Abstract
Background
Tuberculous meningitis (TBM) is the most devastating manifestation of extra-pulmonary tuberculosis. About 33% of TBM patients die due to very late diagnosis of the disease. Conventional diagnostic methods based on signs and symptoms, cerebrospinal fluid (CSF) smear microscopy or liquid culture suffer from either poor sensitivity or long turnaround time (up to 8 weeks). Therefore, in order to manage the disease efficiently, there is an urgent and unmet need for a rapid and reliable diagnostic test.
Methods
In the current study, to address the diagnostic challenge of TBM, a highly rapid and sensitive structural switching electrochemical aptasensor was developed by combining the electrochemical property of methylene blue (MB) with the molecular recognition ability of a ssDNA aptamer. To demonstrate the clinical diagnostic utility of the developed aptasensor, a blinded study was performed on 81 archived CSF specimens using differential pulse voltammetry.
Results
The electrochemical aptasensor developed in the current study can detect as low as 10 pg HspX in CSF background and yields a highly discriminatory response (P<0.0001) for TBM and not-TBM categories with ~95% sensitivity and ~97.5% specificity and has the ability to deliver sample-to-answer in ≤30 minutes.
Conclusion
In summary, we demonstrate a new aptamer-based electrochemical biosensing strategy by exploiting the target-induced structural switching of H63 SL-2 M6 aptamer and electroactivity of aptamer-tagged MB for the detection of HspX in CSF samples for the diagnosis of TBM. Further, the clinical utility of this sensor could be extended for the diagnosis of other forms of tuberculosis in the near future.
Keywords: aptamers, Mycobacterium tuberculosis, TB meningitis, clinical diagnostic, electrochemical sensing
Introduction
Tuberculosis (TB) is a highly infectious disease that causes significant morbidity and mortality worldwide with an estimated 10.4 million new cases and over 1.7 million deaths in 2016. TB continues to pose an enormous threat to public health management systems across the globe, especially in the developing world.1
Tuberculous meningitis (TBM) is the most lethal manifestation of extrapulmonary tuberculosis that causes irreversible damage to the central nervous system (CNS) and has an estimated mortality of 1.5 per 100,000 population in India.2,3 Children and immunocompromised adults are at higher risk of acquiring TBM infection. TBM leads to delayed CNS neurological complications including, but not limited to, cerebral abscess, neurological sequelae, hydrocephalus, etc, and ultimately to death.4 About 33% of TBM patients die due to the delayed diagnosis of the disease. Therefore, in order to manage the disease efficiently, there is an urgent and unmet need for a rapid and reliable diagnostic test.
Conventional diagnostic methods for TBM mainly rely on presentation of signs and symptoms that are often nonspecific and can resemble other meningoencephalitis, which could misguide the clinician. Other conventional approaches for TBM diagnosis include microbiological analysis of cerebrospinal fluid (CSF).5 However, microscopy exhibits poor sensitivity due to the paucibacillary nature of the disease, while CSF culture has a long turnaround time (up to 8 weeks).6 Furthermore, collection of an adequate volume of CSF sample from pediatric patients for culture is a major challenge for clinicians.3,7 A test that has gained some popularity in recent years for diagnosing TBM is Gene Xpert MTB/RIF (Xpert). Xpert is a rapid and automated PCR-based molecular test with a minimal requirement of trained workforce. However, only 55% sensitivity with 84% negative predictive value (NPV) was obtained according to a recently performed meta-analysis of Xpert for TBM when a clinical gold standard was used. This NPV suggests that 1 in 6 TBM patients is being missed in high prevalence TBM settings when tested by Xpert.8–11 A newer advanced version ie, Xpert Ultra has shown a higher sensitivity but its applicability has been demonstrated only in HIV patients.12 Therefore, Xpert as a sole diagnostic test has not been recommended considering the current evidence,8 as delay in the treatment of TBM can lead to fatal consequences. Moreover, the high cost of the test and dependence on proprietary equipment and reagents limits the application of Xpert in resource-limited settings.3,13–15 Therefore, to address the aforementioned challenges, there is an urgent need of a facile, cost-effective, rapid and sensitive test for TBM diagnosis.
HspX is a Mycobacterium tuberculosis antigen (also known as α-crystallin) which is abundantly expressed under hypoxia3,14 and in both active and latent forms of TB.3,14,16 The utility of this antigen in the antibody-based diagnosis of pulmonary TB has already been established.17–19 In another study, the detection of HspX messenger RNA was used to detect viable M. tuberculosis directly in sputum specimens from clinically suspected TB patients.20 Notably, HspX antigen was considered as a reliable biomarker for TBM diagnosis in a polyclonal antibody-based ELISA using CSF.3 While this ELISA evinced a high diagnostic accuracy, a polyclonal antibody-based test has inherent problems that include batch-to-batch variation in antibody titer, limited stability and requirement of a cold chain. These limitations pose a significant challenge during translation and upscaling of laboratory findings into a point-of-care (POC) test for TBM. In recent years, functional nucleic acids known as aptamers (DNA or RNA) have demonstrated the ability to potentially overcome the limitations associated with antibodies.21–26 Owing to these attributes, aptamers are regarded as a strong chemical rival of antibodies, with an ability to replace antibodies in all possible diagnostic formats.22,27 Furthermore, in the last few years, aptamers have demonstrated their utility in the detection of diverse analytes ranging from small molecules to antigens to enzymes to hormones and even whole bacterial or cancer cells.28–30 The unique ability of aptamers to adapt on various diagnostic platforms without compromising their affinity and specificity make them diagnostic reagents of choice.
An ultrasensitive diagnostic platform is highly desirable to detect trace amounts of biomarker present in clinical specimens for the accurate diagnosis of disease.31–33 In recent years, the electrochemical detection platform has generated considerable interest owing to its ease of operation, high sensitivity (up to femtomole), rapid detection, negligible or no requirement of sample processing and the ability to provide both qualitative and quantitative output.32,34 In particular, methylene blue (MB) is useful as a redox probe in aptamer-based electrochemical sensors owing to its remarkable ability to participate in electron transfer, ease in modification of the aptamer by MB, high electroactivity and stability.35
In the current study, we utilize the electron transfer property of MB in combination with target-induced structural switching ability of an in-house generated HspX specific DNA aptamer (H63 SL2-M6)36 for the detection of HspX in CSF samples for the diagnosis of TBM. The secondary structure of this aptamer is shown in Figure S1.
Methods
Reagent and materials
Gold (III) chloride (HAuCl4·3H2O), 2-Mercaptoethanol (β-ME), Tris-HCl, KCl, MgCl2, NaCl and other reagents used in this study were procured from Sigma-Aldrich (Sigma-Aldrich Co., St Louis, MO, USA). Potassium ferrocyanide (K4Fe(CN)6·3H2O) and potassium ferricyanide K3Fe(CN)6 were obtained from HiMedia laboratories (Mumbai, India). HspX specific aptamer (H63 SL2 M6) was custom synthesized from LGC Biosearch (Petaluma, CA, USA) with 5′ MB and 3′ C6 thiol modification. A screen-printed electrode system (SPE) comprising carbon as working electrode (WE; Φ=3 mm), platinum as auxiliary electrode and Ag/AgCl as a reference electrode was procured from ItalSens (PalmSens BV, Houten, the Netherlands) and was used to build an aptamer-based electrochemical sensor. Hexa-His tagged HspX protein was expressed and purified in recombinant from Escherichia coli DH5α using standard Ni-NTA chromatography.
Instrumentation
Electrochemical measurements were carried out for characterizing the fabrication of the sensing interface using an EmstatBlue (PalmSens) equipped with PSTrace software. The aforementioned SPE system was used for electrochemical sensing assays after electrodeposition of gold nanoparticles (GNPs) cyclic voltammetry (CV) were performed in a solution of 5 mm [Fe(CN)63−/4−] and differential pulse voltammetry (DPV) readings were recorded in binding buffer (10 mM Tris-HCl supplemented with 50 mM KCl, 25 mM NaCl and 10 mM MgCl2).
Fabrication and characterization of sensing platform
The electrodeposition of GNPs was carried out for providing a platform for immobilization of thiol-labelled aptamer on carbon SPE electrode using an aqueous solution of 1.0 mM HAuCl4 in the presence of 0.5 M H2SO4 at a constant potential of −0.2 V for 200 seconds. After washing and drying of GNP-deposited SPE, the electrode was characterized using a scanning electron microscope (SEM) and CV. The morphology of the GNP-modified electrodes were observed using SEM (Zeiss evo 18; Carl Zeiss Microscopy, LLC, Thornwood, NY, USA). Further, the screen-printed electrode was characterized before and after deposition of GNPs by CV for electrochemical characterization in 5 mM [Fe(CN)63−/4−]. A potential ranging from −0.4 V to 0.8 V was applied to record the cyclic voltammogram. Following electrochemical deposition of GNPs, 5.0 µL of thiol-activated aptamer (10 µM) in binding buffer was placed on the modified SPE and was incubated at 30°C overnight. Then the aptamer-modified electrode was thoroughly rinsed with binding buffer followed by distilled water to remove the weakly adsorbed aptamer and dried. The aptamer-modified electrode was further exposed to 10 mM 2-mercaptoethanol (β-ME) for 20 minutes to block the uncovered electrode surface. Finally, the modified electrode was rinsed thoroughly with binding buffer followed by double distilled water.
Electrochemical detection of HspX
The prepared SPE-based sensing interface was incubated with 100 ng HspX protein for 15 minutes at room temperature. The sensing interface was incubated with nine differentantigens (HspX, ESAT-6, CFP-10, MPT-64, MPT-51, Ag85complex, GroES, CFP and LAM of M. tuberculosis) and incubated for 15 minutes for determination of selectivity of the sensor. After incubation, the electrodes were washed with binding buffer carefully to remove the unbound protein. In order to detect bound protein, DPV was performed.
Limit of detection
In order to determine the low-end detection limit, first a range (0.01–500 ng) of HspX was spiked in binding buffer and DPV was performed. Further, to study the effect of sample matrix on aptamer–HspX interaction, pooled CSF samples obtained from noninfectious neurological disorder subjects (NIND) was diluted (1:10 in binding buffer) and then it was spiked with HspX (0.01–500 ng). Following this, DPV was performed to determine the lowest possible amount of HspX that can be detected in the CSF.
Application of the aptasensor in clinical assay
To detect HspX level in clinical samples, archived CSF samples were used as described in classification of CSF samples section (Supplementary materials). These CSF samples were diluted to 1:10 in binding buffer (10 mM Tris pH 7.5, 25 mM NaCl, 10 mM MgCl2, 50 mM KCl) before performing the experiment (DPV).
Ethical statement
Ethical clearance for evaluating the antigen-based test to detect TBM was taken from the Institutional Ethics Committee (AIIMS and RMLH). Since the current study was conducted on pediatric CSF samples, all samples were collected after obtaining a written informed consent from parents of the subjects. Patient information was collected in a confidential manner as per ethical principles of the Declaration of Helsinki of 1964 revised by the 59th WMA General Assembly, Seoul, in October 2008.
Results and discussion
Principle of structural switching aptasensor
The schematic in Figure 1 illustrates the principle of the aptamer-based electrochemical biosensor. A dual-labelled aptamer (H63 SL2-M6 with 5′MB and 3′ thiol label having C6 spacer) is immobilized on to the disposable SPE having gold-coated carbon WE. In the absence of HspX protein, the MB-labelled end of the aptamer is in the close proximity of the WE, resulting in high electron transfer. However, upon encounter with HspX in the CSF sample, a target-induced structural change in the aptamer occurs, which increases the distance between MB and WE, causing a rapid decrease in electron transfer, thereby reducing the current. Thus, in presence of the cognate target HspX, a turn-off response is achieved, which forms the basis of electrochemical sensing of HspX.
Figure 1.
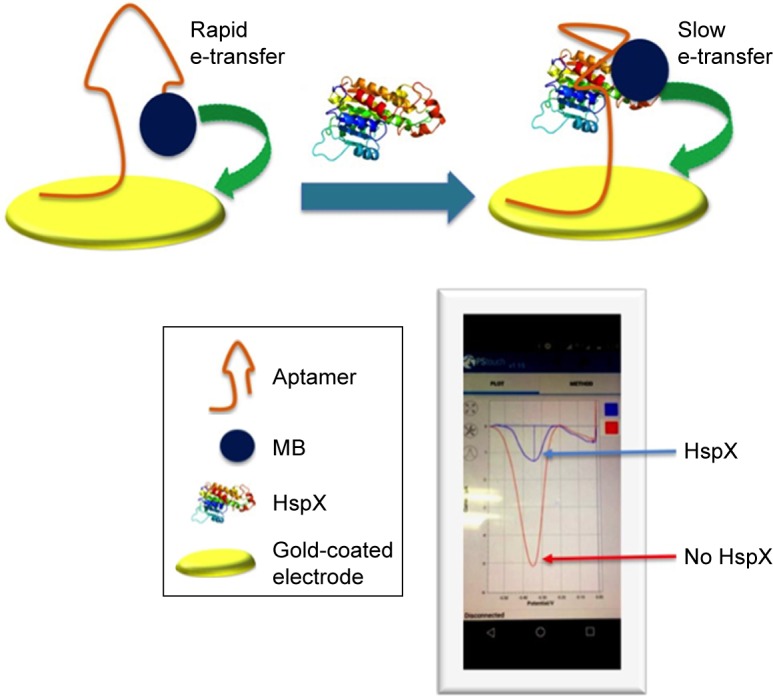
Schematic representation of the aptamer-based electrochemical sensor for the detection of HspX antigen in CSF.
Notes: The aptamer is thiol-labelled at the 3′ end that allows self-assembly of aptamer on a gold-coated electrode. The redox label (MB) is attached at the 5′ of the aptamer and it is in close proximity to the electrode surface, thereby allowing rapid electron transfer. The presence of HspX induces a structural change in the aptamer which increases the distance between MB and the electrode, and electron transfer is thus reduced. The differences in faradaic current before and after addition of HspX were quantified using DPV.
Abbreviations: MB, methylene blue; CSF, cerebrospinal fluid; DPV, differential pulse voltammetry.
Circular dichroism (CD) study
At first, the HspX-induced structural change in the aptamer was monitored using circular dichroism (CD). A negative peak at 246 nm and two positive peaks at 220 and 272 nm were recorded (Figure 2), a typical signature of stem loop/ hairpin type B-DNA structure.37 In the presence of HspX, a structural change is indicated by the change in molecular ellipticity suggesting that the aptamer can work as a molecular switch for the detection of HspX.
Figure 2.
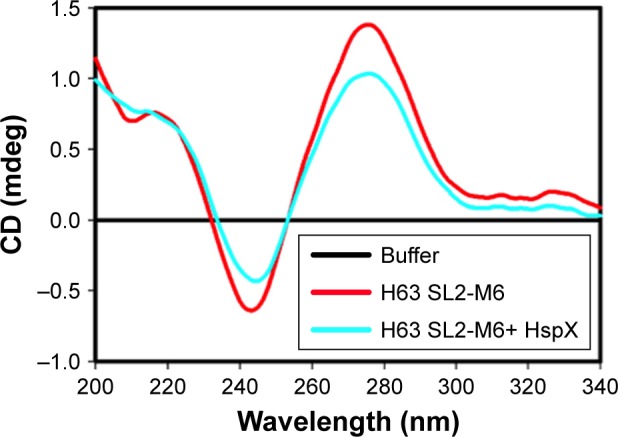
CD spectrum for free aptamer (10 µM) and aptamer in the presence of HspX (20 µM).
Note: In the presence of HspX, the aptamer undergoes target-induced structural change as evident by change in ellipticity.
Abbreviation: CD, circular dichroism.
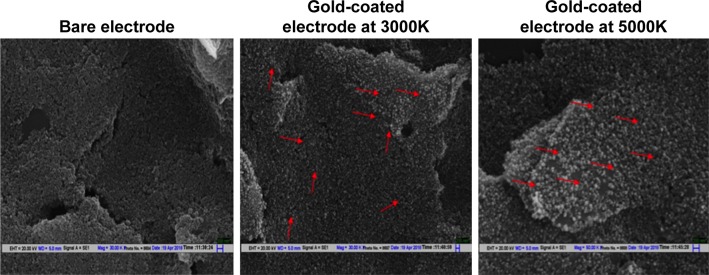
Electrode characterization
The SEM characterization of the bare SPE shows a rough surface, while electrodeposition of gold chloride leads to coating of spherical GNPs on the electrode surface (Figure 3). CV further confirmed the deposition of GNPs on the electrode; an increase in current was recorded on the GNP-coated electrode in comparison to the bare electrode due to the increased conductivity of gold (Figure 4). These results are in concordance with the previously published reports.38
Figure 3.
Scanning electron micrograph of SPE, before and after electrodeposition of gold (at 3000K and 5000K magnification).
Notes: After electrodeposition, spherical GNPs are visible on the electrode surface while bare electrode presents a rough surface. Red arrows point to representative GNPs deposited on the SPE surface.
Abbreviations: SPE, screen-printed electrodes; GNPs, gold nanoparticles.
Figure 4.
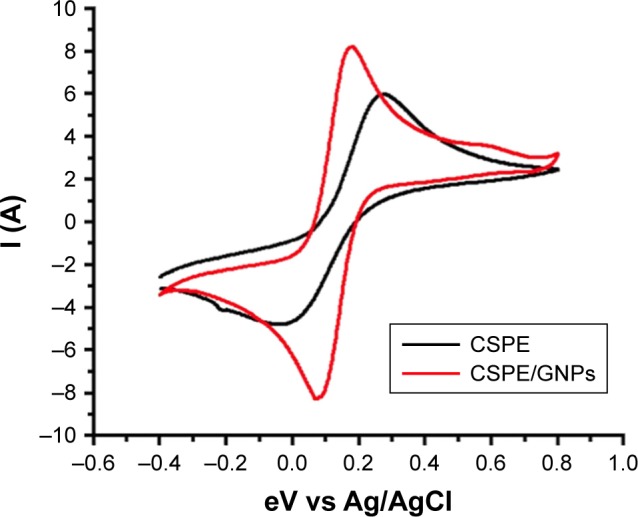
Cyclic voltammogram of CSPE before (black) and after (red) electrodeposition of GNPs.
Note: Electrodeposition of gold makes the electrode surface more conductive, resulting in an increase in current response.
Abbreviations: CSPE, carbon-screen-printed electrode; Ag/AGCl, silver/silver chloride; GNPs, gold nanoparticles.
Evaluation of aptamer specificity
Thereafter, the dual-labelled (MB- and thiol-labelled) H63 SL2-M6 aptamer was immobilized on the gold-coated electrode and used to evaluate the specificity of the sensor. The sensor response in the presence of various M. tuberculosis antigens (HspX, ESAT-6, CFP-10, MPT-64, MPT-51, Ag85complex, GroES, CFP and LAM) was compared and is represented as a three-color gradient heat map (Figure 5). The sensor response is highly selective for HspX and a sharp decrease in current was observed only in presence of HspX. Such a change was not observed in presence of the other antigens that are expressed during TB infection, such as MPT-51, GlcB, CFP-10 and ESAT-6, or secretory proteins such as Ag85C and culture filtrate proteins, or GroES heat shock protein or LAM, a major component of bacterial cell wall.
Figure 5.
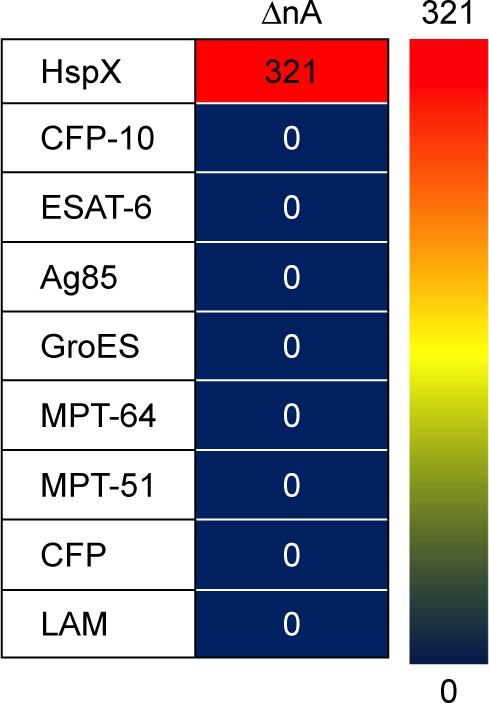
A three-color gradient heat-map representation of sensor response.
Notes: Red color represents highest sensor response while blue indicates the weakest sensor response. The current difference values are also displayed for each antigen. The current difference values obtained in negatives are considered as ‘0’.
Determining limit of HspX detection
The sensitivity of HspX detection using the electrochemical sensing approach was determined next. The DPV of M6 aptamer-modified electrode was recorded in the presence and absence of HspX (500 ng) (Figure S2). The aptamer-based electrochemical sensor was highly sensitive and able to detect as low as 10 pg of HspX (Figure S3). Following this, a range of HspX protein (0.01–500 ng) spiked into CSF (obtained from not-TBM subjects belonging to noninfectious neurological disorders category, described under “Classification of CSF samples” in Supplementary materials) was evaluated to assess the effect of clinical sample matrix on sensor sensitivity. Notably, the aptasensor exhibited a similar low-end detection limit of 10 pg HspX in CSF background (Figure S4). However, the signal strength was marginally reduced in the CSF background, suggesting the possibility of mild quenching of the signal by the sample matrix.
Evaluation of aptasensor in clinical specimens
Finally, to demonstrate the clinical diagnostic utility of the developed aptasensor, its performance was assessed in a blinded manner in 81 archived CSF specimens from pediatric subjects (Supporting information Table S1) belonging to TBM and not-TBM disease control categories, as described in the Supplementary materials section under “Classification of CSF samples”. An ethical permission was obtained from the institutional ethics committee for the use of CSF samples in the current study. A receiver operating characteristic (ROC) curve was constructed using ΔI/mA values (current difference before and after adding the sample) obtained with CSF from definite (true positive) and nontuberculous infectious meningitis (NTIM) group (true negative) samples. The area under the curve (AUC) was 1.0 (Figure S5) and established that the assay is highly robust. The NTIM category comprised of 16 cases of pyogenic bacterial meningitis that included 14 cases that were diagnosed on the basis of response to appropriate antibiotics, clinical presentation along with symptoms and culture confirmed cases of E. coli (N=1), and Acinetobacter sp. (N=1). Based on the cut-off derived from the ROC curve, the performance of the electrochemical sensor was evaluated in all 81 samples (Figure 6). The test yielded a highly discriminatory response (P<0.0001) for TBM and not-TBM category with ~95% sensitivity and ~97.5% specificity. A recent assessment of Xpert on CSF samples showed its performance to be modest with a sensitivity of ~55%.39 While a newer version of Xpert MTB/RIF ie, Xpert Ultra exhibited 70% sensitivity for probable or definite TBM subjects having HIV compared to a uniform case definition.12 In comparison, the aptamer-based electrochemical sensor developed in this study was significantly superior with ~95% sensitivity. This high sensitivity of our diagnostic test is likely due to the high affinity of the aptamer for the target, electrochemical sensing platform and high HspX load present is CSF of TMB subjects. A recent study from our group suggests that HspX is present in a range of 3×106 fg (3 ng) to 2.8×108 fg (280 ng) in 5 µL CSF sample.3 A comparison of the performance of the aptamer-based test reported here with previously reported TBM diagnostic tests is shown in Table 1. Moreover, owing to its simple format, the developed electrochemical sensor meets the requirements of a rapid portable bedside diagnostic test for TBM. Importantly, the electrochemical sensor designed in the current study requires a very small volume of CSF (~4 µL) and addresses the challenge of collecting a large volume of CSF for various diagnostic investigations. In comparison to the existing methods, the test developed in the current study sample-to-answer time of ≤30 minutes with high selectivity and sensitivity. Further, the developed assay can be operated through a mobile phone that makes it a truly POC assay and can also help in rapid dissemination of results and clinical decision-making.
Figure 6.
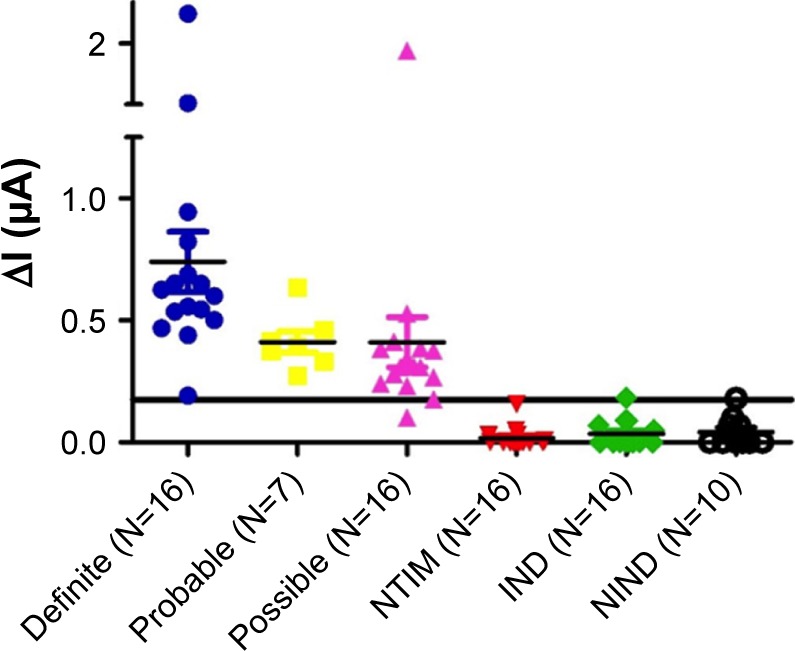
Performance of H63 SL-2 M6 aptamer-based electrochemical sensor on archived CSF samples obtained from TBM patients (N=39; Definite N=16, Probable N=7, Possible N=16) and Not-TBM patients (N=42; NTIM N=16, IND N=16 and NIND N=10).
Abbreviations: CSF, cerebrospinal fluid; IND, infectious neurological disorders; NIND, nonIND; TBM, tuberculous meningitis.
Table 1.
Performance of developed tests in diagnosis of TBM
| Assays for TBM diagnosis | Sensitivity (%) | Specificity (%) | Comments |
|---|---|---|---|
| TB culture40 | 27 | Poor sensitivity | |
| Smear microscopy41 | 10–20 | Poor sensitivity | |
| NAATs | |||
| Xpert MTB/RIF assay39 | 55.1 | 94.8 | Moderate sensitivity, but an excellent rule-in test |
| Nucleic acid commercial amplification tests42 meta-analysis | 56 | 98 | Suboptimal sensitivity, time-consuming, requires DNA isolation and amplification steps |
| Commercial NAATs43 meta-analysis | 64 | 98 | |
| In-house qPCR44 | 87.6 (filtrate) 53.1 (sediment) |
92 | Time-consuming, requires DNA isolation and amplification steps Time-consuming, requires DNA isolation and amplification steps |
| In-house qPCR3 | 100 (definite TBM) 98 (probable and possible TBM) |
96–97 (definite TBM) 98 (probable and possible TBM) |
|
| Antigen detection tests | |||
| This study | 95 | 97.5 | Excellent diagnostic accuracy, laboratory-based format adaptable to POC, only 30 minutes from sample to answer |
| Aptamer-based ALISA HspX36 | 100 | 91 | Excellent diagnostic accuracy, laboratory-based format |
| Antibody-based ELISA3 GlcB, HspX and PstS1 GlcB, HspX and MPT51 |
100 (definite TBM) 92–95 (probable and possible TBM) |
96–97 (definite TBM) 93–96 (probable and possible TBM) |
Excellent diagnostic accuracy, laboratory-based format, time-consuming, uses antibody which might have batch to batch variation |
| Antibody-based ELISA LAM, Ag85 complex, 65 kDa and ESAT-645 | 87 | 84 | Laboratory-based format, time-consuming, uses antibody which might have batch to batch variation |
Abbreviations: TBM, tuberculosis meningitis; TB, tuberculosis; NAATs, nucleic acid amplification tests; POC, point-of-care; ALISA, aptamer-linked immobilized sorbent assay.
Conclusion
In summary, we demonstrate a new aptamer-based electrochemical biosensing strategy by exploiting the target-induced structural switching of H63 SL-2 M6 aptamer and electroactivity of aptamer-tagged MB for the detection of HspX in CSF samples for the diagnosis of TBM. This sensor exhibits high sensitivity and specificity (~95% and ~97.5%, respectively) and sample-to-answer time of ≤30 minutes. One limitation of this study was that the ROC derived cut-off was based on the studied sample size (N=81). However, in order to derive a more robust cut-off we propose to conduct an independent validation of the developed assay on blinded sample sets from geographically distinct populations of TBM patients in the near future. The findings of this proof of concept study clearly indicate the translational potential of the developed electrochemical aptasensor for the detection of TBM, and in future, its application may also be extended for the diagnosis of other forms of tuberculosis.
Supplementary materials
Classification of cerebrospinal fluid (CSF) samples
A subset of 81 pediatric CSF samples that was derived from an archived set of samples described previously by Haldar et al1 was used in the present study. The samples were categorized according to a universal case definition for tuberculous meningitis (TBM).2 Culture/smear/commercial nucleic acid amplification test (NAAT) positive/ acid-fast bacillus (AFB) seen on autopsy were categorized as “definite” TBM. “Probable” and “possible” TBM groups include subjects negative by the above criteria but satisfying the defined clinical criteria, CSF biochemistry and cytology, cerebral imaging criteria and evidence of extraneural TB having a score of ≥10–12 (“probable” TBM) and a score of ≥6–11 (“possible” TBM). In this CSF sample subset (N=81), samples were classified as “definite” TBM on the basis of M. tuberculosis culture positivity only (N=16) and as “probable” TBM (N=7, score range: 10–18), “possible” TBM (N=16, score range: 6–9) and “not-tuberculous meningitis” (not-TBM) with an alternative diagnosis established (N=42). The “not-TBM” category was further sub-classified into nontuberculous infectious meningitis (NTIM, N=16), infectious neurological disorders (IND, N=16) and noninfectious neurological disorders (NIND, N=10). The median age and range of the children in each diagnostic category was quite comparable (Table S1). The NTIM comprised of cases of pyogenic bacterial meningitis that included 2 culture-confirmed cases of E. coli (N=1), and Acinetobacter sp. (N=1). Another 14 cases were diagnosed on the basis of response to appropriate antibiotics, clinical presentation and symptoms. The IND category included 5 cases of meningoencephalitis, 4 cases of enteric encephalopathy, 3 cases of sepsis, 2 cases of cerebral malaria and 1 case each of pneumonia and post diphtheritic polyneuritis. The NIND category included 5 cases of neurodegenerative disorders, 2 cases each of hypocalcemic seizures and transverse myelitis, 1 case of Guillain–Barré syndrome.
Circular dichroism (CD) study
CD spectra were collected as described recently using Jasco J-815 Spectropolarimeter (Jasco Hachioji, Tokyo, Japan) equipped with Peltier junction temperature controller and continuous nitrogen flow was maintained to avoid water condensation around the cuvette as described recently.3 A cuvette with path length of 0.2 cm was used, each spectra was scanned at interval of 0.1 nm at scan rate of 20 nm/min in binding buffer (10 mM Tris pH 7.5,10 mM MgCl2, 50 mM KCl, 25 mM NaCl). Blank spectrum was recorded (binding buffer) and subtracted from the CD spectrum obtained from the aptamer or aptamer + HspX containing solution. Three scans were recorded for each sample in a range of 200–340 nm and their average was plotted to examine the target-dependent change in aptamer structure.
Statistical analysis
The diagnostic potential of the aptamer-based electrochemical sensor was evaluated by obtaining current difference (before and after adding the sample) values in CSF from “definite” TBM (true positives) and NTIM group (true negatives) as described recently.3 The obtained values were used to plot receiver operating characteristic curve (ROC) using GraphPad Prism version 5.00 for Windows, (GraphPad Software, Inc., La Jolla, CA, USA, www.graphpad.com). The cut-off values were obtained to achieve test sensitivity (≥95%) coherent with recent recommendations for diagnostic tests targeting childhood TB. These cut-off values were then applied to the “probable” and “possible” TBM groups, “IND” and “NIND” of not-TBM subjects to evaluate assay performance.
Secondary structure of H63 SL-2 M6 aptamer.
H63 SL-2 M6 Aptamer (black) and H63 SL-2 M6+HspX (red).
Sensor response as a function of protein (HspX) data was fitted using sigmoidal fitting equation in Origin Pro 8 software.
Note: Image inset depicts the limit of detection in the lower antigen range.
Sensor response as a function of protein (HspX) spiked in pooled CSF (NIND).
Notes: Samples and data were fitted using sigmoidal fitting equation in Origin Pro 8 software. Image inset depicts the limit of detection in the lower antigen range. Abbreviations: CSF, cerebrospinal fluid; NIND, noninfectious neurological disorders.
Scatter plot showing sensor response for CSF samples from definite (true positive) and NTIM (true negative) subjects (A). ROC curve derived from sensor response for definite (true positive) and NTIM (true negative) samples (B) showing AUC as 1.0 evincing robust performance of the sensor.
Abbreviations: NTIM, nontuberculous infectious meningitis; AUC, area under the curve; CSF, cerebrospinal fluid; ROC, receiver operating characteristic.
Table S1.
Comparison of various CSF parameters (N=81)a
| Parameters | TBMb (N=39) | NTIM (N=16) | IND (N=16) | NIND (N=10) | TBM vs NTIM | TBM vs IND | TBM vs NIND |
|---|---|---|---|---|---|---|---|
| Age (years) | 7 (2.75–11) | 6 (1.31–10.35) | 7.5 (4.5–13.25) | 5.5 (1.75–10.5) | – | – | – |
| Cells in CSF (per µL) | 30 (5.5–100) | 55 (15–175) | 0.5 (0–2.75) | 0 (0–2) | P=0.55 | <0.05 P=0.0003 | <0.0001 |
| Number of samples with cells (≥10 per µL) | 28 | 12 | 2 | 3 | – | – | – |
| Lymphocyte % | 75 (24.5–100) | 5 (0–65) | 25 (0–100) | 0 (0–100) | <0.05 P=0.02 |
P=0.25 | P=0.22 |
| Number of samples with lymphocytes (>50%) | 25 | 5 | 7 | 7 | – | – | – |
| Neutrophils | 0 (0–30) | 30 (0–92.5) | 0 (0–0) | 0 (0–0) | P=0.17 | P=0.29 | <0.05 P=0.003 |
| CSF sugar (mg%) | 53 (34.5–66.5) | 54 (25–79.75) | 69.5 (60–77.5) | 62 (48.75–82.25) | P=0.83 | <0.05 P=0.013 |
P=0.14 |
| Concomitant blood sugar | 108 (95.5–121.5) | 111 (108–121) | 100 (91–116.5) | 99.5 (92.75–110.25) | P=0.34 | P=0.51 | P=0.41 |
| CSF to blood sugar ratio | 0.47 (0.32–0.58) | 0.49 (0.19–0.7) | 0.7 (0.58–0.728) | 0.69 (0.43–0.82) | P=0.99 | <0.05 P=0.002 |
<0.05 P=0.02 |
| Number of samples with low CSF to blood sugar ratio (<0.5) | 19 | 8 | 3 | 5 | – | – | – |
| CSF protein (mg%) | 97 (75–136) | 75 (43.5–105.5) | 52 (34.5–77.5) | 32.5 (25.5–62.5) | P=0.06 | <0.05 P=0.002 |
<0.05 P=0.0004 |
| Number of samples with raised CSF proteins (>100 mg/dL) | 18 | 4 | 3 | 2 | – | – | – |
Notes:
Median values are indicated; values in brackets indicate the interquartile range. This study was performed on an archived sample from a previous study by Haldar et al.1
Includes “definite”, “probable” and “possible” TBM groups.
Abbreviations: TBM, tuberculosis meningitis; NTIM, nontuberculosis infectious meningitis; IND, infectious neurological disorders; NIND, nonIND; CSF, cerebrospinal fluid.
References
- 1.Haldar S, Sankhyan N, Sharma N, et al. Detection of Mycobacterium tuberculosis GlcB or HspX antigens or devR DNA impacts the rapid diagnosis of tuberculous meningitis in children. PLoS One. 2012;7(9):e44630. doi: 10.1371/journal.pone.0044630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marais S, Thwaites G, Schoeman JF, et al. Tuberculous meningitis: a uniform case definition for use in clinical research. Lancet Infect Dis. 2010;10(11):803–812. doi: 10.1016/S1473-3099(10)70138-9. [DOI] [PubMed] [Google Scholar]
- 3.Dhiman A, Haldar S, Mishra SK, et al. Generation and application of DNA aptamers against HspX for accurate diagnosis of tuberculous meningitis. Tuberculosis. 2018;112:27–36. doi: 10.1016/j.tube.2018.07.004. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This work was supported through a Biotechnology Ignition Grant (BIRAC/IKP0366/BIG-08/16) from the Biotechnology Research Assistance Council (BIRAC) to AptaBharat Innovation Pvt. Ltd. and funding support through the National Biode sign Alliance Project from the Department of Biotechnology, All India Institute of Medical Sciences, which the Government of India awarded to JST (BT/PR14638/ MED/12/483/2010). AD acknowledges the Indian Council of Medical Research (ICMR) for providing Senior Research Fellowship. The excellent technical assistance of Sanjay Kumar and Jitendra Kumar is duly acknowledged. Sarin Prakash and Sunil Kumar are sincerely thanked for their active involvement in data management and sample collection.
Footnotes
Disclosure
The aptamer used in this study is a proprietary reagent and is deposited in the Indian Patent Office as part of Indian Patent application number 201611001550; the patent was transferred to a Translational Health Science and Technology Institute (THSTI) spinoff, AptaBharat Innovation Pvt. Ltd. (ABIPL), for further development of aptamer-based diagnostics. RD is employed by AptaBharat Innovation Pvt. Ltd (ABIPL) and TKS is employed by THSTI. TKS is also the founder of THSTI spinoff, ABIPL and owns 87% stakes in ABIPL. The authors report no other conflicts of interest in this work.
References
- 1.World Health Organization [homepage on the Internet] Global tuberculosis report 2017. [Accessed August 30, 2018]. Available from: http://www.who.int/tb/publications/global_report/en/
- 2.Chakraborty AK. Estimating mortality from tuberculous meningitis in a community: use of available epidemiological parameters in the Indian context. Ind J Tub. 2000;47:9–14. [Google Scholar]
- 3.Haldar S, Sankhyan N, Sharma N, et al. Detection of Mycobacterium tuberculosis GlcB or HspX antigens or devR DNA impacts the rapid diagnosis of tuberculous meningitis in children. PLoS One. 2012;7(9):e44630. doi: 10.1371/journal.pone.0044630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin JH. Tuberculous meningitis: diagnostic and therapeutic challenges. Neurol Clin Pract. 2014;4(3):199–205. doi: 10.1212/CPJ.0000000000000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdon R, Chevret S, Laissy JP, Wolff M. Tuberculous meningitis in adults: review of 48 cases. Clin Infect Dis. 1996;22(6):982–988. doi: 10.1093/clinids/22.6.982. [DOI] [PubMed] [Google Scholar]
- 6.Andersen P, Munk ME, Pollock JM, Doherty TM. Specific immune-based diagnosis of tuberculosis. The Lancet. 2000;356(9235):1099–1104. doi: 10.1016/s0140-6736(00)02742-2. [DOI] [PubMed] [Google Scholar]
- 7.Marais S, Thwaites G, Schoeman JF, et al. Tuberculous meningitis: a uniform case definition for use in clinical research. Lancet Infect Dis. 2010;10(11):803–812. doi: 10.1016/S1473-3099(10)70138-9. [DOI] [PubMed] [Google Scholar]
- 8.Bahr NC, Marais S, Caws M, et al. GeneXpert MTB/RIF to diagnose tuberculous meningitis: perhaps the first test but not the last. Clin Infect Dis. 2016;62(9):1133–1135. doi: 10.1093/cid/ciw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobotka O, Endlicher R, Drahota Z, et al. Impaired mitochondrial functions contribute to 3-bromopyruvate toxicity in primary rat and mouse hepatocytes. J Bioenerg Biomembr. 2016;48(4):363–373. doi: 10.1007/s10863-016-9674-8. [DOI] [PubMed] [Google Scholar]
- 10.Patel VB, Theron G, Lenders L, et al. Diagnostic accuracy of quantitative PCR (Xpert MTB/RIF) for tuberculous meningitis in a high burden setting: a prospective study. PLoS Med. 2013;10(10):e1001536. doi: 10.1371/journal.pmed.1001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nhu NT, Heemskerk D, Thu DDA, et al. Evaluation of GeneXpert MTB/RIF for diagnosis of tuberculous meningitis. J Clin Microbiol. 2014;52(1):226–233. doi: 10.1128/JCM.01834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahr NC, Nuwagira E, Evans EE, et al. Diagnostic accuracy of Xpert MTB/RIF ultra for tuberculous meningitis in HIV-infected adults: a prospective cohort study. Lancet Infect Dis. 2018;18(1):68–75. doi: 10.1016/S1473-3099(17)30474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashyap RS, Kainthla RP, Satpute RM, et al. Demonstration of IgG antibodies to 30 kD protein antigen in CSF for diagnosis of tuberculous meningitis by antibody-capturing ELISA. Neurol India. 2004;52(3):359–362. [PubMed] [Google Scholar]
- 14.Chandramuki A, Bothamley GH, Brennan PJ, Ivanyi J. Levels of antibody to defined antigens of Mycobacterium tuberculosis in tuberculous meningitis. J Clin Microbiol. 1989;27(5):821–825. doi: 10.1128/jcm.27.5.821-825.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med. 2012;9(9):e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro-Garza J, García-Jacobo P, Rivera-Morales LG, et al. Detection of anti-HspX antibodies and HspX protein in patient sera for the identification of recent latent infection by Mycobacterium tuberculosis. PLoS One. 2017;12(8):e0181714. doi: 10.1371/journal.pone.0181714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laal S, Samanich KM, Sonnenberg MG, Zolla-Pazner S, Phadtare JM, Belisle JT. Human humoral responses to antigens of Mycobacterium tuberculosis: immunodominance of high-molecular-mass antigens. Clin Diagn Lab Immunol. 1997;4(1):49–56. doi: 10.1128/cdli.4.1.49-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh KK, Dong Y, Belisle JT, Harder J, Arora VK, Laal S. Antigens of Mycobacterium tuberculosis recognized by antibodies during incipient, subclinical tuberculosis. Clin Diagn Lab Immunol. 2005;12(2):354–358. doi: 10.1128/CDLI.12.2.354-358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori T, Sakatani M, Yamagishi F, et al. Specific detection of tuberculosis infection: an interferon-gamma-based assay using new antigens. Am J Respir Crit Care Med. 2004;170(1):59–64. doi: 10.1164/rccm.200402-179OC. [DOI] [PubMed] [Google Scholar]
- 20.Lakshmipathy D, Kulandai LT, Ramasubban G, Hajib Narahari Rao M, Rathinam S, Narasimhan M. Nested reverse transcriptase–polymerase chain reactions targeting the messenger RNA of ICL2, hspx, and rRNAP1 genes to detect viable Mycobacterium tuberculosis directly from clinical specimens. Int J Mycobacteriol. 2015;4(4):312–317. doi: 10.1016/j.ijmyco.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 21.You M, Chen Y, Peng L, et al. Engineering DNA aptamers for novel analytical and biomedical applications. Chem Sci. 2011;2(6):1003. [Google Scholar]
- 22.Dhiman A, Kalra P, Bansal V, Bruno JG, Sharma TK. Aptamer-based point-of-care diagnostic platforms. Sensors Actuators B Chem. 2017;246:535–553. [Google Scholar]
- 23.Seo HB, Gu MB. Aptamer-based sandwich-type biosensors. J Biol Eng. 2017;11:11. doi: 10.1186/s13036-017-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumari P, Lavania S, Tyagi S, et al. A novel aptamer-based test for the rapid and accurate diagnosis of pleural tuberculosis. Anal Biochem. 2019:564–565. 80–87. doi: 10.1016/j.ab.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Lavania S, Das R, Dhiman A, et al. Aptamer-based TB antigen tests for the rapid diagnosis of pulmonary tuberculosis: potential utility in screening for tuberculosis. ACS Infect Dis. 2018;4(12):1718–1726. doi: 10.1021/acsinfecdis.8b00201. [DOI] [PubMed] [Google Scholar]
- 26.Kaur H, Bhagwat SR, Sharma TK, Kumar A. Analytical techniques for characterization of biological molecules – proteins and aptamers/ oligonucleotides. Bioanalysis. 2019;11(2):103–117. doi: 10.4155/bio-2018-0225. [DOI] [PubMed] [Google Scholar]
- 27.Sharma TK, Bruno JG, Dhiman A. ABCs of DNA aptamer and related assay development. Biotechnol Adv. 2017;35(2):275–301. doi: 10.1016/j.biotechadv.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Rashid F, Shah A, et al. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc Natl Acad Sci U S A. 2015;112(32):10002–10007. doi: 10.1073/pnas.1502159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10(3):282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Kaur H, Bruno JG, Kumar A, Sharma TK. Aptamers in the therapeutics and diagnostics pipelines. Theranostics. 2018;8(15):4016–4032. doi: 10.7150/thno.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong P, Li W, Li J. Applications of aptasensors in clinical diagnostics. Sensors. 2012;12(2):1181–1193. doi: 10.3390/s120201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer F, Mayer G. Selection and biosensor application of aptamers for small molecules. Front Chem. 2016;4:25. doi: 10.3389/fchem.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Tuleouva N, Ramanculov E, Revzin A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal Chem. 2010;82(19):8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Xu J, Liu J, Wang X, Chen B. Disease-related detection with electrochemical biosensors: a review. Sensors. 2017;17(10):2375. doi: 10.3390/s17102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catanante G, Mishra RK, Hayat A, Marty JL. Sensitive analytical performance of folding based biosensor using methylene blue tagged aptamers. Talanta. 2016;153:138–144. doi: 10.1016/j.talanta.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Dhiman A, Haldar S, Mishra SK, et al. Generation and application of DNA aptamers against HspX for accurate diagnosis of tuberculous meningitis. Tuberculosis. 2018;112:27–36. doi: 10.1016/j.tube.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Kalra P, Dhiman A, Cho WC, Bruno JG, Sharma TK. Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity. Front Mol Biosci. 2018;14(5):41. doi: 10.3389/fmolb.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das R, Sharma MK, Rao VK, et al. An electrochemical genosensor for Salmonella typhi on gold nanoparticles-mercaptosilane modified screen printed electrode. J Biotechnol. 2014;188:9–16. doi: 10.1016/j.jbiotec.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Rufai SB, Singh A, Singh J, et al. Diagnostic usefulness of Xpert MTB/ RIF assay for detection of tuberculous meningitis using cerebrospinal fluid. J Infect. 2017;75(2):125–131. doi: 10.1016/j.jinf.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Thakur R, Goyal R, Sarma S. Laboratory diagnosis of tuberculous meningitis – is there a scope for further improvement? J Lab Physicians. 2010;2(1):21–24. doi: 10.4103/0974-2727.66705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thwaites G, Chau TT, Mai NT, Drobniewski F, McAdam K, Farrar J. Tuberculous meningitis. J Neurol Neurosurg Psychiatry. 2000;68(3):289–299. doi: 10.1136/jnnp.68.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pai M, Flores LL, Pai N, Hubbard A, Riley LW, Colford JM. Diagnostic accuracy of nucleic acid amplification tests for tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect Dis. 2003;3(10):633–643. doi: 10.1016/s1473-3099(03)00772-2. [DOI] [PubMed] [Google Scholar]
- 43.Solomons RS, van Elsland SL, Visser DH, et al. Commercial nucleic acid amplification tests in tuberculous meningitis-a meta-analysis. Diagn Microbiol Infect Dis. 2014;78(4):398–403. doi: 10.1016/j.diagmicrobio.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Haldar S, Sharma N, Gupta VK, Tyagi JS. Efficient diagnosis of tuberculous meningitis by detection of Mycobacterium tuberculosis DNA in cerebrospinal fluid filtrates using PCR. J Med Microbiol. 2009;58(5):616–624. doi: 10.1099/jmm.0.006015-0. [DOI] [PubMed] [Google Scholar]
- 45.Flores LL, Steingart KR, Dendukuri N, et al. Systematic review and meta-analysis of antigen detection tests for the diagnosis of tuberculosis. Clin Vaccine Immunol. 2011;18(10):1616–1627. doi: 10.1128/CVI.05205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secondary structure of H63 SL-2 M6 aptamer.
H63 SL-2 M6 Aptamer (black) and H63 SL-2 M6+HspX (red).
Sensor response as a function of protein (HspX) data was fitted using sigmoidal fitting equation in Origin Pro 8 software.
Note: Image inset depicts the limit of detection in the lower antigen range.
Sensor response as a function of protein (HspX) spiked in pooled CSF (NIND).
Notes: Samples and data were fitted using sigmoidal fitting equation in Origin Pro 8 software. Image inset depicts the limit of detection in the lower antigen range. Abbreviations: CSF, cerebrospinal fluid; NIND, noninfectious neurological disorders.
Scatter plot showing sensor response for CSF samples from definite (true positive) and NTIM (true negative) subjects (A). ROC curve derived from sensor response for definite (true positive) and NTIM (true negative) samples (B) showing AUC as 1.0 evincing robust performance of the sensor.
Abbreviations: NTIM, nontuberculous infectious meningitis; AUC, area under the curve; CSF, cerebrospinal fluid; ROC, receiver operating characteristic.
Table S1.
Comparison of various CSF parameters (N=81)a
| Parameters | TBMb (N=39) | NTIM (N=16) | IND (N=16) | NIND (N=10) | TBM vs NTIM | TBM vs IND | TBM vs NIND |
|---|---|---|---|---|---|---|---|
| Age (years) | 7 (2.75–11) | 6 (1.31–10.35) | 7.5 (4.5–13.25) | 5.5 (1.75–10.5) | – | – | – |
| Cells in CSF (per µL) | 30 (5.5–100) | 55 (15–175) | 0.5 (0–2.75) | 0 (0–2) | P=0.55 | <0.05 P=0.0003 | <0.0001 |
| Number of samples with cells (≥10 per µL) | 28 | 12 | 2 | 3 | – | – | – |
| Lymphocyte % | 75 (24.5–100) | 5 (0–65) | 25 (0–100) | 0 (0–100) | <0.05 P=0.02 |
P=0.25 | P=0.22 |
| Number of samples with lymphocytes (>50%) | 25 | 5 | 7 | 7 | – | – | – |
| Neutrophils | 0 (0–30) | 30 (0–92.5) | 0 (0–0) | 0 (0–0) | P=0.17 | P=0.29 | <0.05 P=0.003 |
| CSF sugar (mg%) | 53 (34.5–66.5) | 54 (25–79.75) | 69.5 (60–77.5) | 62 (48.75–82.25) | P=0.83 | <0.05 P=0.013 |
P=0.14 |
| Concomitant blood sugar | 108 (95.5–121.5) | 111 (108–121) | 100 (91–116.5) | 99.5 (92.75–110.25) | P=0.34 | P=0.51 | P=0.41 |
| CSF to blood sugar ratio | 0.47 (0.32–0.58) | 0.49 (0.19–0.7) | 0.7 (0.58–0.728) | 0.69 (0.43–0.82) | P=0.99 | <0.05 P=0.002 |
<0.05 P=0.02 |
| Number of samples with low CSF to blood sugar ratio (<0.5) | 19 | 8 | 3 | 5 | – | – | – |
| CSF protein (mg%) | 97 (75–136) | 75 (43.5–105.5) | 52 (34.5–77.5) | 32.5 (25.5–62.5) | P=0.06 | <0.05 P=0.002 |
<0.05 P=0.0004 |
| Number of samples with raised CSF proteins (>100 mg/dL) | 18 | 4 | 3 | 2 | – | – | – |
Notes:
Median values are indicated; values in brackets indicate the interquartile range. This study was performed on an archived sample from a previous study by Haldar et al.1
Includes “definite”, “probable” and “possible” TBM groups.
Abbreviations: TBM, tuberculosis meningitis; NTIM, nontuberculosis infectious meningitis; IND, infectious neurological disorders; NIND, nonIND; CSF, cerebrospinal fluid.