Abstract
The widespread adoption of electronic health record (HERs) in healthcare systems has created a vast and continuously growing resource of clinical data and provides new opportunities for population-based research. In particular, the linking of EHRs to biospecimens and genomic data in biobanks may help address what has become a rate-limiting study for genetic research: the need for large sample sizes. The principal roadblock to capitalizing on these resources is the need to establish the validity of phenotypes extracted from the EHR. For psychiatric genetic research, this represents a particular challenge given that diagnosis is based on patient reports and clinician observations that may not be well-captured in billing codes or narrative records. This review addresses the opportunities and pitfalls in EHR-based phenotyping with a focus on their application to psychiatric genetic research. A growing number of studies have demonstrated that diagnostic algorithms with high positive predictive value can be derived from EHRs, especially when structured data are supplemented by text mining approaches. Such algorithms enable semi-automated phenotyping for large-scale case-control studies. In addition, the scale and scope of EHR databases have been used successfully to identify phenotypic subgroups and derive algorithms for longitudinal risk prediction. EHR-based genomics are particularly well-suited to rapid look-up replication of putative risk genes, studies of pleiotropy (phenomewide association studies or PheWAS), investigations of genetic networks and overlap across the phenome, and pharmacogenomic research. EHR phenotyping has been relatively under-utilized in psychiatric genomic research but may become a key component of efforts to advance precision psychiatry.
Keywords: EHR, phenotyping, electronic medical records, psychiatric genetics, PheWAS
The implementation of electronic health records (EHRs) in US healthcare systems has expanded dramatically over the past decade, fueled in part by passage of the federal HITECH Act (2009) that provided funding incentives for “meaningful use” of EHRs.[Blumenthal 2011] Comprehensive, longitudinal data captured in EHRs provide a continuously growing repository of clinical and phenotypic data that can enable low-cost population-based studies on a large scale. In particular, the linking of EHR data with biorepositories provides a new platform for psychiatric genetic research. In addition to the use of structured codified data (e.g. demographics, diagnostic codes, medications, laboratory and procedure codes), text mining by natural language processing (NLP) allows the accrual and analysis of detailed, longitudinal clinical data for research purposes.
EHRs provide a unique opportunity for psychiatric research in a variety of domains including epidemiologic studies of risk and resilience factors, pharmacoepidemiology, and genomic and biomarker research. The major advantages of EHR-based research are the large scale and low cost of data collection. The vast repositories of longitudinal health data being collected in health systems across the United States provide ample power for a variety of research applications. Because these data are collected through ongoing clinical care, they are available at much lower cost than would be required for traditional clinical research. However, these advantages come with parallel challenges. The fact that EHR data are generated to serve clinical care and facilitate billing for that care means that they may not be optimal for research purposes. For example, diagnostic data captured in billing codes (typically based on ICD-9 or ICD-10) may have questionable validity. Relevant outcomes and covariates (e.g. lifestyle, family history, and environmental variables) may not be routinely available in the EHR. A patient’s care may be fragmented across different healthcare organizations so that a single EHR may not adequately capture relevant data. The timing of data collection depends on an individual’s encounters with the healthcare provider. The allocation of diagnostic testing and treatments may be confounded by other covariates of interest (“confounding by indication”). Each of these features can complicate causal inferences in EHR-based research. Thus, while EHR-based research has already proven to be a useful resource for biomedical and genetic research, investigators must be thoughtful in their methodologic approach and mindful of its limitations.
In this review, I will highlight key features of EHR research with particular attention the opportunities and challenges involved in psychiatric phenotyping. I will focus on the utility of EHRs for genomic studies and discuss applications that our group and others have explored using electronic medical records for epidemiologic and precision medicine research.
The Phenotypic Landscape of Electronic Health Records.
The widespread use of EHRs is a relatively recent phenomenon. For example, in 2004, only 21% of US office-based physicians had adopted an EHR, but by 2015, this had risen to 87%, and 96% of acute-care hospital had a certified EHR system. [Technology. December 2016; Technology. May 2016] The use of EHRs for research purposes typically involves additional software platforms that can provide an interface for querying, aggregating, and extracting clinical data. Phenotypic data in the EHR are generally derived from two sources: structured, coded variables (including diagnostic and procedure codes, laboratory values, and prescription data) and unstructured material (including narrative notes and test interpretations) produced by clinicians in the course of providing care.
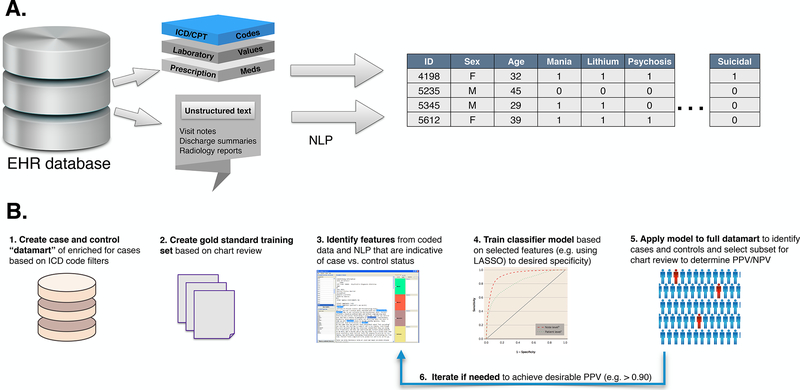
Exploiting these data for research purposes typically requires software platforms that include the creation of relational databases that extract EHR data into a platform that allows users to query the EHR for phenotypic data from these multiple data streams. (see Figure 1A) The use of unstructured material requires more sophisticated computational methods of which NLP is the most widely used. NLP combines computer science and linguistic methods to parse free text (natural language) into parts of speech that can be mapped to standardized concepts or structured data. NLP systems also recognize linguistic context and negation terms (e.g. “not” “no history of”) that could otherwise result in misclassification of a target concept. A number of databases and ontologies (e.g. the Unified Medical Language System, UMLS) are available to map related textual terms and “regular expressions” to structured concepts. [Jensen and others 2012; Liao and others 2015]. In recent years, the integration and sharing of EHR data across healthcare systems has become possible through the implementation of “common data models” (e.g. PCORnet and OMOP) that organize information from different systems into a common standardized format.
Figure 1:
Workflows for leveraging phenotypic data from HER. A) Extraction of clinical data into a research-ready database. Unstructured text can be transformed into standardized coded format through natural language processing (NLP); B) Stages in development of a phenotyping algorithm for case–control analyses. (1) An enriched datamart of cases or controls for the target phenotype is constructed using structured data filters followed by (2) selection of a subset for clinician chart review to establish gold-standard instances. (3) Potential predictors of case (or control) status are extracted from structured and text features in a subset of charts. (4) Using these selected features, a model is trained to predict the gold-standard cases/controls and model metrics are calculated to desired performance. (5) The model is applied to the full datamart and a chart review of a subset of cases (or controls) is conducted to determine PPV and NPV. (6) If desired performance is not achieved, the model can be adjusted until adequate performance (e.g. PPV > .90) is obtained.
Phenotyping algorithms: Validity and Accuracy
Perhaps the most fundamental question facing investigators utilizing the EHR is: how valid are the phenotypes? Imagine a common scenario in which one would like to identify cases and controls for an epidemiologic or genomic study of, say, depression. A simple solution to selecting cases might be to query the EHR using a rule-based algorithm that selects all patients with an ICD-10 code of major depressive disorder (296.2x– 296.3x). Controls could be chosen from among those who never received a depression-related diagnosis. It is perhaps obvious that such a scheme would be likely to misclassify a substantial proportion of cases and controls. For one thing, ICD codes in the clinical record are primarily used for billing rather than research purposes. In some cases, these codes are assigned by non-clinical staff involved in billing and reimbursement rather than clinicians themselves. Or the diagnosis may have been given by a primary care doctor who felt that it was “close enough” for the purposes of justifying prescription of an SSRI to a patient seeking relief from distress. The diagnostic rigor and precision that would be desired for a research study may not apply. In addition, a single instance of a billing code for major depressive disorder (MDD) may have been assigned to a patient who presented with a depressive episode but later experienced a manic episode—that is, a patient who ultimately was determined to have bipolar disorder. Alternatively, the patient’s presumed diagnosis of MDD might have later been felt to be an adjustment disorder or an anxiety disorder.
To address these concerns, investigators utilizing EHR data have typically used one (or both) of two approaches to enhance phenotypic precision. The first utilizes phenotypic algorithms that use filters and rule-based algorithms based on codified, structured data. For example, cases of MDD might be required to have at least two ICD codes for MDD as well as treatment with one or more antidepressants. From this pool, one might exclude individuals with any diagnosis of bipolar disorder and other disorders that might be confused with MDD. Controls might be chosen to have never received a diagnosis of MDD and to have passed through the typical age of risk (e.g. age at least 30 years). The second approach is to incorporate information from unstructured text in the EHR, typically using NLP. The advantage of NLP is that it can incorporate more information relevant to valid phenotypes including symptoms and outcomes reported in narrative notes. For example, an NLP-based algorithm to identify MDD cases might require text documenting depressive symptoms (e.g. depressed mood, anhedonia, neurovegetative symptoms) that are not captured by coded variables. The disadvantage is that it can be methodologically more complicated and time-consuming to derive and validate algorithms that incorporate free text. A recent systematic review of EHR-based case ascertainment for 41 conditions found that incorporating narrative text improved case detection above and beyond coded data alone.[Ford and others 2016]
Regardless of the method used to derive phenotyping algorithms or diagnostic rules, their utility can be gauged by several performance metrics. (Table 1)[see [Wassertheil-Smoller and Smoller 2015] for review of these concepts). The sensitivity of the algorithm refers to its ability to correctly identify individuals with the phenotype of interest. In the case of our algorithm for diagnosing MDD, this would be the proportion of true cases detected by the algorithm among all cases of MDD. The specificity refers to the algorithm’s ability to correctly identify those without the phenotype (e.g. the proportion of true negatives among all individuals without MDD). Sensitivity is the complement of the false negative rate (1- sensitivity) and specificity is the complement of the false positive rate (1- specificity). Thus, for example, as the specificity of an algorithm increases, the false positive rate decreases. Typically, there is also a trade-off between sensitivity and specificity so that increasing one entails a decrease in the other. For example, we might increase the specificity of MDD cases by a rule requiring that cases have a history of treatment with electroconvulsive therapy (ECT). This would be a stringent requirement since ECT would rarely be used in patients without MDD, resulting in a low false positive rate. However, because most individuals with MDD don’t receive ECT, the sensitivity (proportion of true cases detected) would be correspondingly lower.
Table 1.
Key metrics in evaluating the performance of a phenotyping algorithm. P(T+|D+) = probability of test (algorithm) positive, given disease is present. Prev = prevalence.
| Metric | Definition | Formula |
|---|---|---|
| Sensitivity | Proportion of true cases detected |
P(T+|D+) |
| Specificity | Proportion of those without disease who test negative | P(T−|D−) |
| Positive Predictive Value | Probability of disease given positive test |
|
| Negative Predictive Value | Probability of disease-free given negative test |
For algorithms designed to make diagnoses, the optimal diagnostic threshold as a function of this sensitivity and specificity tradeoff can be quantified using a receiver operating characteristic (ROC) curve which plots sensitivity vs. the false positive rate (1-specificity). The goal is to select a threshold that maximizes the area under this curve (AUC). The AUC is also a measure of the classifier’s discrimination performance—how well it correctly classifies those with and without the diagnosis of interest. A classifier that results in an AUC of 0.5 is no better than a coin flip, while an AUC of 1.0 is ideal. In practice, an AUC of 0.80 or higher is considered good.
Importantly, sensitivity and specificity are properties of the algorithm itself and not dependent on the prevalence of the phenotype in the population to which the algorithm is applied. However, the actual predictive value of the algorithm can depend strongly on the prevalence of the disease. In applying the algorithm, we are usually more interested in two measures of predictive value: 1) the positive predictive value (PPV) or the probability that a case defined by the algorithm is a true case, and the negative predictive value (NPV) or the probability that the phenotype is truly absent when the algorithm says it is absent. As shown in Table 1, the PPV and the NPV depend on the sensitivity, specificity and the prevalence. Thus, for a given sensitivity and specificity, when the phenotype is rare, most cases defined by the algorithm will be false positives. In such a scenario, the PPV can be increased when the algorithm is applied to a population that is enriched for the phenotype of interest. For example, if our MDD algorithm has a sensitivity of 90% and a specificity of 90%, its PPV would be 50% in an unselected population where the prevalence is 10%. However, if we enrich the sample by first filtering on those who have at least two ICD codes for MDD, the PPV will be higher. If the true prevalence of MDD in this selected population is, say, 60%, the PPV is now 93%.
Application: Assembling Case-Control Cohorts for Genomic Studies
In a range of studies, we have pursued psychiatric phenotyping using data from the Partners HealthCare system’s EHR. [Barak-Corren and others 2016; Blumenthal and others 2014; Castro and others 2013; Castro and others 2012; Castro and others 2015; Castro and others 2016b; Clements and others 2014; Gallagher and others 2012; Hoogenboom and others 2013; Hoogenboom and others 2014; O’Dushlaine and others 2014; Perlis and others 2012] The Partners system comprises patients at two major academic medical centers in Boston: Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH). An interface called the Research Patient Data Repository (RPDR) provides researchers with a real-time, longitudinal and queryable repository of the Partners EHR including more than 4 million patient records. [Nalichowski and others 2006] In an early application of EHR phenotyping for psychiatric phenotypes, we developed case and control cohorts using NLP for genomic and biomarker studies of treatment resistant depression (TRD). The procedure used provides an illustration of the steps involved in deriving phenotypic algorithms. (see Figure 1B) We first constructed a “datamart” of potential cases from among the millions of patient EHRs in the RPDR by selecting those with at least one ICD code for MDD (296.2–296.3). To derive an NLP classifier of treatment response, we utilized the i2b2 suite of software. [Murphy and others 2009] From the datamart, 5198 patients with MDD and treatment with at least one antidepressant were selected for study. Three board-certified clinical psychiatrists reviewed a set of 724 randomly selected visit notes to determine whether the patient was depressed or well (euthmic/remitted) at the time of each visit. This “clinical gold-standard” was used for training the NLP classifier described below. While reviewing the narrative notes, the psychiatrists also identified words or phrases that related to the clinical status. The i2b2 tools were then used to extract these text elements (“regular expressions”) for use in building the classifier. Next, LASSO regression models were trained to predict clinical status (depressed, well or other) at clinical visits. For comparison, models were trained using three types of predictors: billing codes only; NLP only; or all available data from billing codes and NLP. A rule-based algorithm was used to classify patients as having TRD (cases) or treatment-responsive MDD (controls) based on their longitudinal clinical status after one or more trials of antidepressants and standard definitions of outcome. And, finally, to validate the phenotype definitions, an expert psychiatrist blinded to the algorithmic classification reviewed a random sample of patients and classified them based on standard outcome definitions.
When performance metrics were calculated, the results showed the clear advantages of using NLP-based phenotyping over billing codes alone. For NLP models with 95% specificity, the PPV exceeded 0.85, while those based on billing codes alone did not exceed 0.55. To enable genomic studies, we utilized a laboratory application that links phenotypic data to discarded blood samples obtained through routine clinical care as they become available. [Murphy and others 2009] The case/control sets and their corresponding specimens are then deidentified for use in genomic research. In a proof-of-concept study, we used this method to conduct genomewide analyses of rare copy number variants (CNVs) in TRD. [O’Dushlaine and others 2014] This approach also proved useful in examining neural correlates of depression using brain images obtained during clinical practice (“legacy” scans). We extracted MRI images from the EHR for patients with and without depression (including TRD) whose scans indicated no observable pathology and applied automated and manual segmentation procedures to derive volumetric and white matter structural phenotypes. [Hoogenboom and others 2013; Hoogenboom and others 2014] Consistent with prior evidence, MDD was associated with reduced rostral anterior cingulate volume and treatment resistance was further associated with reduced hippocampal volume. Failure to achieve remission from MDD was also associated with reduced white matter integrity in limbic tracts (fornix and cingulum bundle) and with accelerated age-related decline in white matter integrity. While these analyses provided a proof-of-concept for conducting psychiatric neuroimaging research based on EHR data, they also illustrated some important challenges. Most importantly, the strict quality-control measures needed to identify informative patients and reduce the risk of selection bias in ascertaining clinical MRIs can substantially restrict sample size. A small fraction of patients with billing diagnoses of MDD and concurrent MRI data had suitable narrative notes and scans to allow NLP phenotyping of treatment response and analyses of brain structure.
Nevertheless, these and other studies have demonstrated the utility of using EHR-based phenotyping for psychiatric research. [Lingren and others 2016] A chief advantage of this approach is the ability to rapidly perform analyses that would otherwise be prohibitively costly to perform or require years to assemble adequate study cohorts. It has become clear that genomic studies of both common diseases (typically by GWAS) and rare disease (predominantly DNA sequencing studies) require large sample sizes. In the case of common variant studies of polygenic complex diseases, tens of thousands of cases and controls are needed to provide adequate power to detect loci of individually modest effect. Studies of rare variants and rare disease by definition require similar or larger samples to capture sufficient instances of genotypes and phenotypes of interest.
Still, EHR-based phenotyping for genetic studies will only be valuable if we can be confident in the validity of the derived phenotypes. Fortunately, a large and growing body of evidence in other areas of medicine has supported the validity of EHR-based genomics. For example, GWAS of rheumatoid arthritis, diabetes, asthma, cardiovascular disease and other complex phenotypes have successfully identified loci that were previously established in case-control studies using traditional, in-person clinical assessment. [Hoffmann and others 2017; Kurreeman and others 2011; Namjou and others 2014; Ritchie and others 2010; Roden and Denny 2016; Shen and others 2015; Verma and others 2016; Xu and others 2011] However, psychiatric disorders present a unique challenge in that diagnosis relies on symptoms, behavior and clinical judgment with no pathognomonic laboratory or pathologic findings. As a result, psychiatric genetic studies have traditionally relied on lengthy structured or semi-structured diagnostic interviews administered by trained clinicians or research staff. Validation of EHR-based diagnoses have typically relied on retrospective chart reviews to estimate how well a phenotypic algorithm predicts a true case or control. So how would EHR-derived diagnoses perform relative to the gold-standard of a psychiatric diagnostic interview?
We addressed this question in the context of a large scale study of bipolar disorder genetics, as part of the International Cohort Collection for Bipolar Disorder (ICCBD). [Castro and others 2015] The ICCBD was formed as psychiatric genetic studies began to make clear that sample size had become the rate-limiting step in gene discovery. The ICCBD is a consortium of four sites that aimed to collect clinical data and DNA samples for 19,000 bipolar cases and 19,000 controls over a 4 year period. To achieve this ambitious goal, we turned to high-throughput phenotyping methods. At MGH, we used the i2b2 platform to ascertain 4500 cases and 4500 controls from the Partners HealthCare system. After creating a datamart of potential cases and controls based on ICD billing codes, expert clinicians manually reviewed 612 notes from 209 randomly selected patients in the bipolar mart to develop a gold standard set of cases (by DSM-IV criteria) and to identify relevant terms relevant to be extracted by NLP. In addition to training an NLP-based classifier, we derived additional rule-based classifiers for bipolar disorder using narrow and broad diagnostic criteria as well as a rule-based classifier to identify controls. To evaluate the validity of these classifiers, we applied them to the EHR and identified patients predicted to be cases or controls by the EHR algorithms. Patients were then invited by mail to participate in an in-person validation study during which they underwent semistructured diagnostic interviews (SCID-IV) conducted by experienced doctoral-level clinicians blinded to classifier diagnosis. To further preserve clinician blinding, we recruited individuals from MGH clinics who reported a previous diagnosis of schizophrenia or major depression, disorders commonly considered in the differential diagnosis of bipolar disorder. Thus, this study provided a direct test of EHR phenotyping vs. the accepted gold-standard of direct psychiatric interview.
Based on the direct interview results (N = 190), PPV for the NLP-defined classifier for bipolar disorder was 0.86 with a specificity of 95%. The PPV for controls was 1.0—that is, none of the patients predicted to be controls were found to be cases at direct interview. In addition, we derived NLP algorithms for eight relevant subphenotypes: bipolar subtype, family history of bipolar disorder, age at onset, psychosis, suicide attempt, alcohol abuse, substance abuse, and panic/agoraphobia. When compared to the results of the diagnostic interview the PPVs ranged from .72 (for psychosis) to 0.94 (for age at onset). With these algorithms in hand, we obtained discarded blood samples to accrue DNA from 4500 cases (over 3 years) and 5000 controls (in 10 weeks). We have recently completed genomewide genotyping for the majority of this sample and calculated preliminary heritability estimates from SNP data (h2SNP). The EHR-based phenotypes were significantly heritable and comparable to those observed in prior studies of traditionally ascertained samples (Chia-Yen Chen et al. unpublished). Thus, this study provides evidence for both the clinical and genetic validity of semi-automated EHR-based phenotyping of bipolar disorder.
At the same time, challenges to establishing valid phenotypes remain, beyond the labor- and time-intensive nature of the informatic analyses and clinical validation procedures. Data available through the EHR are limited, by definition, to those captured through interactions with the healthcare system. Thus, features of illness that occur outside this system may not be documented. This can be particularly problematic for so-called “open” systems where patients may receive some but not all of their care. For example, individuals classified as controls may have had an episode of the case phenotype for which they received care outside the system, creating a problem of false negative classification. Similarly, episodes of illness may have occurred within the healthcare system but prior to the implementation of the EHR. Alternative sources of information such as claims data may mitigate these problems by capturing all reimbursed healthcare encounters, but these data do not include the kind of detailed information that would allow NLP based models, for example. Another emerging solution is the availability of interoperable networks of EHR data from multiple institutions, such as the Shared Health Information Network (SHRINE) [McMurry and others 2013], the Scalable Collaborative Infrastructure for a Learning Health System (SCILHS) [Mandl and others 2014], and the Electronic Medical Records and Genomics (eMERGE) [Gottesman and others 2013] networks. Another obvious limitation of EHR resources is the ascertainment bias inherent in the fact that phenotypic data are recorded based on the particular clinical presentation and diagnostic odyssey that apply to individual patients. Relatedly, clinicians may differ in the precision and comprehensiveness with which they record diagnostic codes or narrative text, creating challenges for applying phenotypic algorithms across providers and health systems.
Application: Phenotypic Clusters and Subtyping
Training algorithms for diagnostic phenotyping is related to the concept of “supervised learning” –that is, EHR models are trained against a known gold standard. Another application of interest is the use of large datasets to find phenotypic structure within or across categories—a kind of “unsupervised learning”. Related data-driven approaches to parsing phenotypic complexity include clustering and principal component analyses that can extract phenotypic signatures and dimensions from a heterogeneous mixture of clinical syndromes. Lyalina and colleagues [Lyalina and others 2013] used text mining approaches to annotate records from more than 7000 patients who carried diagnostic codes for autism, schizophrenia or bipolar disorder. They extracted terms related to medications and clinical features and then identified terms that were enriched in the clinical records. Using network and data reduction (principal component analysis) approaches, they were able to identify “phenotypic signatures” of the three disorders. The results revealed distinctive features characterizing autism but significant overlap between schizophrenia and bipolar disorder. In patient-level analyses, they situated individuals in the high-dimensional phenotypic landscape defined by these signatures. Such an approach could be useful in parsing heterogeneity for genomic studies of psychopathology, though its portability to other health systems remains to be established.
Kohane and colleagues [Doshi-Velez and others 2014; Kohane 2015] used the i2b2 platform to assemble a cohort of nearly 5000 individuals, age 15 years or older, with autism spectrum disorder (ASD) diagnoses from the Boston Children’s Hospital longitudinal EHR. Using hierarchical clustering, they identified four distinct subgroups based on clinical trajectories of ASD comorbidities. These comprised: 1) a subtype with a high prevalence of seizures (77.5%); 2) a subtype enriched for multisystem comorbidities (especially gastrointestinal disorders, ear infections and auditory disorders) and otologic comorbidities); 3) a subtype characterized by a high rate of psychiatric disorders; and 4) a residual group with no distinctive comorbidity pattern. The subgroups also differed by age of ASD diagnosis (earliest in the psychiatric comorbidity group) and prevalence of intellectual disability (highest in the subtype enriched for seizure disorders). In a subsequent larger analysis, investigators in the eMERGE network [Lingren and others 2016] were largely able to recapitulate these clusters using an NLP-derived algorithm applied across multiple institutions in more than 20,000 ASD patients. The identification of novel data-driven subtypes provides an enticing opportunity for genomic studies of psychopathology where the heterogeneity of clinical syndromes is widely assumed. If these subtypes reflect more homogeneous etiologies, they may well enhance the power of common and rare variant genomic analyses.
Application: Prediction Algorithms
The vast repository of phenotypic data captured in EHRs provides a powerful platform for predictive modeling of psychiatric disorders and related outcomes, including the use of machine learning approaches. Huang and colleagues [Huang and others 2014] used a combination of structured EHR data and text elements in two health systems to train prediction models for a diagnosis of depression, response to depression treatment, and depression severity. The diagnosis of depression was defined as an ICD-9 code for depression and presence of an antidepressant medication term in the clinical text. The assessment of treatment response and severity was facilitated by the inclusion of PHQ-9 scores in the EHR of one of the health systems. LASSO regression models were trained and validated for each phenotype of interest and results were promising, despite the fact that notes from mental health professionals were unavailable. The model AUC was 0.71 for predicting an EHR diagnosis of depression 6 months in advance of that diagnosis. As Huang et al note, their models achieve sensitivities and specificities comparable to those previously reported for diagnosing depression by primary care physicians. [Mitchell and others 2009]. Models for predicting response to treatment achieved AUCs of 0.66 and 0.75 for response to medication and psychotherapy, respectively. However, the strongest predictor of response was baseline PHQ-9 score, suggesting that these models would not be portable to EHR systems that do not include this measure.
Perhaps no psychiatric phenotype is a more important and challenging target for prediction than suicidal behavior. Suicide is one of the leading causes of death worldwide, and the second leading cause of death among young people. Despite extensive research on the risk factors associated with suicide attempts and death by suicide, the prevalence of suicide has remained virtually unchanged for decades. Most people who die by suicide are seen by clinicians in the year prior to their death, making healthcare systems an important setting for suicide risk prediction and prevention. Unfortunately, there is no accepted algorithm for risk prediction that clinicians can use to integrate the multiple risk factors that may indicate that a patient is likely to make a suicide attempt in the near future. A recent comprehensive meta-analysis found that the predictive ability of known risk factors is weak and has not improved over the past 50 years. [Franklin and others 2016] A number of investigators have used EHR data to identify individuals at risk for suicidal behavior, but studies have been limited by relatively small samples [Baca-Garcia and others 2006; Poulin and others 2014], short follow-up times [Tran and others 2014], specialized populations [Kessler and others 2015], or have not reported performance metrics. [Ilgen and others 2009] We recently developed risk prediction models for suicidal behavior by leveraging the high-dimensional clinical data available in the EHR and using time-varying data across an extended period of follow-up. [Barak-Corren and others 2016] This effort was motivated by an earlier report in which Reis and colleagues [Reis and others 2009] developed naïve Bayesian classifier (NBC) models using structured EHR data that predicted whether a patient was at risk of domestic abuse an average of two years in advance. NBCs are machine learning models that have the advantage of being highly scalable and easily interpretable. We first identified candidate ICD codes for suicidal behavior and manually reviewed more than 2700 notes from 520 patients to select codes with a PPV of > 0.70 based on expert clinician consensus. We then queried the Partners Healthcare RPDR to identify approximately 20,000 cases and 1.7 million controls comprising 8.9 million person-years of follow-up. Cases were supplemented with Massachusetts state death certificates indicating death by suicide. The model was trained in 2/3 of the sample and validated in the remaining 1/3. The model AUC was 0.77 and detected 45% of suicidal behaviors with a specificity of 90% an average of 4 years in advance of their being diagnosed. However, given the low base rate of suicidal behavior, the algorithm PPV was <10%. Efforts are now ongoing to enhance this PPV by applying NBC models to subpopulations enriched for suicidality (e.g. patients being treated for psychiatric disorders). The NBC model assigns weight for the contribution of all variables included from the health record, making it possible to estimate the predictive effect of each of the tens of thousands of EHR features. Although, as would be expected, mental health and substance abuse-related features were among the strongest predictors, certain infections and injury codes were also highly predictive. Thus, the ability to incorporate the full phenotypic breadth of the EHR provides information beyond what would be feasible for individual clinicians to evaluate. In another recent analysis, McCoy and colleagues [McCoy and others 2016] focused on predicting risk of suicide or accidental death among patients discharged from inpatient hospitalization at Partners Healthcare. Models using coded variables with or without the addition of NLP, achieved an AUC of approximately 0.74.
An alternative approach to predicting phenotypes from EHR data, based on unsupervised machine learning approaches, was recently reported by Miotto et al. [Miotto and others 2016] to identify future risk of 78 diseases. Their “deep patient” method begins with an unsupervised neural network pre-processing of patient features to identify reduced dimensional representations of patient profiles. These were then subjected to random forests analysis to predict disease states. Among the conditions for which this deep learning approach performed well in terms of 1 year prediction of diagnosis were psychiatric disorders: schizophrenia (AUC = 0.85) and attention deficit and disruptive behavior disorders (AUC = 0.86).
In sum, EHR data appear to be useful in predicting psychiatric diagnosis and related outcomes using a variety of strategies. Combining these clinical risk models with genomic data may facilitate more advanced precision medicine applications, though this remains to be tested.
Combining EHR data and Genomic Data
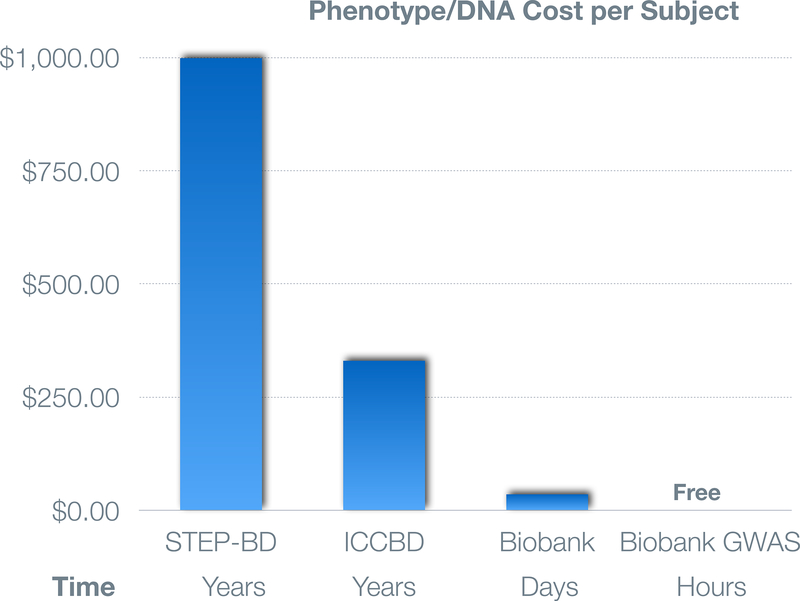
The real promise of advancing psychiatric genomics by leveraging EHR-based data resources derives from the linking of these phenotypic data with genomic data. In recent years, this promise has been progressively realized by the availability of EHR-linked biobanks at healthcare systems in the U.S. and internationally (also see other papers in this special issue). For example, the Partners Healthcare Biobank is a repository of patient blood samples linked to the EHR and supplemented by survey data capturing data not readily available in the EHR (e.g. sun exposure, family history, and sleep habits) [Karlson and others 2016]. At this writing more than 60,000 patients have enrolled using face-to-face and electronic web-based informed consent [Boutin and others 2016] with approximately 2000 added monthly at Partners hospitals including Massachusetts General Hospital, Brigham and Women’s Hospital, and McLean Hospital. In addition to phenotypic data from the full longitudinal EHR (spanning approximately 20 years), we have used a method of unbiased automated feature extraction to build NLP-derived phenotypic algorithms that have been trained and validated by clinician chart review.[Yu and others 2015] These “curated” phenotypes have demonstrated excellent PPVs (≥.90) and NPVs (> 0.95), including those for schizophrenia, bipolar disorder, and major depressive disorder. Genomewide genotyping of 50,000 biobank participants is underway with 15,000 completed thus far. These provide a ready resource for rapid validation (“lookups”) of prior genetic findings, the construction of polygenic risk scores, and phenomewide association studies (described below), many of which are underway. [Smoller and others 2016]. The cost- and time-efficiency of deriving biospecimens and genomic data from EHR-linked biobanks is illustrated in Figure 2 which shows the cost to investigators of obtaining phenotypic data and biospecimens in three studies of bipolar disorder which we have participated in: the STEP-BD genetic repository [Fan and others 2010], the ICCBD (described above), and the Partners Biobank.
Figure 2:
Comparison of cost and time needed to obtain phenotypic data and samples based on experience in studies of bipolar disorder at Partners Healthcare. Approximately 700 samples were collected over a period of years at a cost of approximately $1000/subject in the STEP-BD study. At one-third of this cost, an even larger sample collected as part of the ICCBD (see text). Finally, the Partners Biobank enables investigators to rapidly obtain validated EHR phenotypes and samples at minimal cost and genomewide GWAS data at no cost.
Large-scale U.S. biobanks, including BioVu at Vanderbilt University, MyCode at the Geisinger Health system, and the Kaiser Permanente Biobank have successfully used EHR and genomic data to identify novel disease associations [Denny and others 2011; Hoffmann and others 2017], evaluate the penetrance of putative high-penetrance mutations [Van Driest and others 2016], and identify allelic series that can validate targets for drug development. [Dewey and others 2016]. In 2007, the National Human Genome Research Institute (NHGRI) organized the Electronic Medical Records and Genomics (eMERGE) network (https://emerge.mc.vanderbilt.edu) [Gottesman and others 2013]. At present, eMERGE comprises nine clinical sites, a coordinating center, and two central genotyping and sequencing centers. The eMERGE network has also compiled validated algorithms for EHR phenotyping in a centralized database (PheKB) and demonstrated that these algorithms are portable across healthcare systems. [Kirby and others 2016] Since its inception, the eMERGE network has played a major role in developing best practices for EHR-based genomic research, advancing methods for extracting and validating phenotypic data using semi-automated algorithms, and demonstrating the value of phenomewide association studies (PheWAS). [Crawford and others 2014; Gottesman and others 2013]
The PheWAS approach capitalizes on the broad spectrum of phenotypes captured in the EHR, enabling the discovery of pleiotropic genetic associations. Effectively, a PheWAS is the mirror image of a typical GWAS design, though both take an unbiased approach to high-dimensional association analysis. Where the GWAS typically examines a limited set of target phenotypes and their association with a large number of genetic markers across the genome, the PheWAS typically examines a limited set of target genotypes and their association with a large number of phenotypes across the phenome. [Bush and others 2016; Denny and others 2016] PheWas have been successfully used to identify novel disease loci, replicate GWAS findings and identify pleiotropic effects of loci identified from GWAS of specific phenotypes. [Denny and others 2016] To reduce the dimensionality of the phenotypic space, related diagnostic codes are often grouped into a smaller set of “phecodes”. There is likely to be a trade-off in which collapsing phenotypes may reduce the burden of multiple testing but at the same might obscure association signals by combining potentially heterogeneous phenotypes. Prior research suggests that requiring two or more temporally separate instances of a diagnostic code can improve the PPV of PheWAS phenotypes. [Denny and others 2016] In light of increasing evidence for pleiotropic associations among psychiatric disorders and other phenotypes [Solovieff and others 2013], PheWAS may be particularly useful for psychiatric genetic research.
To date, the application of EHR resources to psychiatric genetics has been relatively limited, but this has begun to change. In one innovative analysis, Simonti and colleagues [Simonti and others 2016] analyzed phenotypic and genomewide data across the eMERGE network for ~28,000 individuals to determine whether SNPs derived from Neanderthal admixture are associated with a range of diseases. They observed modest but significant heritability attributable to Neanderthal variants for eight phenotypes including mood disorders. Further, Neanderthal loci were enriched for associations with neurologic and psychiatric diseases and for brain eQTLs (as indexed by expression in cerebellum and temporal cortex).
Another recent analysis [Prieto and others 2016] examined the phenotypic effects of loci previously identified in GWAS of bipolar disorder using data on 7316 patients with EHR data and genomewide genotypes in the Mayo Clinic’s Mayo Genomic Consortium. ICD-9 codes were used to identify individuals with 19 disease phenotypes that are often comorbid with bipolar disorder. Evidence was found for pleiotropic associations of SNPs in CACNA1C with cardiac dysrhythmias and SVEP1 with essential hypertension, though these did not survive correction for multiple testing.
In addition to enabling studies of pleiotropy, EHR-linked biobanks provide a valuable resource for rapid look-up and replication of putative risk loci. For example, loci implicated in GWAS and rare variant studies of a given disorder can be readily queried in biobanks to validate the association in a clinical setting.
Future Directions and Opportunities
The coming years will see important opportunities to advance the application of EHR-based psychiatric genomic research. For example, investigators in the eMERGE network have recently launched an effort (PsycheMERGE) to harmonize phenotypic data for a range of psychiatric disorders and conduct cross-disorder and functional genomic analyses. Psychiatric diagnoses derived from ICD codes are also available in the UK Biobank (N = 500,000), providing another large resource for genetic studies. And, the U.S. Department of Veteran Affairs’ Million Veteran Program (http://www.research.va.gov/MVP/default.cfm) has already enrolled more than 500,000 veterans, with genomic studies of substance use and PTSD underway. Perhaps the most eagerly anticipated resource for large-scale deeply-phenotyped genomic research is the NIH’s Precision Medicine Initiative (PMI) [Collins and Varmus 2015; Precision Medicine Initiative (PMI) Working Group 2015]. The PMI’s All of Us program aims to enroll a diverse cohort of a million or more participants across the United States with multiple data streams including EHRs, physical assessments, participant surveys, mobile health technologies, and a range of biospecimens. Participants will be followed for ten years or more, providing a unique longitudinal program for examining the role of genetic, environmental, and lifestyle factors on health outcomes and treatment response. The inclusion of behavioral and psychiatric phenotypes should enable unprecedented opportunities for psychiatric genetic and pharmacogenomic research.
The tens of thousands of phenotypes available in EHRs and biobanks provide clear advantages but also challenges: given the high-dimensional nature of these resources, which phenotypes are most fruitful for genomic analyses? Typically, research studies have focused on a small fraction of the available clinical variables. We have developed computational tools that allow the prioritization of phenotypes based on their SNP-heritability (h2SNP). [Ge and others 2016; Ge and others 2015] In a recent effort [Ge and others 2016], we introduced a computationally efficient “moment-matching method” capable of handling very large samples sizes and high-dimensional phenotypes. To illustrate its utility, we conducted a phenome-wide heritability analysis of 551 traits derived from the interim data release (N > 152,000) of the large-scale, population-based UK Biobank, comprising both quantitative phenotypes and EHR disease codes. We further demonstrated the moderating effect of three sociodemographic variables (age, sex, and socioeconomic status) on heritability. The results provide a ranking of heritability across the phenome, highlighting phenotypes that may warrant priority for genetic association studies. We also observed significant moderation of heritability by sociodemographic variables or numerous traits, underscoring the importance of considering population characteristics in interpreting heritability. Of note, heritability estimates for 14 pairs of self-reported illness and ICD-10 codes that represent the same or closely matched diseases were largely consistent and had a Pearson correlation of 0.78, indicating that both phenotypic approaches captured useful and comparable variations in these phenotypes. Approaches like these may allow future genetic studies to focus on the most informative phenotypes within the immense landscape of EHR data.
The phenotypic diversity captured in EHRs also provides an opportunity to explore or validate phenotypic and genetic relationships across the “disease-ome.” For example, Rzhetsky et al.[Rzhetsky and others 2007] developed a probabilistic modeling approach to estimate phenotypic relationships among 161 disorders by mining 1.5 million patient records from a clinical database at the Columbia University Medical Center. Using a set of modeling assumptions including genetic penetrance functions, they were able to create networks of correlated phenotypes and infer genetic overlaps among disorders. Of note, neuropsychiatric disorders figured prominently in these networks. In particular, autism, schizophrenia, and bipolar disorder were predicted to have substantial genetic overlap with each other and with a range of neurologic and medical disorders. In a subsequent analysis that included billing and claims data from more than 100 million patients, Rzhetsky and colleagues [Blair and others 2013] identified novel comorbidity relationships between Mendelian and complex diseases, suggesting that Mendelian disease genes have pleiotropic effects on sets of complex common diseases. In support of this, GWAS results for these complex disease sets were enriched in the linked Mendelian disease genes. Of relevance to psychiatric genetics, their analysis predicted the relationship of specific loci to multiple psychiatric disorders as previously observed in cross-disorder GWAS. [Cross-Disorder Group of the Psychiatric Genomics 2013]
Future research may also address some of the inherent limitations of EHR data. As mentioned earlier, the fact that health data are recorded during particular episodes of clinical care by providers who differ in their approach to diagnosis and documentation creates a potential for ascertainment bias in the types and extent of available phenotypic information. In one sense, unascertained phenotypes could be seen as a “missing data problem” for which imputation might be possible. Newer phenotype imputation methods that capitalize on both phenotypic correlations and genetic relatedness in high-dimensional datasets [Dahl and others 2016] may be particularly relevant for studies combining EHR and genomic data. Another limitation to EHR datasets that is relevant to genomic research is the frequent lack of information on family history. In an innovative recent analysis, Polubriaginof, Tatonetti and colleagues [Polubriaginof and others 2016] mined emergency contact data from the EHR to infer 4.7 million familial relationships among patients in two large hospital systems. Using an algorithm that enabled the reconstruction of extended pedigrees, they estimated the heritability and familial recurrence rates of more than 700 phenotypes. Surprisingly, the phenotype with the largest observed heritability estimate was “victim of child abuse” (h2 = 0.90, 95% CI: 0.73–1.0) and among psychiatric diagnoses the most heritable phenotype was “adjustment disorder with mixed emotional features” (h2 = 0.43, 95% CI: 0.30–0.59).
In addition to their potential value for identifying disorder risk loci, EHR-based genomic resources may accelerate pharmacogenetic research. We and others have used the EHR in pharmacoepidemiology studies to identify novel adverse effects of psychotropic medications. [Blumenthal and others 2014] [Castro and others 2013; Castro and others 2012; Castro and others 2016a; Castro and others 2016b; Clements and others 2014; Gallagher and others 2012; Iqbal and others 2015; Tatonetti and others 2011]. To date, however, few studies have capitalized on the availability of psychotropic drug response data in the EHR for genetic research. Success in this area will need to address the challenges of extracting valid treatment outcomes from electronic records. These include incomplete documentation of medication compliance and imprecise capture of symptom change or adverse events over time. Nevertheless, EHR-based pharmacogenomic research may be particularly informative for “precision psychiatry” efforts to identify subgroups of patients who may be at risk for serious adverse events such as clozapine-induced agranulocytosis, lithium-induced renal failure or antidepressant-induced QT prolongation.
In sum, EHR databases linked to biospecimens and genomic data provide important, as yet untapped, opportunities for psychiatric genetic research. The principal challenges involve the extraction of valid, interoperable phenotyping algorithms, but a growing literature has documented numerous approaches to addressing the challenge and successful use cases for a broad range of medical and, increasingly, neuropsychiatric phenotypes. With the growing availability of large-scale biobanks, the impending implementation of genomic data into medical care, and massive research efforts such as the Million Veteran Program and All Of Us Research Program, the prospects for scientific discovery in this arena are substantial.
Acknowledgments
Supported in part by NIH awards R01MH085542 and K24MH094614 and by support from the Tommy Fuss Fund and the Demarest Lloyd, Jr. Foundation.
Dr. Smoller is a Tepper Family MGH Research Scholar. Presented in part at the “Leveraging Electronic Medical Records for Psychiatric Research Workshop”, NIMH, September 15, 2016. “All of Us” is a service mark of the U.S. Department of Health and Human Services.
Footnotes
Disclosures:
Dr. Smoller is an unpaid member of the Scientific Advisory Board of PsyBrain Inc. and the Bipolar/Depression Research Community Advisory Panel of 23andMe.
References
- Baca-Garcia E, Perez-Rodriguez MM, Basurte-Villamor I, Saiz-Ruiz J, Leiva-Murillo JM, de Prado-Cumplido M, Santiago-Mozos R, Artes-Rodriguez A, de Leon J. 2006. Using data mining to explore complex clinical decisions: A study of hospitalization after a suicide attempt. J Clin Psychiatry 67(7):1124–1132. [DOI] [PubMed] [Google Scholar]
- Barak-Corren Y, Castro VM, Javitt S, Hoffnagle AG, Dai Y, Perlis RH, Nock MK, Smoller JW, Reis BY. 2016. Predicting Suicidal Behavior From Longitudinal Electronic Health Records. Am J Psychiatry:appiajp201616010077. [DOI] [PubMed] [Google Scholar]
- Blair DR, Lyttle CS, Mortensen JM, Bearden CF, Jensen AB, Khiabanian H, Melamed R, Rabadan R, Bernstam EV, Brunak S, Jensen LJ, Nicolae D, Shah NH, Grossman RL, Cox NJ, White KP, Rzhetsky A. 2013. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell 155(1):70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal D 2011. Wiring the health system--origins and provisions of a new federal program. N Engl J Med 365(24):2323–2329. [DOI] [PubMed] [Google Scholar]
- Blumenthal SR, Castro VM, Clements CC, Rosenfield HR, Murphy SN, Fava M, Weilburg JB, Erb JL, Churchill SE, Kohane IS, Smoller JW, Perlis RH. 2014. An electronic health records study of long-term weight gain following antidepressant use. JAMA Psychiatry 71(8):889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin NT, Mathieu K, Hoffnagle AG, Allen NL, Castro VM, Morash M, O’Rourke PP, Hohmann EL, Herring N, Bry L, Slaugenhaupt SA, Karlson EW, Weiss ST, Smoller JW. 2016. Implementation of Electronic Consent at a Biobank: An Opportunity for Precision Medicine Research. J Pers Med 6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Oetjens MT, Crawford DC. 2016. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 17(3):129–145. [DOI] [PubMed] [Google Scholar]
- Castro VM, Clements CC, Murphy SN, Gainer VS, Fava M, Weilburg JB, Erb JL, Churchill SE, Kohane IS, Iosifescu DV, Smoller JW, Perlis RH. 2013. QT interval and antidepressant use: a cross sectional study of electronic health records. BMJ 346:f288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro VM, Gallagher PJ, Clements CC, Murphy SN, Gainer VS, Fava M, Weilburg JB, Churchill SE, Kohane IS, Iosifescu DV, Smoller JW, Perlis RH. 2012. Incident user cohort study of risk for gastrointestinal bleed and stroke in individuals with major depressive disorder treated with antidepressants. BMJ Open 2(2):e000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro VM, Kong SW, Clements CC, Brady R, Kaimal AJ, Doyle AE, Robinson EB, Churchill SE, Kohane IS, Perlis RH. 2016a. Absence of evidence for increase in risk for autism or attention-deficit hyperactivity disorder following antidepressant exposure during pregnancy: a replication study. Transl Psychiatry 6:e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro VM, Minnier J, Murphy SN, Kohane I, Churchill SE, Gainer V, Cai T, Hoffnagle AG, Dai Y, Block S, Weill SR, Nadal-Vicens M, Pollastri AR, Rosenquist JN, Goryachev S, Ongur D, Sklar P, Perlis RH, Smoller JW, International Cohort Collection for Bipolar Disorder C. 2015. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am J Psychiatry 172(4):363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro VM, Roberson AM, McCoy TH, Wiste A, Cagan A, Smoller JW, Rosenbaum JF, Ostacher M, Perlis RH. 2016b. Stratifying Risk for Renal Insufficiency Among Lithium-Treated Patients: An Electronic Health Record Study. Neuropsychopharmacology 41(4):1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements CC, Castro VM, Blumenthal SR, Rosenfield HR, Murphy SN, Fava M, Erb JL, Churchill SE, Kaimal AJ, Doyle AE, Robinson EB, Smoller JW, Kohane IS, Perlis RH. 2014. Prenatal antidepressant exposure is associated with risk for attention-deficit hyperactivity disorder but not autism spectrum disorder in a large health system. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Varmus H. 2015. A new initiative on precision medicine. N Engl J Med 372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG, Denny JC, Bush WS, Haines JL, Roden DM, McCarty CA, Jarvik GP, Ritchie MD. 2014. eMERGEing progress in genomics-the first seven years. Front Genet 5:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C. 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381(9875):1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl A, Iotchkova V, Baud A, Johansson A, Gyllensten U, Soranzo N, Mott R, Kranis A, Marchini J. 2016. A multiple-phenotype imputation method for genetic studies. Nat Genet 48(4):466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Bastarache L, Roden DM. 2016. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annu Rev Genomics Hum Genet 17:353–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de Andrade M. 2011. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89(4):529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, Gusarova V, O’Dushlaine C, Gottesman O, Trejos J, Hunt C, Van Hout CV, Habegger L, Buckler D, Lai KM, Leader JB, Murray MF, Ritchie MD, Kirchner HL, Ledbetter DH, Penn J, Lopez A, Borecki IB, Overton JD, Reid JG, Carey DJ, Murphy AJ, Yancopoulos GD, Baras A, Gromada J, Shuldiner AR. 2016. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med 374(12):1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi-Velez F, Ge Y, Kohane I. 2014. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics 133(1):e54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ionita-Laza I, McQueen MB, Devlin B, Purcell S, Faraone SV, Allen MH, Bowden CL, Calabrese JR, Fossey MD, Friedman ES, Gyulai L, Hauser P, Ketter TB, Marangell LB, Miklowitz DJ, Nierenberg AA, Patel JK, Sachs GS, Thase ME, Molay FB, Escamilla MA, Nimgaonkar VL, Sklar P, Laird NM, Smoller JW. 2010. Linkage disequilibrium mapping of the chromosome 6q21–22.31 bipolar I disorder susceptibility locus. Am J Med Genet B Neuropsychiatr Genet 153B(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. 2016. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 23(5):1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin JC, Ribeiro JD, Fox KR, Bentley KH, Kleiman EM, Huang X, Musacchio KM, Jaroszewski AC, Chang BP, Nock MK. 2016. Risk Factors for Suicidal Thoughts and Behaviors: A Meta-Analysis of 50 Years of Research. Psychol Bull. [DOI] [PubMed] [Google Scholar]
- Gallagher PJ, Castro V, Fava M, Weilburg JB, Murphy SN, Gainer VS, Churchill SE, Kohane IS, Iosifescu DV, Smoller JW, Perlis RH. 2012. Antidepressant response in patients with major depression exposed to NSAIDs: a pharmacovigilance study. Am J Psychiatry 169(10):1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen C-Y, Neale BM, Sabuncu MR, Smoller JW. 2016. Phenome-wide Heritability Analysis of the UK Biobank. BiorXiv 10.1101/070177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Sabuncu MR, Smoller JW. 2015. Massively expedited genome-wide heritability analysis (MEGHA). Proc Natl Acad Sci U S A 112(8):2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, Sanderson SC, Kannry J, Zinberg R, Basford MA, Brilliant M, Carey DJ, Chisholm RL, Chute CG, Connolly JJ, Crosslin D, Denny JC, Gallego CJ, Haines JL, Hakonarson H, Harley J, Jarvik GP, Kohane I, Kullo IJ, Larson EB, McCarty C, Ritchie MD, Roden DM, Smith ME, Bottinger EP, Williams MS, e MN. 2013. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 15(10):761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. 2017. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet 49(1):54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom WS, Perlis RH, Smoller JW, Zeng-Treitler Q, Gainer VS, Murphy SN, Churchill SE, Kohane IS, Shenton ME, Iosifescu DV. 2013. Feasibility of studying brain morphology in major depressive disorder with structural magnetic resonance imaging and clinical data from the electronic medical record: a pilot study. Psychiatry Res 211(3):202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom WS, Perlis RH, Smoller JW, Zeng-Treitler Q, Gainer VS, Murphy SN, Churchill SE, Kohane IS, Shenton ME, Iosifescu DV. 2014. Limbic system white matter microstructure and long-term treatment outcome in major depressive disorder: a diffusion tensor imaging study using legacy data. World J Biol Psychiatry 15(2):122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, LePendu P, Iyer SV, Tai-Seale M, Carrell D, Shah NH. 2014. Toward personalizing treatment for depression: predicting diagnosis and severity. J Am Med Inform Assoc 21(6):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgen MA, Downing K, Zivin K, Hoggatt KJ, Kim HM, Ganoczy D, Austin KL, McCarthy JF, Patel JM, Valenstein M. 2009. Exploratory data mining analysis identifying subgroups of patients with depression who are at high risk for suicide. J Clin Psychiatry 70(11):1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal E, Mallah R, Jackson RG, Ball M, Ibrahim ZM, Broadbent M, Dzahini O, Stewart R, Johnston C, Dobson RJ. 2015. Identification of Adverse Drug Events from Free Text Electronic Patient Records and Information in a Large Mental Health Case Register. PLoS One 10(8):e0134208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PB, Jensen LJ, Brunak S. 2012. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13(6):395–405. [DOI] [PubMed] [Google Scholar]
- Karlson EW, Boutin NT, Hoffnagle AG, Allen NL. 2016. Building the Partners HealthCare Biobank at Partners Personalized Medicine: Informed Consent, Return of Research Results, Recruitment Lessons and Operational Considerations. J Pers Med 6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Warner CH, Ivany C, Petukhova MV, Rose S, Bromet EJ, Brown M 3rd, Cai T, Colpe LJ, Cox KL, Fullerton CS, Gilman SE, Gruber MJ, Heeringa SG, Lewandowski-Romps L, Li J, Millikan-Bell AM, Naifeh JA, Nock MK, Rosellini AJ, Sampson NA, Schoenbaum M, Stein MB, Wessely S, Zaslavsky AM, Ursano RJ, Army SC. 2015. Predicting suicides after psychiatric hospitalization in US Army soldiers: the Army Study To Assess Risk and rEsilience in Servicemembers (Army STARRS). JAMA Psychiatry 72(1):49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, Pacheco JA, Tromp G, Pathak J, Carrell DS, Ellis SB, Lingren T, Thompson WK, Savova G, Haines J, Roden DM, Harris PA, Denny JC. 2016. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 23(6):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane IS. 2015. An autism case history to review the systematic analysis of large-scale data to refine the diagnosis and treatment of neuropsychiatric disorders. Biol Psychiatry 77(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreeman F, Liao K, Chibnik L, Hickey B, Stahl E, Gainer V, Li G, Bry L, Mahan S, Ardlie K, Thomson B, Szolovits P, Churchill S, Murphy SN, Cai T, Raychaudhuri S, Kohane I, Karlson E, Plenge RM. 2011. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 88(1):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, Gainer VS, Shaw SY, Xia Z, Szolovits P, Churchill S, Kohane I. 2015. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingren T, Chen P, Bochenek J, Doshi-Velez F, Manning-Courtney P, Bickel J, Wildenger Welchons L, Reinhold J, Bing N, Ni Y, Barbaresi W, Mentch F, Basford M, Denny J, Vazquez L, Perry C, Namjou B, Qiu H, Connolly J, Abrams D, Holm IA, Cobb BA, Lingren N, Solti I, Hakonarson H, Kohane IS, Harley J, Savova G. 2016. Electronic Health Record Based Algorithm to Identify Patients with Autism Spectrum Disorder. PLoS One 11(7):e0159621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyalina S, Percha B, LePendu P, Iyer SV, Altman RB, Shah NH. 2013. Identifying phenotypic signatures of neuropsychiatric disorders from electronic medical records. J Am Med Inform Assoc 20(e2):e297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl KD, Kohane IS, McFadden D, Weber GM, Natter M, Mandel J, Schneeweiss S, Weiler S, Klann JG, Bickel J, Adams WG, Ge Y, Zhou X, Perkins J, Marsolo K, Bernstam E, Showalter J, Quarshie A, Ofili E, Hripcsak G, Murphy SN. 2014. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): architecture. J Am Med Inform Assoc 21(4):615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy TH Jr., Castro VM, Roberson AM, Snapper LA, Perlis RH. 2016. Improving Prediction of Suicide and Accidental Death After Discharge From General Hospitals With Natural Language Processing. JAMA Psychiatry 73(10):1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry AJ, Murphy SN, MacFadden D, Weber G, Simons WW, Orechia J, Bickel J, Wattanasin N, Gilbert C, Trevvett P, Churchill S, Kohane IS. 2013. SHRINE: enabling nationally scalable multi-site disease studies. PLoS One 8(3):e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto R, Li L, Kidd BA, Dudley JT. 2016. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci Rep 6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AJ, Vaze A, Rao S. 2009. Clinical diagnosis of depression in primary care: a meta-analysis. Lancet 374(9690):609–619. [DOI] [PubMed] [Google Scholar]
- Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, Zeng Q, Dubey A, Gainer V, Mendis M, Glaser J, Kohane I. 2009. Instrumenting the health care enterprise for discovery research in the genomic era. Genome research 19(9):1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalichowski R, Keogh D, Chueh HC, Murphy SN. 2006. Calculating the benefits of a Research Patient Data Repository. AMIA Annu Symp Proc:1044. [PMC free article] [PubMed] [Google Scholar]
- Namjou B, Marsolo K, Caroll RJ, Denny JC, Ritchie MD, Verma SS, Lingren T, Porollo A, Cobb BL, Perry C, Kottyan LC, Rothenberg ME, Thompson SD, Holm IA, Kohane IS, Harley JB. 2014. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to Eosinophilic Esophagitis. Front Genet 5:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dushlaine C, Ripke S, Ruderfer DM, Hamilton SP, Fava M, Iosifescu DV, Kohane IS, Churchill SE, Castro VM, Clements CC, Blumenthal SR, Murphy SN, Smoller JW, Perlis RH. 2014. Rare copy number variation in treatment-resistant major depressive disorder. Biol Psychiatry 76(7):536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, Cai T, Goryachev S, Zeng Q, Gallagher PJ, Fava M, Weilburg JB, Churchill SE, Kohane IS, Smoller JW. 2012. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med 42(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polubriaginof F, Quinnies K, Vanguri R, Yahi A, Simmerling M, Ionita-Laza I, Salmasian H, Bakken S, Hripcsak G, Goldstein D, Kiryluk K, Vawdrey DK, Tatonetti NP. 2016. Estimate of disease heritability using 4.7 million familial relationships inferred from electronic health records. BiorXiv 10.1101/066068. [DOI] [Google Scholar]
- Poulin C, Shiner B, Thompson P, Vepstas L, Young-Xu Y, Goertzel B, Watts B, Flashman L, McAllister T. 2014. Predicting the risk of suicide by analyzing the text of clinical notes. PLoS One 9(1):e85733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precision Medicine Initiative (PMI) Working Group. 2015. The Precision Medicine Initiative Cohort Program – Building a Research Foundation for 21st Century Medicine.
- Prieto ML, Ryu E, Jenkins GD, Batzler A, Nassan MM, Cuellar-Barboza AB, Pathak J, McElroy SL, Frye MA, Biernacka JM. 2016. Leveraging electronic health records to study pleiotropic effects on bipolar disorder and medical comorbidities. Transl Psychiatry 6:e870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis BY, Kohane IS, Mandl KD. 2009. Longitudinal histories as predictors of future diagnoses of domestic abuse: modelling study. BMJ 339:b3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, Basford MA, Brown-Gentry K, Balser JR, Masys DR, Haines JL, Roden DM. 2010. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 86(4):560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Denny JC. 2016. Integrating electronic health record genotype and phenotype datasets to transform patient care. Clin Pharmacol Ther 99(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Wajngurt D, Park N, Zheng T. 2007. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A 104(28):11694–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Hoffmann TJ, Melles RB, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Jorgenson E. 2015. Differences in the Genetic Susceptibility to Age-Related Macular Degeneration Clinical Subtypes. Invest Ophthalmol Vis Sci 56(8):4290–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, Li R, Pathak J, Ritchie MD, Roden DM, Verma SS, Tromp G, Prato JD, Bush WS, Akey JM, Denny JC, Capra JA. 2016. The phenotypic legacy of admixture between modern humans and Neandertals. Science 351(6274):737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Karlson EW, Green RC, Kathiresan S, MacArthur DG, Talkowski ME, Murphy SN, Weiss ST. 2016. An eMERGE Clinical Center at Partners Personalized Medicine. J Pers Med 6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 14(7):483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatonetti NP, Denny JC, Murphy SN, Fernald GH, Krishnan G, Castro V, Yue P, Tsao PS, Kohane I, Roden DM, Altman RB. 2011. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 90(1):133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technology. OotNCfHI. December 2016. Office-based Physician Electronic Health Record Adoption, Health IT Quick-Stat #50.
- Technology. OotNCfHI. May 2016. ‘Non-federal Acute Care Hospital Electronic Health Record Adoption,’ Health IT Quick-Stat #47.
- Tran T, Luo W, Phung D, Harvey R, Berk M, Kennedy RL, Venkatesh S. 2014. Risk stratification using data from electronic medical records better predicts suicide risks than clinician assessments. BMC Psychiatry 14:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driest SL, Wells QS, Stallings S, Bush WS, Gordon A, Nickerson DA, Kim JH, Crosslin DR, Jarvik GP, Carrell DS, Ralston JD, Larson EB, Bielinski SJ, Olson JE, Ye Z, Kullo IJ, Abul-Husn NS, Scott SA, Bottinger E, Almoguera B, Connolly J, Chiavacci R, Hakonarson H, Rasmussen-Torvik LJ, Pan V, Persell SD, Smith M, Chisholm RL, Kitchner TE, He MM, Brilliant MH, Wallace JR, Doheny KF, Shoemaker MB, Li R, Manolio TA, Callis TE, Macaya D, Williams MS, Carey D, Kapplinger JD, Ackerman MJ, Ritchie MD, Denny JC, Roden DM. 2016. Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records. JAMA 315(1):47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Verma SS, Pendergrass SA, Crawford DC, Crosslin DR, Kuivaniemi H, Bush WS, Bradford Y, Kullo I, Bielinski SJ, Li R, Denny JC, Peissig P, Hebbring S, De Andrade M, Ritchie MD, Tromp G. 2016. eMERGE Phenome-Wide Association Study (PheWAS) identifies clinical associations and pleiotropy for stop-gain variants. BMC Med Genomics 9 Suppl 1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassertheil-Smoller S, Smoller J. 2015. Chapter 5: Mostly about screening Biostatistics and epidemiology: A primer for health and biomedical professionals, 4th Ed: Springer; p 133–142. [Google Scholar]
- Xu H, Jiang M, Oetjens M, Bowton EA, Ramirez AH, Jeff JM, Basford MA, Pulley JM, Cowan JD, Wang X, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. 2011. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc 18(4):387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Liao KP, Shaw S, Gainer VS, Churchill SE, Szolovits P, Murphy SN, Kohane IS, Cai T. 2015. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. AMIA. [DOI] [PMC free article] [PubMed] [Google Scholar]