Abstract
In this Review, we highlight some recent developments in the discovery and application of broadly neutralizing antibodies (bnAbs) to human immunodeficiency virus (HIV); i.e., antibodies able to neutralize diverse isolates of HIV. We consider the characterization of novel bnAbs, recent data on the effects of bnAbs in vivo in humans and animal models, and the importance of both kinds of data for the application of Abs to prophylaxis and therapy and to guide vaccine design. We seek to place newly discovered bnAbs in the context of existing bnAbs, and we explore the various characteristics of the antibodies that are most desirable for different applications.
In virology, no antibody (Ab) responses have been as extensively studied as those to HIV. Responses to natural infection with HIV have been investigated in many people, hundreds of monoclonal antibodies (mAbs) to the surface envelope (Env) glycoprotein of HIV have been isolated, and immunization of diverse animals and humans with Env proteins has been extensively explored. The results have provided a compelling picture of two titans of evolution, the virus and the humoral immune system, driving each other to complex solutions to maintain their functionality1. Some of the Abs are now being pursued directly as prophylactic or therapeutic reagents and, more indirectly, to help guide the design of vaccines against HIV. The impact of the studies has extended to many other pathogens, in terms of both the utility of Abs as anti-pathogen agents (‘super-antibodies’) and strategies for vaccine development2.
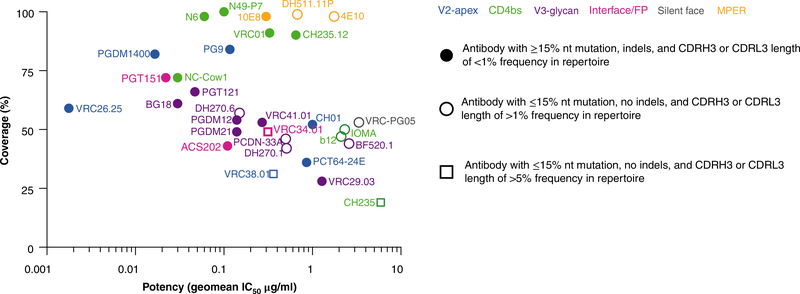
In this Review, we do not aim to repeat information that is available in many excellent reviews already published3–9; instead, we seek to highlight some very recent developments. As will be seen, there is tension between the qualities of Abs that might best serve their different uses. The best Abs for prophylaxis and therapy are probably those with the greatest potency and breadth. Such Abs typically have high levels of somatic hypermutation (SHM), insertions and/or deletions (indels) and, often, unusually long CDRH3 complementarity-determining regions, which all have disadvantages as templates for vaccine design. Abs with less SHM, no indels and CDRH3 regions of closer to average length might be a more realistic starting point for vaccine design than the former Abs (Fig. 1 and Table 1). The latter Abs are typically less potent and broad in neutralization. Put simply, one might argue that ‘outstanding Abs’ (such as those with a half-maximal inhibitory concentration (IC50) of <1 μg/ml) are best for prophylaxis and therapy, but ‘realistic Abs’ might be best for vaccine design (Fig. 1 and Table 1). Notably, these are qualitative assessments of Ab ‘elicitability’ that can be compared with a previously reported quantitative analysis called ‘Antibody Features Frequency’10. However, it should be borne in mind that there is not a definitive measure of the levels of bnAbs that will protect humans against exposure to HIV, so it remains unknown how potent the Abs induced by vaccine design need to be. Some estimates can be made from studies of animal models, as discussed below, but this remains a major uncertainty in the field.
Fig. 1 |. Neutralization coverage of large panels of global isolates by bnAbs as a function of neutralization potency.
Neutralization was measured through the use of pseudoviruses in a TZM-bl assay101 and is presented as potency (geometric mean (geomean) IC50, in μg/ml) of newly discovered bnAbs (bold) and earlier bnAbs (less intense), grouped by epitope targeted (colors in key, top right), as in Table 1. Symbols (key, right) indicate ‘elicitability’: Abs with ≥15% overall nucleotide mutation in the heavy and light chain combined, indels or CDR3 length frequencies (CDRH3 or CDRL3) <1% that of the Ab repertoire are least favorably elicited; Abs with ≤15% overall nucleotide mutation in the heavy and light chain combined, no indels and CDR3 length frequencies >1% that of the Ab repertoire are more favorable (‘realistic Abs’); Abs with ≤15% overall nucleotide mutation in the heavy and light chain combined, no indels and CDR3 length frequencies >5% that of the Ab repertoire are most favorable (‘realistic Abs’; this includes only three bnAbs here). The CDRH3 length-frequency cut-offs for comparison with those above are as follows. About 97% of Abs in the human repertoire have a CDRH3 length between 8 and 23 amino acids, and each length is represented at least 1% or more in the repertoire. About 73% of Abs in the repertoire range between 11 and 18 amino acids, and each length is represented at least 5% or more in the repertotire. Correspondingly, about 99% of the λ-CDRL3 lengths range between 8 and 13 amino acids, with each length represented at 1% or more, and 96% range between 9 and 12 amino acids, with each length represented at 5% or more. For the κ-CDRL3, 98% range between 8 and 11 amino acids, with each length represented at 1% or more, and 94% range between 8 and 10 amino acids, with each length represented at 5% or more.
Table 1|.
Breadth, potency and select characteristics of bnAbs
| Epitope | Ab | IC50 geometric mean (μg/ml) | Coverage | Number of viruses tested | HC V gene | lC V gene | CDRH3 length (aa) | CDRl3 length (aa) | VHHJ + VκHJ or VλHJ (%nt) | Insertions (+) or deletions (−) | Refs. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| V2 apex | VRC26.25 | 0.002 | 59% | 174 | HV3-30*03 | λV1-51*02 | 38 | 12 | 10 | 22 | |
| V2 apex | PCT64–24E | 0.911 | 33% | 115 | HV3-15*01 | κV3-20*01 | 25 | 8 | 8 | 25 | |
| V2 apex | VRC38.01 | 0.361 | 31% | 210 | HV3-13*01 | κV2-28*01 | 18 | 10 | 13 | 24 | |
| V2 apex | PG9 | 0.118 | 84% | 200 | HV3-33*05 | λV2-14*01 | 30 | 10 | 11 | 17 | |
| V2 apex | PGDM1400 | 0.017 | 81% | 194 | HV1-8*01 | κV2-14*01 | 34 | 9 | 18 | 12 | |
| V2 apex | CH01 | 1.007 | 52% | 195 | HV3-20*01 | κV3-20*01 | 26 | 9 | 13 | 102 | |
| V3 glycan | BG18 | 0.032 | 62% | 119 | HV4-4*02 | λV3-25*03 | 23 | 11 | 18 | 35 | |
| V3 glycan | DH270.1 | 0.510 | 42% | 179 | HV1-2*02 | λV2-23*02 | 20 | 10 | 6 | 34 | |
| V3 glycan | DH270.6 | 0.151 | 57% | 179 | HV1-2*02 | λV2-23*02 | 20 | 10 | 11 | 34 | |
| V3 glycan | PGDM12 | 0.134 | 57% | 111 | HV3-11*03 | κV2-24*01 | 21 | 9 | 16 | CDRH1 (−2 aa) | 12 |
| V3 glycan | VRC41.01 | 0.275 | 53% | 107 | HV4-39*07 | κV3-20*01 | 21 | 9 | 16 | 103 | |
| V3 glycan | PGDM21 | 0.139 | 50% | 111 | HV4-34*08 | κV3-20*01 | 20 | 9 | 18 | CDRH2 (+4 aa) | 12 |
| V3 glycan | PCDN-33A | 0.410 | 49% | 125 | HV4-34*01 | κV3-20*01 | 22 | 8 | 11 | 104 | |
| V3 glycan | BF520.1 | 2.600 | 44% | 34 | HV1-2*02 | κV3-15*01 | 20 | 11 | 6 | 32 | |
| V3 glycan | VRC29.03 | 1.276 | 28% | 179 | HV4-59*08 | κV3-20*01 | 20 | 9 | 12 | CDRH2 (+6 aa) | 31 |
| V3 glycan | PGT121 | 0.048 | 66% | 200 | HV4-59*01 | λV3-21*01 | 26 | 12 | 17 | L-FR3 (+3 aa), L-FR1 (−7 aa) | 109 |
| V3 glycan | 10–1074 | 0.039 | 68% | 200 | HV4-59*01 | λV3-21*01 | 26 | 12 | NA | L-FR3 (+3 aa), L-FR1 (−4 aa) | 105 |
| CD4bs | N49-P7 | 0.100 | 100% | 117 | HV1-2*02 | λV2-11*01 | 21 | 5 | 19 | CDRL1 (−6 aa) | 41 |
| CD4bs | N6 | 0.058 | 98% | 327 | HV1-2*02 | κV1-33*01 | 15 | 5 | 25 | 40 | |
| CD4bs | NC-Cow1 | 0.028 | 72% | 117 | HV1-7*01 | - | 62 | - | - | 53 | |
| CD4bs | IOMA | 2.320 | 50% | 118 | HV1-2*02 | λV2-23*02 | 19 | 8 | 10 | 52 | |
| CD4bs | CH235 | 5.927 | 19% | 176 | HV1-46*01 | κV3-15*01 | 15 | 8 | 6 | 106 | |
| CD4bs | CH235.12 | 0.650 | 90% | 199 | HV1-46*01 | κV3-15*01 | 15 | 8 | 19 | 51 | |
| CD4bs | b12 | 2.204 | 47% | 200 | HV1-3*01 | κV3-20*01 | 20 | 9 | 12 | 48 | |
| CD4bs | VRC01 | 0.329 | 91% | 196 | HV1-2*02 | κV3-20*01 | 14 | 5 | 23 | CDRL1 (−3 aa) | 110 |
| CD4bs | 3BNC117 | 0.097 | 85% | 198 | HV1-2*02 | κV1-33*01 | 12 | 5 | 19 | H-FR3 (+4 aa), CDRL1 (−4 aa) | 56 |
| CD4bs | CH103 | 0.699 | 85% | 150 | HV4-61*08 | λV3-1*01 | 15 | 10 | 15 | CDRL1 (−3 aa) | 50 |
| Silent face | VRC-PG05 | 3.714 | 53% | 78 | HV3-7*01 | κV4-1*01 | 19 | 8 | 9 | 15 | |
| Interface- FP | VRC34.01 | 0.310 | 49% | 179 | HV1-2*02 | κV1-9*01 | 15 | 9 | 11 | 59 | |
| Interface- FP | ACS202 | 0.140 | 44% | 81 | HV3-30*03 | κV1-33*01 | 24 | 9 | 15 | CDRH2 (+1 aa) | 60 |
| Interface- FP | PGT151 | 0.023 | 72% | 200 | HV3-30*03 | κV2D-29*02 | 28 | 9 | 16 | 55 | |
| Interface- FP | 35O22 | 0.151 | 56% | 200 | HV1-18*03 | λV2-14*02 | 16 | 10 | 22 | H-FR3 (+8 aa) | 57 |
| Interface- FP | 8ANC195 | 1.115 | 66% | 200 | HV1-3*03 | κV1-5*03 | 22 | 9 | 21 | CDRH1 (+1 aa), H-FR3 (+4 aa), CDRH2 (−2 aa), HCDRL1 (+1 aa) | 56 |
| MPER | DH511.11P | 0.674 | 99% | 180 | HV3-15*01 | κV1-39*01 | 23 | 11 | 15 | 65 | |
| MPER | 4E10 | 1.765 | 98% | 200 | HV1-69*17 | κV3-20*01 | 20 | 9 | 10 | 107 | |
| MPER | 10E8 | 0.299 | 98% | 199 | HV3-15*05 | λV3-19*01 | 22 | 12 | 17 | 111 |
Neutralization breadth and potency were captured mainly from the tool CATNAP100 or, in some cases, the primary manuscript (far right column). Overall nucleotide mutation (%nt) was calculated as the mutation frequency given the genes encoding the heavy-chain variable (V) region and joining (J) region (HC V gene), as well as those encoding the light-chain V and J regions (LC V gene). The neutralization IC50 values reported are Ab inhibitory concentrations that reduce viral infectivity by 50% relative to neutralization in the absence of Ab (measured on TZM-bl cells). Geometric mean neutralization IC50 was calculated by exclusion of resistant viruses. Neutralization breadth was calculated on the basis of a neutralization IC50 cut-off of between 10 μg/ml and 50 μg/ml.
Novel bnAbs
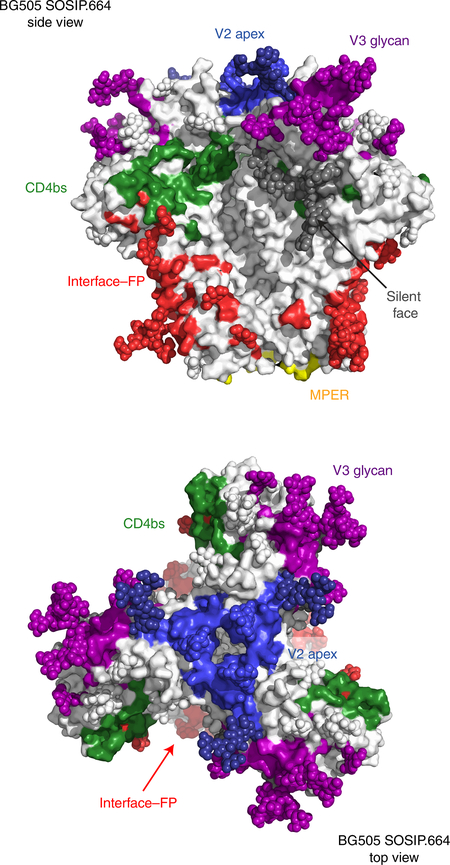
bnAbs are generally considered to puncture the glycan-shield defenses of the HIV Env trimer in five regions (Fig. 2), each of which is probably involved in Env function. Thus, the V2 apex region is believed to be involved in maintaining the metastability of the Env spike, so that after ligation of the co-receptor CD4, it rearranges to fully expose the binding site for the co-receptor CCR511. The V3 glycan site is formed partly by a section of the CCR5 co-receptor site and partly by the surrounding camouflaging glycans12. The CD4-binding site (CD4bs) is of course associated with binding of the primary receptor for HIV, CD4. The interface region for glycoproteins gp120 and gp41 includes the fusion peptide (FP) and the site for cleavage of gp160 into gp120 and gp41. Unusually for a viral Env structure, the hydrophobic FP is partially exposed on virions13. The membrane-proximal external region (MPER) is part of the fusion machinery14. To this list of sites, we can now add a sixth site, defined by the bnAb VRC-PG0515, which binds to the center of the ‘silent face’ of gp120 (so called because when the structure of gp120 was first determined, Abs that recognize residues in this region had not been described and therefore it was believed to be immunologically silent16). VRC-PG05 recognizes glycans at residues Asn295, Asn262 and Asn448, and 85% of VRC-PG05’s interacting binding surface is to glycans, which makes it the most heavily glycan-dependent bnAb after 2G12, which recognizes glycans exclusively. The principal glycan involved in VRC-PG05’s binding is the highly conserved (99%) glycan at Asn262, which buries almost 750 Å2 of the surface area during interaction with the Ab. VRC-PG05 is relatively modest in potency (geometric mean, ~4 μg/ml), which is more characteristic of the first generation rather than the second generation of bnAbs5, but it has relatively substantial breadth (53%).
Fig. 2 |. The bnAb epitopes on the HIV env trimer.
Amino-acid residues and glycans comprising epitope regions of SOSIP.664 gp140 trimers of the HIV isolate BG505 (BG505 SOSIP.664) that is targeted by bnAbs, in a side view (top) and top view (bottom); colors indicate epitope regions: blue, V2 apex; purple, V3 glycan; green, CD4bs; dark gray, silent face; red, interface–FP; and yellow, MPER.
bnAbs to the V2 apex
bnAbs to the V2 apex epitope target the apex of the HIV Env trimer. The key feature of these bnAbs is a long protruding CDRH3 region (≥24 residues) in a β-hairpin or hammerhead conformation that penetrates between V2 glycans, particularly those at Asn160, and has an anionic tip, including sulfated tyrosines that contact basic residues on β-strand C of the V2 region17–19. The bnAbs to the V2 apex are among the most potent Abs described, although they rarely achieve more than 70% breadth and tend to show incomplete neutralization (<100%)20,21. New members of the CAP256 family of bnAbs have been described through the use of B cell culture22, including the bnAb CAP256-VRC26.25, which neutralizes 59% of a large panel of global isolates of HIV and about 70% of a large panel of clade C isolates of HIV with a IC50 (geometric mean) of around 2 ng/ml. These numbers probably make this Ab the most potent bnAb described so far. In addition, for the CAP256 family, sialic acid–bearing glycans are recognized by germline-reverted Abs and serve as ‘anchors’ for the evolution of nAb breadth23.
A novel bnAb lineage, N90-VRC38, was discovered by affinity sorting of B cells on virus-like particles24. These Abs recognize the V2 apex without a long protruding CDRH3. A largely neutral CDRH3 of 16 residues makes multiple interactions between Ab side chains and Env side chains, whereas the prototypical bnAbs to the V2 apex, such as PG9, make extensive interactions between the CDRH3 main-chain parallel β-strand and Env strand C. Serum fingerprinting studies have shown that VRC38 specificities are relatively common responses in HIV-infected people, suggestive of possible advantages in targeting this specificity through vaccine design. However, the dependence of the Abs on a glycan at Asn130, which is present in less than half of HIV isolates, reduces the breadth of neutralization to around 30%. The IC50 (geometric mean) is of the order of 0.4 μg/ml.
A new bnAb lineage (PCT64) was identified by longitudinal studies of a donor from International AIDS Vaccine Initiative (IAVI) Protocol C25. These PCT64 Abs have a classic phenotype of bnAbs to the V2 apex in terms of a long, anionic, 25-residue CDRH3 that structurally mimics the long rod of bnAb PGT145 (discussed below), rather than the hammerhead structure of bnAbs PG9 and PG16. This Ab lineage is attractive from a vaccine-design standpoint, in that mature Abs have relatively little SHM (around 10%) and no indels. However, again, potency (IC50 = ~0.9 μg/ml) and breadth (33%) are modest for antibody PCT64–23E, a representative somatic variant.
Structural information on the interaction between V2 apex–targeting bnAbs and Env has relied on information obtained from complexes of the prototypical Abs PG9, PG16 and CH01–04 with epitope scaffolds. The first trimer structure of a bnAb to the V2 apex was provided in the form of a cryo-electron-microscopy construction of Ab PGT145 bound to the stabilized recombinant Env trimer from the HIV isolate BG50511. The protruding CDRH3 of PGT145 binds along the threefold axis of the trimer between Asn160 glycans and makes asymmetric contacts with the three glycans and the anionic tip of the CDRH3 contacts residues from three gp120 protomers. The trimer specificity of the Ab, typically used for trimer purification, was thus explained. In addition, SHM in the other CDR regions of PGT145 are crucial for stabilizing the elongated CDRH3 structure. Structures of bnAbs to the V2 apex from early and late in the evolution of the lineage have shown that the CDRH3 loop rigidifies, the bnAb angle of approach steepens, and surface charges are altered to accommodate glycan changes during the development of neutralization breadth26.
bnAbs to the V3 glycan site
bnAbs to the V3 glycan site are the most common of all Abs found in HIV infection27–29. Several new lineages of these bnAbs have been discovered, many with features that distinguish them from the prototypical Abs5 described so far, such as PGT121, PGT128 and 10–1074. A donor from IAVI Protocol C was the source for the isolation of bnAbs of the PCDN lineage30. The Abs show moderate potency (an IC50 of ~0.4 μg/ml) and breadth (~49%), relatively little SHM and no indels. Another study has described three different lineages of bnAbs to the V3 glycan site from donor N170, with moderate potency (about 1 μg/ml) and breadth (up to 28%)31. However, these Abs have features typical of bnAbs to the V3 glycan site, including the use of variable heavy-chain region 4 (VH4), an insertion in CDRH2 and a long CDRH3. bnAb BF520.1, isolated from an infant only 1 year after infection, had low levels of SHM, as expected, as well as no indels and substantial breadth (~44%), but moderate potency (an IC50 of ~3 μg/ml)32. Infants can develop bnAbs more rapidly than adults do, and multiple specificities have been described in some children33. More potent bnAbs (with an IC50 as low as 0.15 μg/ml) with substantial breadth (of up to 57%) have been isolated from the DH270 lineage34. DH270 had limited SHM and no indels, which stimulated interest in this Ab as a vaccine template. The most recent bnAb to the V3 glycan site with the best combination of potency and breadth described is BG1835, which has breadth as great as that of the prototypical bnAbs to the V3 glycan site but is somewhat more potent. The lack of indels in this Ab make it an attractive template for vaccine design and, given the success of the induction of bnAbs in the PGT121 germline-reverted Ab knock-in mouse model through the use of tailored immunogens36,37, there is particularly great interest in immunogen design for BG18 from a vaccine standpoint. SHM in BG18 is high, but Ab engineering has shown many of these mutations are not required for function (W.R. Schief, unpublished observations). Structural studies30,32,38 in the cases noted above have helped to define how the Abs navigate the glycan shield in the vicinity of the Asn332 glycan and access the GDIR motif (at residues 324–327) associated with binding of CCR512.
bnAbs to the CD4bs
The bnAbs to the CD4bs probably provide the best combination of breadth and potency among all bnAb specificities. They typically take longer than any other specificity to develop in natural infection and have, on average, the highest level of SHM. One class of bnAbs to the CD4bs are known as ‘CD4-mimic Abs’; their paratopes are characterized by the use of one of the two VH segments, VH1–2 or VH1–46, that mimic the immunoglobin-like amino-terminal domain of CD4 in interacting with the CD4bs. The most potent of these Abs invariably use a five-residue CDRL3 and are called ‘VRC01-class Abs’, for the prototypical Ab in this category39. An important development has been the discovery of two bnAbs of the VRC01 class with essentially pan–strain-neutralizing ability (98–100% breadth) at a median IC50 of ~0.1 μg/ml (refs 40,41). The bnAb N6 is able to avoid steric clashes with glycans40, which had restricted the breadth of some other Abs of the VRC01 class. N49P7 was isolated through the use of information from a proteomics study of the plasma response of an elite neutralizer (a person with outstanding serum neutralization breadth, on the basis of an alogrithm that compares cross-clade neutralization titers108) and was shown to access conserved residues within the inner domain of gp120, distinct from other Abs of the VRC01 class41. Both Abs (N6 and N49P7) would seem to be promising candidates for prophylactic and/or therapeutic applications. Because of their combined potency and breadth, Abs of the VRC01 class have also been a favorite for vaccine targeting through the use of sequential immunization strategies42–46, and the germline-targeting immunogen eOD-GT8 nanoparticle42 has now entered phase I clinical trials. Several different subclasses of VRC01 class Ab–expressing naive B cells were identified through the use of eOD-GT8 affinity selection47.
Another class of Abs to the CD4bs are known as ‘loop dependent Abs’; their paratope relies on the CDRH3 generated by recombination of variable-diversity-joining (V(D)J) gene segments for recognition of the CD4bs. Previously identified Abs that target the CD4bs epitope in this manner include b1248, HJ1649, and the CH103 Ab lineage50. The development of CH103 was tracked longitudinally, and the appearance of a VH1–46 ‘helper’ lineage (CH235) was associated with the development of neutralization breadth. With added time, this helper lineage also developed notable breadth and potency (90% breadth at a potency of 0.6 μg/ml)51. Finally, a novel bnAb, IOMA52, uses the VH1–2 germline segment but lacks the short CDRL3 characteristic of the VRC01 class. This Ab has modest breadth and potency. A high-resolution crystal structure of IOMA in complex with natively glycosylated Env has revealed how the Ab breaches the glycan shield52 and has paved the way for the design of vaccines targeting IOMA.
The most unusual bnAbs to the CD4bs described so far, and the only such Abs generated by vaccination, are those generated by the immunization of cows with recombinant HIV Env53. A subset of Abs in cows have an ultralong CDRH3 of up to 70 residues, and cows respond to immunization with SOSIP with rapid and potent serum bnAb responses. A mAb isolated from one of the cows showed potent and broad neutralization comparable to that of many bnAbs isolated from humans infected with HIV. This study showed that, although repeated immunization with trimer does not induce bnAbs in most animals, the task is straightforward, given an appropriate naive Ab repertoire, as in cows53. Unsurprisingly, Abs with ultra-long CDRH3 regions are not found in humans, but antibodies with CDRH3 regions 20–30 residues in length, which are a common feature of naturally occurring bnAbs, are typically present at measurable if low frequencies in the naive repertoire54.
Abs to the interface region
The interface region between gp120 and gp41 is the target of many bnAbs, the prototypes of which have been PGT15155, 8ANC19556 and 35O2257. PGT151 binds selectively to fully cleaved trimer and is often used to report on native Env configuration. It also stabilizes the Env trimer and had been used in that context, particularly in cryo-electron-microscopy studies58. Recently some interest has focused on bnAbs directed to the interface region and reactive with FP. The FP-targeting bnAb VRC3459 neutralizes ~49% of viruses with an IC50 of ~0.3 μg/ml, and bnAb ACS20260 neutralizes ~44% of viruses with an IC50 of ~0.1 μg/ml. Because of the proximity of heterogeneous complex glycans on gp41, ACS202 incompletely neutralizes a subset of viruses. Both Abs bind to amino-terminal residues of FP and the gp41 glycan at Asn88. The discovery of these FP-targeting bnAbs has stimulated attempts to elicit such Abs via immunization. mAbs that neutralize up to ~30% of global isolates can be isolated via the immunization of mice, guinea pigs and nonhuman primates (NHPs)61. Escape from vaccine-elicited FP-targeting Abs in mice seems to be mediated largely by mutations in the FP, in contrast to escape from VRC34 in humans, which occurs via mutations in the FP and distal interacting sites of the Env trimer62. The data suggest that vaccines that incorporate some diversity in the FP might assist in countering variation in viruses to which vaccinees are exposed.
bnAbs to the mPeR
bnAbs to the MPER are typically very broad but have moderate potency. They also often display some degree of autoreactivity or polyreactivity. The latter is probably associated with the hydrophobic character of CDRH3 regions used by many of these Abs to interact with membranes and allow close approach of the Abs to the membrane, as required for recognition of their protein epitopes in the MPER63. The most potent of the Abs to the MPER, 10E8, does not seem to have substantial autoreactivity or polyreactivity. An Ab-surface mutational strategy combined with neutralization studies was used to generate a version of 10E8 with potency about tenfold higher than that of the parental Ab, with retained nearly ‘pan’ breadth64. Variants with added hydrophobic residues or positively charged residues positioned to interact with viral membrane lipids or viral sialic acid moieties, respectively, were investigated in particular. For DH511, a novel MPER-targeting lineage, the most effective Ab showed nearly ‘pan’ breadth at an IC50 of ~1 μg/ml (ref. 65). The Abs were isolated from both memory B cells and plasma. Interestingly, the unmutated common ancestor of DH511 and several other lineage members were polyreactive, suggestive of possible hurdles in vaccine design strategies templated on this Ab lineage.
An important aspect of the interaction of bnAbs with HIV Env is that essentially all the Abs incorporate features to interact directly with glycans and/or to accommodate glycans, so there is considerable interest in fully elucidating the glycans on Env. Emerging data indicate that there are some differences between the glycosylation at specific sites on recombinant stabilized trimers and that on trimers on the virus surface26,66,67. Of note, glycan-array data indicate that several bnAbs bind with greatest avidity to certain hybrid N-glycans, which suggests that such less-common glycans might serve as components of immunogens designed to elicit bnAbs68.
bnAbs in prophylaxis, therapy and vaccine design
The value of new bnAbs, ranging from those with outstanding activity but unusual features to those with more-modest activity but more-normal profiles, in prophylaxis and in vaccine design depends on the Ab levels required to provide protection against exposure to HIV. There have been many excellent studies and reviews on protection against viral challenge by passive transfer of bnAbs to NHPs and mouse models7,69–72. Accordingly, here we will focus instead on recent updates in this area and also offer a discussion about defining the titer of Ab that would most likely offer protection against HIV.
Until recently, only a handful of chimeric simian–human immunodeficiency virus (SHIV) isolates were available for use in passive-transfer experiments, as only select HIV Env sequences establish robust infection with AIDS-like progression in NHPs. A major milestone in this area was the finding that substitution of bulky hydrophobic or basic amino acids at residue 375 of HIV Env enhances its affinity for rhesus CD4, which leads to improved entry of the virus into target rhesus CD4+ T cells and corresponding improvements in replicative capacity73. This approach is nearly universal for the generation of SHIVs from HIV Env sequences of different clades, including A, B, C and D. Through the use of an alternative approach to generate SHIVs, two new clade C isolates, SHIV-325c and SHIV327c74,75, were generated by cloning of an Env-encoding sequence from an acutely HIV clade C–infected donor in South Africa into the SHIV KB9-AC backbone and subsequent passage in vitro in primary human peripheral blood mononuclear cells. Ultimately, the generation of diverse SHIV isolates brings researchers closer to recapitulating the diversity of global HIV isolates in controlled in vivo models for comprehensive evaluation of the advantages and shortfalls of different bnAbs in providing protection against HIV.
Historically, passive-transfer experiments in NHPs focused on evaluating the protection provided by mAbs against single SHIV molecular clones. Such studies have now expanded to explore protection against more diverse challenge viruses, such as protection against a mixture of two different SHIV isolates76,77. One such study focused on the development of a new Ab construct for prophylaxis and therapy by combining three Ab specificities into a single trispecific Ab construct76. A tri-specific Ab containing fragments of Abs VRC01, PGDM1400 and 10E8v4 was compared with single administration of either VRC01 or PGDM1400 in passive-transfer experiments in rhesus macaques76. Macaques were then challenged with a mixture of SHIV-BaLP4, which is sensitive to VRC01 but is resistant to PGDM1400, and SHIV-325c, which is resistant to VRC01 but is sensitive to PGDM1400. Following challenge, 75% of macaques in the VRC01-treated group and 62% of macaques in the PGDM1400-treated group became infected. In contrast, the group treated with the tri-specific Ab was completely protected against infection. Similarly, the efficacy of a combination of PGT121 and PGDM1400, as well as that of each mAb alone, against a mixture of isolates of SHIV-SF162P3 (clade B; resistant to PGDM1400 but sensitive to PGT121) and SHIV-325c (clade C; sensitive to PGDM1400 but resistant to PGT121) was tested77. 100% of macaques that received a single mAb were infected, but 100% of macaques that received both mAbs together were protected77.
The vast majority of Ab passive-transfer experiments in NHPs utilize cell-free virus (i.e., virions in suspension). Cell-associated viruses are derived from the transfer of infected cells delivered through semen or other infectious bodily fluids, which leads to cell–cell transmission of HIV. Some studies have reported reduced efficacy of bnAbs for cell–cell transmission of HIV78–83, while others have shown effective activity84,85. One study compared the ability of bnAb PGT121 to provide protection against cell-free virus versus its ability to provide protection against cell-associated virus86. PGT121 was delivered intravenously into pig-tailed macaques (at a dose of 1 mg per kg body weight (mg/kg)) that were subsequently challenged with a single high dose of SHIV-SF162P3-infected splenocytes. In this experiment, PGT121 demonstrated partial protection efficacy, with three of the six macaques (50%) showing complete protection, two macaques showing a 1-week delay in viremia and one macaque showing a 7-week delay in viremia. In comparison, PGT121 protected 100% of macaques against cell-free virus. Another study evaluating the efficacy of protection following different challenge routes in rhesus macaques found that PGT126 was able to provide protection similarly regardless of whether the challenge was delivered by a vaginal or rectal route87.
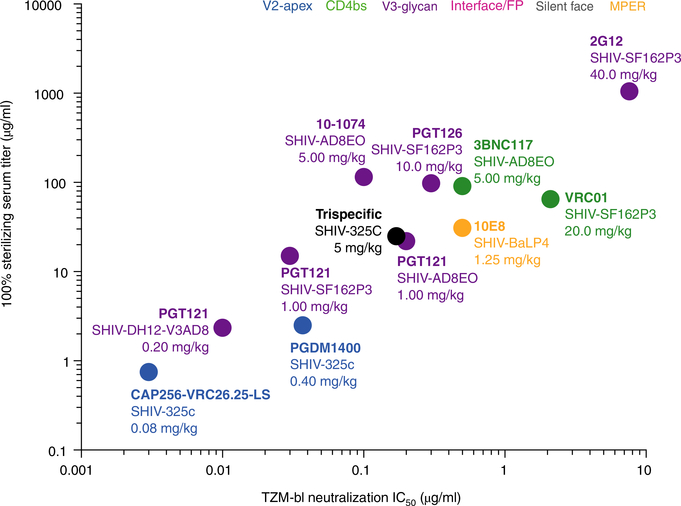
While there has been clear progress in the NHP model of passive transfer, perhaps the most important aspect of work in this area is identifying the minimal serum titers of Abs needed to provide protection against exposure to HIV. A comparison of previous bnAb passive-transfer experiments has reinforced the concept that potent in vitro neutralizing activity against challenge virus on TZM-bl cells translates to potent in vivo protection in rhesus macaques70,74,76,77,88–91 (Fig. 3). The TZM-bl cell line is an engineered HeLa human cervical cancer cell line that expresses CD4 and the chemokine receptors CXCR4 and CCR5 and also encodes firefly luciferase under control of a Tat promoter that is activated after entry of HIV. Typically, the serum concentration of a bnAb that provides protection in vivo far exceeds the in vitro IC50 neutralizing titer. Comparison of in vitro neutralization with in vivo protection across different studies has indicated that an in vivo concentration roughly 200-fold above the neutralization IC50 measured in vitro on TZM-bl cells is generally needed to ensure 100% protection against viral challenge in passive-transfer NHP studies (Fig. 3). Notably, however, this value can range quite appreciably from 1:30 (in the case of protection provided by VRC01 against SHIV-SF162P3) to 1:1,150 (in the case of protection provided by 10–1074 against SHIV-AD8EO), which could be due to differences in biodistribution of the Ab, different characteristics of the challenge virus, limited numbers of animals in the challenge experiment or other unknown factors.
Fig. 3 |. Serum concentration of bnAbs needed to provide complete protection against viral challenge.
Serum concentrations of bnAbs (measured immediately before the time of mucosal challenge) plotted against neutralization (measured using challenge virus in a TZM-bl assay) for various challenge SHIVs and Ab doses (in plot; key), showing that the serum concentration of bnAbs required for complete protection against viral challenge in 100% of animals (vertical axis) in the SHIV–macaque model is directly related to the in vitro neutralizing titer (horizontal axis).
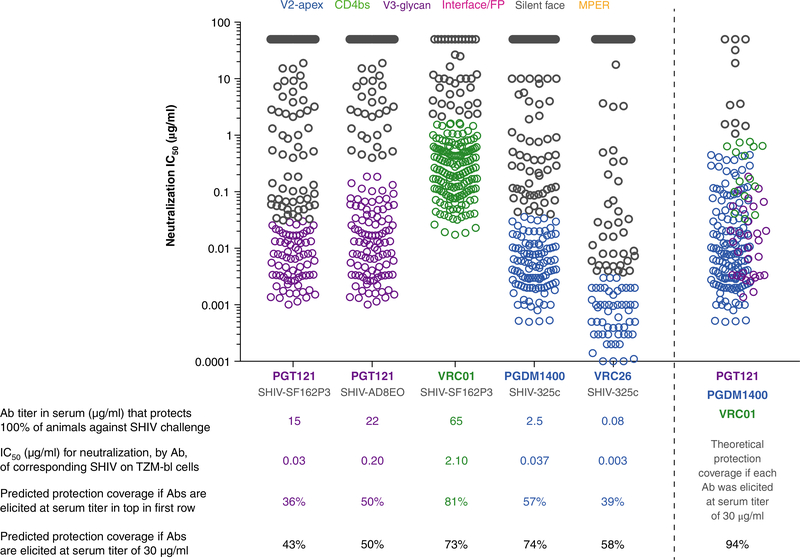
The bnAb, SHIV and macaque-protection data and the breadth and potency of bnAbs directed against global isolates can be extrapolated for estimation of the proportion of HIV strains a human would be protected against at a given serum concentration of a bnAb (Fig. 4). For example, a serum concentration of PGDM1400 of 2.5 μg/ml was reported to protect 100% of macaques against the virus SHIV-325c74. The neutralization IC50 of PGDM1400 against SHIV-325c in TZM-bl cells is 0.037 μg/ml. About 57% of a large panel of global HIVs are neutralized by PGDM1400 at an IC50 value of 0.037 μg/ml or less; i.e., about 57% of global isolates are as sensitive as or more sensitive than SHIV-325c is to PGDM1400. With the assumption that the SHIV–macaque model is predictive of protection in humans, then the extrapolated data suggest that PGDM1400 at a serum concentration of 2.5 μg/ml would provide protection against 57% of global isolates. These extrapolations are of course gross estimates; for example, the macaque-protection data are based on challenge with a high dose of SHIV, probably much higher than would ever be encountered during human exposure, and evidence suggests that lower Ab titers are effective against lower challenge doses in the SHIV–macaque model92. The extrapolations are also generally based on the relationship between in vitro neutralization sensitivity and protection against a single SHIV isolate. Different SHIVs might result in somewhat different estimates. For example, PGT121 provides protection against SHIV-SF162P3 and SHIV-AD8EO at a serum concentration of 15 μg/ml and 22 μg/ml, respectively. On TZM-bl cells, PGT121 neutralizes those challenge viruses at an IC50 value of 0.03 μg/ml or 0.20 μg/ml, respectively. Accordingly, on the basis of the SHIV-SF162P3 challenge data, we estimate PGT121’s protection coverage to be 36%. The protection coverage against global isolates, on the basis of the SHIV-AD8EO challenge data, however, is estimated to be 50%. We next sought to estimate the protection coverage of a vaccine that is capable of eliciting PGT121, PGDM1400 and VRC01. The concentration of the immunoglobulin IgG in human serum is approximately 10 mg/ml, and a serum concentration of 30 μg/ml, which is an approximate titer of 1:300, corresponds to a reasonable sustained vaccine-induced titer achieved via standard immunization protocols93. At this concentration, the protection coverage by most bnAbs increases, and if PGT121, PGDM1400 and VRC01 could be elicited at a serum concentration of 30 μg/ml for each Ab, then we project approximately 94% protection coverage (Fig. 4).
Fig. 4 |. Predicted protection coverage in humans by bnAbs, determined from neutralization coverage and titers and available SHIV protection data.
Protection coverage (% value; bottom two rows below plot) against global isolates was estimated on the basis of the ability of the bnAb (horizontal axis) to protect 100% of rhesus macaques at a given serum concentration (first row below plot) and the neutralization potency of the bnAb against the challenge virus. For PGT121, a serum concentration of 15 μg/ml or 22 μg/ml was required for sterilizing immunity against virus challenge with SHIV-SF162P3 or SHIV-AD8EO, respectively. The neutralization IC50 of PGT121 against these challenge SHIVs on TZM-bl cells was measured to be 0.03 μg/ml for SF162P3 and 0.20 μg/ml for AD8EO. On the basis of neutralization breadth and potency on cross-clade virus panels, PGT121 can neutralize 36% of viruses at an IC50 of 0.03 μg/ml or less and can neutralize 50% of viruses at an IC50 of 0.20 μg/ml or less. Accordingly, we estimate PGT121 would provide 36% protection coverage if it were elicited by vaccination at a serum concentration of 15 μg/ml (on the basis of data for SHIV-SF162P3) and to provide 50% protection coverage if elicited at a concentration of 22 μg/ml (data for SHIV-AD8EO data). We also provide estimated protection coverage if each mAb or combinations of mAbs were elicited at a serum concentration of 30 μg/ml, as a reasonable sustained concentration of Abs elicited by vaccination93. In the plot, each symbol represents a unique SHIV isolate (194–200 isolates tested per Ab), with colors (key) indicating the epitope targeted by the Ab (along horizontal axis; SHIV below), and each symbol position represents the neutralization IC50 in the TZM-bl assay (vertical axis).
Clearly, a key piece of information missing from the field is the protection efficacy of bnAbs against viral exposure in humans in vivo. To remedy this, the National Institute of Allergy and Infectious Diseases has launched two parallel phase IIb multinational clinical trials, HVTN703/HPTN081 and HVTN704/HPTN085, to evaluate Ab-mediated prevention against HIV, with target enrollment of 27,00 men (or transgender people who have sex with men) and 1,900 women in sub-Saharan Africa, respectively94. The trials will assess the mAb VRC01, which will be administered by intravenous infusion every 8 weeks in two dose groups: one at 10 mg/kg and another at 30 mg/kg. The participants will be tested for HIV infection at every study visit and will be monitored for 12 weeks after the final administration of Ab at week 80. Data from these trials will mark an important milestone in the HIV-prevention field, as they will show, for the first time, the concentration of nAbs required to afford protection against HIV in humans. These data will be highly informative for ‘next-generation’ bnAb-prophylaxis product-development programs to determine appropriate bnAb dosage needed to provide protection, as well as for the broader field of the design of vaccines against HIV, to determine the average nAb titers that will be needed to afford immunity to HIV. Finally, this information can be used to ‘benchmark’ old and new passive-transfer studies in NHPs, which will still be an important animal model in the future for testing different constructs and conditions, as these studies are much more affordable and much less resource-intensive than is a human clinical trial.
In parallel to the development of bnAbs for use as prophylaxis, equivalent progress has been made for the use of bnAbs in therapy and cure. Following the administration of PGT121 to rhesus macaques infected with SHIV-SF162P3, tissues that tested positive for HIV RNA at day 7 were cleared at day 10 (ref. 95), which suggested that the immune system is able to clear infected cells in the presence of bnAbs. Similarly, a mother-to-child transmission model of NHPs showed that macaques given bnAbs 24 h after oral inoculation with SHIV-SF162P3 were protected89. Notably, bnAbs are now being evaluated in humans. In one study, multiple infusions of 3BNC117 (an antibody to the CD4bs), at a dose of 30 mg/kg, into 13 patients with viremia resulted in average time to rebound of 8.4 weeks (ref. 96). These participants were pre-screened for viral sensitivity to 3BNC117, and resistant viruses were found for six of eight participants after their rebound. In another study, administration of VRC01 to eight patients resulted in a decrease of viral load between 1.1 log10 and 1.8 log10 in six of the eight subjects, with a mean nadir of 9 days (ref. 97). After their rebound, all subjects showed evidence of resistance to VRC01. Finally, the Ab 10–1074 (to V3 glycan) was administered, at a dose 30 mg/kg, to 13 patients with viremia. 11 of the 13 subjects who had 10–1074-sensitive viruses showed rapid decrease in viremia, by an average of 1.52 log10. Emergence of 10–1074-resistant viruses occurred in the first weeks after Ab infusion, with evidence of different escape variants98. These results were obtained with single mAbs, which indicates that combinations of bnAbs targeting different epitopes might have even-more-potent effects. Much remains to be explored for the use of bnAbs in therapy and cure, but these data highlight the promise of bnAbs.
Conclusions
The discovery of new bnAbs to HIV is still a very active field. For prophylaxis and therapy, the emphasis is on Abs with even greater neutralization potency and breadth than those described so far. In addition, and not discussed in this Review, there is much ongoing work on optimizing bnAbs in terms of Ab effector function and Ab half-life in serum2,69,99. For vaccine design, there has been increased interest in what we have called ‘realistic Abs’ as templates for design. However, the serum concentrations of nAbs that provide protection in humans remain unknown and therefore it remains unknown if ‘realistic Abs’, which are typically less potent than many of the best bnAbs, would be effective at concentrations attainable by vaccination. We await the results of the Ab-mediated prevention trial for important indications.
Acknowledgements
We thank I. Wilson and W. Schief for comments on the manuscript; and L. Hangartner, C. Corbaci and B. Briney for assistance with figure preparation. Supported by the US National Institute of Allergy and Infectious Diseases (D.R.B.), IAVI (D.R.B.), the Bill and Melinda Gates Foundation (D.R.B.), the Ragon Institute (D.R.B.), the Bill & Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (D.S.) and the United States Agency for International Development (D.S.). A full list of IAVI donors is available at https://www.iavi.org/what-we-do/partner/donors. Contributions from the International AIDS Vaccine Initiative do not necessarily reflect the views of USAID or the United States government.
Footnotes
Competing interests
The authors declare no competing interests.
online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41590-018-0235-7.
References
- 1.Burton DR, Poignard P, Stanfield RL & Wilson IA Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker LM & Burton DR Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol 18, 297–308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escolano A, Dosenovic P & Nussenzweig MC Progress toward active or passive HIV-1 vaccination. J. Exp. Med 214, 3–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD & Mascola JR HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Burton DR & Hangartner L Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol 34, 635–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCoy LE & Burton DR Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev 275, 11–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegu A, Hessell AJ, Mascola JR & Haigwood NL Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev 275, 296–312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris L & Mkhize NN Prospects for passive immunity to prevent HIV infection. PLoS Med. 14, e1002436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subbaraman H, Schanz M & Trkola A Broadly neutralizing antibodies: what is needed to move from a rare event in HIV-1 infection to vaccine efficacy? Retrovirology 15, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jardine JG et al. Minimally mutated HIV-1 broadly neutralizing antibodies to guide reductionist vaccine design. PLoS Pathog. 12, e1005815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46, 690–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sok D et al. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45, 31–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancera M et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molinos-Albert LM, Clotet B, Blanco J & Carrillo J Immunologic insights on the membrane proximal external region: a major human immunodeficiency virus type-1 vaccine target. Front. Immunol 8, 1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T et al. A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48, 500–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyatt R & Sodroski J The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Walker LM et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejchal R et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. USA 107, 11483–11488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancera M et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84, 8098–8110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy LE et al. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLoS Pathog. 11, e1005110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb NE, Montefiori DC & Lee B Dose-response curve slope helps predict therapeutic potency and breadth of HIV broadly neutralizing antibodies. Nat. Commun. 6, 8443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose NA et al. New member of the V1V2-directed CAP256VRC26 lineage that shows increased breadth and exceptional potency. J. Virol 90, 76–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrabi R et al. Glycans function as anchors for antibodies and help drive HIV broadly neutralizing antibody development. Immunity 47, 524–537.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cale EM et al. Virus-like particles identify an HIV V1V2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity 46, 777–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landais E et al. HIV envelope glycoform heterogeneity and localized diversity govern the initiation and maturation of a V2 apex broadly neutralizing antibody lineage. Immunity 47, 990–1003.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rantalainen K et al. Co-evolution of HIV envelope and apex-targeting neutralizing antibody lineage provides benchmarks for vaccine design. Cell Rep. 23, 3249–3261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker LM et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landais E et al. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog. 12, e1005369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgiev IS et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756 (2013). [DOI] [PubMed] [Google Scholar]
- 30.MacLeod DT et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the env high-mannose patch. Immunity 44, 1215–1226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longo NS et al. Multiple antibody lineages in one donor target the glycan-V3 supersite of the HIV-1 envelope glycoprotein and display a preference for quaternary binding. J. Virol. 90, 10574–10586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonich CA et al. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell 166, 77–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ditse Z et al. HIV-1 subtype C-infected children with exceptional neutralization breadth exhibit polyclonal responses targeting known epitopes. J. Virol. 92, e00878–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonsignori M et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med 9, eaai7514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freund NT et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med 9, eaal2144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steichen JM et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Escolano A et al. Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166, 1445–1458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes CO et al. Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun 9, 1251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sajadi MM et al. Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173, 1783–1795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jardine JG et al. HIV-1 vaccines. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briney B et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166, 1459–1470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sok D et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dosenovic P et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian M et al. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166, 1471–1484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havenar-Daughton C et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med 10, eaat0381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burton DR et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Corti D et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5, e8805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao HX et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonsignori M et al. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165, 449–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gristick HB et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol 23, 906–915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sok D et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548, 108–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Briney BS, Willis JR & Crowe JE Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One 7, e36750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falkowska E et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheid JF et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JH, Ozorowski G & Ward AB Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong R et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Gils MJ et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol 2, 16199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu K et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med 24, 857–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dingens AS et al. Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV. PLoS Pathog. 14, e1007159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Irimia A et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog. 13, e1006212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwon YD et al. Surface-matrix screening identifies semi-specific interactions that improve potency of a near pan-reactive HIV-1-neutralizing antibody. Cell Reports 22, 1798–1809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams LD et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol 2, eaal2200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao L et al. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 9, 3693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Struwe WB et al. Site-specific glycosylation of virion-derived HIV-1 Env is mimicked by a soluble trimeric immunogen. Cell Rep. 24, 1958–1966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shivatare VS et al. Unprecedented role of hybrid N-glycans as ligands for HIV-1 broadly neutralizing antibodies. J. Am. Chem. Soc 140, 5202–5210 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Gautam R et al. A single injection of crystallizable fragment domain-modified antibodies elicits durable protection from SHIV infection. Nat. Med 24, 610–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moldt B et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA 109, 18921–18925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Gils MJ & Sanders RW In vivo protection by broadly neutralizing HIV antibodies. Trends Microbiol. 22, 550–551 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Hessell AJ & Haigwood NL Animal models in HIV-1 protection and therapy. Curr. Opin. HIV AIDS 10, 170–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA 113, E3413–E3422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Julg B et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med 9, eaal1321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang HW et al. Generation and evaluation of clade C simian-human immunodeficiency virus challenge stocks. J. Virol. 89, 1965–1974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu L et al. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358, 85–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Julg B et al. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci. Transl. Med 9, eaao4235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abela IA et al. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 8, e1002634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duncan CJ et al. High-multiplicity HIV-1 infection and neutralizing antibody evasion mediated by the macrophage-T cell virological synapse. J. Virol 88, 2025–2034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gombos RB et al. Inhibitory effect of individual or combinations of broadly neutralizing antibodies and antiviral reagents against cell-free and cell-to-cell HIV-1 transmission. J. Virol 89, 7813–7828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schiffner T, Sattentau QJ & Duncan CJ Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine 31, 5789–5797 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Li H, Zony C, Chen P & Chen BK Reduced potency and incomplete neutralization of broadly neutralizing antibodies against cell-to-cell transmission of HIV-1 with transmitted founder Envs. J. Virol 91, e02425–e02416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reh L et al. Capacity of broadly neutralizing antibodies to inhibit HIV-1 cell-cell transmission is strain- and epitope-dependent. PLoS Pathog. 11, e1004966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malbec M et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J. Exp. Med. 210, 2813–2821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCoy LE et al. Neutralisation of HIV-1 cell-cell spread by human and llama antibodies. Retrovirology 11, 83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parsons MS et al. Partial efficacy of a broadly neutralizing antibody against cell-associated SHIV infection. Sci. Transl. Med 9, eaaf1483 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Moldt B et al. Neutralizing antibody affords comparable protection against vaginal and rectal simian/human immunodeficiency virus challenge in macaques. AIDS 30, 1543–1551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pegu A et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci. Transl. Med 6, 243ra88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hessell AJ et al. Early short-term treatment with neutralizing human monoclonal antibodies halts SHIV infection in infant macaques. Nat. Med. 22, 362–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shingai M et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 211, 2061–2074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hessell AJ et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5, e1000433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hessell AJ et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med 15, 951–954 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinn CP et al. Humoral and cell-mediated immune responses to alternate booster schedules of anthrax vaccine adsorbed in humans. Clin. Vaccine Immunol 23, 326–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gilbert PB et al. Basis and statistical design of the passive HIV-1 antibody mediated prevention (AMP) test-of-concept efficacy trials. Stat. Commun. Infect. Dis 9, 20160001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J et al. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science 353, 1045–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scheid JF et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bar KJ et al. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med 375, 2037–2050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Caskey M et al. Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat. Med 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bournazos S & Ravetch JV Anti-retroviral antibody FcγR-mediated effector functions. Immunol. Rev. 275, 285–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoon H et al. CATNAP: a tool to compile, analyze and tally neutralizing antibody panels. Nucleic Acids Res. 43(W1), W213–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarzotti-Kelsoe M et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bonsignori M et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and Their inferred unmutated common ancestors. J. Virol. 85, 9998–10009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Munir Alam S et al. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci. Transl. Med 9, eaai7521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.MacLeod DT et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44, 1215–1226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mouquet H et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA 109, E3268–E3277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao F et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 158, 481–491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buchacher A et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10, 359–369 (1994). [DOI] [PubMed] [Google Scholar]
- 108.Simek MD et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-Throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Walker LM et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu X et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang J et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]