Abstract
Background: Real-time knowledge of the somatic genome can influence management of patients with metastatic castration-resistant prostate cancer (mCRPC). While routine metastatic tissue biopsy is challenging in mCRPC, plasma circulating tumor DNA (ctDNA) has emerged as a minimally invasive tool to sample the tumor genome. However, no systematic comparisons of matched “liquid” and “solid” biopsies have been performed that would enable ctDNA profiling to replace the need for direct tissue sampling.
Methods: We performed targeted sequencing across 72 clinically relevant genes in 45 plasma cell-free DNA (cfDNA) samples collected at time of metastatic tissue biopsy. We compared ctDNA alterations with exome sequencing data generated from matched tissue and quantified the concordance of mutations and copy number alterations using the Fisher exact test and Pearson correlations.
Results: Seventy-five point six percent of cfDNA samples had a ctDNA proportion greater than 2% of total cfDNA. In these patients, all somatic mutations identified in matched metastatic tissue biopsies were concurrently present in ctDNA. Furthermore, the hierarchy of variant allele fractions for shared mutations was remarkably similar between ctDNA and tissue. Copy number profiles between matched liquid and solid biopsy were highly correlated, and individual copy number calls in clinically actionable genes were 88.9% concordant. Detected alterations included AR amplifications in 22 (64.7%) samples, SPOP mutations in three (8.8%) samples, and inactivating alterations in tumor suppressors TP53, PTEN, RB1, APC, CDKN1B, BRCA2, and PIK3R1. In several patients, ctDNA sequencing revealed robust changes not present in paired solid biopsy, including clinically relevant alterations in the AR, WNT, and PI3K pathways.
Conclusions: Our study shows that, in the majority of patients, a ctDNA assay is sufficient to identify all driver DNA alterations present in matched metastatic tissue and supports development of DNA biomarkers to guide mCRPC patient management based on ctDNA alone.
Circulating cell-free tumor DNA (ctDNA) is an emerging biomarker across a range of solid malignancies, including metastatic castration-resistant prostate cancer (mCRPC). Cell-free DNA (cfDNA) is shed into the bloodstream by nonmalignant and cancer cells, but in mCRPC patients the proportion of tumor-derived cfDNA (ctDNA fraction) is frequently greater than 1%, enabling comprehensive tumor genome profiling (1–4). Several recent studies have demonstrated that mutations and copy number changes in the plasma cfDNA of mCRPC patients are consistent with somatic landscapes previously established through metastatic tissue profiling (1–3,5–9). However, no systematic comparisons of matched liquid and solid biopsies have been performed.
mCRPC is a lethal disease and an inevitable consequence of managing castrate-sensitive prostate cancer with androgen deprivation therapy. The treatment landscape of mCRPC is increasingly complex and confounded by partial and unpredictable cross-resistance between androgen receptor (AR) pathway-directed agents (eg, enzalutamide and abiraterone) and a shifting consensus on when to utilize taxane-based chemotherapies (10). Molecular subtyping breakthroughs have spurred clinical trials of targeted agents aimed at distinct genotypes, including poly (ADP-ribose) polymerase (PARP) inhibitors in patients with biallelic defects in genes involved in homologous recombination (11) and PI3K/AKT pathway inhibition in patients with PTEN deletions (12). Thus, there is an urgent need for real-time and practical tumor biomarkers to guide therapy selection.
Tissue biopsies remain the gold standard for biomarker development, with contemporary clinical trials frequently mandating collection of solid tumor material for patient stratification or eligibility. However, routine biopsy of CRPC metastases may not be feasible because of its cost, morbidity, and low yield due to the bone-predominant metastatic spread of prostate cancer. This prevents the enrollment of patients with nonaccessible metastatic lesions and suffers from a biopsy failure rate of 25% to 75% in some CRPC case series (13–17). By contrast, ctDNA profiling is theoretically available to any patient able to provide a blood sample and can be sequentially performed on the same patient with minimal morbidity and modest cost. Nevertheless, until ctDNA is proven to represent the genomic information detected in solid tissue, it cannot replace the need for metastatic tissue biopsy of patients. To address this, we profiled 45 matched metastatic tissue and liquid biopsies from mCRPC patients and quantified the concordance of somatic alterations across key driver genes.
Methods
Clinical Cohort
Patients with mCRPC and evidence of progression on the Prostate Cancer Clinical Trials Working Group (PCWG2) criteria were prospectively enrolled in a tissue acquisition protocol at one of five investigational sites (18,19). Approval for this study was granted by the local ethics review board. Written informed consent was obtained from all participants prior to enrollment. A computerized tomography–guided core needle biopsy was obtained from an accessible metastatic lesion in bone or soft tissue. Patients underwent paired collection of blood (for cfDNA) on the day of tumor biopsy.
Targeted cfDNA Sequencing
Aliquots of plasma and buffy coat were isolated from blood collected in EDTA tubes and stored at –80 °C prior to DNA extraction, performed as previously described (1,20). We employed a targeted sequencing strategy capturing the exonic regions of 72 genes (total = 950 kb). For each sample, 10 to 100 ng of DNA was used for library preparation and quantification according to our established protocols (20). Pools of up to 25 purified libraries were hybridized to the capture panel for a minimum of 16 hours at 47 °C. The subsequent wash, recovery, and amplification of the captured regions were performed according to the NimbleGen SeqCap EZ system protocols (Roche, Basel, Switzerland). Final libraries were purified with Agencourt AMPure beads (Beckman Coulter, CA) and quantitated with the KAPA quantitative polymerase chain reaction (qPCR) kit (Roche, Basel, Switzerland). Pools were diluted to 20 pM and were sequenced on Illumina MiSeq (V3 600 cycle kit) or HiSeq 2500 (V4 250 cycle kit) machines. Protocols for data analyses and comparisons with matched tissue data are provided in the Supplementary Methods (available online).
Tissue Exome Sequencing
Metastatic tissue biopsies were verified for tumor content and formalin-fixed for pathology assessment. DNA was isolated from formalin-fixed paraffin-embedded tumor sample cores under Clinical Laboratory Improvement Amendments (CLIA) conditions. For each sample, 200 ng of DNA was used for library preparation using KAPA library preparation kits (Roche, Basel, Switzerland) with Illumina adapters (San Diego, CA). Library quantification was carried out with an Agilent Bioanalyzer (Santa Clara, CA) before and after PCR amplification (12 cycles). Target capture was performed with the Nimblegen SeqCap EZ Exome kit (Roche, Basel, Switzerland). Hybridized libraries were washed and amplified for standardized input, and paired-end 2×75 bp sequencing was performed using an Illumina HiSeq 4000.
Statistical Analysis
Somatic mutations unique to one biopsy type were identified using two-sided Fisher exact tests. P values of less than .01 were considered statistically significant. Concordance of copy number calls between solid and liquid biopsies was quantified using Pearson correlation of coverage log ratios. cfDNA yield and prostate-specific antigen (PSA) concentration were compared between patients with and without detectable ctDNA using the rank-sum test. All statistical tests were two-sided.
Results
ctDNA Fractions and Somatic Landscape
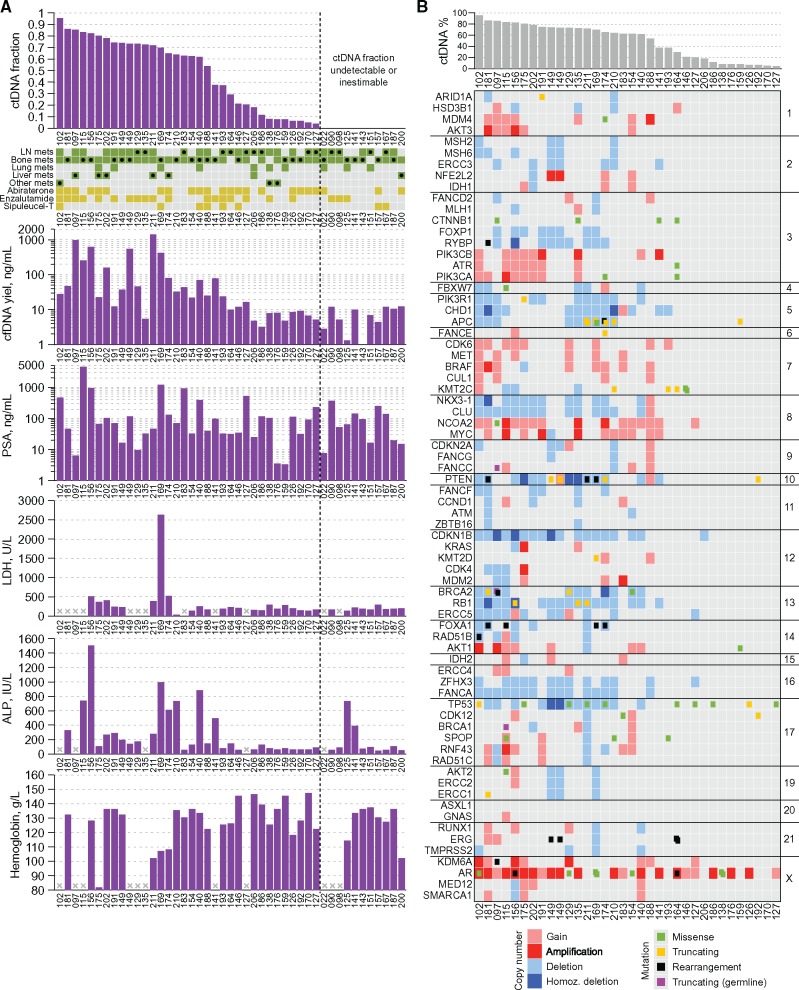
The Stand Up 2 Cancer/Prostate Cancer Foundation West Coast Dream Team (WCDT) is an ongoing prospective study collecting mCRPC patient metastatic tissue biopsies and analyzing histologic, genomic, and transcriptomic variables in the context of clinical outcomes (18). We acquired plasma samples collected at time of metastatic tissue biopsy from patients enrolled in the WCDT study considering only patients (n = 42) where 1) tissue biopsy was successful and 2) exome sequencing data of tumor tissue was available. Three patients provided matched biopsies at two time points, meaning a total of 45 paired solid and liquid biopsies were available for study. Thirty-five of 45 samples (77.8%) were collected from patients who had developed progressive disease following at least one line of AR-targeted therapy (Table 1, Figure 1A;Supplementary Table 1, available online).
Table 1.
Summary of clinical characteristics in patients at time of cfDNA collection (n = 45)*
| Characteristics | No. (%) |
|---|---|
| Median age (range), y | 70 (45–90) |
| ECOG Performance status | |
| 0 | 26 (57.8) |
| 1 | 16 (35.6) |
| 2 | 2 (4.4) |
| NA | 1 (2.2) |
| Median PSA (range), ng/mL | 70.2 (3.4–4478) |
| Median LDH (range), U/L | 200 (31–2643) |
| Median ALP (range), U/L | 107 (46–1506) |
| Median hemoglobin (range), g/L | 130 (82–147) |
| Median cfDNA yield (range), ng/mL | 12.2 (1.0–1380) |
| Bone metastases | 41 (91.1) |
| Lung metastases | 10 (22.2) |
| Liver metastases | 6 (13.3) |
| Prior abiraterone | 22 (48.9) |
| Prior enzalutamide | 21 (46.7) |
| Prior abiraterone + enzalutamide | 10 (22.2) |
ALP = alkaline phosphatase; cfDNA = cell-free DNA; ECOG = Eastern Cooperative Oncology Group; LDH = lactate dehydrogenase; NA = not applicable; PSA = prostate-specific antigen.
Figure 1.
Somatic alterations detected in plasma cell-free DNA. A) Schematic showing the proportion of cell-free DNA that was tumor derived (the circulating cell-free tumor DNA [ctDNA] fraction) and the relationship of this variable to select clinical characteristics. The grid provides an overview of metastatic locations in each patient at the time of sampling (green), with filled black circles indicating the region that was subjected to tissue biopsy concomitant to plasma collection. Orange squares denote prior exposure to (and progression on) three major systemic therapies for metastatic castration-resistant prostate cancer (mCRPC) at the time of paired sample collection. B) Matrix of mutations and copy number alterations detected in independent analysis of plasma cell-free DNA (cfDNA) samples. All 72 mCRPC driver genes included in the cfDNA sequencing panel are shown (sorted by chromosome and position). Note that sensitivity of copy number calling is diminished in samples with less than 35% ctDNA. ALP = alkaline phosphatase; cfDNA = cell–free DNA; ctDNA = circulating cell-free tumor DNA; LN = lymph node; PSA = prostate-specific antigen.
We subjected all 45 cfDNA samples to targeted sequencing (median depth = 839×) of 72 mCRPC genes (Supplementary Figure 1, available online). Thirty-four of 45 (75.6%) cfDNA samples had a quantifiable ctDNA fraction greater than 2% (median = 62.7%, range = 4.0%–95.3%) (Figure 1A;Supplementary Figure 2, available online). An additional five cfDNA samples exhibited an AR amplification but were not amenable to ctDNA fraction estimation because of a lack of detectable somatic mutations or autosomal copy number changes across the genes in our panel (Figure 1A). Samples with a quantifiable ctDNA fraction had higher cfDNA yields than those without (median = 24.4 vs 6.8 ng/mL, P = .002, rank-sum test) (Figure 1A). PSA concentrations did not differ between patients with and without detectable ctDNA (P = .71, rank-sum test), although the seven patients with highest PSA levels (>370 ng/mL) all had detectable ctDNA. The cohort included six patients with liver metastases, most of whom displayed high cfDNA yields (median = 118 ng/mL, range = 4.8–1380 ng/mL) and high ctDNA fractions (median = 75.0%, range = 18.1%–85.3%) (Figure 1A). The cohort also included seven patients whose metastatic biopsy tissue harbored a histological component of small cell neuroendocrine carcinoma. All cfDNA samples from these patients had a quantifiable ctDNA fraction (median = 65.0%, range = 7.8%–82.3%). Collectively, these data are consistent with previous reports associating high ctDNA fractions with metrics of tumor burden in mCRPC (2,21).
Detected alterations across the 34 samples with quantifiable ctDNA greater than 2% included AR amplifications in 22 (64.7%) samples, SPOP mutations in three (8.8%) samples, and inactivating alterations in tumor suppressors TP53, PTEN, RB1, APC, CDKN1B, BRCA2, and PIK3R1 (Figure 1B;Supplementary Figure 3, available online). In the five patients with small cell neuroendocrine histology in their metastatic tissue biopsy and a liquid biopsy with ctDNA fractions above 35%, there was evidence for biallelic loss of TP53 and/or RB1 in all but one liquid biopsy (Supplementary Table 2, available online), consistent with tissue-based studies of this disease variant where these aberrations are common (22,23).
Representation of Tissue Biopsy Mutations in ctDNA
We identified a total of 109 somatic mutations among the 34 cfDNA samples with quantifiable ctDNA (Supplementary Table 3, available online). This amounted to an average mutation rate of 3.4 mutations per megabase, similar to prior studies of metastatic tissue biopsies (5,6). This estimate may be inflated because of recurrent driver mutations in the target regions.
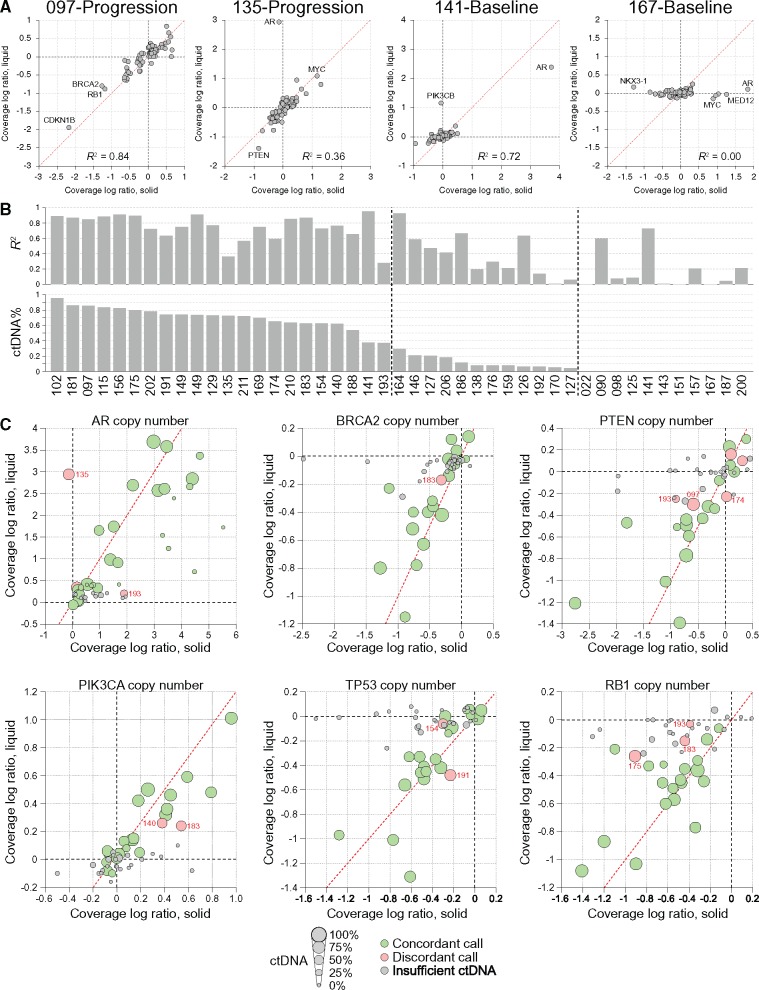
We compared our results with exome sequencing data (median depth 78×) from the 34 matched metastatic tissue biopsies (Supplementary Figure 4, available online). A mutation analysis restricted to the same regions as the cfDNA panel revealed 78 somatic mutations, a remarkable 73 (93.6%) of which were also identified in our independent analysis of the cfDNA samples (Figure 2A;Supplementary Table 3, available online). Of the five mutations detected in a solid biopsy but not in matched cfDNA, only two were protein altering (in CDK12 and MLH1). All five missed mutations were present in their respective cfDNA samples at allele fractions between 0.5% and 1.3%, so although they did not pass our signal-to-noise thresholds, they would likely be robustly detected with ultra-deep sequencing (incorporating unique molecular identifiers) or digital droplet PCR (4,24). Four of the missed mutations were from samples with less than 10% ctDNA. Among the 19 paired samples with more than one shared somatic mutation, the hierarchy of variant allele fractions for shared mutations was highly concordant between the liquid and solid biopsy (Figure 2B).
Figure 2.
Concordance of mutation calls between solid and liquid biopsies. A) Bar plot showing the variant allele frequencies for driver mutations in selected clinically relevant metastatic castration-resistant prostate cancer (mCRPC) genes. For each mutation, allele frequencies are provided for the solid tissue biopsy (S) and the cell–free DNA (cfDNA) liquid biopsy (L). The circulating cell-free tumor DNA (ctDNA) fraction corresponding to each mutation is shown in the lower panel. B) Variant allele frequencies for somatic mutations shared between matched liquid and solid biopsies, showing broad conservation of mutant allele fraction hierarchy. ctDNA fraction for each liquid biopsy is provided at the bottom. For patient 149, two cfDNA samples obtained at different time points are shown. AF = allele frequency; ctDNA = circulating cell-free tumor DNA.
Missense mutations in the ligand binding domain of AR are present in 10% to 25% of mCRPC patients and can have implications for patient response to AR-directed therapy (1,2,5,6,25). Seven patients (16.7%) in our study harbored AR mutations (exclusively L702H and/or H875Y). All AR mutations identified in solid biopsies were also detected in the corresponding liquid biopsy, with the exception of two patients (022 and 187), whose liquid biopsies lacked detectable ctDNA (Figure 2A;Supplementary Table 3, available online). Patients 138 and 169 each harbored double AR L702H and H875Y mutations. These mutations had supporting evidence in both biopsy types, although the L702H mutation did not pass our detection thresholds in the solid biopsy of patient 169 because of low sequencing depth and the presence of two L702H reads in the paired germline sample (potentially from ”contaminating” circulating tumor cells [CTCs] in the leukocyte fraction). AR mutations are typically mutually exclusive to high-level gene amplifications (1,2,5). This was true in six of seven patients in our cohort. However, patient 102 carried an AR amplification and an H875Y mutation at 95.3% and 94.5% allele fraction in liquid and solid biopsy, respectively, suggesting that the mutation occurred prior to amplification (Supplementary Figure 5, available online).
Mutational Heterogeneity Unique to ctDNA
Performing the reverse analysis, 36 of 109 (33.0%) somatic mutations detected in liquid biopsies were not detected in the corresponding solid biopsy. Twenty-nine of these were explained by insufficient coverage in the solid biopsy (median 10×), but seven mutations displayed a robust and statistically significant lack of evidence in the solid biopsy (P < .01, Fisher exact test) (Supplementary Table 3, available online). All seven were protein altering, and four were clear cancer drivers, including a TP53 missense, RB1 frameshift, PTEN frameshift, and APC stopgain mutation (Figure 2A;Supplementary Table 3, available online). In patient 174, ctDNA analysis revealed deleterious mutations in APC, TP53, and PTEN at allele fractions of 31.9%, 28.2%, and 12.9%, for which there were no supporting reads in the solid biopsy despite corresponding read depths of 38×, 37×, and 44× (P < .01 for all three comparisons, Fisher exact test) (Supplementary Figure 6, available online). In this patient, who had developed disease progression following enzalutamide and abiraterone therapy at the time of sample collection, the site of tissue biopsy was a liver metastasis, but the patient also had multiple bone metastases that were not biopsied. These results demonstrate the potential for liquid biopsy to capture clinically relevant mutational heterogeneity that may be missed or underestimated by tissue biopsy of a single metastatic site.
Concordance of Copy Number Calls Between Liquid and Solid Biopsies
Twenty-two of 45 (48.9%) cfDNA samples had a sufficient ctDNA fraction (>35%) for detecting copy number changes based on coverage log ratio (Supplementary Table 4, available online). For those samples, there was a very high correlation between the coverage log ratios of matched liquid and solid biopsies (median R2 = 0.76, range = 0.28–0.94) (Figure 3A;Supplementary Figure 7, available online). For samples with ctDNA fractions of 1% to 35%, coverage log ratios were still correlated (median R2 = 0.35), albeit to a lesser degree because of the reduced dynamic range of signal. Unsurprisingly, samples with undetectable or unquantifiable ctDNA showed no correlation (median R2 = 0.08), with the exception of two samples carrying strong AR amplifications (Figure 3A).
Figure 3.
Concordance of genome copy number between liquid and solid biopsies. A) Shows representative scatter plots showing the correlation for coverage log ratio across the 72 genes in our targeted panel. Each gene is represented as a single circle. B) Shows the R2 value for this correlation across all samples in the cohort and the relationship to circulating cell-free tumor DNA (ctDNA) fraction. C) Bubble plots showing the concordance for copy number calls across individual genes, and how concordance depends on ctDNA fraction. Discordant calls are indicated in red, with patient ID annotated. ctDNA = circulating cell-free tumor DNA.
AR amplifications were detected in 15 of 22 liquid biopsies with ctDNA fractions greater than 35% (Supplementary Figure 5, Supplementary Table 4, available online). Thirteen of these amplifications were detected in the matched solid biopsy, suggesting high concordance for this key mCRPC driver event (Figure 3B). However, in one notable exception (patient 135), the liquid biopsy identified a 10-fold AR amplification that was not detected in the corresponding lymph node metastatic biopsy. Furthermore, while this pair shared a CTNNB1 p.D32N hotspot mutation, the liquid biopsy carried an RB1 frameshift mutation that was also absent in the matched lymph node metastasis (Supplementary Table 3, available online). Patient 135 also had bone metastases, so it is plausible that the tumor clones with AR amplification were confined to the bone disease at time of biopsy collection. Among the 23 liquid biopsies in our cohort with ctDNA fractions of less than 35% (or inestimable), we identified a further 11 AR amplifications, all of which were corroborated in matched solid biopsies (Figure 3B). Although low ctDNA fractions preclude statements about the absence of copy number alterations, they do not preclude detection of strong copy number changes when they are present.
Across the clinically informative mCRPC driver genes AR, BRCA2, ATM, PTEN, PIK3CA, PIK3CB, PIK3R1, TP53, and RB1, we observed a concordance of 88.9% for individual gene copy number calls between evaluable liquid and solid biopsies (Figure 3, C–G; Supplementary Figure 8, available online). Among the rare discordant calls in the aforementioned genes, the median absolute coverage log ratio difference between paired samples was 0.28, suggesting only minor or subclonal differences. Furthermore, in 72.7% of discordant calls, there was weak evidence (absolute coverage log ratio > 0.1) for the corresponding coverage log ratio change in the noncalled member of the pair, but not sufficient evidence to pass our thresholds. It is plausible that the majority of discordant calls would be found concordant by a more sensitive assay capturing additional genomic territory across target genes or through algorithmic improvements. This hypothesis is supported by the fact that copy number calls were more discordant for short genes with few target regions such as MYC, FOXA1, CDKN1B, and FANCF (66.9% concordance for genes with less than 10 target regions compared with 83.9% concordance for genes with 10 or more target regions; P = 4.3 × 10-7, Fisher exact test) (Supplementary Figure 8, available online).
Four patients with quantifiable ctDNA fractions were subsequently treated with platinum-based chemotherapy. One (patient 174) had a sustained tumor response to carboplatin, with a greater than 50% reduction in size of multiple liver metastases and decline in serum PSA from a baseline of 471 ng/mL to a nadir of 26 ng/mL, and a response duration over six months. This patient had a biallelic deletion of BRCA2 detected in baseline ctDNA and matched metastatic biopsy (Supplementary Figure 3 and Supplementary Table 4, available online).
Genomic Rearrangements Detected in Liquid and Solid Biopsies
Although most large structural rearrangements fall within introns and are undetectable by exon-focused sequencing approaches, we detected a total of 19 somatic genomic rearrangements among the 34 liquid biopsies with a quantifiable ctDNA fraction (Supplementary Table 5, available online). No somatic rearrangements were detected in the 11 liquid biopsies that lacked quantifiable ctDNA. Detected rearrangements included three PTEN disrupting rearrangements, one TMPRSS2-ERG fusion, one APC truncating deletion, four FOXA1 rearrangements (including one apparent insertion of an Alu element from 14q into the middle of the FOXA1 coding region), and two in-frame deletions in the BRCA2 coding region. Each rearrangement was unique to one patient, no supporting reads were observed in other patients. Nine of the 19 (47.4%) rearrangements displayed sequence-level evidence in the paired solid biopsy (Supplementary Table 5, available online). The sensitivity of rearrangement detection was limited in the solid whole exome sequencing biopsies because of their shorter read length (75 base pairs) and limited coverage. In two patients (156 and 164), we identified rearrangements that juxtaposed the AR 3’-UTR with other locations in chromosome X, but these likely represent passenger events accompanying the AR amplifications observed in both samples.
Discussion
The interrogation of metastatic tissue biopsies has suggested that real-time knowledge of the CRPC somatic genome can influence clinical management. A “liquid” biopsy using plasma ctDNA offers a minimally invasive and practical tool to access such information. Our study shows that a ctDNA-based assay is, in the majority of patients, sufficient to identify all driver DNA alterations that are present in matched metastatic tissue. Importantly, this supports the development of DNA biomarkers to guide mCRPC patient management based on ctDNA alone.
Several prior studies have demonstrated high plasma ctDNA fractions in mCRPC patients progressing on systemic therapy (1–3,26,27). Bolstered by these results, we aimed to survey a large proportion of the clinically actionable mCRPC genome by using a 72-gene panel. With sequencing data covering approximately 950 kb of the genome at approximately 800× coverage, 75% of patients examined had informative cfDNA samples. We and others have shown that high ctDNA fractions are associated with poor prognosis and are correlated with indices of overall tumor burden in mCRPC patients (2,21). Therefore, the patients missed by our approach may represent those with a more favorable prognosis and perhaps most likely to benefit from standard of care (and least likely to require personalized interventions). Nevertheless, these patients may benefit from digital droplet PCR or ultra-deep sequencing incorporating unique molecular identifiers, thereby achieving greater sensitivity at specific mutation hotspots (4). Similarly, assays that survey large numbers of heterozygous SNPs can inform on monoallelic deletions with better sensitivity than those focused solely on exonic regions. Commercial screens for fetal aneuploidy in maternal plasma do not routinely provide results below 4% fetal cfDNA fractions (28), suggesting that even chromosome-wide copy number changes in cancer patients will be challenging to resolve in low ctDNA fractions. Ultimately, although a cfDNA assay will sometimes fail at detecting a somatic alteration if the ctDNA fraction is very low, a well-designed panel allows differentiation between absence of knowledge (ie, insufficient ctDNA) and a negative result (no alteration in the ctDNA). We envisage a scenario where obtaining metastatic tissue biopsies could serve as a fallback whenever ctDNA fractions are insufficient.
Our study included only patients with a positive metastatic tissue biopsy that yielded sufficient material for genomic analysis. Success rates with ctDNA approaches must be compared with those achievable with tissue biopsy. In patients with mCRPC, bones are often the only site of metastatic spread. Success rates for obtaining tumor tissue from bone in prostate cancer vary from 25% to 75% and (similar to ctDNA fraction) are positively correlated with metrics of overall tumor burden (13–17). Furthermore, considerable selection bias exists in identification of patients eligible for tissue biopsy as many metastases are inaccessible or too small to biopsy. In a large series of biopsies such as ours, bone metastases are underrepresented relative to their clinical prevalence, given the relative ease of soft tissue biopsy.
An important advantage of ctDNA is its ability to integrate somatic information from more than one metastatic lesion and thereby survey intrapatient tumor heterogeneity. Our data suggest that important drivers of therapy resistance (AR amplification, WNT pathway disruption) robustly detected in ctDNA can be missed by a single metastatic tissue biopsy. Conversely, we did not observe discordance between liquid and solid biopsies for established “truncal” events such as SPOP and FOXA1 mutations (29). A recent study of tissue biopsies from 54 men with mCRPC has suggested high concordance between multiple metastatic sites for the majority of cancer drivers (7), and despite some notable exceptions, our study is consistent with this hypothesis. A major limitation of the current study was the use of different sequencing approaches for the solid and liquid biopsies and the lower sequencing depth of the solid biopsy data. Although we corrected for this issue by using statistics to determine which mutations were most likely specific to liquid biopsies, it is possible that higher sequencing depth would have enabled the detection of more subclonal mutations in the solid biopsies.
A large proportion of patients in this study had progression of their disease following at least one line of AR-targeted therapy such as abiraterone or enzalutamide at the time of biopsy, and consistent with this, we observed a high frequency of AR alterations. The postabiraterone and enzalutamide setting is currently where most targeted agents are tested, and therefore represents the patient population poised to benefit the most from the development of prospective biomarkers. We recently demonstrated that ctDNA analysis is sufficient to demonstrate somatic loss-heterozygosity in patients with germline BRCA2 mutations (20). Similarly, we and others have shown that PTEN loss is detected in ctDNA from mCRPC patients (1,2). Patients with these respective genotypes may benefit from PARP inhibitors or PI3K/AKT pathway blockade (11,12). Indeed, a patient in our study who had a BRCA2 homozygous deletion identified through ctDNA sequencing had an objective response to a DNA-damaging agent. Here, we show that a well-designed ctDNA assay will not miss important actionable alterations, such as BRCA2 or PTEN loss, that are present in matched metastatic tissue biopsies, the current gold standard for mCRPC tumor genotyping. The remarkable concordance of ctDNA and metastatic tissue biopsies in mCRPC suggests that ctDNA assays could be confidently used to molecularly stratify patients for prognostic and predictive purposes.
Funding
This work was supported by a Stand Up to Cancer Prostate Cancer Foundation Prostate Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0812). Additional support was provided by Prostate Cancer Canada through the Movember Rising Star in Prostate Cancer research program (AW), the Emil Aaltonen Foundation (MA), and a Terry Fox New Frontiers Program Project (grant No. TFF116129 to MG, AW, KC).
Notes
The study sponsors had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
References
- 1. Wyatt AW, Azad AA, Volik SV, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2(12):1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer.Sci Transl Med. 2015;7(312):312re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulz P, Belic J, Graf R, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer.Nat Commun. 2016;7:12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Volik S, Alcaide M, Morin RD, Collins C.. Cell-free DNA (cfDNA): Clinical significance and utility in cancer shaped by emerging technologies.Mol Cancer Res. 2016;14(10):898–908. [DOI] [PubMed] [Google Scholar]
- 5. Robinson D, Van Allen EM, Wu Y-M, et al. Integrative clinical genomics of advanced prostate cancer.Cell. 2015;161(5):1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer.Nature. 2012;487(7406):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer.Nat Med. 2016;22(4):369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer.Nature. 2015;520(7547):353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong MKH, Macintyre G, Wedge DC, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer.Nat Commun. 2015;6:6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi K, Hotte SJ, Joshua AM, et al. Treatment of mCRPC in the AR-axis-targeted therapy-resistant state.Ann Oncol. 2015;26(10):2044–2056. [DOI] [PubMed] [Google Scholar]
- 11. Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer.N Engl J Med. 2015;373(18):1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Bono JS, De Giorgi U, Massard C, et al. PTEN loss as a predictive biomarker for the Akt inhibitor ipatasertib combined with abiraterone acetate in patients with metastatic castration-resistant prostate cancer (mCRPC).Ann Oncol. 2016;27(suppl_6):7180. doi:10.1093/annonc/mdw372.02. [Google Scholar]
- 13. Ross RW, Halabi S, Ou S-S, et al. Predictors of prostate cancer tissue acquisition by an undirected core bone marrow biopsy in metastatic castration-resistant prostate cancer—a Cancer and Leukemia Group B study.Clin Cancer Res. 2005;11(22):8109–8113. [DOI] [PubMed] [Google Scholar]
- 14. Efstathiou E, Titus M, Wen S, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer.Eur Urol. 2015;67(1):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKay RR, Zukotynski KA, Werner L, et al. Imaging, procedural and clinical variables associated with tumor yield on bone biopsy in metastatic castration-resistant prostate cancer.Prostate Cancer Prostatic Dis. 2014;17(4):325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spritzer CE, Diana Afonso P, Vinson EN, et al. Bone marrow biopsy: RNA isolation with expression profiling in men with metastatic castration-resistant prostate cancer—factors affecting diagnostic success. Radiology. 2013:121782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lorente D, Omlin A, Zafeiriou Z, et al. Castration-resistant prostate cancer tissue acquisition from bone metastases for molecular analyses.Clin Genitourin Cancer. 2016;14(6):485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aggarwal R, Beer TM, Gleave M, et al. Targeting adaptive pathways in metastatic treatment-resistant prostate cancer: Update on the Stand Up 2 Cancer/Prostate Cancer Foundation–Supported West Coast Prostate Cancer Dream Team.Eur Urol Focus. 2016; 2(5):469–471. [DOI] [PubMed] [Google Scholar]
- 19. Scher HI, Halabi S, Tannock I, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group.J Clin Oncol. 2008;26(7):1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Annala M, Struss WJ, Warner EW, et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol. 2017; in press. [DOI] [PubMed] [Google Scholar]
- 21. Wyatt AW, Annala M, Beja K, et al. Genomic alterations in circulating tumor DNA (ctDNA) are associated with clinical outcomes in treatment-naive metastatic castration-resistant prostate cancer (mCRPC) patients commencing androgen receptor (AR)-targeted therapy.Ann Oncol. 2016;27(suppl_6):59P. doi:10.1093/annonc/mdw363.08. [Google Scholar]
- 22. Akamatsu S, Wyatt AW, Lin D, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer.Cell Rep. 2015;12(6):922–936. [DOI] [PubMed] [Google Scholar]
- 23. Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer.Nat Med. 2016;22(3):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joseph JD, Lu N, Qian J, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509.Cancer Discov. 2013;3(9):1020–1029. [DOI] [PubMed] [Google Scholar]
- 25. Lorente D, Mateo J, Zafeiriou Z, et al. Switching and withdrawing hormonal agents for castration-resistant prostate cancer.Nat Rev Urol. 2015;12(1):37–47. [DOI] [PubMed] [Google Scholar]
- 26. Azad AA, Volik SV, Wyatt AW, et al. Androgen receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(10):2315–2324. [DOI] [PubMed] [Google Scholar]
- 27. Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer.Sci Transl Med. 2014;6(254):254ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy.N Engl J Med. 2015;372(17):1589–1597. [DOI] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer.Cell. 2015;163(4):1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.