.17 (1.72–48.86) | 0.009 |
| SETBP1 f | 6.37 (0.76–53.48) | 0.09 | 4.52 (0.48–42.10) | 0.19 |
Hazard ratio > 1 or Hazard ratio < 1 indicate a higher or lower risk. Only variables with a univariable P ≤ 0.20 were included in the multivariable models. Mutations such as IDH1 and TP53 were either not analyzed or detected in our cohort, thus they were not included in this analysis
CI confidence interval, WBC white blood cells, CN-AML cytogenetically normal AML, ITD internal tandem duplication
aLow vs high expression
b > 60 vs ≤ 60 years
c ≥ 30 vs < 30 × 109/L
dAdverse vs intermediate vs favorable
ePresent vs absent
fMutated vs wild type
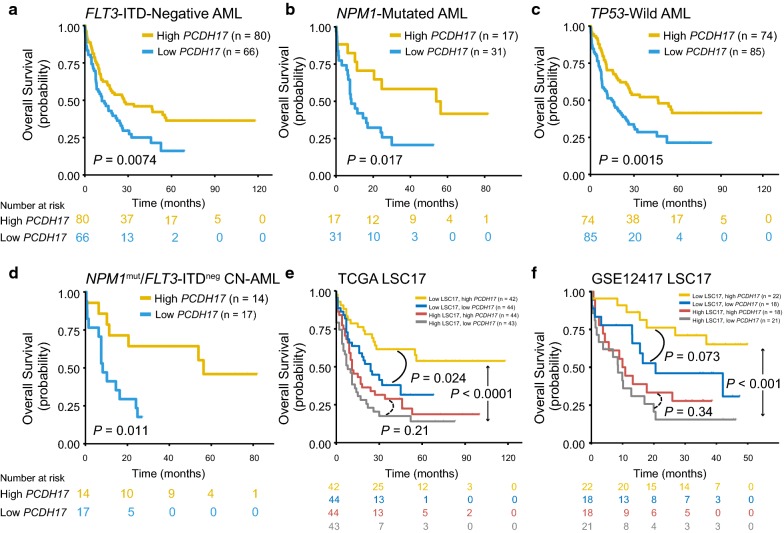
The strong prognostic significance of PCDH17 status led us to hypothesize that it may provide additional prognostic information in the molecularly defined subgroups of AML patients. Indeed, exploratory subgroup analyses showed that the dichotomous stratification of PCDH17 expression was still able to discriminate between shorter and longer OS in the FLT3-ITD absent, NPM1 mutated, and TP53 wild-type patients (P = 0.0074, 0.017, and 0.0015, respectively) (Fig. 6a–c), despite the aforementioned interaction among these alterations. Moreover, PCDH17 status identified a poor-prognostic subset of patients within the FLT3-ITDnegative/NPM1mutated low-risk CN-AML group (P = 0.011, Fig. 6d), although this analysis is limited by a relatively small number of patients (n = 31). For DFS, however, we found that reduced PCDH17 only had significant prognostic value in the FLT3-ITD absent and TP53 wild-type subsets (Additional file 1: Figure S2).
Fig. 6.
Reduced PCDH17 expression predicts a worse outcome in the molecularly defined subgroups of AML patients. a–d Kaplan–Meier curves of OS stratified by PCDH17 in the genetically defined subgroups of AML patients, as indicated. Low PCDH17 expression is associated with poor outcome in all subgroups. e, f Kaplan–Meier curves of OS in the TCGA (e) and the GSE12417 cohort 1 (f) as stratified by the LSC17 signature. The low- and high-risk patients were further dichotomized by PCDH17 expression status. PCDH17 expression status could further stratify survival in the subgroup with low LSC17 score. Patients were separated into high and low PCDH17 group based on the median expression levels of PCDH17
A recently published risk score (LSC17) [13], generated using 17 LSC-specific genes, has proved to be a strong prognostic indicator independent from traditional prognosticators and other reported LSC signatures. We recalculated the LSC17 score (see Additional file 1 for code details) using the TCGA dataset and the GSE12417 dataset (cohort 1) and found it could accurately discriminate between shorter and longer OS in both cohorts (P < 0.0001 and P < 0.001, respectively) (Fig. 6e, f). When applied PCDH17 status to the subsets of patients stratified by the LSC17 score, we found PCDH17 expression further dichotomizes survival within the low-risk group (P = 0.024 for the TCGA cohort and P = 0.073 for the GSE12417 cohort) (Fig. 6e, f). This suggests that PCDH17 could improve risk stratification in patients with a low LSC17 score.
Association between PCDH17 expression and treatment response
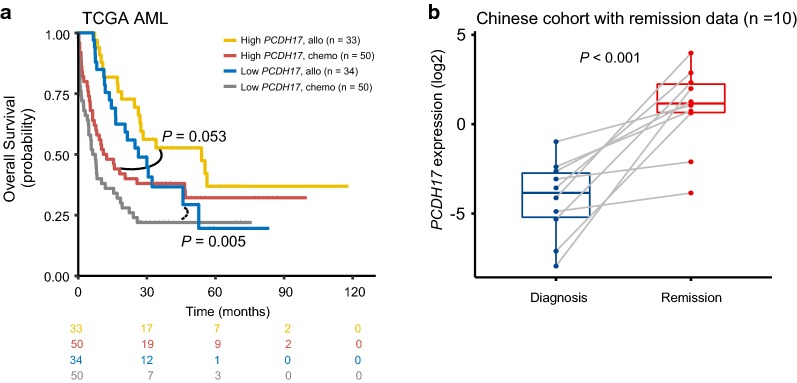
In the discovery cohort, 67 patients received allogeneic stem-cell transplantation (allo-HSCT); this therapy, though effective, is generally reserved for high-risk patients because of significant treatment-related mortality [35]. Thus, we asked whether patients with low PCDH17 expression would benefit from allo-HSCT. We separately analyzed the prognostic impact of allo-HSCT in patients with low and high PCDH17 expression. Results show that the benefit from allo-HSCT was more significant in patients with low PCDH17 expression (P = 0.005), as compared with those with high expression (P = 0.053) (Fig. 7a). This suggests that patients with low PCDH17 expression would potentially be candidates for allo-HSCT.
Fig. 7.
PCDH17 expression may help predict treatment response in AML patients. a Kaplan–Meier curves of OS for patients with high or low PCDH17 expression as stratified by treatment options (allo versus chemo) in the TCGA cohort. Allo, allogeneic transplantation; chemo, chemotherapy. b PCDH17 expression changes in 10 follow-up patients with paired BM-MNC samples from diagnosis to remission in the Chinese cohort. Each grey line represents one patient. PCDH17 expression was up-regulated in all patients after remission. The P value was calculated with a Wilcoxon rank-sum test
To further investigate PCDH17 expression changes in the surveillance of AML, we assessed its expression in 10 patients with paired BM-MNC samples from diagnosis to complete remission (CR). Our data demonstrated that PCDH17 expression was significantly increased in CR after induction therapy (P < 0.001, Fig. 7b).
Distinct gene- and microRNA-expression signatures associated with PCDH17 expression in AML
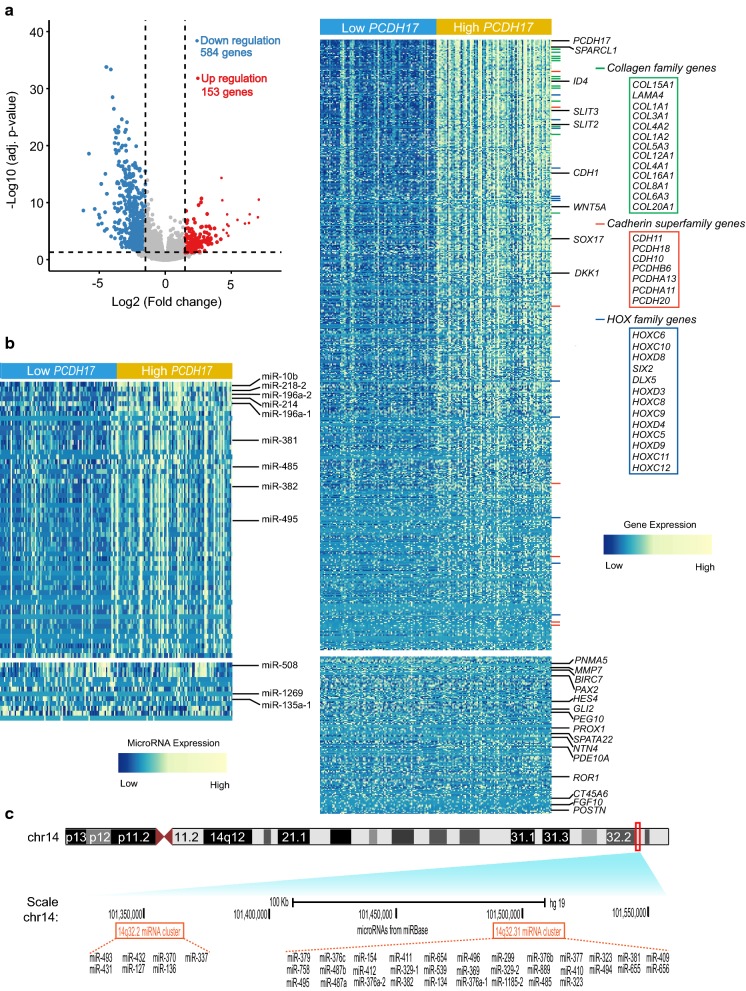
To extend the observation that clinical features differ between AML samples with low versus high PCDH17 expression, we first compared the transcriptomes of low PCDH17 expressed group with those of high expression. This comparison yielded 739 differentially expressed genes (FDR < 0.05, |log2 FC| > 1.5; Fig. 8a; Additional file 5), in which 584 genes were positively correlated with PCDH17 expression levels, and 153 were negatively correlated.
Fig. 8.
Gene/microRNA signatures associated with PCDH17 expression. a Left: volcano plot showing gene expression differences between patients with low and high PCDH17 expression. Significantly down-regulated genes (blue points) and up-regulated genes (red points) in patients with low PCDH17 expression are indicated. Right: heatmap showing the gene expression signature associated with PCDH17 expression (FDR < 0.05, |log2 FC| > 1.5). Up-regulated and down-regulated genes mentioned in the text are labeled on the rows or in the boxes. b Heatmap showing the microRNA expression signature associated with PCDH17 expression (FDR < 0.05, |log2 FC| > 1.5). Up-regulated and down-regulated microRNAs mentioned in the text are indicated. c UCSC browser screenshot of the microRNAs located on the 14q32 region. Top shows schematic representation of chromosome 14, with the 14q32 locus indicated as a red rectangle. Bottom shows the relative positions of the 39 microRNAs that are positively associated with PCDH17 expression (left, the 14q32.2 microRNA cluster; right, the 14q32.31 microRNA cluster)
Among the positively associated genes, the PCDH17 gene itself was ranked as the highest, as expected; other members of the cadherin superfamily, the HOXC and HOXD gene family, and the collagen family were also identified within this signature. Importantly, several tumor suppressors were found. Among them were negative regulators of epithelial-mesenchymal transition (EMT)—SLIT2, ID4, and CDH1—all three genes were known to be regulated by methylation- and EZH2-mediated repression in cancers, and the Wnt antagonists, such as DKK1, WNT5A, and SOX17. One of the most differentially expressed genes was SPARCL1, whose repression has been reported as a negative prognostic indicator in a number of cancers [36, 37]. SPARCL1 is best known for its role as an inhibitor of metastatic-invasion in solid tumors [38]; whether this gene has an antileukemic effect, however, remains to be identified.
As distinct from the tumor suppressive signatures that were positively associated with PCDH17 expression, those negatively correlated with PCDH17 expression consists mainly of oncogenes. Among them were known cancer/testis antigen genes (PNMA5, SPATA22, ROR1, and CT45A6) and some new candidates (PAX2, HES4, PEG10, NTN4, PDE10A, and POSTN), these genes could be useful tumor markers and potential therapeutic targets. Another noteworthy feature was an enrichment of EMT-inducing factors, including MMP7, BIRC7, GLI2, PROX1, and FGF10. Three of them—MMP7, BIRC7, and PROX1—were known as transcriptional targets of Wnt/β-catenin signaling [39–41]. Gli2, a transcription factor of the Hedgehog pathway, has recently been reported as a negative prognostic factor in AML patients, and its expression was significantly correlated with FLT3 mutation status [42].
We also derived a PCDH17 associated microRNA expression signature comprising 69 microRNAs (FDR < 0.05, |log2 FC| > 1.5; Fig. 8b; Additional file 6). Of the 69 microRNAs, 56 (fifty-six) microRNAs were down-regulated and 13 up-regulated in patients with low PCDH17 expression. To our surprise, more than two-thirds (n = 39) of the microRNAs in the former group were encoded within a microRNA cluster at 14q32 (Fig. 8c). This microRNA cluster, originally described as a human imprinted locus, were frequently deleted or epigenetically silenced in various cancers [43, 44], highlighting its tumor suppressive roles. Specifically, reduced expression of three microRNAs in this cluster (miR-381, miR-485, and miR-495) was associated with multidrug resistance in leukemia [45], and miR-495 has been reported as a tumor suppressor in MLL-rearranged AML [46]. We also identified three microRNAs embedded in the HOX cluster (miR-10b, miR-196a-1 and miR-196a-2): while miR-10b was shown to be negatively correlated with BAALC expression in AML [47], miR-196a was reported as a negative ERG regulator [48]. In addition, EMT-related microRNAs were noted, including miR-218-2, which was frequently repressed and co-methylated with its host gene SLIT3 [49]; miR-214, which targets EZH2, β-catenin, and BIRC7 [50]—all were genes implicated in cancer metastasis and invasion; miR-382, which targets ROR1 [51]. It is worth noting that SLIT3, BIRC7, and ROR1 were all among the aforementioned gene expression signatures. As for those negatively associated with PCDH17, only 3 of the 8 genes have previously been associated with carcinogenesis: miR-1269 was shown to mediate tumor metastasis by targeting HOXD10 in colorectal cancer [52]; the two other microRNAs (miR-508 and miR-135a-1) were mainly implicated in drug resistance.
GSEA analyses for PCDH17 expression associated transcriptomes
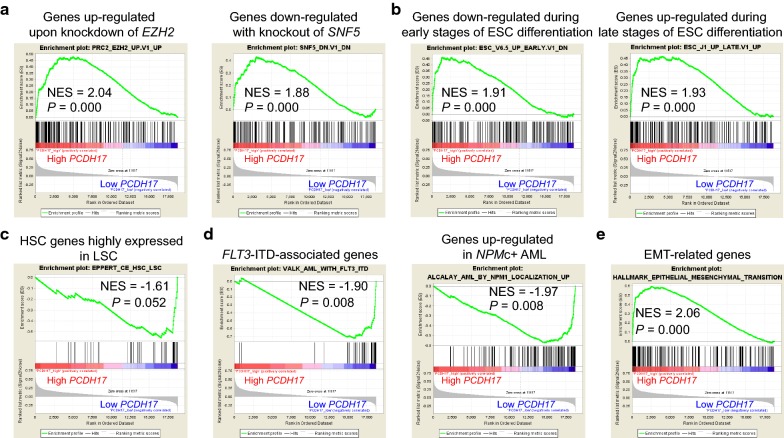
Considering the above differential analysis might lose some genes of potential biological meanings, we performed a gene set enrichment analysis (GSEA) by comparing transcriptomes of low PCDH17 expressers with that of high PCDH17 expressers. Our results confirmed and extended the above findings: (1) high PCDH17 expressers showed significant enrichment of genes up-regulated upon knockdown of EZH2 and genes down-regulated with depletion of SNF5, an established antagonist gene of EZH2 (Fig. 9a). It was found that EZH2, a component of the Polycomb repressive complex 2 (PRC2), could predispose certain TSGs to cancer-associated promoter hypermethylation during the early stages of stem cell development [53], consistent with our observation that PCDH17 was significantly hypermethylated in LSCs; (2) transcriptomes of high PCDH17 expressers are enriched in genes down-regulated during early differentiation of embryonic stem cells (ESCs) and those up-regulated in late stage (Fig. 9b), which agreed favorably with the dynamic expression pattern of PCDH17 during hematopoiesis; (3) low PCDH17 expressers tend to express HSC genes that are highly expressed in LSC (Fig. 9c), indicating that the gene itself might be down-regulated in LSC; (4) two earlier gene sets previously correlated with FLT3-ITD and NPM1 mutation were found enriched in the low PCDH17 signature (Fig. 9d); and (5) as expected, PCDH17-associated genes were significantly correlated with the EMT gene signature (Fig. 9e).
Fig. 9.
PCDH17 expression is associated with distinct gene signatures in AML. a GSEA plots show enrichment of EZH2 and SNF5 gene sets in PCDH17 high expressers versus low expressers. b GSEA comparison of PCDH17 high expressers versus low expressers shows enrichment of genes down-regulated during early stages of ESC differentiation and genes up-regulated during late stages of ESC differentiation. c GSEA plots show up-regulation of AML HSC/LSC signature genes in PCDH17 low expressers versus high expressers. d GSEA plots show enrichment of FLT3-ITD and NPM1 gene sets in PCDH17 low expressers versus high expressers. e GSEA plot shows enrichment of EMT-related signature in PCDH17 high expressers versus low expressers. The normalized enrichment scores (NES) and P values are indicated in each plot. ESC, embryonic stem cell
Together, these results further demonstrate that low PCDH17 expression is associated with distinct molecular features in AML.
Discussion
Our results showed that PCDH17 was significantly down-regulated in the bulk AML as well as in the LSC populations, as compared to their normal counterparts. This repressed expression, mediated mainly by DNA methylation, was correlated with distinct clinical and biological features. Most importantly, we found PCDH17 repression to be an independent prognostic factor for shorter OS and DFS in AML; it could add additional prognostic value by stratifying molecularly defined patients into more homogeneous groups.
Although reduced PCDH17 expression has been reported in a number of tumor types [19], it has, thus far, not been demonstrated in the context of AML. Our study shows that PCDH17 was repressed in AML and, in particular, in AML stem cells (LSCs). This repression was observed in four microarray profiling datasets of AML and confirmed by RT-qPCR, suggesting down-regulation of PCDH17 expression as a common alteration in AML. We also tracked PCDH17 expression during differentiation of major hematopoietic lineages. Despite the well-established role of PCDH17 in neural development, its involvement in hematopoiesis has, to our knowledge, been rarely documented. Our analysis shows that PCDH17 transcript levels are highest in the HSC and decrease sharply as HSC differentiate into multipotent progenitor (MPP). This observation was consistent with the findings that adhesion molecules tend to be up-regulated in HSCs and down-regulated upon early commitment [54, 55]. It is possible that high levels of PCDH17 may facilitate retention of HSCs in the stem cell niche, whereas decreased PCDH17 expression might result in destabilized cell–matrix interactions, thereby allowing HSCs to mobilize and differentiate. In pathological settings, however, PCDH17 inactivation might confer upon HSCs/pre-LSCs an enhanced proliferation activity, which cooperates with other phenotypic changes, eventually leading to fully transformed LSCs. Interestingly, we found PCDH17 expression was increased in the mature cell populations, possibly because PCDH17 may participate in the homing process of peripheral blood leukocyte.
Several prognostically relevant LSC gene signatures have been reported thus far [10–13]; these signatures consist of genes that are highly expressed not only in LSC but also in HSC, leaving the prognostic value of tumor suppressor genes in LSC largely undetermined. Given that PCDH17 expression was significantly repressed in LSC, we sought further to investigate its clinical implications. Our results showed that decreased PCDH17 expression was associated with female sex, higher median WBC counts, as well as higher median PB and BM blast counts, indicative of a high proliferative potential of PCDH17 repressed leukemia cells. With respect to the prognostic relevance, we found low PCDH17 expression predicted inferior OS and DFS in the whole TCGA cohort as well as in the subgroup of cytogenetically normal patients; the predictive value for OS was further validated in three independent cohorts. In addition, multivariate analysis demonstrated that PCDH17 expression was an independent prognostic factor for worse OS and DFS. In the subset of patients with CN-AML, however, PCDH17 expression was only independently associated with poorer OS, but not with poorer DFS.
Interestingly, we found patients with low PCDH17 expression harboring FLT3-ITD and NPM1 mutations (the two most common mutations in CN-AML) more often than those with PCDH17 high expression; GSEA analysis further confirmed this observation. In contrast, low PCDH17 expression was found to be negatively associated with TP53 mutations (a mutation occurred frequently in AML with complex aberrant karyotype). These results indirectly reflect the fact that FLT3-ITD mutation was often detected together with NPM1 mutation, whereas TP53 mutation never or rarely co-occurred with the former two mutations [3, 56]. They also indicate a possible pathogenic link between these mutations and PCDH17 alteration. It should be noted that, however, these mutations, which themselves have well-recognized prognostic impacts, are potential confounding factors for outcome prediction. We therefore analyzed the prognostic value of PCDH17 within the mutationally defined subgroups. Low PCDH17 expression still negatively impacts survival in the FLT3-ITD wild-type, NPM1 mutated, and TP53 absent patient subsets, suggesting that PCDH17 expression status could further refine the molecular classification of subsets of AML patients with or without specific mutations.
Finally, we derived PCDH17-associated gene/microRNA expression signatures in AML. One prominent feature was the identification of several EMT-related genes, including E-cadherin repressors (i.e., Gli2, CNTN1, and CTNND2), epithelial markers (CDH1), matrix metalloproteinases (MMP7), and type IV collagens (i.e., COL4A1 and COL4A2). Notably, three EMT-related genes from the signatures (CDH1, BIRC7, and Gli2) were reported by us and others to have profound prognostic significances in AML patients [42, 57, 58]. It was somewhat surprising, however, that EMT modulators could have such implications in leukemia, which, unlike carcinomas, is already endowed with metastatic potential. Recent studies have started to shed some light on this puzzle. For example, ZEB2, a classic EMT regulator, has been shown to regulate early hematopoiesis [59]; its depletion caused aberrant differentiation and reduced proliferation of AML cells, indicating cell context-dependent activities of EMT regulators [60]. Moreover, using an MLL-AF9-induced murine model, Stavropoulou et al. [61] have identified an EMT-like gene signature that was linked to short latency, high aggressiveness, and poor prognosis of AML. This might be explained by the proinvasive and stem-like properties associated with deregulated EMT-like transcriptomes. Future research should further investigate their distinct roles in the context of hematological malignancies.
Another interesting finding was the deregulated expression of Wnt signaling pathway genes, including Wnt antagonists (i.e., DKK1, WNT5A, and SOX17) and Wnt targets (i.e., MMP7, BIRC7, and PROX1). Indeed, PCDH17 was shown to be an inhibitor of the Wnt/β-catenin signaling in breast cancer [30]; the same might also hold true in AML. Thus, it is possible that repressed PCDH17 contributes Wnt pathway activation and enhanced proliferation of leukemic cells, which is consistent with our observation of higher WBC counts and percentages of BM and PB blasts in patients with low PCDH17 expression. In addition to PCDH17, our earlier studies have established the prognostic significances of some other Wnt antagonists in AML, such as SOX17, NKD2, and SFRP genes [31–33], highlighting the importance of deregulated Wnt signaling in leukemogenesis.
For microRNAs co-expressed with PCDH17, some of them were previously reported to be negatively associated with the unfavorable prognostic factors in AML-that is, MDR1 (i.e., miR-381, miR-485, and miR-495) [45], BAALC (miR-10b) [47], and ERG (miR-196a) [48], further supporting the negative prognostic significance of deregulated PCDH17.
Conclusions
In summary, low PCDH17 expression is an independent, poor prognostic factor in AML and has additional prognostic value in stratifying molecularly defined subgroups. Furthermore, low PCDH17 expressers are characterized by a gene-expression signature that reflects the deregulations of EMT- and Wnt pathway-related genes. It is possible that these deregulations contribute to the adverse outcome observed in low PCDH17 patients. Future studies are warranted to confirm this hypothesis.
Additional files
Additional file 1. This file contains Additional Figures (Figures S1–S2) and Additional Tables (Tables S1–S3). It also includes Additional Methods information and R code for regenerating the LSC17 score.
Additional file 2. List of 443 down-regulated probes/genes in LSCs versus normal HSCs.
Additional file 3. List of 863 hypermethylated CpGs in LSCs versus normal HSCs.
Additional file 4. Methylation levels of PCDH17 as measured by targeted bisulfite sequencing.
Additional file 5. Differentially expressed genes between AML patients with low and high PCDH17 expression.
Additional file 6. Differentially expressed microRNAs between AML patients with low and high PCDH17 expression.
Authors’ contributions
JL, JQ, and Z-QD designed the study; Z-JX performed all bioinformatic analyses and wrote the manuscript; JL, JQ, and Z-QD advised Z-JX in bioinformatic analyses and commented on the manuscript; Z-JX, J-CM, J-DZ, and X-MW performed the cellular and molecular experiments; D-MY designed all the primers used in this study; WZ, R-BJ, D-HW, and L-JT contributed to the collection of clinical samples. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets analyzed in this study are available in the following open access repositories:
TCGA, http://www.cbioportal.org, http://gdac.broadinstitute.org.
GEO, https://www.ncbi.nlm.nih.gov/geo/ (GEO accession numbers: GSE12417, GSE24006, GSE30029, GSE42519, GSE63270 and GSE63409).
BloodSpot, http://servers.binf.ku.dk/bloodspot/.
The in-house methylation data was provided as Additional file 4. Other datasets are available from the corresponding author upon reasonable request.
Consent for publication
Written informed consents were obtained from all enrolled individuals prior to their participation.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee and Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81270630), Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project of Jiangsu Province (2015-WSN-115), Zhenjiang Clinical Research Center of Hematology (SS2018009), China Postdoctoral Science Foundation Funded Project (2016M601748), Youth Medical Talents Project of “Ke Jiao Qiang Wei” Project of Jiangsu Province (QNRC2016450), Social Development Foundation of Zhenjiang (SH2016045, SH2017040), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX17_182), Social Development Foundation of Kunshan (KS1624), Clinical Medical Science, Development Foundation of Jiangsu University (JLY20160011), and the Key Medical Talent Program of Zhenjiang City.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- LSC
leukemia stem cell
- AML
acute myeloid leukemia
- TSG
tumor suppressor gene
- HSC
hematopoietic stem cell
- TCGA
The Cancer Genome Atlas
- PCDH17
protocadherin17
- BM
bone marrow
- PB
peripheral blood
- OS
overall survival
- ESCC
esophageal squamous cell carcinoma
- RT-qPCR
real-time quantitative PCR
- FACS
fluorescence-activated cell sorting
- FLASH
fast length adjustment of short reads
- PCA
principal component analysis
- s
5-aza-2′-deoxycytidine
- RQ-MSP
real-time methylation-specific polymerase chain reaction
- DFS
disease-free survival
- FDR
false discovery rate
- GSEA
gene set enrichment analysis
- MNC
mononuclear cell
- PMN
polymorphonuclear cell
- WBC
white blood cell
- CN
cytogenetically normal
- allo-HSCT
allogeneic stem-cell transplantation
- CR
complete remission
- EMT
epithelial-mesenchymal transition
- PRC2
polycomb repressive complex 2
- ESC
embryonic stem cell
- MPP
multipotent progenitor
Contributor Information
Zi-jun Xu, Email: xuzijun1989@hotmail.com.
Ji-chun Ma, Email: majichun606@sohu.com.
Jing-dong Zhou, Email: zhoujingdong1989@qq.com.
Xiang-mei Wen, Email: wenxiangmei@126.com.
Dong-ming Yao, Email: yaodongming1990@163.com.
Wei Zhang, Email: zhangweizwmz@126.com.
Run-bi Ji, Email: haidi_2000@163.com.
De-hong Wu, Email: durwardwu@163.com.
Li-juan Tang, Email: 1121647294@qq.com.
Zhao-qun Deng, Phone: +86-511-88915586, Email: zqdeng2002@163.com.
Jun Qian, Phone: +86-511-88915586, Email: qianjun0007@hotmail.com.
Jiang Lin, Phone: +86-511-88915586, Email: linjiangmail@sina.com.
References
- 1.Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 2.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 3.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, Baty JD, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, Dohner H, Pollack JR. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 6.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Lowenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 7.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, (Eds) Thiele J. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. IARC: Lyon; 2017. p. 141–145.
- 9.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 10.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 12.Jung N, Dai B, Gentles AJ, Majeti R, Feinberg AP. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun. 2015;6:8489. doi: 10.1038/ncomms9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 14.Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 2012;26:344–349. doi: 10.1101/gad.184341.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, Mehra R, Wang X, Ghosh D, Shah RB, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adh Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14:121–134. doi: 10.1038/nrc3647. [DOI] [PubMed] [Google Scholar]
- 18.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Hajj N, Dittrich M, Haaf T. Epigenetic dysregulation of protocadherins in human disease. Semin Cell Dev Biol. 2017;69:172–182. doi: 10.1016/j.semcdb.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T, Inazawa J. Frequent silencing of protocadherin17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31:1027–1036. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 21.Hu X, Sui X, Li L, Huang X, Rong R, Su X, Shi Q, Mo L, Shu X, Kuang Y, et al. Protocadherin17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229:62–73. doi: 10.1002/path.4093. [DOI] [PubMed] [Google Scholar]
- 22.Costa VL, Henrique R, Danielsen SA, Eknaes M, Patricio P, Morais A, Oliveira J, Lothe RA, Teixeira MR, Lind GE, Jeronimo C. TCF21 and PCDH17 methylation: an innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6:1120–1130. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 23.Uyen TN, Sakashita K, Al-Kzayer LF, Nakazawa Y, Kurata T, Koike K. Aberrant methylation of protocadherin17 and its prognostic value in pediatric acute lymphoblastic leukemia. Pediatr Blood Cancer. 2017 doi: 10.1002/pbc.26259. [DOI] [PubMed] [Google Scholar]
- 24.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou JD, Zhang LC, Zhang TJ, Gu Y, Wu DH, Zhang W, Ma JC, Wen XM, Guo H, Lin J, Qian J. Dysregulation of miR-200s clusters as potential prognostic biomarkers in acute myeloid leukemia. J Transl Med. 2018;16:135. doi: 10.1186/s12967-018-1494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He PF, Xu ZJ, Zhou JD. Methylation-associated DOK1 and DOK2 down-regulation: potential biomarkers for predicting adverse prognosis in acute myeloid leukemia. J Cell Physiol. 2018;233:6604–6614. doi: 10.1002/jcp.26271. [DOI] [PubMed] [Google Scholar]
- 27.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST + : architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:1023–1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin X, Xiang T, Mu J, Mao H, Li L, Huang X, Li C, Feng Y, Luo X, Wei Y, et al. Protocadherin17 functions as a tumor suppressor suppressing Wnt/beta-catenin signaling and cell metastasis and is frequently methylated in breast cancer. Oncotarget. 2016;7:51720–51732. doi: 10.18632/oncotarget.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang CY, Lin J, Qian W, Yang J, Ma JC, Deng ZQ, Yang L, An C, Wen XM, Zhang YY, Qian J. Low SOX17 expression: prognostic significance in de novo acute myeloid leukemia with normal cytogenetics. Clin Chem Lab Med. 2014;52:1843–1850. doi: 10.1515/cclm-2014-0487. [DOI] [PubMed] [Google Scholar]
- 32.Li XX, Zhou JD, Zhang TJ, Yang L, Wen XM, Ma JC, Yang J, Zhang ZH, Lin J, Qian J. Epigenetic dysregulation of NKD2 is a valuable predictor assessing treatment outcome in acute myeloid leukemia. J Cancer. 2017;8:460–468. doi: 10.7150/jca.16914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Zhang TJ, Wen XM, Zhou JD, Ma JC, An C, Zhang W, Xu ZJ, Lin J, Qian J. Hypermethylation of secreted frizzled-related proteins predicts poor prognosis in non-M3 acute myeloid leukemia. Onco Targets Ther. 2017;10:3635–3644. doi: 10.2147/OTT.S136502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapin N, Bagger FO, Jendholm J, Mora-Jensen H, Krogh A, Kohlmann A, Thiede C, Borregaard N, Bullinger L, Winther O, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood. 2014;123:894–904. doi: 10.1182/blood-2013-02-485771. [DOI] [PubMed] [Google Scholar]
- 35.Cornelissen JJ, Gratwohl A, Schlenk RF, Sierra J, Bornhauser M, Juliusson G, Racil Z, Rowe JM, Russell N, Mohty M, et al. The European LeukemiaNet AML Working Party consensus statement on allogeneic HSCT for patients with AML in remission: an integrated-risk adapted approach. Nat Rev Clin Oncol. 2012;9:579–590. doi: 10.1038/nrclinonc.2012.150. [DOI] [PubMed] [Google Scholar]
- 36.Yu SJ, Yu JK, Ge WT, Hu HG, Yuan Y, Zheng S. SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are prognosis-related in colorectal cancer. World J Gastroenterol. 2011;17:2028–2036. doi: 10.3748/wjg.v17.i15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurley PJ, Marchionni L, Simons BW, Ross AE, Peskoe SB, Miller RM, Erho N, Vergara IA, Ghadessi M, Huang Z, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc Natl Acad Sci USA. 2012;109:14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagliardi F, Narayanan A, Mortini P. SPARCL1 a novel player in cancer biology. Crit Rev Oncol Hematol. 2017;109:63–68. doi: 10.1016/j.critrevonc.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Dey N, Young B, Abramovitz M, Bouzyk M, Barwick B, De P, Leyland-Jones B. Differential activation of Wnt-beta-catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PLoS ONE. 2013;8:e77425. doi: 10.1371/journal.pone.0077425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan D, Liu L, Gu D. Transcriptional regulation of livin by beta-catenin/TCF signaling in human lung cancer cell lines. Mol Cell Biochem. 2007;306:171–178. doi: 10.1007/s11010-007-9567-6. [DOI] [PubMed] [Google Scholar]
- 41.Karalay O, Doberauer K, Vadodaria KC, Knobloch M, Berti L, Miquelajauregui A, Schwark M, Jagasia R, Taketo MM, Tarabykin V, et al. Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proc Natl Acad Sci USA. 2011;108:5807–5812. doi: 10.1073/pnas.1013456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellbrock J, Latuske E, Kohler J, Wagner K, Stamm H, Vettorazzi E, Vohwinkel G, Klokow M, Uibeleisen R, Ehm P, et al. Expression of hedgehog pathway mediator GLI represents a negative prognostic marker in human acute myeloid leukemia and its inhibition exerts antileukemic effects. Clin Cancer Res. 2015;21:2388–2398. doi: 10.1158/1078-0432.CCR-14-1059. [DOI] [PubMed] [Google Scholar]
- 43.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 44.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y, Mao Z, Shannon MF, Fan JY. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS ONE. 2013;8:e82062. doi: 10.1371/journal.pone.0082062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X, Huang H, Li Z, He C, Li Y, Chen P, Gurbuxani S, Arnovitz S, Hong GM, Price C, et al. MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2012;109:19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwind S, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Becker H, Whitman SP, Wu YZ, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116:5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coskun E, von der Heide EK, Schlee C, Kuhnl A, Gokbuget N, Hoelzer D, Hofmann WK, Thiel E, Baldus CD. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res. 2011;35:208–213. doi: 10.1016/j.leukres.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Guan H, Wei G, Wu J, Fang D, Liao Z, Xiao H, Li M, Li Y. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1334–E1344. doi: 10.1210/jc.2013-1053. [DOI] [PubMed] [Google Scholar]
- 50.Penna E, Orso F, Taverna D. miR-214 as a key hub that controls cancer networks: small player, multiple functions. J Invest Dermatol. 2015;135:960–969. doi: 10.1038/jid.2014.479. [DOI] [PubMed] [Google Scholar]
- 51.Tan H, He Q, Gong G, Wang Y, Li J, Wang J, Zhu D, Wu X. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 2016;48:181–190. doi: 10.3892/ijo.2015.3241. [DOI] [PubMed] [Google Scholar]
- 52.Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 54.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 55.Forsberg EC, Prohaska SS, Katzman S, Heffner GC, Stuart JM, Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 57.Zhang TJ, Zhou JD, Ma JC, Deng ZQ, Qian Z, Yao DM, Yang J, Li XX, Lin J, Qian J. CDH1 (E-cadherin) expression independently affects clinical outcome in acute myeloid leukemia with normal cytogenetics. Clin Chem Lab Med. 2017;55:123–131. doi: 10.1515/cclm-2016-0205. [DOI] [PubMed] [Google Scholar]
- 58.El-Mesallamy HO, Hegab HM, Kamal AM. Expression of inhibitor of apoptosis protein (IAP) livin/BIRC7 in acute leukemia in adults: correlation with prognostic factors and outcome. Leuk Res. 2011;35:1616–1622. doi: 10.1016/j.leukres.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 59.Goossens S, Janzen V, Bartunkova S, Yokomizo T, Drogat B, Crisan M, Haigh K, Seuntjens E, Umans L, Riedt T, et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 2011;117:5620–5630. doi: 10.1182/blood-2010-08-300236. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Mar BG, Zhang H, Puram RV, Vazquez F, Weir BA, Hahn WC, Ebert B, Pellman D. The EMT regulator ZEB2 is a novel dependency of human and murine acute myeloid leukemia. Blood. 2017;129:497–508. doi: 10.1182/blood-2016-05-714493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stavropoulou V, Kaspar S, Brault L, Sanders MA, Juge S, Morettini S, Tzankov A, Iacovino M, Lau IJ, Milne TA, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30:43–58. doi: 10.1016/j.ccell.2016.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. This file contains Additional Figures (Figures S1–S2) and Additional Tables (Tables S1–S3). It also includes Additional Methods information and R code for regenerating the LSC17 score.
Additional file 2. List of 443 down-regulated probes/genes in LSCs versus normal HSCs.
Additional file 3. List of 863 hypermethylated CpGs in LSCs versus normal HSCs.
Additional file 4. Methylation levels of PCDH17 as measured by targeted bisulfite sequencing.
Additional file 5. Differentially expressed genes between AML patients with low and high PCDH17 expression.
Additional file 6. Differentially expressed microRNAs between AML patients with low and high PCDH17 expression.
Data Availability Statement
The datasets analyzed in this study are available in the following open access repositories:
TCGA, http://www.cbioportal.org, http://gdac.broadinstitute.org.
GEO, https://www.ncbi.nlm.nih.gov/geo/ (GEO accession numbers: GSE12417, GSE24006, GSE30029, GSE42519, GSE63270 and GSE63409).
BloodSpot, http://servers.binf.ku.dk/bloodspot/.
The in-house methylation data was provided as Additional file 4. Other datasets are available from the corresponding author upon reasonable request.