Abstract
The placental epigenome plays a vital role in regulating mammalian growth and development. Aberrations in placental DNA methylation are linked to several disease states, including intrauterine growth restriction and preeclampsia. Studying the evolution and development of the placental epigenome is critical to understanding the origin and progression of such diseases. Although high-resolution studies have found substantial variation between placental methylomes of different species, the nature of methylome variation has yet to be characterized within any individual species. We conducted a study of placental DNA methylation at high resolution in multiple strains and closely related species of house mice (Mus musculus musculus, Mus m. domesticus, and M. spretus), across developmental timepoints (embryonic days 15–18), and between two distinct layers (labyrinthine transport and junctional endocrine). We observed substantial genome-wide methylation heterogeneity in mouse placenta compared with other differentiated tissues. Species-specific methylation profiles were concentrated in retrotransposon subfamilies, specifically RLTR10 and RLTR20 subfamilies. Regulatory regions such as gene promoters and CpG islands displayed cross-species conservation, but showed strong differences between layers and developmental timepoints. Partially methylated domains exist in the mouse placenta and widen during development. Taken together, our results characterize the mouse placental methylome as a highly heterogeneous and deregulated landscape globally, intermixed with actively regulated promoter and retrotransposon sequences.
Keywords: DNA methylation, placenta, junctional zone, labyrinthine zone, epigenome
Introduction
The placenta forms the crucial link between mother and developing offspring during mammalian pregnancy. It is responsible for anchoring the fetus to the uterine wall, secretes hormones that adapt maternal physiology, prevents immunological rejection of the fetus, and exchanges substrates between fetal and maternal blood spaces. These functions are strongly conserved across mammals, despite extraembryonic tissues displaying remarkable morphological variation (Furukawa et al. 2014) and placenta-specific genes accumulating a high rate of nonsynonymous mutations (Hughes et al. 2000; Chuong et al. 2010).
To effectively perform its wide array of functions, the placenta is composed of multiple trophoblast cell types which in the mouse are organized into specialized zones. The junctional zone lies proximal to the uterine wall and is composed of invasive endocrine trophoblast cells: the spongiotrophoblast, glycogen and giant cells. These cell types are important in promoting maternal immune tolerance, decidual vascularization and maternal metabolic adjustments that favor fetal nutrient delivery (Hu and Cross 2010). The other main layer of the mouse placenta is called the labyrinthine zone, and lies proximal to the developing embryo. It is composed of a dense network of fetal capillaries and maternal blood spaces that are lined with syncytiotrophoblast cells, which exchange nutrients, gases and waste between mother and fetus (Coan et al. 2005; Sferruzzi-Perri et al. 2009). Genomic studies have provided insight into placentation and diversification of placental morphology (Cross 2000; Roberts and Cooper 2001), and the recent maturation of assays designed to explore epigenetic features allows us to study these phenomena at unprecedented resolution.
DNA methylation occurs primarily on cytosines of CpG dinucleotides in mammals (Bird 1985). Across the genome, most CpGs are methylated. In somatic cells, hypomethylated intervals have an average size of up to a few kilobases, although larger intervals exist. These regions tend to co-locate with promoters and enhancers, and methylation through these intervals is associated with gene silencing and restriction of regulatory activity (Jones 2012). Retrotransposons are methylated in most cell types, and this phenomenon is one form of genomic defense against their expression (Walsh et al. 1998). DNA methylation changes in measurable and consistent ways as tissues differentiate (Seisenberger et al. 2012) and can be compared across species (Molaro et al. 2011; Pai et al. 2011), enabling a precise characterization of cellular and species identity.
In mammals, ∼70% of the cytosines in CpG dinucleotides are methylated in somatic cells (Song et al. 2013), compared with closer to 50% methylated in the placenta (Ehrlich et al. 1982; Razin et al. 1984). A large body of evidence suggests that the placental methylome plays a critical functional role, and targeted assays have implicated aberrant DNA methylation in several placental disease phenotypes, including pre-eclampsia (Yuen et al. 2010; Kulkarni et al. 2011; Hogg et al. 2013) and growth restriction (Banister et al. 2011; Lambertini et al. 2011). The recent advent of whole genome bisulfite sequencing (WGBS) has provided higher resolution maps of DNA methylation in the placenta that have confirmed this lower methylation and detected the presence of partially methylated domains (PMDs), long stretches of the genome where methylation levels drop below the background, primarily hypermethylated state (Schroeder et al. 2013). PMD presence is correlated with changes in gene expression, and outside of placenta they have only been observed in cancer and cultured cell lines (excluding ESCs and iPSCs) (Lister et al. 2009; Hansen et al. 2011).
A recent study found that despite widespread morphological changes, the placenta’s globally lowered methylation state is a common feature across mammalian species (Schroeder et al. 2015). Reduced levels of methylation may allow for species-specific endogenous retroviral activity in the placenta, and could represent regulatory variation that is typically silenced in other tissues (Chuong et al. 2013). Prior work also observed similarity between trophoblast methylomes and oocytes, suggesting minimal de novo methylation in extraembryonic-derived tissues following fertilization (Schroeder et al. 2015). Knocking out de novo methyltransferases Dnmt3a/3b in trophoblasts resulted in few defects compared with wild type at embryonic day 9.5, further supporting a reduction or lack of de novo methylation in placenta to that timepoint (Branco et al. 2016).
Little is known about how the placental epigenome varies within an individual species. A full understanding of this within-species heterogeneity must precede the identification of meaningful between-species differences. In this study, we explored the placental methylome from six strains of three closely related mouse species and report a globally deregulated epigenome relative to other differentiated tissues. Species-specific methylation patterns primarily existed inside retrotransposon subfamilies, particularly in RLTR10 and RLTR20 subfamilies. Regulatory regions such as CpG islands and promoter regions displayed conservation across species. Promoter methylation levels showed a unique distribution wherein highly methylated promoters displayed intermediate methylation levels, rather than near-complete methylation as observed in other tissues. We used differential expression between placenta and other tissues to show that this intermediate methylation remains associated with gene repression.
Additionally, we produced the first purified methylomes of the functionally distinct placental junctional and labyrinthine zones at two developmental timepoints. We identified a subtle but consistent hypomethylation of the junctional zone globally, as well as many concentrated differences at gene promoters. Promoter differences were enriched on the X chromosome, and most differentially methylated promoters were hypermethylated in the junctional zone relative to the labyrinthine. Differential methylation between developmental timepoints uncovered evidence for progressive PMD formation in the placenta as well as widespread de novo methylation, suggesting a role for the epigenome in mediating differentiation of the layers towards term. In spite of earlier studies suggesting the methylation in the placenta is static, our studies demonstrate a dynamic methylation program that varies by species, genetic strain, layer, and developmental timepoint.
Results
Our study relied upon two data sets, which we will refer to as the “interspecific” and “intraspecific” data sets, respectively. The interspecific data set included 12 WGBS methylomes from three species, including four samples from each of Mus musculus musculus, M. m. domesticus, and M. spretus. We reduced the potential confounds of inbreeding and litter effects by intercrossing two strains per species. We sequenced to an average depth of 6.2× per covered CpG per sample, and surveyed an average of 77% of CpGs genome-wide per placenta.
The intraspecific data set concentrated on a single genetic strain, C57BL/6J, the genome reference. We produced 24 WGBS methylomes from the two main placental layers (labyrinthine zone [LZ] and junctional zone [JZ]), from two developmental timepoints (embryonic days 15 [E15] and 18 [E18]), and from male and female siblings collected from three different litters. In this intraspecies data set, each sample was sequenced to an average depth of 1.45× per covered CpG, and surveyed on an average 50% of CpGs genome-wide. Replicates for each factor allowed us to combine these methylomes for high coverage where necessary and increase statistical power to detect differences across factors. Due to poor quality, one sample was thrown out.
The full experimental design and quality control statistics for all methylomes produced for this study can be found in supplementary figure 1 and tables 1 and 2, Supplementary Material online, and descriptions of the mouse strains used can be found in the “Materials and Methods” section.
Placenta DNA Methylation Is Globally Heterogeneous but Highly Conserved at Regulatory Regions
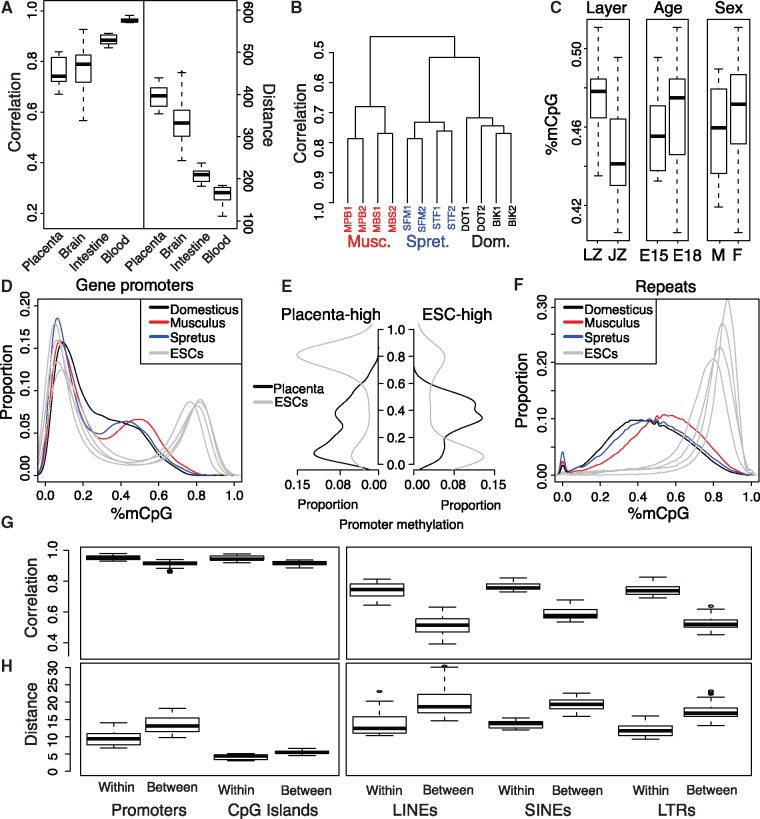
We observed global hypomethylation of the mouse placenta relative to other tissues: genome-wide methylation levels across interspecific samples ranged from 43.3% to 53.8%, and varied by species (ANOVA, P < 0.015). Comparison of placental methylation levels in whole placental samples at single CpG resolution also revealed significantly higher within-tissue heterogeneity when compared with other fully differentiated tissues. To illustrate this, we computed the Pearson correlation and Euclidean distance between all pairs of whole placenta samples from the same species. We plotted these as a boxplot, together with boxplots of pairwise correlation and distance for three other tissues: brain (Lister et al. 2013), intestine (Hon et al. 2013; Kaaij et al. 2013; Sheaffer et al. 2014) and blood (Kieffer-Kwon et al. 2013) (fig. 1A). Despite this within-tissue variability, genome-wide methylation levels clustered reasonably well by strain and species in the placenta (fig. 1B), although they did not precisely capture the true species-level evolutionary relationship (Tucker 2006; Sarver et al., in press). Intra-species samples clustered well by layer, however both comparisons of single-CpG heterogeneity and pairwise binned correlations suffered from substantially lower sequencing depth (supplementary fig. 2A and B, Supplementary Material online). In the intraspecific data set, the junctional zone was less methylated than the labyrinthine zone (P < 0.017) (fig. 1C), but there was no significant difference in global levels of CpG methylation by developmental timepoint or sex.
Fig. 1.
DNA methylation is variable in the placenta, except at regulatory regions. (A) Pearson correlation and Euclidean distance between pairs of whole placenta samples, compared with pairwise correlation and distance between samples of other tissues at single CpG resolution. (B) Hierarchical clustering of pairwise binned correlation for whole placental samples in 1-kb bins. Three-letter codes indicate genetic strain, number indicates individual. (C) Global methylation between layers, timepoints, and sex. (D) Promoter methylation density plot comparing interspecies placenta and ESCs. (E) Promoter methylation distributions in placenta and ESC for genes upregulated in placenta (left) and ESC (right). (F) Retrotransposon methylation density plot. (G and H) Within- and between-species pairwise correlation and distance by genomic feature.
DNA methylation is an important component of transcriptional regulation, with methylation of retrotransposons and gene promoters strongly correlated with their repression (Boyes and Bird 1991). Figure 1D presents methylation levels in gene promoters in the placenta of each mouse species and high quality WGBS embryonic stem cell (ESC) methylomes from four separate projects curated in MethBase (Song et al. 2013; Li et al. 2015; Lu et al. 2014; Harten et al. 2015; Yearim et al. 2015). In the placenta, promoter methylation remained bimodal, but with the high mode typically associated with transcriptional repression shifted from the near-complete methylation seen in other tissues to intermediate levels.
To explore how this reduced promoter methylation level might relate to transcriptional regulation, we identified differentially expressed (DE) genes between 5 E14.5 C57Bl6/J x FVB/n mouse placenta RNA-seq experiments (Mould et al. 2013) and ESC RNA-seq data derived from two studies with matched methylation and expression data (Lu et al. 2014; Yearim et al. 2015). We identified differentially expressed genes (see “Materials and Methods” section) and plotted them by their normalized counts and log fold change between ESC and placenta (supplementary fig. 3A, Supplementary Material online). We filtered for DE genes with a log-fold change of >5 and counts per million (CPM) of <5, leaving us 797 and 733 DE genes expressed higher in placenta and ESC, respectively, and plotted the promoter methylation distributions for these genes (fig. 1E). For genes that were higher expressed in placenta, we observed nearly complete methylation of the promoter in ESCs and hypomethylation of the promoter in placenta. In genes higher expressed in ESCs however, the promoter methylation levels of placenta reached only intermediate levels. This pattern was conserved when considering the promoter methylation distributions of differentially expressed genes between placenta and other tissues, including brain (Lister et al. 2013), intestine (Sheaffer et al. 2014), and blood (Kieffer-Kwon et al. 2013) (supplementary figs. 3 and 4 and all DE genes included in supplementary table 3, Supplementary Material online). These observations indicate that epigenomic repression of transcription in the placenta does not require methylation levels as high as seen in other tissues.
Another unique feature of the placental methylome is the hypomethylation of retrotransposons. Almost all retrotransposons are methylated in most other tissues, but show a relaxed methylation state in the placenta (fig. 1F). Of the 2,191,618 annotated retrotransposons overlapping at least one CpG, we sequenced to a depth of at least five observations per copy in 96.2%, 95.0%, and 81.9% of all retrotransposons in M. m. domesticus, M. m. musculus, and M. spretus, respectively (see “Materials and Methods” section). Comparisons of these interspecies samples revealed uniformly high correlation and low Euclidean distance for promoters and CpG islands, indicating conservation of epigenomic state at these regulatory regions. In contrast, we observed strong species-specific patterns in all classes of retrotransposons (fig. 1G and H). These patterns are consistent with an arms race hypothesis (Crespi and Nosil 2013), where methylation divergence is driven by conflict with genomic parasites.
Quantifying Species- and Layer-Specific Methylation Changes in the Placenta
To identify the biologically meaningful and statistically significant epigenetic differences driven by species, layer, and developmental timepoints, we used RADMeth to find differentially methylated (DM) CpGs, combine the P values of neighboring (within 100 bp) CpGs, and perform false discovery rate correction according to Benjamini and Hochberg (1995).
To compare species, we identified DM CpGs for each species relative to the other two species combined. This allows us to identify features specific to each species.
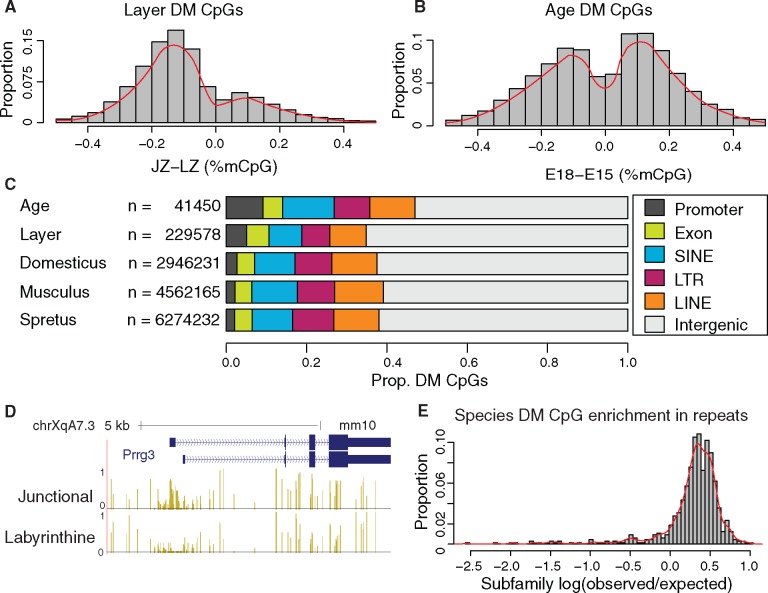
To compare layers and developmental timepoints, we accounted for the full experimental design to avoid confounding by other factors. Importantly, we removed one female sample from the junctional zone (M1043-F5-F15-JZ) from the same developmental timepoint as the missing LZ sample to ensure equal numbers of male and female samples while calling differential methylation. Principle component analysis (Wold et al. 1987) using our identified DM CpGs as the feature set for each factor showed a clear segregation by factor status (supplementary fig. 5, Supplementary Material online). Most of the significantly DM CpGs between layers are hypomethylated in the junctional zone relative to the labyrinthine zone (fig. 2A). We also detected a sizeable number of CpG sites whose methylation level increased between E15 and E18 (fig. 2B), suggesting that while previous studies on Dnmt3a/b knockout mice revealed normal trophoblast formation during early development, de novo methylation likely plays a role in the late stage development and differentiation of the placental layers.
Fig. 2.
Methylomes differ between layers, developmental timepoints, and species. (A and B) Directional DM CpG methylation distributions between layer and age. (C) DM CpG quantity and location by genomic region, with N equal to the total number of DM CpGs for each factor. (D) Example promoter that is differentially methylated between layers. (E) Distribution of enrichment of Mus musculus-specific DM CpGs in retrotransposon subfamilies, showing enrichment (log O/E > 0) of differential methylation in almost every subfamily.
To identify the sources of species-, layer-, and age-specific variation, we investigated DM CpG occupancy inside various genomic regions (fig. 2C). We observed an order of magnitude more differences by species than by layer or age, and DM CpGs seem to be uniformly distributed throughout the genome. To identify the DM CpGs that are most likely to drive meaningful differences in transcriptional regulation, we focused on those located in gene promoters. For each promoter, we counted the number of significantly DM CpGs with a methylation difference of at least 30% between levels for each factor. Between layers, this yielded five genes with at least 10 DM promoter CpGs, with the top two (Srrt and Zmym3) having 45 and 25 DM CpGs, respectively. Three of these genes were on the X-chromosome, and analysis of male and female samples separately show conservation of magnitude and directionality of this differential methylation between sexes (supplementary fig. 6A–C, Supplementary Material online). Subject to the same analysis, differences between ages yielded only two genes with at least 10 DM CpGs (table 1). Of note, the next highest difference between ages (8 DM CpGs) was Tjp1, a human ortholog of which was previously associated with trophoblast cell differentiation and whose promoter was methylated in E18 samples (Pidoux et al. 2010). No gene set was enriched for placenta-related gene ontology terms.
Table 1.
Top Differentially Methylated Promoters between Placental Layers and Developmental Timepoints.
| Rank | Layer | # DM CpGs | Age | # DM CpGs |
|---|---|---|---|---|
| 1 | Srrt | 45 | Cdc42 | 16 |
| 2 | Zmym3a | 25 | Picalm | 12 |
| 3 | Stag2a | 25 | Tjp1 | 8 |
| 4 | Prrg3a | 11 | ||
| 5 | 1810009A15Rik | 10 |
Between layers, there is an enrichment for X-chromosome genes.
Interestingly, differentially methylated gene promoters between layers were enriched on the X-chromosome and primarily hypermethylated in the junctional zone, despite most DM CpGs showing junctional hypomethylation. An example of junctional zone promoter hypermethylation is shown in figure 2D.
An analysis of the X-chromosome revealed global hypomethylation relative to autosomes (, supplementary fig. 7A, Supplementary Material online) in females but similar methylation levels in males. We also observed elevated methylation levels in CpG islands of both male and female X chromosomes relative to autosomes, but with greater levels in female placentas (, supplementary fig. 7B, Supplementary Material online), suggesting CpG islands have elevated methylation levels on the inactive X. The male CpG island methylation increase is slightly enriched in the junctional layer (P < 0.03, supplementary fig. 7C, Supplementary Material online).
To better understand the strong species-specific retrotransposon signals we observed, we utilized the RepeatMasker annotation of retrotransposons in the LINE, SINE, and LTR classes (removing all nonretrotransposons from the annotation). We computed the enrichment of species-specific DM CpGs in each retrotransposon subfamily given each subfamily’s total CpG density. The distribution of observed over expected (O/E) ratios of DM CpG occupancy inside retrotransposon subfamilies is notably shifted to the right, indicating that almost all retrotransposon subfamilies are more differentially methylated between species than expected by chance (fig. 3E). By filtering for subfamilies with at least 50 DM CpGs and O/E ratio of at least 2×, we saw almost exclusive enrichment in RLTR10 and RLTR20 subfamilies (supplementary table 4, Supplementary Material online), notably in the same broad group of ERVs (ERV2) as the retrotransposons bearing species-specific enhancers identified previously (Chuong et al. 2013).
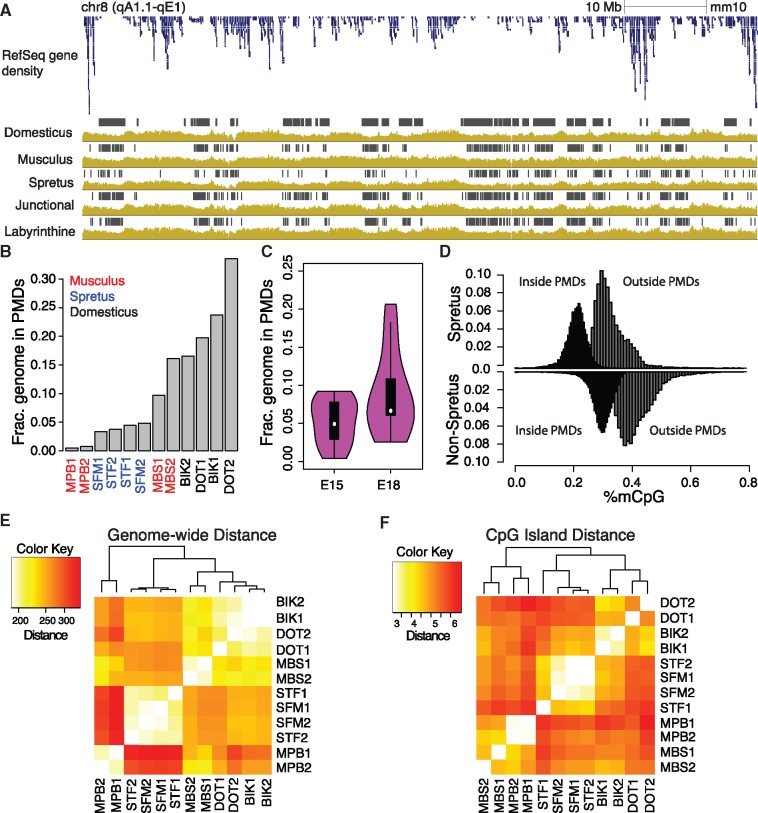
Fig. 3.
Partially methylated domains exist in mouse placenta and are spatially conserved across species. (A) Methylation levels (yellow bar plots) and identified PMD locations (gray boxes) in a selected genomic interval; genes shown in blue. (B) Genomic fraction covered by PMDs in whole placenta samples. (C) Genomic fraction covered by PMDs at E15 and E18 in intra-specific samples. (D) Distributions of CpG methylation levels inside and outside PMDs in Mus spretus and non-M. spretus samples. (E) Pairwise distances between genome-wide methylation profiles (average level in 1-kb bins). (F) Pairwise distance by species in CpG islands.
Progressive PMD Formation in the Mouse Placenta
PMDs are megabase-scale stretches of the genome with consistently low methylation relative to the background of genome-wide equilibrium methylation level. They were first observed in human immortalized cell lines (Lister et al. 2009) and later found to be present in cancer (Hansen et al. 2011; Berman et al. 2012) and then observed in human placenta methylomes (Schroeder et al. 2013). In contrast to a prior study that reported the absence of PMDs in mouse placenta (Schroeder et al. 2015), we identified a highly reproducible segmentation of the methylome in all three mouse species into background and PMD regions using the HMM approach described in Song et al. (2013) (fig. 3A). This method segments the methylome based on consecutive observations of weighted average methylation levels inside 1-kb bins, reducing the effect of local hypermethylation introduced by regulatory regions such as gene promoters or enhancers. We compared the previously reported mouse placenta methylome to our own data and found PMDs covering 26.5% of the genome and reaching similar in-PMD methylation levels to our own using the same identification technique, with a bin size of 20 kb to compensate for substantially lower coverage than our own whole placental samples (supplementary fig. 8A, Supplementary Material online).
Placental PMDs are located in gene poor regions and exist in both layers of the placenta (fig. 3A). Taking the union of PMDs across all interspecific samples, we observed an overlap with 8,828 gene promoters compared with an expected 12,025, given the size of the genome and assuming a hypergeometric distribution of the overlaps. While PMD locations stayed generally constant, the overall fraction of the genome inside PMDs varied substantially, from nearly absent in the MPB strain to extremely prevalent in the DOT strain (fig. 3B). For this interspecific data, the collection method for the whole placenta samples allowed the age of the embryo at dissection to vary by up to 5 days (see “Materials and Methods” section), and we observed an order-of-magnitude decrease in embryonic weight in the MPB strain that suggests an earlier developmental stage and absent PMDs (supplementary table 1, Supplementary Material online). Therefore, we hypothesized that PMDs gradually appear over developmental time in the placenta. In the intraspecific data, analysis of PMD size between developmental timepoints in late gestation revealed an increase in the size of PMDs between E15 and E18 (P < 0.032) (fig. 3C). While PMDs widened over time, the methylation level inside PMDs did not show significant differences between timepoints (P < 0.47). The methylation levels outside of PMDs increased slightly but not significantly, consistent with the observed global methylation levels of the two timepoints. Corroborating the observations reviewed in Novakovic and Saffery (2013), the Dnmt1 promoter is hypomethylated in mouse placenta at all timepoints, in all layers, and in all species, suggesting that PMD formation in the mouse placenta is likely not driven by differential expression of Dnmt1.
The globally lower methylation level in M. spretus compared with the other two species led to lower average methylation inside PMDs (P < 0.0006) (fig. 3D) but similar PMD depth. Interestingly, despite the lack of PMDs in MPB leading to large overall methylation differences between those and the other M. musculus samples (fig. 3E), CpG island methylation remained extremely close regardless of PMD presence. This suggests that CpG islands remain under direct regulation even inside PMDs (fig. 3F). Average methylation levels for CpG islands inside PMDs were slightly elevated in our mouse placenta samples, corroborating the finding in (Schroeder et al. 2013). However, few CpG islands inside placental PMDs displayed methylation >80% (supplementary fig. 8B, Supplementary Material online), while methylation of CpG islands inside cancer PMDs regularly exceeds 80% (Toyota et al. 1999; Hansen et al. 2011).
Discussion
The placenta is a rapidly evolving organ that plays a temporary but essential role in mammalian development. In this study, we explored the role of the placental epigenome on its function and evolution. We characterized the placental epigenome as globally deregulated, possessing within-tissue methylation variance substantially higher than in other differentiated tissues. This noisy and globally hypomethylated state, relative to other tissues, remains a fundamentally distinct and poorly understood feature of placental cells. Recent studies have shown that this low methylation likely originated very early and persists through the trophoblast lineage (Branco et al. 2016). In addition, the globally low methylation appears to be a common feature across distant species of mammal (Schroeder et al. 2015; Branco et al. 2016). We showed that global variation in placental methylation is conserved in differentiated placental layers and across later timepoints. The source of this variability could be rooted in the placenta’s transient nature, allowing its epigenome to erode during development without much harm to the overall success of the embryo. Another possibility is that the placenta’s adaptive response to its environment (Fowden and Moore 2012; Sferruzzi-Perri and Camm 2016) manifests in global methylation changes in subpopulations of cells not yet detectable without applying single-cell methods.
Despite globally heterogeneous methylation patterns, we observed similar levels of methylation across species at regulatory regions, including gene promoters and CpG islands both inside and outside of promoters (as annotated in the UCSC table browser, see “Materials and Methods” section). While methylation in placental promoters showed within-tissue consistency, the actual distribution was placenta-specific, showing a shift in the “high” methylation range that remained correlated with repressive effects on gene expression. This opens up an interesting question: if the background methylation level in differentiated somatic cells is higher than needed for its role in gene expression, why does it remain at consistently high levels with such small variation between somatic cell types?
We produced the first methylomes of the junctional and labyrinthine zones and observed substantial de novo methylation between E15 and E18. Our results suggest that while Dnmt3a/3b may not be necessary in the early development of the trophoblast (Branco et al. 2016), it could play a role in later maturation of the placental layers. Using male and female placental methylomes, we indirectly explored methylation on the inactivated paternal X chromosome in females. We identified globally lower methylation levels in females, but elevated CpG island methylation on the X chromosome, which contributes to X inactivation (Csankovszki et al. 2001).
The impact of the globally lowered methylation state on retrotransposon activity in the placenta also remains poorly understood. Our analysis revealed that retrotransposon families were more likely to show species-specific methylation than expected, with differentially methylated CpGs especially enriched in members of the RLTR10 and RLTR20 subfamilies. This pattern stands in stark contrast to nonplacental tissues, where retrotransposons are usually methylated (Bestor 2000). This elevated tolerance to retro-element hypomethylation and expression may be required for placenta-specific phenomena, such as previously identified exaptation events of specific retrotransposons by the placenta to evade the maternal immune system (Mi et al. 2000; Feschotte and Gilbert 2012) or co-option of certain retrotransposon subfamilies as placenta-specific enhancer elements (Chuong et al. 2013). In contrast to the study by Chuong et al. (2013), which identified placenta-specific enhancer elements at mouse-specific retrotransposons not present in the rat, we identified differential methylation in retrotransposons that are present in all species studied. This could represent more recent adaptations of the placental regulatory program, although further study is needed.
Though selection to maintain genome integrity in extra-embryonic tissues will certainly be lower than in the embryo or its germline, too much retro-element expression still represents a potential danger to genome integrity. Retro-element hypomethylation may be possible due to the redundant nature of mechanisms for retrotransposon silencing (Aravin et al. 2007; Reichmann et al. 2013), allowing their sequences to act as an enhancers for nearby genes while limiting their transcriptional activity. In turn, differentially methylated subfamilies may help fuel the rapid diversification of placenta-specific regulatory networks. Although the mechanisms of retrotransposon-derived enhancers have been studied previously (McDonald et al. 1997; Ruda et al. 2004), further study is needed to explore the direct impact of differential retrotransposon methylation state between species on the transcription of nearby orthologous genes, and to understand what, if any, role retrotransposon-mediated transcription may play in the human placenta.
To function properly, the placenta must invade and integrate with maternal tissues, a process that shares some similarities to the invasive behaviors of some cancers (Novakovic and Saffery 2013). Partially methylated domains exist in the mouse placenta, are absent in our smallest, developmentally young embryos, and widen between E15 and E18, suggesting that they arise as a function of developmental time. These PMDs share conserved locations across species, layer, and are found in the same gene-poor regions as in cancers. PMDs correlate with late replicating domains in human (Berman et al. 2012), and therefore may arise in both cancer and placental cells as a consequence of rapid cell division outpacing the maintenance of methylation in these regions. CpG island hypermethylation is a hallmark of cancer methylomes and is enriched within cancer PMDs. Mouse placenta PMDs show no CpG island hypermethylation of the type reported in the PMDs of cancer methylomes (Berman et al. 2012). Further studies are required to determine if PMDs have any significance in placental function. However, this shared feature of placental and cancer methylomes is striking and any model to explain PMDs will be more appealing if its explanatory power extends to both cancer and placenta epigenomes.
Materials and Methods
Interspecies Whole Placenta Tissue Collection
All animal husbandry, experimental procedures, and personnel were approved by the University of Southern California’s Institutional Animal Care and Use Committee, protocol #11394. Mice were housed under a 14:10 hour light cycle with food and water ad libitum. To investigate species differences in placental methylation, crosses between wild derived inbred strains were established, developed and distributed by François Bonhomme and colleagues (U. Montpellier). For M. domesticus, we made reciprocal crosses between strains BIK (originally isolated from Kefar Galim, Israel) and DOT (Tahiti); for M. musculus, MPB (Bialowieza, Poland) and MBS (Sokolovo, Bulgaria); and for M. spretus, STF (Fondouk Djedid, Tunisia) and SFM (Montpellier, France). For all crosses, a single stud and dam were housed together for four and a half days, and then split. 10 days later, females were euthanized, uteri were collected and the number of viable conceptuses counted, leading to gestational ages between E11 and E16. The embryos and placenta were dissected and weighed. Two placentae were sampled from a single litter in each cross direction and snap frozen in liquid nitrogen. Placenta are named according to their maternal strain. We attempted to sample only female placenta, through two replicated attempts to PCR-amplify two Y- and one X-linked region (Kunieda et al. 1992), but for logistical reasons we had to include two male placentae.
Intraspecies Junctional and Labyrinthine Zone Tissue Collection
All experiments were carried out under the UK Home Office Animals (Scientific Procedures) Act 1986. C57Bl/6 females were housed under dark:light 12:12 conditions with free access to water and the standard diet used in the University of Cambridge Animal Facility. At 8–10 weeks, females were mated with C57Bl/6 males and the day a copulatory plug was found was denoted as embryonic day 1 of pregnancy (term = 20.5 days). On embryonic days 15 or 18 of pregnancy (days correspond to the periods of rapid placental and fetal growth, respectively), mouse dams were schedule 1 killed by cervical dislocation. Uteri were collected and the number of viable conceptuses counted. Embryos and placentas were dissected and weighed. Each placenta from the litter was rapidly separated into the functionally distinct zones, the labyrinthine transport and junctional endocrine zones (Sferruzzi-Perri et al. 2009), in ice cold sterile PBS before rapid snap freezing in liquid nitrogen. Fetal tails were kept for sexing using standard genotyping methods including using primers to detect the SRY gene (5′-CCCAGCATGCAAAATACAGA-3′ and 5′-TCAACAGGCTGCCAATAAAA-3′), an internal control gene (5′-AGTGGCTAACGCTGAGTGGT-3′ and 5′-GTGCCTGTCGGAGGAGAAC-3′) and with agarose gel electrophoresis. From each litter, the male and female placenta with its weight closest to the litter mean was used for further analysis.
DNA Extraction and Methylation Assay
Using a Qiagen DNA extraction kit, DNA was extracted and purified for all inter- and intraspecies samples. DNA was fragmented to 100- to 300-bp fragments by sonication and end repaired before ligation of methylated sequencing adapters. Bisulfite treatment was performed using the Zymo EZ DNA Methylation Gold kit. Following bisulfite treatment, DNA fragments were de-salted and size selected to produce a 200- to 300-bp short-insert library, subjected to PCR, and size selected again before 100-bp paired-end reads were sequenced using an Illumina Hiseq4000.
Data Analysis
Reads were mapped to the mm10 reference genome using WALT (Chen et al. 2016). Calculation of methylation levels, bisulfite conversion rate, and identification of PMDs was performed as described in (Song et al. 2013), and all values are available in supplementary tables 1 and 2, Supplementary Material online. Weighted methylation levels as defined by Schultz et al. (2012) were used to calculate average methylation levels in genomic regions. All browser plots were created using the UCSC genome browser tool (Kent et al. 2002). Promoters were defined as ±1 kb from the mm10 RefSeq TSS based on the observation that hypomethylation frequently occurs there on the kilobase scale (Molaro et al. 2011). CpG islands were identified as described in Gardiner-Garden and Frommer (1987). Retrotransposon copies were annotated by RepeatMasker (Smit et al. 1996) and downloaded from the UCSC Table Browser (Karolchik et al. 2004). Pearson correlation and Euclidean distance at single CpG resolution in whole placenta samples were calculated from the 2,801,446 CpG sites with 5× or greater sequencing depth across all samples. Pairwise comparisons between whole placental samples included only within-species comparisons. Intraspecies single CpG site correlations and distances were computed from 8,180 CpG sites with 3× or greater sequencing depth across all samples. Boxplots for brain, intestine, and blood were produced with a random set of CpG sites downsampled to the number covered in placental samples. Pearson correlation, Euclidean distance, and distributions were produced using only promoters, CpG islands, and retrotransposons with at least five CpG observations to reduce the discretizing effect of low-coverage observations.
ANOVA between global methylation levels with species as the factor was performed in R as a one-way analysis of variance. Differentially methylated CpGs between species were called using (Dolzhenko and Smith 2014) with species-specific methylation signatures identified using the other two species as background samples. Multiple testing correction of combined DM CpG P values was done as described in (Benjamini and Hochberg 1995) using an alpha level of 0.05. Observed over expected (O/E) ratios used for enrichment and depletion analysis of subfamilies were calculated as follows:
| (1) |
Public RNA-seq reads were mapped using STAR (Dobin et al. 2013). BAM files were converted to read counts using HT-seq (Anders et al. 2014) and differentially expressed genes were identified using edgeR (Robinson et al. 2010). We placed an upper bound on the CPM of differentially expressed genes analyzed to focus on those genes that were nearly silenced in one cell type relative to the other.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to members of the Smith lab for their helpful discussion and comments. Brent Young assisted with mouse husbandry and experiments. Funding was provided by an EU COST ACTION grant and Erasmus Traineeship (J.L.T.), a Royal Society Dorothy Hodgkin Fellowship (A.N.S.P.), and National Insitutes of Health grants GM098536 (M.D.D.) and HG005238 (A.D.S.). The authors declare no conflict of interest.
Author Contributions
B.E.D., A.D.S., and M.D.D. designed the study. Bioinformatic analysis was performed by B.E.D. and A.D.S. Whole placental sample dissection and library preparation was performed by M.D.D. Placental layer dissection was performed by J.L.T. and A.N.S.P. The manuscript was written by B.E.D., M.D.D. and A.D.S. with input from A.N.S.P. All authors read and approved the final manuscript.
References
- Anders S, Pyl PT, Huber W.. 2014. Htseq: a python framework to work with high-throughput sequencing data. Bioinformatics 312:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J.. 2007. The piwi-pirna pathway provides an adaptive defense in the transposon arms race. Science 3185851:761–764. [DOI] [PubMed] [Google Scholar]
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ.. 2011. Infant growth restriction is associated with distinct patterns of dna methylation in human placentas. Epigenetics 67:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 571:289–300. [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. 2012. Regions of focal dna hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 441:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. 2000. The dna methyltransferases of mammals. Hum Mol Genet. 916:2395–2402. [DOI] [PubMed] [Google Scholar]
- Bird AP. 1985. Cpg-rich islands and the function of dna methylation. Nature 3216067:209–213. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A.. 1991. Dna methylation inhibits transcription indirectly via a methyl-cpg binding protein. Cell 646:1123–1134. [DOI] [PubMed] [Google Scholar]
- Branco MR, King M, Perez-Garcia V, Bogutz AB, Caley M, Fineberg E, Lefebvre L, Cook SJ, Dean W, Hemberger M, et al. 2016. Maternal dna methylation regulates early trophoblast development. Dev Cell 362:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith AD, Chen T.. 2016. Walt: fast and accurate read mapping for bisulfite sequencing. Bioinformatics 3222:3507–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MK, Soares MJ, Baker JC.. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 453:325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Tong W, Hoekstra HE.. 2010. Maternal–fetal conflict: rapidly evolving proteins in the rodent placenta. Mol Biol Evol. 276:1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan P, Ferguson-Smith A, Burton G.. 2005. Ultrastructural changes in the interhaemal membrane and junctional zone of the murine chorioallantoic placenta across gestation. J Anat. 2076:783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi B, Nosil P.. 2013. Conflictual speciation: species formation via genomic conflict. Trends Ecol Evol. 281:48–57. [DOI] [PubMed] [Google Scholar]
- Cross JC. 2000. Genetic insights into trophoblast differentiation and placental morphogenesis In Seminars in cell & developmental biology. volume 11(2). Academic Press. Elsevier; p. 105–113. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R.. 2001. Synergism of xist rna, dna methylation, and histone hypoacetylation in maintaining x chromosome inactivation. J Cell Biol. 1534:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. Star: ultrafast universal rna-seq aligner. Bioinformatics 291:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, Smith AD.. 2014. Using beta-binomial regression for high-precision differential methylation analysis in multifactor whole-genome bisulfite sequencing experiments. BMC Bioinformatics 151:215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, Gehrke C.. 1982. Amount and distribution of 5-methylcytosine in human dna from different types of tissues or cells. Nucleic Acids Res. 108:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C.. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 134:283–296. [DOI] [PubMed] [Google Scholar]
- Fowden A, Moore T.. 2012. Maternal-fetal resource allocation: co-operation and conflict. Placenta 33:e11–e15. [DOI] [PubMed] [Google Scholar]
- Furukawa S, Kuroda Y, Sugiyama A.. 2014. A comparison of the histological structure of the placenta in experimental animals. J Toxicol Pathol 271:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M.. 1987. Cpg islands in vertebrate genomes. J Mol Biol. 1962:261–282. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 438:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harten SK, Oey H, Bourke LM, Bharti V, Isbel L, Daxinger L, Faou P, Robertson N, Matthews JM, Whitelaw E.. 2015. The recently identified modifier of murine metastable epialleles, rearranged l-myc fusion, is involved in maintaining epigenetic marks at cpg island shores and enhancers. BMC Biol. 131:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Blair JD, McFadden DE, von Dadelszen P, Robinson WP.. 2013. Early onset pre-eclampsia is associated with altered dna methylation of cortisol-signalling and steroidogenic genes in the placenta. PloS One 85:e62969.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B.. 2013. Epigenetic memory at embryonic enhancers identified in dna methylation maps from adult mouse tissues. Nat Genet. 4510:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Cross JC.. 2010. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol. 542:341.. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Garbayo JM, Roberts RM.. 2000. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc Natl Acad Sci. 977:3319–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of dna methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 137:484–492. [DOI] [PubMed] [Google Scholar]
- Kaaij LT, van de Wetering M, Fang F, Decato B, Molaro A, van de Werken HJ, van Es JH, Schuijers J, de Wit E, de Laat W, et al. 2013. Dna methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 145:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ.. 2004. The ucsc table browser data retrieval tool. Nucleic Acids Res. 32(Suppl 1):D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.. 2002. The human genome browser at ucsc. Genome Res. 126:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Tang Z, Mathe E, Qian J, Sung M-H, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 1557:1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Chavan-Gautam P, Mehendale S, Yadav H, Joshi S.. 2011. Global dna methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol. 302:79–84. [DOI] [PubMed] [Google Scholar]
- Kunieda T, Xian M, Kobayashi E, Imamichi T, Moriwaki K, Toyoda Y.. 1992. Sexing of mouse preimplantation embryos by detection of y chromosome-specific sequences using polymerase chain reaction. Biol Reprod. 464:692–697. [DOI] [PubMed] [Google Scholar]
- Lambertini L, Lee T-L, Chan W-Y, Lee M-J, Diplas A, Wetmur J, Chen J.. 2011. Differential methylation of imprinted genes in growth-restricted placentas. Reprod Sci. 1811:1111–1117. [DOI] [PubMed] [Google Scholar]
- Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z.. 2015. Distinct roles of dnmt1-dependent and-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol. 161:115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 3416146:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human dna methylomes at base resolution show widespread epigenomic differences. Nature 4627271:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y.. 2014. Role of tet proteins in enhancer activity and telomere elongation. Genes Dev. 2819:2103–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JF, Matyunina LV, Wilson S, Jordan IK, Bowen NJ, Miller WJ.. 1997. Ltr retrotransposons and the evolution of eukaryotic enhancers In Evolution and impact of transposable elements. Netherlands: Springer; p. 3–13. [PubMed] [Google Scholar]
- Mi S, Lee X, Li X-P, Veldman GM, Finnerty H, Racie L, Lavallie E, Tang X-Y, Edouard P, Howes S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 4036771:785–789. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD.. 2011. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 1466:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AW, Pang Z, Pakusch M, Tonks ID, Stark M, Carrie D, Mukhopadhyay P, Seidel A, Ellis JJ, Deakin J, et al. 2013. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for x inactivation. Epigenet Chromatin 61:19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Saffery R.. 2013. Placental pseudo-malignancy from a dna methylation perspective: unanswered questions and future directions. Front Genet. 4285:10–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y.. 2011. A genome-wide study of dna methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 72:e1001316.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux G, Gerbaud P, Gnidehou S, Grynberg M, Geneau G, Guibourdenche J, Carette D, Cronier L, Evain-Brion D, Malassiné A, et al. 2010. Zo-1 is involved in trophoblastic cell differentiation in human placenta. Am J Physiol Cell Physiol. 2986:C1517–C1526. [DOI] [PubMed] [Google Scholar]
- Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, Sciaky-Gallili N, Cedar H.. 1984. Variations in dna methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci. 818:2275–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J, Reddington JP, Best D, Read D, Öllinger R, Meehan RR, Adams IR.. 2013. The genome-defence gene tex19. 1 suppresses line-1 retrotransposons in the placenta and prevents intra-uterine growth retardation in mice. Hum Mol Genet. 229:1791–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J, Cooper D.. 2001. Pathogenesis and genetics of pre-eclampsia. Lancet 3579249:53–56. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 261:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruda V, Akopov S, Trubetskoy D, Manuylov N, Vetchinova A, Zavalova L, Nikolaev L, Sverdlov E.. 2004. Tissue specificity of enhancer and promoter activities of a herv-k (hml-2) ltr. Virus Res. 1041:11–16. [DOI] [PubMed] [Google Scholar]
- Sarver B, Keeble S, Cosart T, Tucker P, Dean MD.. in press. Phylogenomic insights into genome evolution and speciation in mice. Genome Biol Evol. 93:726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HOK, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, et al. 2013. The human placenta methylome. Proc Natl Acad Sci. 11015:6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Jayashankar K, Douglas KC, Thirkill TL, York D, Dickinson PJ, Williams LE, Samollow PB, Ross PJ, Bannasch DL, et al. 2015. Early developmental and evolutionary origins of gene body dna methylation patterns in mammalian placentas. PLoS Genet. 118:e1005442.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR.. 2012. levelingthe playing field for analyses of single-base resolution dna methylomes. Trends Genet. 2812:583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W.. 2012. The dynamics of genome-wide dna methylation reprogramming in mouse primordial germ cells. Mol Cell 486:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sferruzzi-Perri AN, Camm EJ.. 2016. The programming power of the placenta. Front Physiol. 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sferruzzi-Perri AN, Macpherson AM, Roberts CT, Robertson SA.. 2009. Csf2 null mutation alters placental gene expression and trophoblast glycogen cell and giant cell abundance in mice. Biol Reprod. 811:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schübeler D, Kaestner KH.. 2014. Dna methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 286:652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley R, Green P.. 1996. Repeatmasker. Available from: http://www.repeatmasker.org.
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD.. 2013. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PloS One 812:e81148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa J-PJ.. 1999. Cpg island methylator phenotype in colorectal cancer. Proc Natl Acad Sci. 9615:8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK. 2006. Systematics of the genus mus. Mouse Biomed Res Hist Wild Mice Genet. 1:13–21. [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH.. 1998. Transcription of iap endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 202:116–117. [DOI] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P.. 1987. Principal component analysis. Chemometrics Intell Lab Syst. 2(1–3):37–52. [Google Scholar]
- Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm J-P, Nissim-Rafinia M, Cohen A-HS, Rippe K, Meshorer E, et al. 2015. Hp1 is involved in regulating the global impact of dna methylation on alternative splicing. Cell Rep. 107:1122–1134. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Peñaherrera MS, von Dadelszen P, McFadden DE, Robinson WP.. 2010. Dna methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 189:1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.