Abstract
Purpose:
Ras-GTPase activating proteins (RasGAPs), notably NF1 and RASA1, mediate negative control of the RAS/MAPK pathway. We evaluated clinical and molecular characteristics of NSCLC with RASA1 mutations in comparison with NF1-mutated cases.
Experimental Design:
Large genomic datasets of NSCLC [MSK-IMPACT™ dataset at MSKCC (n=2004), TCGA combined lung cancer dataset (n=1144)] were analyzed to define concurrent mutations and clinical features of RASA1-mutated NSCLCs. Functional studies were performed using immortalized human bronchial epithelial cells (HBECs) and NSCLC lines with RasGAP truncating mutations in RASA1, NF1, or both.
Results:
Overall, approximately 2% of NSCLCs had RASA1 truncating mutations, and this alteration was statistically, but not completely, mutually exclusive with known activating EGFR (p=0.02) and KRAS (p=0.02) mutations. Unexpectedly, RASA1 truncating mutations had a strong tendency to co-occur with NF1 truncating mutations (p<0.001). Furthermore, all patients (16/16) with concurrent RASA1/NF1 truncating mutations lacked other known lung cancer drivers. Knockdown of RASA1 in HBECs activated signaling downstream of RAS and promoted cell growth. Conversely, restoration of RASA1 expression in RASA1-mutated cells reduced MAPK and PI3K signaling. While growth of cell lines with inactivation of only one of these two RasGAPs showed moderate and variable sensitivity to inhibitors of MEK or PI3K, cells with concurrent RASA1/NF1 mutations were profoundly more sensitive (IC50: 0.040μM trametinib). Finally, simultaneous genetic silencing of RASA1 and NF1 sensitized both HBECs and NSCLC cells to MEK inhibition.
Conclusions:
Cancer genomic and functional data nominate concurrent RASA1/NF1 loss of function mutations as a strong mitogenic driver in NSCLC which may sensitize to trametinib.
Keywords: RASA1, NF1, RasGAP, non-small cell lung carcinoma, MEK inhibitor
INTRODUCTION
Lung cancer is the most frequently diagnosed cancer and a leading cause of cancer related mortality worldwide, accounting for nearly 1.4 million deaths each year (1, 2). It has become standard practice to profile non-small cell lung carcinomas (NSCLCs), especially adenocarcinomas, for recurrent targetable alterations such as mutant EGFR, MAP2K1, BRAF, NRAS, or rearranged ALK, RET, ROS1, NTRK1, NTRK2, NTRK3, or amplified ERBB2 or MET exon 14 skipping (3–9), which, as strong mitogenic drivers, are mutually exclusive. Routine clinical genomic profiling of lung adenocarcinoma to comprehensively detect all known targetable alterations has been recently shown to result in use of matched therapy in a substantial proportion of patients (10). Despite these encouraging data, not all mitogenic drivers are readily targetable (e.g. KRAS, NRAS) and there remain at least 12% of patients with lung adenocarcinoma with no clear mitogenic driver (10).
RAS proteins are predominantly localized to the inner surface of the plasma membrane and act as binary switches cycling between an inactive RAS-GDP and an active RAS-GTP. GTP-bound RAS is able to bind and activate effector pathways, including the RAF/mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT signaling cascades, which play central roles in cell growth and survival. Ras-GTPase activating proteins (RasGAPs), notably NF1 and RASA1, terminate the active state of RAS by stimulating GTP hydrolysis, thereby mediating negative control of the RAS pathway (11). NF1 and RASA1 are ubiquitously expressed; the former is abundant in neurons, Schwann cells, astrocytes, and leukocytes (12); and the latter plays an important role as a regulator of blood and lymphatic vessel growth (13). Accordingly, germline mutations of NF1 or RASA1 cause neurofibromatosis type I and capillary malformation-arteriovenous malformation, respectively. Currently, various RasGAPs with overlapping patterns of tissue distribution but with nonredundant functions have been identified (14, 15), and the tumor-suppressive functions of RasGAPs have been shown in several cancer types such as neuroblastoma, glioblastoma, melanoma, prostate cancer, and breast cancer (16–24). With respect to lung cancer, NF1 inactivating mutations are enriched in NSCLCs lacking KRAS alterations; however, they are not completely mutually exclusive (25, 26). Furthermore, the clinical and molecular characteristics of NSCLC with RASA1 mutations have not been defined. The Cancer Genome Atlas (TCGA) studies showed that RASA1 and NF1 mutations are frequently detected in both lung adenocarcinoma (1.7% for RASA1; 11% for NF1) and squamous cell carcinoma (4% for RASA1; 11% for NF1) (25, 26).
The link between RasGAPs and RAS signaling, and clinical genomic data suggest that RASA1 and NF1 loss of function mutations may be mitogenic drivers in NSCLCs. Recent studies have demonstrated that inhibition of MAPK signaling can be effective in RAF/MAPK activated cancers, such as BRAF-mutant melanomas. Trametinib, which acts downstream of RAS to suppress signaling through the MAPK cascade, is approved by the FDA for the treatment of BRAF V600E-mutant melanomas. Trametinib has superior pharmacological properties compared with other MEK inhibitors because it impairs feedback reactivation of ERK (27), suggesting that trametinib may have greater clinical activity in RAF/MAPK activated tumors. Importantly, NSCLCs with RASA1 or NF1 inactivating mutations may also be sensitive to this strategy of downstream inhibition because of the negative regulation of RAS activity associated with RasGAPs.
Here, we evaluate the clinical and molecular characteristics of NSCLCs with RASA1 mutations in comparison with NF1-mutated cases. Our findings identify concurrent RASA1/NF1 loss of function mutations as a strong mitogenic driver in NSCLC, and suggest that therapeutic strategies for RAF/MAPK activated tumors may also have efficacy in patients whose tumors show this distinctive genotype.
MATERIALS AND METHODS
MSK-IMPACT assay
The MSK-IMPACT assay was conducted as previously described (28, 29). In brief, DNA from formalin-fixed paraffin-embedded primary or metastatic lung tumors and patient-matched normal blood samples was extracted and sheared. Barcoded libraries from tumor and normal samples were captured, sequenced, and subjected to a custom pipeline to identify somatic mutations. Sequencing consisted of deep sequencing of all exons and selected introns of a custom 341 cancer-associated gene panel, with an updated panel containing 410 genes. All exons tested had a minimum depth of coverage of 100×. The alterations detected were all confirmed to be somatic, because mutations were called against the patient’s matched normal sample.
Cell lines and cell culture
EPLC272H and LCLC103H were purchased from German Resource Center for Biological Materials (DSMZ). RERFLCKJ and RERFLCAI were purchased from RIKEN BRC Cell Bank, and ABC-1 was purchased from JCRB Cell Bank. The human bronchial epithelial cells were immortalized with CDK4 and hTERT (HBEC3KT cell line) and were obtained from Dr. John Minna (UT South Western) (30). Other cells were purchased from the American Type Culture Collection. HBEC3KT cells were cultured in keratinocyte serum free media supplemented with EGF and 2% bovine pituitary extract. All cell lines were cultured in RPMI-1640 supplemented with 10% FBS, 1% penicillin-streptomycin in 5% CO2 atmosphere at 37°C. All cell lines were routinely tested for mycoplasma and were found to be negative and the NF1 and RASA1 status of all cell lines was confirmed to be consistent with the data from the CCLE.
Plasmids
The lentiviral pLKO.1 MISSION shRNA constructs targeting RASA1, KRAS or non-target control (NT) were purchased from Sigma-Aldrich. The lentiviral full-length RASA1 expressing construct (#Ex-T0725-Lv102) and empty vector control were purchased from GeneCopoeia. For packaging, we used psPAX2 (Addgene) and VSV-G/pMD2 (Addgene).
Western-blot assays
RAS-GTP levels were detected using an Active Ras detection kit, following the manufacturer instructions (Cell Signaling). Cells were lysed in RIPA lysis buffer (ThermoFisher Scientific). Lysates were denatured in 2× sample buffer at 55 °C for 5 min, resolved on 4–12% NuPAGE gels (Invitrogen) and transferred onto PVDF (polyvinylidene fluoride) membranes. Membranes were blocked in 3% BSA in TBST buffer (TBS with Tween-20) for 1 h at room temperature and probed with primary antibodies (Table S1). Bound antibodies were detected with peroxidase-labeled goat anti-mouse IgG or goat anti-rabbit IgG (R&D Systems) and developed with ECL western blotting detection reagents (GE Healthcare).
Phospho-kinase array analysis
We used a human Phospho-Kinase Array Kit containing duplicate validated controls and capture antibodies specific for phosphorylation of 43 human kinases (R&D Systems) to simultaneously detect relative kinase phosphorylation levels, according to the manufacturer’s protocol. A total of 5×106 cells were plated in 10 cm dishes, then deprived of serum for 24 hours. In brief, the array membranes were blocked, incubated with each cell lysate overnight at 4 °C, washed, incubated with biotinylated antibodies for 2 hours at room temperature, washed again, incubated with streptavidin-HRP for 30 minutes at room temperature, washed again and developed with ECL western blotting detection reagents. The kinase spots were visualized with X-ray films (Ewen-Parker X-ray). The average pixel densities of duplicate spots were determined using the ImageJ software (http://imagej.nih.gov/ij/).
Phospho-RTK and apoptosis array analyses
A human Phospho-RTK Array Kit and a Human Apoptosis Antibody Array Kit containing duplicate validated controls and capture antibodies specific for phosphorylation of 49 RTKs or 35 apoptosis-related proteins (R&D Systems) were used to simultaneously detect the relative kinase phosphorylation levels and apoptosis-related protein levels, respectively. A total of 5×106 cells were plated in 10 cm dishes, then deprived of serum for 24 hours. In a Human Apoptosis Antibody Array, cells were treated with inhibitors for 96 hours, and after treatment, cells were lysed in lysis buffer. Both phospho-RTK and apoptosis array analyses were performed according to the manufacture’s protocol. The spots were visualized with X-ray films (Ewen-Parker X-ray). The average pixel densities of duplicate spots were determined using the ImageJ software (http://imagej.nih.gov/ij/).
Cell proliferation assay
A total of 2.5×104 cells were plated in six-well plates. Cells were counted the day after plating and at 48 h intervals using a Countess Automated Cell Counter (Invitrogen).
Cell Viability Assay
Cell viability of HBEC3KT with RASA1 knock down or non-target control was assessed using a CellTiter-Blue® Cell Viability Assay (Promega). Cells were plated at a density of 3,000/well in a 96-well plate in medium containing 10% FBS, and incubate for 3 days. Viable cell numbers were determined using the CellTiter-Glo assay kit according to the manufacturer’s protocol (Promega). Each assay consisted of duplicate samples and the experiments were repeated in triplicate at minimum. Data were expressed as relative luciferase activity (percentages relative to control cells).
Growth inhibition Assay
Cells were seeded in white walled 96-well plates at a density of 1,500 to 7,000 cells per well and exposed to drug alone or combination the following day. At 120 hours after drug addition, alamarBlue® Reagent (Promega) was added, and fluorescence was measured on a Spectramax spectrophotometer (Molecular Devices). The data were fitted to dose-response Inhibition and IC50 was calculated using Graphpad Prism® software (version 7.0a). All inhibitors were obtained from Selleckchem.
Statistical analysis
Data are expressed as mean ± SEM unless indicated otherwise. Statistical significance was determined by analysis of variance (ANOVA) using Dunnett’s multiple-comparison post-test with GraphPad Prism® software version 7.0a unless otherwise noted.
RESULTS
RASA1 and NF1 truncating mutations preferentially co-occur in patients with NSCLC
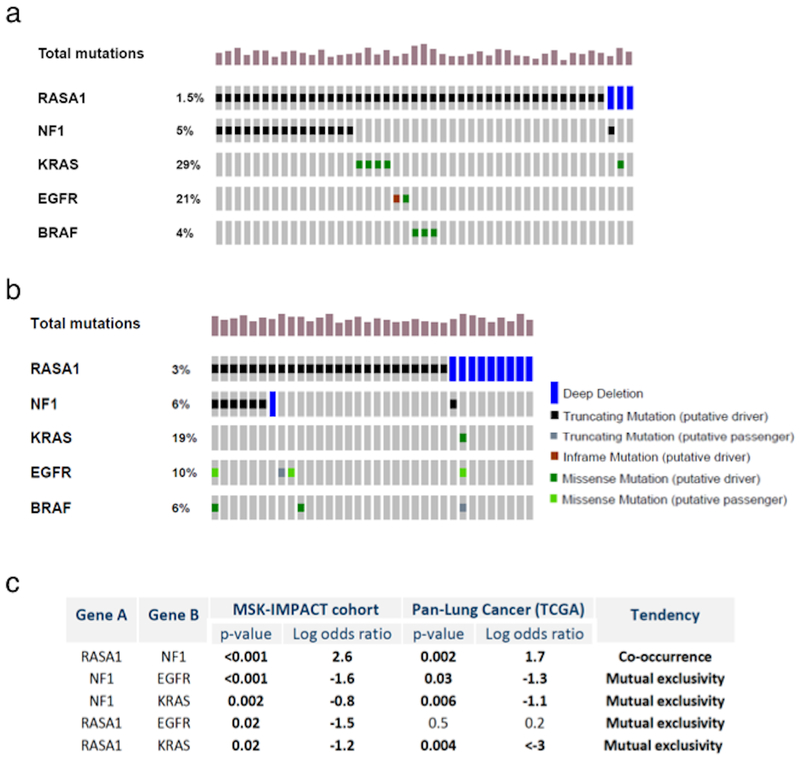
Genomic data from the MSK-IMPACT clinical cohort consisting of 2900 patients with NSCLC (data from 01/01/2014 to 9/24/2017) revealed truncating mutations of RASA1 and NF1 in 1.5% and 5% of cases, respectively (Figure 1a). Analysis performed through the cBioPortal (31) demonstrated a statistically significant tendency toward co-occurrence of RASA1 and NF1 truncating mutations or homozygous deletion (p < 0.001; Figure 1c). In contrast, the presence of these two mutations, seen in 16/2900 patients (0.6%), was completely mutually exclusive with known activating EGFR or KRAS mutations (p<0.001 and 0.002, respectively; Figure 1c), as well as other common lung cancer mitogenic drivers such ALK/RET/ROS1 fusions, ERBB2 exon 20 insertions, BRAF mutations, and MET exon 14 skipping mutations (not shown). Among other common lung cancer tumor suppressors, cases with RASA1 truncating mutations were significantly enriched for TP53 alterations (p<0.001) while STK11 and CDKN2A alterations were not over-represented (not shown). RASA1 and NF1 mutation rates were slightly higher in the TCGA combined squamous carcinoma and adenocarcinoma dataset (32) (n = 1144 patients; Figure 1b), where truncating mutations (or homozygous deletions) were identified in 3% and 6% of NSCLC, respectively (Figure 1b), likely due to a higher proportion of squamous cell carcinoma (42% in the TCGA analysis vs 10% in the MSKCC cohort). Analysis of the TCGA data also confirmed the significant tendency toward co-occurrence of truncating mutations (or homozygous deletions) of RASA1 and NF1, seen in 8/1144 TCGA cases (0.7%), and the mutual exclusivity of their co-inactivation with canonical EGFR and KRAS mutations (Figure 1C) and other known mitogenic drivers (not shown). An integrated analysis of copy number data, mRNA expression data, and mutations was performed using the TCGA squamous carcinoma and adenocarcinoma datasets (25,26). This confirmed that truncating mutations and deep (likely homozygous) deletions of both genes are associated with low mRNA expression levels (Figure S1). The mutation details, smoking histories, and basic demographic and pathologic data for 24 patients (16 MSK-IMPACT, 8 TCGA) with co-inactivation of RASA1 and NF1 are provided in Table S2.
Figure 1. RASA1 truncating mutations display a strong tendency to co-occur with NF1 truncating mutations.
Distribution and frequency of RASA1 and NF1 truncating mutations in non-small cell lung carcinoma (NSCLC) as identified by MSK-IMPACT dataset (a) and TCGA combined lung cancer dataset (b). Summary table of p-values and log odd ratio showing significant tendency for co-occurrence of RASA1 and NF1 mutations and mutual exclusivity with EGFR and KRAS alterations for both genes in the MSK-IMPACT NSCLC cohort (c).
Tumor suppressive function of RASA1 in lung tumorigenesis in vitro
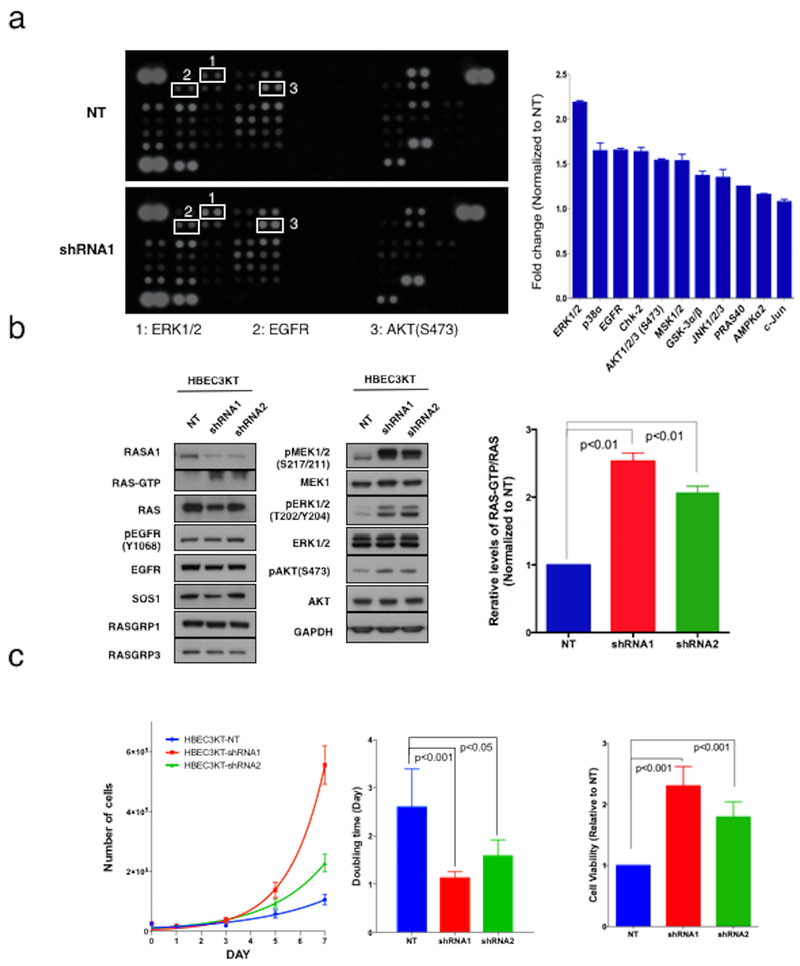
To evaluate the function of RASA1 in human lung cells, we suppressed RASA1 expression with lentiviral shRNA in immortalized human bronchial epithelial cells (HBEC3KT cells). We first analyzed the effect of RASA1 knockdown on the phosphorylation state of 43 proteins and total amounts of 2 related proteins using a human phospho-kinase array, which revealed a significant increase in the phosphorylation of proteins associated with MAPK and PI3K signaling in HBEC3KT cells (Figure 2a). We further tested RAS-GTP levels, phosphorylation of AKT and ERK, as well as expression of various nucleotide exchange factors (GEFs) including SOS1, RasGRP1 and RasGRP3 since the activity of RAS is controlled by the ratio of bound GTP to GDP, and these processes are regulated by RasGEFs and RasGAPs. RASA1 knockdown activated downstream signaling of RAS via an increase in RAS-GTP (Figure 2b), which promoted cell growth in HBEC3KT cells (Figure 2c). Interestingly, there was more prominent elevation in pERK levels compared with pAKT upon RASA1 knockdown in HBEC3KT cells.
Figure 2. Tumor suppressive function of RASA1 in lung tumorigenesis in vitro.
(a) Phospho-Kinase array showing RASA1 knockdown effects on the phosphorylation state of multiple kinases in HBEC3KT cells (left panel). Average of the mean pixel densities in duplicate spots were measured and normalized to non-target shRNA control (NT). The bar graph shows significantly increased (p<0.01) phosphoproteins, such as ERK1/2 and AKT1/2 (right panel). (b) In HBEC3KT cells that retain RASA1 expression, RNAi-mediated suppression of RASA1 (shRNA1 and shRNA2) resulted in the activation of RAS and its downstream signaling (left panel). Average of the mean pixel densities of RAS-GTP in three independent experiments are shown for each shRNAs. The relative levels of RAS-GTP to RAS were normalized to that of non-targeting shRNA (NT). Inactivation of RASA1 induced a statistically significant increase in RAS-GTP level (right panel). (c) Growth curves of HBEC3KT cells with RNAi-mediated suppression of RASA1. Data points show triplicate averages ± SEM (left panel). Inactivation of RASA1 induced statistically significant decreases in doubling times (HBEC3KT-NT: 2.6±0.8 days; HBEC3KT-shRNA1: 1.1±0.1days; HBEC3KT-shRNA2: 1.6±0.83 days) (center panel). Cell viability of HBEC3KT with RASA1 knockdown or non-target control (NT) was assessed using a CellTiter-Blue® Cell Viability Assay. Inactivation of RASA1 induced a statistically significant increase in cell viability (right panel).
Loss of RasGAPs expression in NSCLC lines with RasGAP truncating mutations
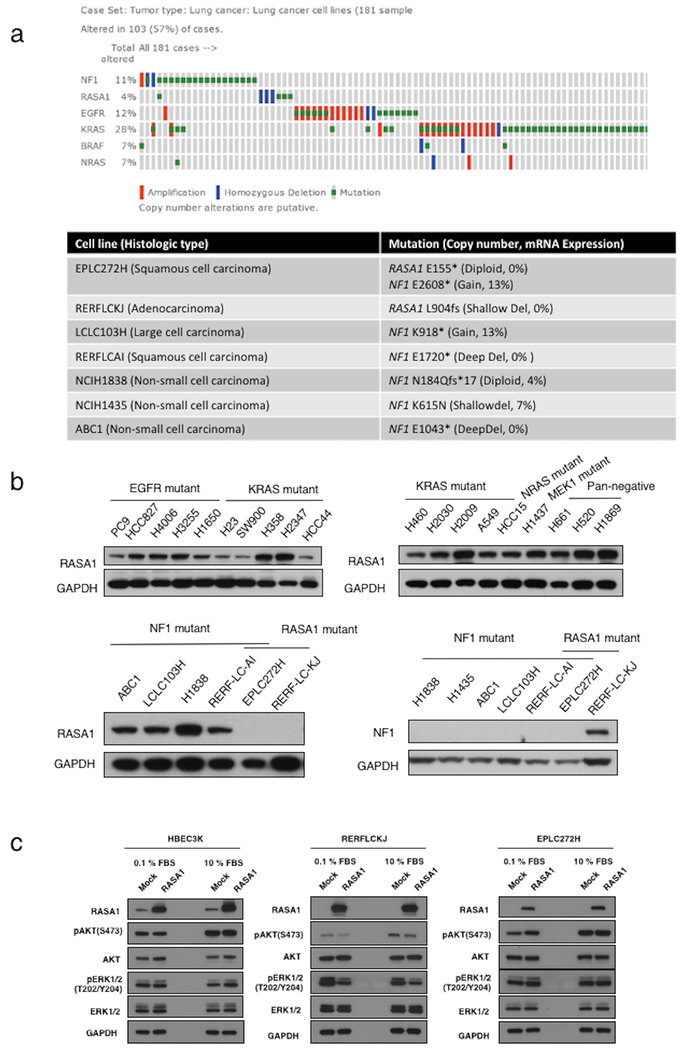
To identify human lung cancer cell lines with endogenous loss of RASA1, we evaluated RASA1 expression in a panel of 26 lung cancer cell lines. Only two cell lines exhibited no RASA1 expression, both of which harbored RASA1 truncating mutations (pE155* in EPLC272H cells and pL904fs in RERFLCKJ cells). EPLC272H cell also contained an NF1 truncating mutation (pE2608*) and exhibited no NF1 expression (as did other cell lines with NF1 truncating mutations) (Figure 3a, b). To confirm a functional role for RASA1 inactivation in human lung cancers, we reintroduced RASA1 in two RASA1-null cell lines, EPLC272H, and RERFLCKJ. Ectopic RASA1 expression suppressed signaling through the MAPK and AKT pathways in RERFLCKJ cells; there were stronger reductions in pERK levels compared with pAKT levels. In contrast, no effect was detected in either HBEC3KT cells that express RASA1 endogenously, suggesting that its supraphysiologic overexpression does not further suppress downstream signaling or EPLC272H cells that lack both RASA1 and NF1 expression, suggesting that restoration of RASA1 function alone may not be sufficient to restore normal downstream signaling in lung cancer cells with co-inactivation of NF1. (Figure 3c). To verify further the dependency of RASA1-mutant cells on RAS signaling, we carried out shRNA-mediated knockdown experiments. As expected, KRAS knockdown with 2 different shRNAs led to growth inhibition in both EPLC272 and RERFLCKJ cells (data not shown).
Figure 3.
Identification of RASA1-mutated lung cancer cell lines in the CCLE dataset. (a) RASA1-mutated lung cancer cell lines and 20 NF1-mutated lung cancer cell lines were identified. (b) RASA1 and NF1 protein levels in a panel of 26 human lung cancer cell lines, as determined by western blotting. Lung cancer cell lines commonly retain RASA1 expression; the exceptions were EPLC272H and RERFLCKJ. (c) Western blots of phosho (p)-AKT and pERK levels in HBEC3KT (left), EPLC272H (center), and RERFLCKJ (right) cells following expression of control vector or RASA1. Restoration of RASA1 expression resulted in reduced signaling of MAPK pathway only in RASA1-mutated cells (RERFLCKJ), but not in RASA1/NF1 co-mutated NSCLC lines (EPLC272H).
Modulation of MAPK and PI3K/AKT signaling pathways upon MEK or PI3K inhibition
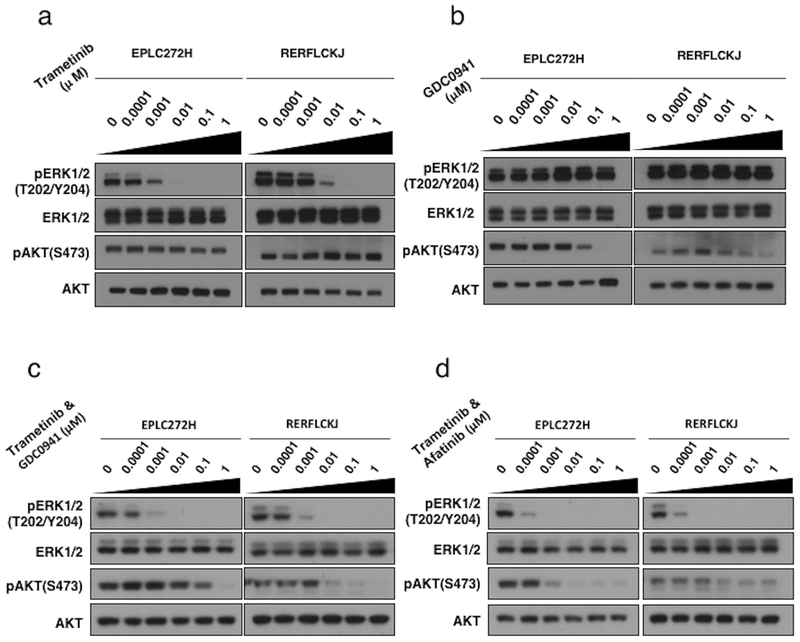
Given that the loss of the RASGAP protein NF1 has been shown to lead to sensitivity to inhibition downstream of RAS (15, 33), we next examined whether RASA1 loss has a similar effect. Specifically, to assess how RASA1 mutated NSCLC lines respond to the MEK inhibitor trametinib (GSK1120212), we analyzed effects of trametinib on the signaling of MAPK and PI3K/AKT pathways by Western blot in comparison with PI3K inhibitor (GDC0941). In both RASA1 mutated NSCLC lines (EPLC272H and RERFLCKJ), trametinib induced a potent dose-dependent decrease of pERK1/2 levels. In contrast, GDC0941 reduced pAKT levels only at higher doses. Furthermore, RASA1/NF1 co-mutated cells (EPLC272H) were more sensitive to trametinib than RASA1-mutated/NF1 wild type cells (RERFLCKJ), showing a 10-fold decrease in pERK levels (Figure 4a, b). We further analyzed the co-administration of MEK inhibitor with inhibitors of PI3K or EGFR (afatinib). This showed that combining trametinib and GDC0941 induced a dose-dependent decrease of both pERK and pAKT levels (Figure 4c). Furthermore, inhibition of pERK levels was more prominent when cells were exposed to a combination of trametinib and afatinib, showing a greater than 10-fold decrease in pERK, compared to trametinib alone. (Figure 4d).
Figure 4. Inhibition of MAPK and PI3K/AKT signaling in RASA1-mutated NSCLC cell lines.
Inhibition of phosphorylation of ERK or AKT in EPLC272H and RERFLCKJ cells by trametinib (a), GDC0941, co-administration of trametinib and GDC0941, and co-administration of trametinib and afatinib (d).
Profound dependence of growth of RASA1/NF1 co-mutated NSCLC lines on MAPK signaling
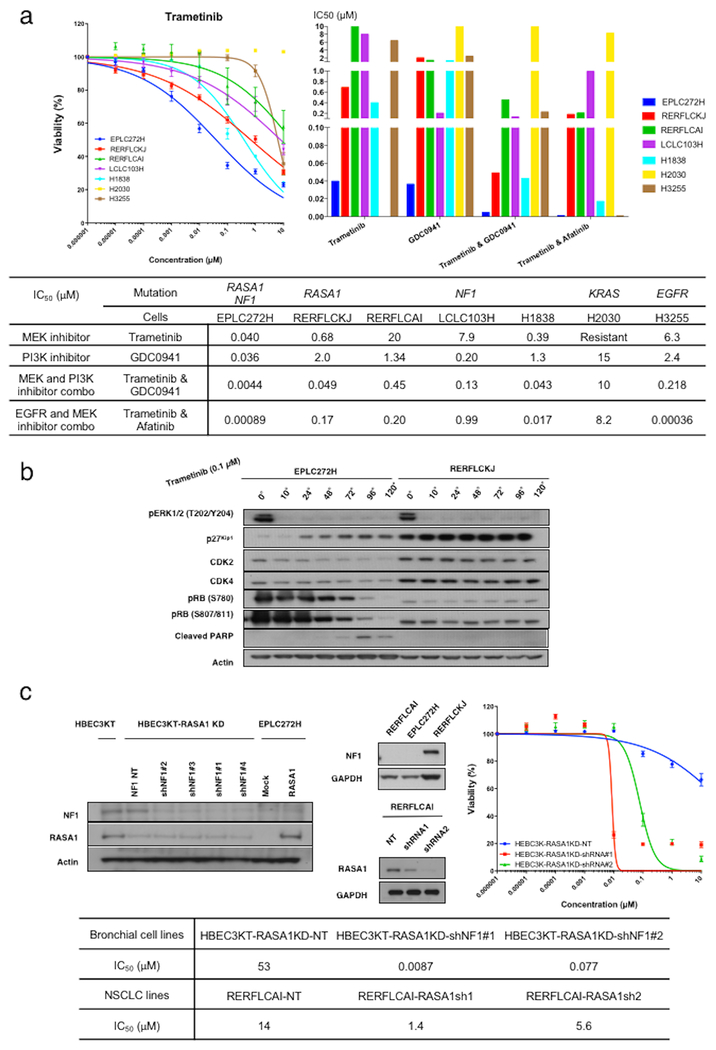
Since previous studies (15–21) and our results show that loss of RasGAPs including RASA1 trigger activation of the MAPK and PI3K/AKT signaling through RAS, we next tested the sensitivity of RASA1 mutated NSCLC cells against variety of kinase inhibitors with in vitro cell growth inhibition assays. Kinase inhibitors included MEK inhibitor (trametinib), PI3K inhibitor (GDC0941), and EGFR inhibitor (afatinib). A panel of NSCLC cells were cultured in vitro in the presence of increasing concentrations of kinase inhibitors for 120 hours to determine the half maximal inhibitory concentration (IC50). The panel included RASA1/NF1 co-mutated (EPLC272H), RASA1-mutated (RERFLCKJ), NF1-mutated (RERFLCAI, LCLC103H, H1838), KRAS mutated (H2030) and EGFR-mutated (H3255) NSCLC lines. KRAS-mutated cells were resistant to all inhibitors examined. EGFR mutated cells were moderately sensitive to combination of MEK and PI3K inhibitors, and sensitive to combined inhibition of MEK and EGFR, as expected. NSCLC lines with only RASA1 or NF1 mutation (but not both) showed moderate and variable sensitivity to all inhibitors examined. Surprisingly, RASA1/NF1 co-mutated cells showed profound sensitivity to the same inhibitors (IC50: 0.040μM trametinib; 0.036μM GDC0941; 0.0044μM combination of GSK1120201/GDC0941; 0.00089 μM combination of trametinib/afatinib) (Figure 5a).
Figure 5. MAPK pathway as a potential target in RASA1/NF1 co-mutated NSCLCs.
(a) A panel of NSCLC lines including RASA1/NF1-co-mutated (EPLC272H), RASA1-mutated (RERFLCKJ), NF1-mutated (RERFLCAI, LCLC103H, and H1838), KRAS-mutated (H2030) and EGFR-mutated (H3233) NSCLC lines were grown in the presence of each drug, and IC50 values were determined using AlamarBlue assay. KRAS and EGFR mutated NSCLC lines were used as control. Error bars denote SEM. The RASA1/NF1-co-mutated cell line EPLC272H was the most sensitive to MEK (trametinib) and PI3K (GDC0941) inhibitors. RERFLCKJ cells were moderately sensitive to MEK inhibitor. Among 3 NF1 mutated NSCLC cells, only H1838 cells were moderately sensitive to MEK inhibitor. NF1 mutated NSCLC cells showed similar sensitivity to PI3K inhibitor. Overall, all cells except KRAS-mutated NSCLC lines showed increased sensitivity to co-administration of trametinib with either a PI3K or EGFR inhibitor. (b) Western blot analyses were performed to analyzed the expression level of key cell-cycle and apoptotic signaling molecules in EPLC272 and RERFLCKJ cells. Elevated p27 and cleaved PARP, and decreased CDK2, CDK4, and pRB were observed only in EPLC272H cells in a time-dependent manner. (c) HBEC3KT cells with RASA1 knockdown were stably infected with non-targeting shRNA (NT) or NF1-targeting sequences (shNF1#1 to #4). Conversely, NF1-mutated RERFLCAI cells were stably infected with non-targeting shRNA (NT) or RASA1-targeting sequences (shRNA1 and shRNA2). Knockdown by each shRNA was confirmed using Western blotting (upper, right and center). The simultaneous inactivation of RASA1 and NF1 induced sensitivity to trametinib in both cell lines (upper, right and lower).
MAPK inhibition induces cell cycle arrest and apoptosis in RASA1/NF1 co-mutated NSCLC lines
To determine whether or not the decrease in cell survival was due to apoptosis and cell cycle arrest, we investigated the expression level of key cell-cycle and apoptotic signaling molecules over time after administration of trametinib (0.1 μM) in both EPLC272H and RERFLCKJ cells by western blot. (Figure 5b) Our experiments provide evidence of cell cycle arrest (e.g. elevated p27, and decreased CDK2 and CDK4, and subsequently decreased pRB) after 48 hours and evidence of apoptosis (e.g. cleaved PARP) after 72 hours in EPLC272H cells, but not in RERFLCKJ cells, suggesting that inhibition of MAPK signaling is sufficient to induce cell-cycle arrest and apoptosis in RASA1/NF1 co-mutated NSCLC lines, but not in RASA1-only mutated NSCLC lines. (Figure 5b) To further investigate the involvement of apoptotic signaling molecules following trametinib and afatinib co-administration in EPLC272H and RERFLCKJ cells, we performed apoptosis array hybridizations using a human apoptosis array from R&D Systems. This showed that a number of apoptotic signaling proteins, such as cleaved caspase-3 are modulated following co-administration of trametinib and afatinib in both EPLC272H and RERFLCKJ cells (Figure S2a). Next, the time course of PARP cleavage after administration of combination of trametinib and afatinib was investigated in both EPLC272H and RERFLCKJ cells by western blot. Our results showed that cleaved PARP was increased in both cell lines after 48 hours (Figure S2b). These data suggest that single agent trametinib may be sufficient to induce cell cycle arrest and apoptosis in RASA1/NF1 co-inactivated NSCLC, but not in lung cancers where one of these two RasGAPs is still functional but this will need to be confirmed if additional RASA1/NF1 co-inactivated NSCLC cell lines become available.
Simultaneous knockdown of RASA1 and NF1 induces sensitivity to trametinib
Since our analysis of large cancer genomic datasets showed that RASA1 mutations tend to co-occur with NF1 mutations in NSCLCs, and our studies of native NSCLC cell lines showed that the RASA1/NF1 co-mutated EPLC272H cells are highly sensitive to MEK inhibition, we further investigated whether experimental inactivation of both RASA1 and NF1 confers sensitivity to trametinib in other cell lines. We first performed cell-based sensitivity profiling using HBEC3KT cells stably expressing both RASA1 and NF1 shRNAs. Trametinib treatment markedly reduced growth of HBEC3KT cells with both RASA1 and NF1 knockdown compared with cells with only RASA1 knockdown (Figure 5c). Furthermore, we found that trametinib could inhibit the growth of NF1 mutated NSCLC cells (RERFLCAI) when RASA1 is also knockdown (Figure 5c). Thus, the simultaneous loss of RASA1 and NF1 induces sensitivity to trametinib both in immortalized bronchial epithelial cells and NSCLC lines.
Clinicopathological features of patients with concurrent RASA1/NF1 truncating mutations
Finally, we reviewed the characteristics of patients whose NSCLCs show co-inactivation of RASA1 and NF1. Patients with truncating mutations in both RASA1 and NF1 (or deep deletion of RASA1 and truncating mutation of NF1, n=1) had a mean and median age of 69 (range 46-81), 50% were female and consisted entirely of former or current heavy smokers (Table S3). RASA1-only mutated tumors were more often squamous cell carcinoma (63%; Chi-square p<0.0001) than tumors with NF1 alterations only which were predominantly adenocarcinomas (81%); 50% of tumors with RASA1/NF1 co-mutations were squamous. Other clinical and pathological features did not differ between these three subgroups (Table S3). Focusing on patients with longer follow up data, survival analysis was performed on the first 109 MSK-IMPACT patients with inactivating alterations in one or both genes, a cohort with a median follow up of 13.3 months. Patients with RASA1, NF1 and RASA1/NF1 co-mutated tumors, had a median overall survival of 64, 39 and 18 months respectively, suggesting that the latter may be an aggressive subset of NSCLC but these differences did not reach statistical significance (Figure S3).
DISCUSSION
NSCLCs, particularly lung adenocarcinomas, have emerged as striking examples of cancers that have been molecularly redefined by the discovery of activating oncogene mutations that serve as strong mitogenic drivers and, as such, are largely mutually exclusive. While inactivating mutations in the NF1 and RASA1 tumor suppressors are expected to increase MAPK signaling and cell proliferation be mitogenic, it has been less clear how they fit into the “molecular pie chart” classification of lung adenocarcinoma as they are, individually, not as consistently mutually exclusive with strong mitogenic drivers such as EGFR and KRAS mutations (10, 25, 26, 32). Approximately 1.5% and 3% of NSCLCs harbor RASA1 truncating mutations in the MSK-IMPACT and TCGA cohorts, respectively (32). Our analysis of these genomic data reveals that RASA1 truncating mutations display a strong tendency to co-occur with NF1 truncating mutations, and that concurrent RASA1/NF1 truncating mutations are completely mutually exclusive with other strong mitogenic drivers, suggesting that their redundant functions as RasGAPs provide a selective pressure for dual inactivation in order to achieve a level of activated RAS signaling that could substitute for other lung cancer drivers such as KRAS or EGFR mutations, as well as other known mitogenic drivers in lung cancer. Strong mitogenic drivers are expected to be early, “truncal” events in studies of NSCLC evolution. Thus, it is notable that a recent analysis of NSCLC evolution in 100 patients found that some RASA1 mutations occurred early in evolution, and one patient (1%) had both RASA1 and NF1 inactivation as early events in the absence of another strong driver (34). Furthermore, there is also evidence for increased co-occurrence of NF1 mutations with alterations of other RasGAPs (including RASA1) and RasGEFs across a wide range of human cancers (35). Finally, whereas T cell-lineage knockouts of NF1 or RASA1 show only a minor phenotype and do not result in T cell acute lymphoblastic leukemia/lymphoma (T-ALL), their combined deletion leads to the development of T-ALL (36), suggesting that, in some cellular settings, loss of both NF1 and RASA1 is needed to produce the high levels of signaling downstream of RAS necessary for malignant transformation. Such data, along with our analysis of the cancer genomic data nominate concurrent RASA1/NF1 truncating mutations as a strong mitogenic driver in NSCLC, a nomination for which we now provide functional data in NSCLC cell lines.
We demonstrate that loss of RASA1 expression activates downstream signaling of RAS via an increase of RAS-GTP, which promotes cell growth in immortalized bronchial epithelial cells. Conversely, restoration of RASA1 expression reduces signaling through the MAPK and PI3K pathways in NSCLC lines harboring RASA1 truncating mutation. Moreover, NSCLC lines with concurrent RASA1/NF1 mutations show more profound sensitivity to inhibitors of MEK, both in western blots and cell growth inhibition assays, as compared to only RASA1 mutated lines. Likewise, simultaneous knockdown of RASA1 and NF1 enhances sensitivity to the MEK inhibitor trametinib, suggesting that more complete cellular loss of RasGAP function induced sensitivity to inhibitors which act downstream of RAS.
In comparison with patients with only RASA1 or NF1 mutated tumors, RASA/NF1 co-mutated patients showed similar demographic features and smoking histories (Table S3). However, there was a significant difference seen in the histologic type observed in each cohort: RASA1 or NF1 truncating mutations were enriched among squamous cell carcinomas and adenocarcinomas, respectively, whereas RASA1/NF1 co-mutated tumors were more evenly distributed. Targeted therapy is commonly used in patients with select driver-positive lung adenocarcinomas, but there remains a dearth of effective targeted agents for molecular subsets of lung squamous cell carcinoma. This study indicates that inhibition of MEK in NSCLC lines harboring concurrent RASA1/NF1 truncating mutations dramatically inhibit cell growth in vitro. Notably, these findings have potential therapeutic implications for patients with not only lung adenocarcinoma but also squamous cell carcinoma with concurrent RASA1/NF1 truncating mutations, providing a preclinical rationale for the evaluation of MEK inhibitors in this molecular subset of NSCLC that lacks other targetable alterations.
The preferential co-inactivation of NF1 and RASA1 that we have identified suggests that partial redundancy between NF1 and RASA1 requires that both be inactivated to obtain an oncogenic level of MAPK pathway activation in lung cancer. Whether this concept of co-inactivation of negative regulators of a pathway leading to striking sensitivity to pathway inhibition extends beyond these RASGAPs is presently unclear, but there are mounting data supporting its broader relevance. For instance, detailed genomic analysis of a bladder cancer patient with exceptional response to an mTORC1 inhibitor revealed nonsense mutations in TSC1 and NF2, both negative regulators of mTORC1 (37). Likewise, a recent analysis in malignant pleural mesothelioma identified a small subset with co-inactivation of LATS2 and NF2 that is associated with increased sensitivity to mTOR inhibition (38). Finally, a recent study found that cell lines with co-inactivation of ARID1A and RNF43, both alterations known to activate beta-catenin signaling, are exquisitely sensitive to aurora kinase inhibition (39). Thus, in several settings or pathways, the specific co-occurrence of loss-of-function mutations may lead to vulnerabilities that can be exploited as potential therapeutic targets.
In summary, our cancer genomic and functional data nominate concurrent RASA1/NF1 loss of function mutations as a strong mitogenic driver in NSCLC. Patients whose tumors show this distinctive genotype should be considered for trials of MEK inhibitors. Because of the relatively high background mutation rate of smoking-associated NSCLC, we have focused our data analyses on truncating mutations of RASA1 and NF1, reasoning that a high proportion of missense mutations in these two genes may be passenger mutations of uncertain functional significance and would add noise to the genomic data analysis. However, it is likely that a subset of missense mutations in these genes, for instance in their RasGAP domains, may be functionally inactivating, potentially expanding the size of this small but targetable molecular subset of NSCLC. Finally, while targetable alterations are generally much more common in never-smokers, it is notable that patients whose NSCLC harbor concurrent RASA1/NF1 loss of function mutations, along with patients with MAP2K1 activating mutations (40), represent strongly smoking-associated subsets that are potentially targetable, opening new therapeutic avenues for some patients with smoking-induced NSCLC.
Supplementary Material
Translational Relevance.
Approximately 2% of non-small cell lung carcinomas (NSCLCs) harbor RASA1 truncating mutations. Here, we show that RASA1 truncating mutations have a strong tendency to co-occur with NF1 truncating mutations, and that alterations of these two negative regulators of RAS signaling, when concurrent, are mutually exclusive with other lung cancer mitogenic drivers. This is notable because the major mitogenic drivers defined to date in NSCLC have been oncogenes activated by mutation or gene fusion, not tumor suppressors. NSCLC cells with concurrent RASA1/NF1 mutations show profound sensitivity to MEK inhibition. Taken together, cancer genomic data and our functional data nominate concurrent RASA1/NF1 loss of function mutations as a strong mitogenic driver in NSCLCs. Patients whose tumors show this distinctive genotype should be considered for clinical trials of MEK inhibitors.
Acknowledgement
We thank Dr. Greg Riely for assistance with smoking data.
Grant Support: NCI grant P01 CA129243 (M.L.), the Lung Cancer Research Foundation (M.L.), and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (NCI P30 CA008748)
Footnotes
Conflict of interest: none
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med. 2013;188:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–9. [DOI] [PubMed] [Google Scholar]
- 10.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maertens O, Cichowski K. An expanding role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv Biol Regul. 2014;55:1–14. [DOI] [PubMed] [Google Scholar]
- 12.Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene. 2007;26:4609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapinski PE, Kwon S, Lubeck BA, Wilkinson JE, Srinivasan RS, Sevick-Muraca E, et al. RASA1 maintains the lymphatic vasculature in a quiescent functional state in mice. J Clin Invest. 2012;122:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King PD, Lubeck BA, Lapinski PE. Nonredundant functions for Ras GTPase-activating proteins in tissue homeostasis. Sci Signal. 2013;6:re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernards A GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. [DOI] [PubMed] [Google Scholar]
- 16.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, et al. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Soong J, Mohanty S, Xu L, Scott G. The neural guidance receptor Plexin C1 delays melanoma progression. Oncogene. 2013;32:4941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arafeh R, Qutob N, Emmanuel R, Keren-Paz A, Madore J, Elkahloun A, et al. Recurrent inactivating RASA2 mutations in melanoma. Nat Genet. 2015;47:1408–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–28. [DOI] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013;123:340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017. [DOI] [PubMed] [Google Scholar]
- 35.Stites EC, Trampont PC, Haney LB, Walk SF, Ravichandran KS. Cooperation between Noncanonical Ras Network Mutations. Cell Rep. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubeck BA, Lapinski PE, Oliver JA, Ksionda O, Parada LF, Zhu Y, et al. Cutting Edge: Codeletion of the Ras GTPase-Activating Proteins (RasGAPs) Neurofibromin 1 and p120 RasGAP in T Cells Results in the Development of T Cell Acute Lymphoblastic Leukemia. J Immunol. 2015;195:31–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tranchant R, Quetel L, Tallet A, Meiller C, Renier A, de Koning L, et al. Co-occurring Mutations of Tumor Suppressor Genes, LATS2 and NF2, in Malignant Pleural Mesothelioma. Clin Cancer Res. 2017;23:3191–202. [DOI] [PubMed] [Google Scholar]
- 39.Mina M, Raynaud F, Tavernari D, Battistello E, Sungalee S, Saghafinia S, et al. Conditional Selection of Genomic Alterations Dictates Cancer Evolution and Oncogenic Dependencies. Cancer Cell. 2017;32:155–68 e6. [DOI] [PubMed] [Google Scholar]
- 40.Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, et al. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clin Cancer Res. 2015;21:1935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.