Abstract
Abdominal aortic aneurysm (AAA) is a local dilatation of the abdominal aortic vessel wall and is among the most challenging cardiovascular diseases as without urgent surgical intervention, ruptured AAA has a mortality rate of >80%. Most patients present acutely after aneurysm rupture or dissection from a previous asymptomatic condition and are managed by either surgery or endovascular repair. Patients usually are old and have other concurrent diseases and conditions, such as, diabetes, obesity and hypercholesterolemia making surgical intervention more difficult. Collectively, these issues have driven the search for alternative methods of diagnosing, monitoring and treating AAA using therapeutics and/or less invasive approaches. Non-coding RNAs—small non-coding RNAs (microRNAs) and long-non-coding RNAs (lncRNAs) - are emerging as new fundamental regulators of gene expression. Researchers and clinicians are aiming at targeting these microRNAs and lncRNAs and exploit their potential as clinical biomarkers and new therapeutic targets for abdominal aortic aneurysms. While the role of miRNAs in AAA is established, studies on lncRNAs are only beginning to emerge, suggesting their important yet unexplored role in vascular physiology and disease. Here, we review the role of noncoding RNAs and their target genes focusing on their role in AAA. We also discuss the animal models used for mechanistic understanding of AAA. Furthermore, we discuss the potential role of microRNAs and lncRNAs as clinical biomarkers and therapeutics.
Keywords: microRNAs, long-noncoding RNAs, abdominal aortic aneurysms (AAAs), aortic dissection, circulating biomarkers
1. Introduction: Pathophysiology of AAA
Abdominal aortic aneurysm (AAA) is defined as a permanent dilation or bulging of the aorta. AAA is relatively common and its rupture leads to a potentially life–threatening condition.1 Ruptured AAA is the 13th-leading cause of death in the United States and is estimated to cause ~ 15,000 deaths per year. The frequency of rupture is 4.4 per 100,000 patients. Despite increased survival following diagnosis, incidence and crude mortality seem to be increasing.2 Globally, the frequency rate of asymptomatic AAA ranges from 4–8% while the frequency of AAA rupture ranges from 4 to 13 cases per 100,000 persons.3 Aging and being male are two strong risk factors of AAA.1 It is important to note that the frequency rate of AAA in males is much higher compared to the females early on which increases with aging. In addition, white men have the highest incidence of AAA (~3.5 times that in African American men).1 Other risk factors include smoking, hypertension, and atherosclerosis in coronary artery or peripheral arteries.1. Most AAAs occur in individuals with advanced atherosclerosis. Atherosclerosis may induce AAA formation by causing mechanical weakening of the aortic wall with loss of elastic recoil, along with degenerative ischemic changes occurring in the adventitial layer. However, it is unclear whether atherosclerosis is causing AAA or conspire with other pro-AAA factors. It is interesting, however, to note that the patients at greatest risk for AAA are men who are older than 65 years and have peripheral atherosclerotic disease.1
The diameter of the aorta with AAA can increases by >50% increase over its normal size.4 Most AAAs begin below the renal arteries and terminate above the iliac arteries. The size, shape, and extent of AAAs vary considerably. Like aneurysms of the thoracic aorta, AAAs may be broadly described as either fusiform (circumferential) or saccular (more localized). AAA develops as the extracellular matrix proteins are degraded and remodeled in the aortic wall, resulting in the loss of the structural integrity and dilation of the wall.5–9 The pathogenic mechanisms underlying AAA include proteolytic degradation of aortic-wall connective tissue6, 10–13, inflammation and immune responses14–18, biomechanical wall stress19–24, molecular genetics25–27, and genetic predisposition5–8, 28–31, as have been reviewed previously. Other less common causes of AAA include cystic medial necrosis, arteritis, trauma, inherited connective-tissue disorders, and anastomotic disruption.32
Most AAAs are asymptomatic, progressive, and often are detected as an incidental finding on diagnostic imaging obtained for other reasons. At present, the only treatment option for AAA is surgical repair via laparotomy or less invasive endovascular stents.32 Unfortunately, there is no therapeutic treatment available to prevent, delay or reverse the pathology of AAA other than reducing the risk factors such as stop smoking, controlling blood pressure and lipid levels. This is in part due to our lack of understanding of the pathophysiological mechanism(s) and key regulators thereof. Efforts have been underway to identify the genomic and epigenetic factors that regulate AAA pathogenesis as they may lead to better mechanistic understanding and therapeutic targets to prevent and treat the disease.
Through transcriptome studies using samples obtained from patients with AAA and animal models of AAA, various genes involved in extracellular matrix degradation, inflammation, and other critical pathophysiology observed in AAA formation have been identified.33–47 The combination of proteolytic degradation of aortic-wall connective tissue, inflammation and immune responses, biomechanical wall stress, and molecular genetics represents a dynamic process that leads to aneurysmal deterioration of aortic tissue. Matrix metalloproteinases (MMPs) and their inhibitors are present in normal aortic tissue and are responsible for vessel-wall remodeling. Aneurysmal tissue are associated with increased MMP activity and decreased inhibitor activity, which favor the degradation of the structural extracellular matrix (ECM) proteins, elastin and collagen. The mechanism that regulates degradation of elastin and collagen and remodeling of ECM in the aortic wall of AAAs is controlled by the balance between proteases including MMPs and protease inhibitors including tissue inhibitor of MMPs (TIMPs).8, 48 The detailed underlying mechanisms that regulate this key process still needs much more studies and involves non-coding RNAs. For these mechanistic studies human tissue samples and animal models, which will be further summarized below, have played crucial roles.
2. Animal models for AAA
Three different models of mouse AAA, elastase infusion, calcium chloride sponge, and Angiontesin II (AngII) infusion are widely used. While each model shows some features of human AAA, no single animal model has been shown to develop all clinical features of AAA. Therefore, many studies use all three mouse models for comprehensive understanding of human AAA.
Elastase Infusion
The infusion of elastase into the infrarenal segment of rat or mouse aortas has been a frequently used model of AAAs.26, 41, 49 Elastase infusion degrades elastin leading to the loss of structural integrity of the abdominal aorta. The procedure involves isolation of a segment of the abdominal aorta and incubation of porcine pancreatic elastase (PPE) for a few minutes before restoration of flow. This causes extensive destruction of the elastic lamellae and the infiltration of macrophages in the adventitial region within 2 weeks. Using this model, several microRNAs have been identified: miR-21, miR-24, and miR-29.50, 51
Calcium Chloride model
Peri-aortic application of calcium chloride leads to structural disruption of the medial layer and inflammatory responses.48, 49. In this model, a gauze soaked in a calcium chloride solution is placed over the aorta. In this model, inflammation occurs on the luminal and medial layers of the aortas, leading to an increase in the aortic diameter >1.5 fold. This model induces luminal dilation without the mechanical effects that develops in the elastase-infused model.48 MicroRNAs identified/studied using this model are miR-33 and miR-19a, miR-19b, miR-132, and miR-221.52, 53
Angiotensin-II (AngII) infusion model
Infusion of AngII into low-density lipoprotein (LDL) receptor−/− (LDLR−/−), apolipoprotein E (ApoE)−/− mice, or C57BL6 mice injected with adeno-associated virus (AAV) encoding for gain-of-function mutant of proprotein convertase subtilisin/kexin type 9 (PCSK9) (AAV-PCSK9) to overexpress PCSK9, which in turn knocks down LDLR in the liver, is another widely used approach to induce AAA or aortic dissection.6, 49, 54, 55 Delivery of a high-dose AngII at doses of 500 to 1000 ng/kg per minute, via subcutaneously implanted osmotic mini-pumps, leads to AAAs in the suprarenal region within the 28-day infusion period. Using this model, following miRNAs have been studied: miR-33, miR-145, miR-103a, miR-712, miR-205, miR-21, miR-24 and miR-29.50, 51, 56–58
Studies from these animal models have provided key insights into the mechanisms of the initiation and progression of AAA.
3. Non-coding RNAs
It has been suggested that more than 97% of genome encodes for noncoding transcripts. It is likely that the differences in organism complexity may arise from the vast differences in noncoding transcripts between higher and lower organisms. Many of these noncoding transcripts are processed to generate small noncoding RNA such as miRNA or lncRNA. Although, microRNAs have gained widespread attention for their role in diverse physiological and pathophysiological processes, the knowledge about the functioning of lnc-RNAs still remains limited. Through their interaction with DNA, RNA and proteins, noncoding RNA have emerged as key regulators of gene expression under both physiological and pathological conditions. The table below summarizes the similarities and differences between miRNAs and lncRNAs.
3.1. MicroRNAs and AAA
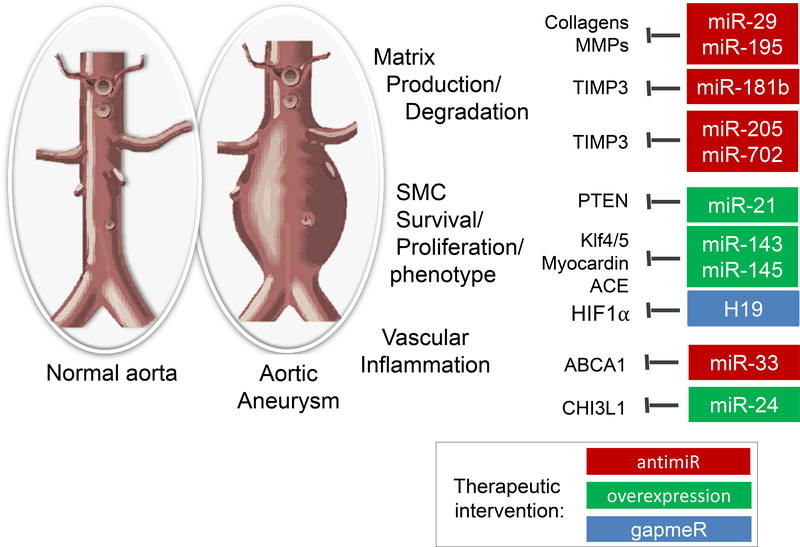
MicroRNAs (miRs) are the best-characterized class of non-coding RNAs. These small single stranded RNAs are 18–21 nucleotides in length and inhibit gene expression by incorporation into a protein complex called RNA-induced silencing complex (RISC) and binding to mRNAs.61 One miR can bind to multiple mRNAs, frequently hundreds, so that miRs can broadly regulate gene expression patterns. Several miRs have been described over the last decade to regulate vascular remodeling.62, 63 Of interest, miRs can also be detected in the circulation and can serve as biomarkers for disease, including aneurysmal disease.64 Multiple cell types including endothelial cells, smooth muscle cells, and immune cells in the vasculature play important role in development and progression of aneurysms. Many miRNAs have been shown to target these cell types, especially vascular smooth muscle cells, in the vasculature to alter the pathophysiology of AAA. Here we review these miRs associated with AAA for potential therapeutic translation based on pre-clinical evidence in animal models (Figure 1).
Figure 1:
MicroRNAs regulating aortic aneurysm formation or progression. Selected targets that were shown to contribute to the effects of the miRs in aneurysm formation are shown. Red labelled miRs promote aneurysm formation and may be targets by antimiRs for therapeutic interventions, whereas green labelled miRs inhibit aneurysm formation.
miR-29:
One of the first described miRs that regulates aneurysm formation is miR-29.51, 65, 66 MiR-29 inhibits expression of several extracellular matrix proteins, as well as anti-apoptotic factors, thereby having an aneurysm-promoting effect on smooth muscle cells. Inhibition of miR-29, using systemic application of anti-miRs in mouse models, blocks the formation and progression of aneurysms50, 65–68, even on a long-term basis.67 miR-29 in synergy with other members of miR-15 family have been implicated in regulating the vascular extracellular matrix.68 Since systemic blockade of miR-29 could potentially induce fibrosis in other organs 69, suggesting a potential off-target effects of the anti-miRs as a long-term therapeutics. Therefore, a targeted, local delivery of anti-miRs seems to be worth considering for therapeutic development.
miR-21:
Another very promising and one of the earliest described miRs to be involved in vascular remodeling is miR-21.50 Overexpression of miR-21 leads to a decrease in PTEN and a subsequent survival of smooth muscle cells, which induces vessel wall stability in a mouse model of elastin degradation-induced aneurysms. Therapeutic overexpression or local delivery of functional miRs is challenging, since modifications that increase the lifetime or uptake by target cells limit functional incorporation of the miR in RISC.70 Alternatively, viral vectors or nanoparticles might be used to deliver precursor microRNAs or mimics. Although some progress has been made with respect to endothelial or vascular targeting nanoparticles 71–75, so far no highly efficient and specific targeted delivery for AAA has been reported. This is the limiting factor for the development of a potential miR-21-based aneurysm therapy. Interestingly, Wang et al, showed that anti-miR-21 delivered as a drug-eluting stent successfully prevented in-stent myointimal hyperplasia in a humanized rat model.76 While not tested in AAA, this result suggests the potential of stent-mediated local delivery of anti-miRs as a novel AAA therapy.
miR-24:
miR-24 overexpression also shows promise as therapeutic strategy for aneurysmal disease.77 The main effects that miR-24 seems to have is on limitation of macrophage survival, cytokine production and macrophage recruitment, thereby limiting vessel wall inflammation and extracellular matrix breakdown. Mechanistically, miR-24 limits the expression of CHI3L1, a secreted glycoprotein, which inhibits the production of the cytokines IL8 and CCL2 by smooth muscle cells and macrophages. For therapeutic development, the same limitations as described above for miR-21 delivery, also apply for miR-24.
miR-33:
Another promising miR that has effects in both smooth muscle cells and macrophages is miR-33.52 This miR, known to inhibit the cholesterol transporter ABCA1, is a natural pro-inflammatory regulator in smooth muscle cells and macrophages.78, which was recently described to inhibit smooth muscle cell proliferation and subsequent vein graft disease.79 However, attenuation of miR-33 leads to a decrease of p38 and JNK signaling, as a result of increased ABCA1 expression, and inhibition of CaCl2 and Angiotensin II-induced aneurysm formation in mice. Interestingly, miR-33 deletion also increases HDL levels as well as anti-inflammatory properties of HDL particles, attributed to effects in the liver. However, unexpectedly, genetic deletion of miR-33 or a systemic long-term treatment with anti-miR-33 changed metabolism leading to loss of anti-atherogenic effects, but macrophage-specific genetic deletion of miR-33 showed the desired anti-atherogenic effects.52, 80 These results demonstrate the importance of considering a tissue- or cell-type targeted delivery of anti-miR-33 as potential therapeutic approach.
miR-143/145:
One of the first-described smooth muscle-enriched miR clusters81, miR-143/145, was also inversely associated with aneurysms in humans.82 Indeed, miR-143 and miR-145 induce a contractile, quiescent, and mature phenotype in smooth muscle cells and its local delivery may induce stabilization of aneurysms. Interestingly, both miR-143 and miR-145 have been described to be transported from one cell type to another. More specifically, miR-143/145 from endothelial cells can be taken up by smooth muscle cells83, but also vice versa.84 This mechanism may be exploited therapeutically, where one can envision that patient-derived vesicles or exosomes containing these miRs may be locally deposited to increase the stability of the vessel wall.
miR-181b:
MiR-181b is increased in human aneurysms and targets TIMP3, an inhibitor of extracellular matrix breakdown, as well as elastin, and inhibition of miR-181b prevents aneurysm formation and even attenuates progression of existing aneurysmal disease.85 The same study also reports a decrease in atherosclerosis after miR-181b inhibition. However, aging, one of the risk factors for aneurysms, reduces the expression of miR-181b and genetic deletion of miR-181b induces vascular stiffness and increases blood pressure.86 Furthermore, miR-181b was also described to decrease inflammation and atherosclerosis (contradicting the study described above 85) by attenuating NFκB nuclear translocation87, so that any strategy based on miR-181b inhibition to prevent aneurysm growth should carefully consider the risk of increasing atherosclerosis or vascular stiffness.
miR-195:
Circulating levels of miR-195 in blood plasma inversely correlates with aortic diameter.68 An independent study also suggested that plasma miR-195 measurements may be of prognostic value, as it described plasma miR-195 as differentially expressed in fast versus slow growth aneurysms value.88 Another study found that miR-195 was also differentially expressed in human aneurysmal tissue.64 Mechanistically, miR-195 targets extracellular matrix (ECM) proteins, but also matrix metalloproteinases (MMPs), so that therapeutic targeting of miR-195 is not likely to be straightforward.68, 89–92 Here, a more specific approach such as use of target-site blockers would be useful to inhibit the miRNA and a selected mRNA interaction.93 For example, Target-site blockers could be designed to block the specific interaction between miR-195 and elastin without affecting the interaction of miR-195 with other mRNAs targets such as MMP2/9.
miR-205 and miR-712:
Finally, miR-205 and its mouse-specific homolog miR-712, can be targeted to inhibit aneurysm formation in mice.57 The mechanism by which miR-205 and miR-712 functions is similar to that described for miR-181 above, namely via the inhibition of TIMP3, so that miR-205 or miR-712 blockade leads to an increase in TIMP3 expression 94, a reduction in ECM degradation and subsequent stabilization of the vessel wall. Inhibition of these miRs also diminishes atherosclerosis, so that, in contrast to miR-181 inhibition strategies, therapeutic targeting of miR-205 in patients may be beneficial for both atherosclerosis and aneurysmal disease.57, 94
3.2. lncRNAs and AAA
Long non-coding RNAs (lncRNAs) are RNAs that are more than 200 nucleotides in length that do not code for proteins. Based on their relative location in reference to the protein-coding genes, lncRNAs have been classified into five categories:96 (Also see Table 1)
antisense RNA (overlaps a protein coding gene on the opposite strand),
lincRNA (long intergenic non-coding RNA),
sense overlapping RNA (contains a coding gene within an intronic sequence)
sense intronic RNA (located within intron of a coding/non-coding gene)
3’ overlapping ncRNA (located within the 3 UTR of larger gene).
Table 1.
Similarities and differences between miRNAs and lncRNAs
| Characteristics | miRNA | lncRNAs |
|---|---|---|
| Size | Short (18 to 21 nucleotides) | >200 nucleotides |
| Primary mode of action | Negatively regulating the expression by complementary binding to target mRNAs and causing their degradation or inhibiting translation | Highly diverse, regulate gene expression by
various mechanisms. Lnc-RNAs may work as 1. Signaling cues 2. Guides 3. Decoys 4. Scaffolds 5. miRNA sponges |
| Genomic location |
-Exonic -Intergenic (between genes) -Intronic (embedded in a gene) |
-Antisense
RNA -LincRNA -Sense overlapping RNA -Sense intronic RNA - 3’ overlapping ncRNA |
| Secondary structure | Usually the precursor miRNA forms a hairpin structure | Forms simple to complex secondary structures 59, 60 |
| Post-transcriptional processing | Typically produced as pri-miRNAs, which are processed by miRNA-processing enzymes (Drosha, DGCR8) and Dicer to generate pre-miR and mature miRNAs | Undergo post-transcriptional processing like mRNAs, i.e. 5’ capping, polyadenylation, and splicing |
| Computational gene target prediction | Relatively easy. Multiple bioinformatics tools/ prediction algorithms available that help to predict target genes | Difficult to predict. Not available. |
| Conservation between species | Relatively well conserved nucleotide sequences across species, although some species-specific miRNAs exist | Poorly conserved between species at the level of primary nucleotide sequences, but may have conserved secondary structures |
| Experimental gain-of-function strategies | miRNA mimics, over-expression plasmids, transgenic overexpression | over-expression plasmids, transgenic overexpression |
| Experimental loss-of-function strategies | Anti-miRs, antagomiRs, and morpholinos, genetic deletions | GapmeRs, genetic deletions or mutations |
Unlike the relatively straightforward mechanisms by which miRNAs regulate their target mRNAs through complimentary binding, much less is known about the role and the mechanism(s) by which lncRNAs regulate gene expression (Table 1). This is in part due to their diverse biogenesis 97, low abundance and poor conservation amongst species.98, 99 LncRNAs have been shown to be three-dimensional regulators of transcription and translation, for example by acting as molecular decoys and scaffolds, or through guiding ribonucleoprotein complexes to their targets via protein binding.100, 101 Furthermore, they can function as host genes for the transcription of miRNAs (in the nucleus) or miRNA sponges (in the cytoplasm).102 In addition, a mere act of lncRNA transcription process itself was known to regulate gene expression, 103 further adding complexity to the action mechanisms of lncRNAs. In contrast to miRNAs, lncRNA expression appears to be highly tissue-specific (approximately 80%)104, which may become important in potential therapeutic and biomarker approaches as the risk for off-target effects and non-specific detection decreases.
To date, few studies have assessed the role of lncRNAs in the context of vascular disease development and progression. Examples include Meg3, which is a crucial regulator of endothelial aging and angiogenesis 105, as well as lincRNA-p21, which mediates SMC survival and macrophage activity in the atherosclerotic process.3, 106 At present, however, the only lncRNA specifically linked to AAA is H19.107 Studies using other lncRNAs have only been observational and speculative for their potential role(s) in aneurysm development and progression. Some studies have revealed a crucial involvement for lncRNAs in mechanisms known to be of key importance to AAA development and expansion (e.g., SMC proliferation and apoptosis, as well as aortic inflammation). However, only few reports cited here were able to provide in vivo evidence for a suspected molecular mechanism of action being mediated through lncRNAs. Future studies will have to answer the question whether other lncRNAs apart from H19 are as effective in mediating the fate of aortic aneurysms as miRNAs. Here, we briefly summarize recent lncRNA studies in the context of aortic aneurysms.
H19:
Recently, the first lncRNA with a functional implication in AAA development and progression was reported. Li et al. identified the lncRNA H19 using RNA sequencing of AngII and PPE-induced AAAs in mice.107 H19 was substantially increased in progressing AAAs, and inhibition of H19 prevented aneurysm growth in both the AngII and PPE models of AAA in mice as well as in the LDLR-depleted Yucatan mini-pigs and human patient samples with AAAs. Increased H19 expression levels negatively correlated with SMC proliferation and migration, while positively correlating with apoptosis in vivo. Mechanistically, hypoxia-inducible factor 1-alpha (HIF1α) was identified as the major downstream target of H19. Increased SMC apoptosis was mediated though cytoplasmic and nucleic interactions between H19 and HIF1α. In the cytoplasm, H19-HIF1α interaction increased p53 levels and activity. In the nucleus, H19 triggered transcription of HIF1α via recruiting the SP1 transcription factor to the promoter region. These results suggest that H19 is a novel therapeutic target of AAA and that H19 LNA-GapmeR is a potential AAA therapy. This study further suggests the potential role of additional lncRNAs in AAA development and their application as diagnostic markers and therapeutics.
SENCR:
Bell et al. used RNAseq of human coronary artery SMCs to discover the vascular cell-enriched lncRNA SENCR (Smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA).108 SENCR is transcribed in antisense to the FLI1 gene, and active mainly in the cytoplasm. Inhibition of SENCR indicated only marginal cis-acting activity of SENCR on FLI1 as well as other nearby genes. However, RNA profiling in SMCs upon SENCR depletion resulted in decreased expression of Myocardin and various other SMC contractile genes and increases in pro-migratory marker genes. Functional assessment of SENCR inhibition utilizing scratch wound and Boyden chamber assays further showed SENCR as a powerful novel mediator of SMC proliferation and migration. However, its role in AAA is yet to be demonstrated.
SMILR:
Another lncRNA with potential implications for AAA disease is the vascular remodeling associated transcript SMILR (Smooth Muscle Cell Enriched Long Non-Coding RNA).109 SMILR was identified by RNAseq in SMCs upon interleukin-1α (IL1α) and platelet derived growth factor (PDGF) stimulation. Knockdown of SMILR significantly reduced cell proliferation rates. Importantly, its expression is increased in unstable atherosclerotic plaques and plasma from patients with elevated C-reactive protein levels.
Lnc-Ang362:
The LncRNA362 was identified in rat aortic SMCs upon AngII stimulation, an important factor in human and experimental AAA development, as well as in atherosclerosis and hypertension.110 LncRNA362 was identified as a host transcript for miRs-221/−222, which are both well-established drivers of SMC proliferation in vascular diseases.111 Its role in AAA is unknown yet.
HOTAIR:
HOTAIR, a well-studied lncRNA in the cancer field, was shown to be decreased in sporadic thoracic abdominal aorta (TAA) specimens, and to negatively correlate with the size of aortic diameter.112 Depletion of HOTAIR was not only capable of reducing proliferation and triggering apoptosis rates, but also to limit the expression of the collagen isoforms type I and III. However, its role in AAA in animal studies is yet to be demonstrated.
Pi15:
Falak et al 113 found the protease inhibitor 15 (Pi15), as well as another non-validated lncRNA, to be implicated in disruptions of elastic laminae in rat aneurysms. Whether these lncRNAs play a role in human AAA development and mouse models of AAA needs to be studied.
lnc-HLTF-5:
Li et al. carried out a microarray-based RNA profiling study using human TAA tissue samples and identified 147 lncRNAs that were differentially regulated compared to the controls.114 They further identified that lnc-HLTF-5 correlates with an increased ascending aortic diameter, AAA-relevant MMP9 expression, and arterial hypertension.114 Interestingly they found that more than half of these deregulated lncRNAs in TAA were sense-overlapping RNAs, containing protein-coding genes within their intronic sequences. These findings implicate the potential importance of these lncRNAs to their corresponding protein-coding genes in aneurysm development.
HIF1α-AS1:
Another study, using sera from patients with thoraco-abdominal aortic aneurysm (TAAA) patients.115, identified the lncRNA HIF1α antisense RNA 1 (HIF1α-AS1) to be increased. Inhibition of HIF1α-AS1 by siRNA decreased the expression of the caspase-3 and −8, while upreguating the levels of Bcl2. Furthermore, inhibition of HIF1α-AS1 limited palmitic acid-induced SMC apoptosis.116 However, the role of HIF1 α -AS1 in animal models of AAA needs to be established.
AK056155:
The lncRNA AK056155 has been reported for being increased in subjects with a subform of TAA, the Loeys-Dietz syndrome (LDS). Interestingly, expression of AK056155 correlated with AKT/PI3K/TGF-β signaling, which are the known key factors contributing to aneurysm development and disease progression.117 They showed that AK056155 expression is stimulated by TGF-β1 in HUVECs. Interestingly, in combination with TGF-β1 stimulation, AK056155 expression can be augmented upon treatment with the PI3K inhibitor LY294002 or the AKT inhibitor GDC-0068. Their role in AAA development in human tissues and animal models needs to be established.
Linc00540:
Finally, a recent meta-analysis of all available AAA-specific genome-wide association studies (GWAS) identified linc00540 as a novel risk locus for AAA.118 Expression data and functional studies in disease relevant cell subtypes as well as in vivo models are currently lacking for this lncRNA. This is further complicated by the fact that no mouse homologue for this lincRNA exists. Identifying its mouse homologues would be helpful in further studying the role of this lncRNA in AAA development.
4. Circular RNAs in aneurysms
Like lncRNAs, circular RNA transcripts (circRNAs) have been a recent addition to the functionally relevant non-coding RNAs in our genomic landscape. Upon their discovery in the 1990s, they have long been disregarded, due in part to a limitation in their detection method. Recently, this was changed through the support of novel bioinformatics in combination with deep RNA sequencing approaches.119 Recent studies suggest thousands of endogenous circRNAs in mammalian cells, with some of them being highly abundant and evolutionarily conserved. CircRNAs have been shown to mediate miRNA function (e.g., via sponging), and to control events important in transcription (e.g., RNA folding, endonuclease protection).120 CircRNAs can be produced by the direct ligation of 5’ and 3’ ends of linear RNAs, by “back-splicing”, in which a downstream 5’ splice site (donor) is linked to an upstream 3’ site (acceptor), or as intermediates in RNA processing reactions.119 Data on circRNAs in aneurysm disease are scarce, but emerging studies have shown evidence of abnormal expression of circRNAs in the disease context. However, the functional role(s) of these circRNAs in AAA development in animal models and their therapeutic potential is yet to be established.
circ000595:
Zheng and colleagues have screened circRNAs in tissue specimens from AAA patients and found increased expression of circ000595 in diseased specimens. A similar pattern was found in hypoxic aortic SMCs, in which knockdown of circ000595 reduced apoptosis. They further showed that miR-19a might serve as a potential downstream target of circ000595.121
circRNA-101238:
Another study utilized a targeted circRNA array to explore differentially expressed circRNAs in human tissue specimens from TAA patients undergoing surgery. Here, Zou et al. identified circRNA-101238 not only as being deregulated with the disease, but also to potentially affect miR-320a expression as well as MMP9 levels.122
circ0021001:
One recent effort identified the circ0021001 in peripheral blood of patients with intracranial aneurysms without showing its potential contribution to aneurysmal expansion.123
5. Summary and perspectives
Emerging studies have clearly established that non-coding RNAs such as microRNAs, lncRNAs and circRNAs are involved in AAA development and progression. While the roles of miRNAs and lncRNAs have been established, the functional role of circRNAs in AAA is yet to be elucidated. The role and therapeutic potential of miRNAs (miR-21, 24, 29, 33, 143/145, 181b, 195, 205 and 712) have been demonstrated in various animal models of AAA with some cross-validations in human aneurysmal tissue samples. In contrast, the only lncRNA that has clearly demonstrated its role in AAA is H19, although several lncRNAs (SENCR, SMILR, lncAng362, HOTAIR, Pi5, lnc-HLTF5, Hif1α-AS1, AK056155 and lnc00540) have shown their potential roles in some pathophysiological processes of AAA development. Therefore, there is a tremendous need for additional studies for miRNAs, while the need to study the roles and mechanisms of lncRNAs and circRNAs in AAA are wide open. This information will lead to better understanding of mechanisms of AAA and development of therapies and diagnostic approaches.
To understand the biological and pathophysiological mechanisms by which these non-coding RNAs regulate the AAA development and progression, it is essential to identify and validate additional miRNAs, lncRNAs and circRNAs that are abnormally expressed in the diseased human tissues and relevant animal models. Given the tissue-specific expression of lncRNAs and potentially other non-coding RNAs, it is important to consider collecting cell-type specific RNA samples to determine the cell-specific expression of non-coding RNAs under various disease conditions. For example, whenever possible, collecting RNAs from key cell types such as endothelial cells, smooth muscle cells and immune cells would be helpful to obtain specific insights into the cell-type specific changes in the noncoding RNAs in AAA development. A combination of cell sorting approaches and in vitro experiments would be further helpful.
Once their expression is validated, the molecular mechanisms of these lncRNAs should be tested in different cell types in vitro and in vivo. The most critical and difficult part of establishing the molecular mechanisms of noncoding RNAs is to identify the genes that are directly regulated by them. Although, several in silico miRNA target gene prediction programs 124 (TargetScan, miR-Walk, miR-TarBase, etc.) are available to help identifying their potential gene targets, it is nevertheless a daunting and challenging task to validate their gene targets in the specific cell types and tissues. Given the diverse and complex three-dimensional regulatory mechanisms of lncRNAs, there are no simple target gene prediction algorithms available. Initial efforts should be aimed at studying the genes which are either embedded, overlapping, or in close proximity to the genomic location the lncRNAs of interest. Although the computational methods for detection and downstream analysis of circRNAs are emerging, more sophisticated tools are needed to identify and study the circRNAs involved in AAAs.125
To study the functional role of these noncoding RNAs in AAA, it is highly advisable to use both gain-of-function and loss-of-function approaches in both in vitro and in vivo models (Table 1). For miRNAs, anti-miRs and miR-mimics, are popular choices, while GapmeRs and CRISPR-Cas9 mediated genome editing are gaining popularity to study the role of lncRNAs in addition to conventional genetic engineering approaches. These tools may also be used for potential therapeutic approaches.
The critical challenges in using these tools for manipulating miRNA and lncRNA levels are specificity, safety and methods of delivery. Since miRNAs can target numerous genes simultaneously in various tissues and cells, it is important to minimize potential off-gene target effects and off-tissue effects by considering the use of targeted-delivery methods and more gene-specific miRNA inhibitors such as target site blockers (TSBs). Currently, these miRNA manipulators are delivered in vivo either systemically or locally. Systemic delivery of miRNA inhibitors or mimics are carried out using either native or modified oligomers such as locked nucleic acid or 2’-O-methyl nucleotide derivatives. Moreover, these oligomers are typically delivered systemically in saline, but can also be delivered using carriers such as lipid nanoparticles126–129, gold nanoparticles130–132, exosomes133–140 with or without additional surface coatings for targeted delivery. For example, naked anti-miRs in saline can be injected systemically to inhibit AAA in mouse models.57 In contrast, lipid nanoparticles coated with VCAM1-targeting peptide carrying anti-miR-712 was used to deliver the payload to the inflamed endothelium to inhibit atherosclerosis in mice with minimum off-target effects.128 Although this approach has not been tested in AAA, it would be worthwhile to test its efficacy and reduced off target effects in the murine AAA models. Another method to deliver anti-miRs is by using drug-eluting stents for local delivery to arteries. 76 This approach could be used to deliver similar inhibitors of miRNAs or lncRNAs using coated endovascular devices locally to the AAA regions to minimize off target effects and enhanced therapeutic effects. Furthermore, using the local delivery approach with a combination strategy targeting multiple miRNAs could also be a promising therapeutic option. For example, treatment with miR-24 mimic and anti-miR-21 cocktail, or anti-miR-29 and anti-miR-181b cocktail may have more potent therapeutic effects on AAA than targeting single miRNA alone. However, it is important to determine whether the potential benefits of targeting multiple miRNAs using a cocktail of anti-miRs outweigh the potentially increased off target effects.
In addition to the therapeutic targeting of miRNAs and lncRNAs, they may serve as biomarkers to monitor progression of AAA in patients. Several studies have identified miRNAs for their diagnostic potential for existing aneurysms.17, 64, 77, 141 Since ultrasound imaging is widely available, economical, sensitive and effective in identifying aortic aneurysms, using miRNAs merely as a biomarker may not add additional significant prognostic value. However, a miRNA-based biomarker may be quite useful to predict aneurysm expansion rates and the risk of future ruptures. It may offer a significant benefit in the stratification and monitoring of patients with small, but growing AAAs. In this regard, Wanhainen and colleagues have identified a panel of 20 miRNAs that are expressed differently in patients with either fast- or slow-growing AAAs.88 Although the number of individuals (169 AAA cases and 48 age-matched controls) included into this study was relatively small, the results from this explorative analysis encourage us to carry out much larger studies to identify ad classes of non-coding RNAs as prognostic biomarkers for AAA progression.
In summary, many noncoding RNAs have been identified as critical regulators of AAA and the list is growing, but more efforts are needed to identify additional players and demonstrate their action mechanisms and their diagnostic and therapeutic potential.
Table 2.
Summary of microRNAs regulating pathophysiology of AAAs
| miRNA | Cell types | Potential biomarker | Animal model | miRNA modulation method | Results | Target genes | References |
|---|---|---|---|---|---|---|---|
| mir-29 | SMC EC FB | No | Elastase, AngII, Marfan | Viral OE Anti-miR |
Inhibition reduces AAA | collagens elastin |
62, 65, 95 |
| miR-21 | SMC EC FB |
No | Elastase, AngII | Viral OE Anti-miR |
OE reduces AAA | PTEN | 50 |
| miR-24 | SMC EC MPh |
Yes | Elastase, AngII | Viral OE Inhibition |
OE reduces AAA | CHI3L1 | 77 |
| miR-33 | SMC MPh HC |
No | AngII, CCl2 | Knockout | Inhibition reduces AAA | ABCA1 | 78,80 |
| miR-143 miR-145 |
SMC FB EC | No* | AngII | Viral OE Knockout | OE induces SMC differentiation | KLF4 PHACTR4 | 83 |
| miR-181b | MPh EC | No | AngII | Anti-miR | Conflicting results | TIMP3 elastin |
85–87 |
| miR-195 | SMC | Yes | AngII | AntagomiR | Inhibition reduces AAA | collagen
proteoglycan elastin SMAD3 |
68, 89–91 |
| miR-205 miR-712 |
EC SMC | No | AngII | Anti-miR | Inhibition reduces AAA | TIMP3 | 57 |
SMC: Smooth muscle cells, EC: Endothelial cells, FB: Fibroblasts, MPh: Macrophages, HC: Hepatocytes, AngII: Angiotensin II, OE: Overexpression. *miR-143/145 may be useful biomarkers for vascular disease, but not specific for aneurysms.
Acknowledgments
Sources of funding:
This work was supported by funding from National Institutes of Health grants HL119798 and HL095070 to HJ. HJ is John and Jan Portman Professor and would like to dedicate this paper in memory of late Mr. Portman for his generous support for the endowed Professorship. SD is funded through the Excellence Cluster Cardiopulmonary Systems (German Research Foundation) and the LOEWE (Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz) Centre for Cell and Gene Therapy (State of Hesse) and the German Research Foundation (SFB834). RAB is funded by the German Research Foundation (DFG, SFB834), the European Research Council (ERC, NOVA), the German Center for Cardiovascular Research (DZHK), the Amsterdam Academic Alliance (AAA), and the Netherlands Organisation for Scientific Research (NWO, Vidi). LM is funded through the European Research Council (ERC NORVAS), the German Center for Cardiovascular Research (DZHK), the German Research Foundation (DFG, Heisenberg Programm), the Swedish Research Council (VR), and the Swedish Heart-Lung-Foundation (HLF).
Nonstandard Abbreviation and Acronyms:
- AAA
Abdominal aortic aneurysm
- AAV
adeno-associated virus
- ABCA1
ATP Binding Cassette Subfamily A Member 1
- AngII
Angiontesin II
- AS
antisense
- Cas9
CRISPR associated protein 9
- CHI3L1
Chitinase 3 Like 1
- circRNA
circular RNA transcript
- CRISPR-Cas9
clustered regularly interspaced short palindromic repeats
- DGCR8
DiGeorge Syndrome Critical Region Gene 8
- EC
Endothelial cells
- ECM
extracellular matrix
- FB
Fibroblasts
- HC
Hepatocytes
- HIF1α
hypoxia-inducible factor 1 alpha
- HLTF5
Helicase Like Transcription Factor 5
- HOTAIR
HOX Transcript Antisense RNA
- IL8
Interleukin 8
- LDL
low-density lipoprotein
- LDLR
low-density lipoprotein receptor
- LDS
Loeys-Dietz syndrome
- LincRNA
long intergenic non-coding RNA
- Lnc-RNA
long-non-coding RNA
- Meg3
Maternally Expressed 3
- MMP
Matrix metalloproteinase
- Mph
Macrophage
- NFκB
Nuclear Factor Kappa B
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PDGF
platelet derived growth factor
- Pi15
protease inhibitor 15
- PPE
porcine pancreatic elastase
- PTEN
Phosphatase and tensin homolog
- RISC
RNA-induced silencing complex
- RNAseq
RNA sequencing
- SENCR
Smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA
- SMC
Smooth muscle cells
- SMILR
Smooth Muscle Cell Enriched Long Non-Coding RNA
- TAA
Thoracic aortic aneurysms
- TIMP
tissue inhibitor of matrix metalloproteinase
- TSB
target site blocker
- UTR
untranslated region
Footnotes
Disclosures:
Reinier Boon and Stefanie Dimmeler have a patent on the use of miR-29 in AAA and Stefanie Dimmeler is a member of Scientific Advisory Board of Miragen Therapeutics. There are no disclosures for Sandeep Kumar, Lars Maegdefessel and Hanjoong Jo.”
References
- 1.Aggarwal S, Qamar A, Sharma V, Sharma A. Abdominal aortic aneurysm: A comprehensive review. Exp Clin Cardiol. 2011;16:11–15 [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P, American Heart Association Council on E, Prevention Statistics C, Stroke Statistics S. Heart disease and stroke statistics-2018 update: A report from the american heart association. Circulation. 2018;137:e67–e492 [DOI] [PubMed] [Google Scholar]
- 3.Howard DP, Banerjee A, Fairhead JF, Handa A, Silver LE, Rothwell PM, Oxford Vascular S. Age-specific incidence, risk factors and outcome of acute abdominal aortic aneurysms in a defined population. Br J Surg. 2015;102:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston KW, Rutherford RB, Tilson MD, Shah DM, Hollier L, Stanley JC. Suggested standards for reporting on arterial aneurysms. Subcommittee on reporting standards for arterial aneurysms, ad hoc committee on reporting standards, society for vascular surgery and north american chapter, international society for cardiovascular surgery. Journal of vascular surgery. 1991;13:452–458 [DOI] [PubMed] [Google Scholar]
- 5.Ghorpade A, Baxter BT. Biochemistry and molecular regulation of matrix macromolecules in abdominal aortic aneurysms. Ann N Y Acad Sci. 1996;800:138–150 [DOI] [PubMed] [Google Scholar]
- 6.Daugherty A, Cassis LA. Mechanisms of abdominal aortic aneurysm formation. Current atherosclerosis reports. 2002;4:222–227 [DOI] [PubMed] [Google Scholar]
- 7.Limet R, Nusgens B, Verloes A, Sakalihasan N. Pathogenesis of abdominal aortic aneurysm (aaa) formation. Acta chirurgica Belgica. 1998;98:195–198 [PubMed] [Google Scholar]
- 8.Rabkin SW. The role matrix metalloproteinases in the production of aortic aneurysm. Progress in molecular biology and translational science. 2017;147:239–265 [DOI] [PubMed] [Google Scholar]
- 9.Thompson RW, Liao S, Curci JA. Vascular smooth muscle cell apoptosis in abdominal aortic aneurysms. Coronary artery disease. 1997;8:623–631 [DOI] [PubMed] [Google Scholar]
- 10.Reilly JM. Plasminogen activators in abdominal aortic aneurysmal disease. Ann N Y Acad Sci. 1996;800:151–156 [DOI] [PubMed] [Google Scholar]
- 11.Samad F, Schneiderman J, Loskutoff D. Expression of fibrinolytic genes in tissues from human atherosclerotic aneurysms and from obese mice. Ann N Y Acad Sci. 1997;811:350–358; [DOI] [PubMed] [Google Scholar]
- 12.Michel JB, Martin-Ventura JL, Egido J, Sakalihasan N, Treska V, Lindholt J, Allaire E, Thorsteinsdottir U, Cockerill G, Swedenborg J, consortium FE. Novel aspects of the pathogenesis of aneurysms of the abdominal aorta in humans. Cardiovascular research. 2011;90:18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eagleton MJ. Inflammation in abdominal aortic aneurysms: Cellular infiltrate and cytokine profiles. Vascular. 2012;20:278–283 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Tang C, Qin Y. Cathepsins: A new culprit behind abdominal aortic aneurysm. Regenerative medicine research. 2013;1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale MA, Ruhlman MK, Baxter BT. Inflammatory cell phenotypes in aaas: Their role and potential as targets for therapy. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:1746–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuivaniemi H, Ryer EJ, Elmore JR, Tromp G. Understanding the pathogenesis of abdominal aortic aneurysms. Expert review of cardiovascular therapy. 2015;13:975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Shang T, Huang C, Yu T, Liu C, Qiao T, Huang D, Liu Z, Liu C. Plasma micrornas serve as potential biomarkers for abdominal aortic aneurysm. Clin Biochem. 2015;48:988–992 [DOI] [PubMed] [Google Scholar]
- 18.Jalalzadeh H, Indrakusuma R, Planken RN, Legemate DA, Koelemay MJ, Balm R. Inflammation as a predictor of abdominal aortic aneurysm growth and rupture: A systematic review of imaging biomarkers. European journal of vascular and endovascular surgery : the official journal of the European Society for Vascular Surgery. 2016;52:333–342 [DOI] [PubMed] [Google Scholar]
- 19.Vorp DA, Vande Geest JP. Biomechanical determinants of abdominal aortic aneurysm rupture. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:1558–1566 [DOI] [PubMed] [Google Scholar]
- 20.Vorp DA. Biomechanics of abdominal aortic aneurysm. Journal of biomechanics. 2007;40:1887–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dua MM, Dalman RL. Hemodynamic influences on abdominal aortic aneurysm disease: Application of biomechanics to aneurysm pathophysiology. Vascular pharmacology. 2010;53:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGloughlin TM, Doyle BJ. New approaches to abdominal aortic aneurysm rupture risk assessment: Engineering insights with clinical gain. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:1687–1694 [DOI] [PubMed] [Google Scholar]
- 23.Raut SS, Chandra S, Shum J, Finol EA. The role of geometric and biomechanical factors in abdominal aortic aneurysm rupture risk assessment. Annals of biomedical engineering. 2013;41:1459–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz-Rixen T, Keese M, Hakimi M, Peters A, Bockler D, Nelson K, Grundmann RT. Ruptured abdominal aortic aneurysm-epidemiology, predisposing factors, and biology. Langenbeck’s archives of surgery. 2016;401:275–288 [DOI] [PubMed] [Google Scholar]
- 25.Waliszewski K, Slomski R, Oszkinis G, Majewski W. [genetic aspects of the pathogenesis of aortic abdominal aneurysms]. Polski merkuriusz lekarski : organ Polskiego Towarzystwa Lekarskiego. 2005;18:111–114 [PubMed] [Google Scholar]
- 26.Thompson RW, Curci JA, Ennis TL, Mao D, Pagano MB, Pham CT. Pathophysiology of abdominal aortic aneurysms: Insights from the elastase-induced model in mice with different genetic backgrounds. Ann N Y Acad Sci. 2006;1085:59–73 [DOI] [PubMed] [Google Scholar]
- 27.Makrygiannis G, Courtois A, Drion P, Defraigne JO, Kuivaniemi H, Sakalihasan N. Sex differences in abdominal aortic aneurysm: The role of sex hormones. Annals of vascular surgery. 2014;28:1946–1958 [DOI] [PubMed] [Google Scholar]
- 28.Halloran BG, Baxter BT. Pathogenesis of aneurysms. Seminars in vascular surgery. 1995;8:85–92 [PubMed] [Google Scholar]
- 29.Thompson RW. Basic science of abdominal aortic aneurysms: Emerging therapeutic strategies for an unresolved clinical problem. Current opinion in cardiology. 1996;11:504–518 [DOI] [PubMed] [Google Scholar]
- 30.van Vlijmen-van Keulen CJ, Pals G, Rauwerda JA. Familial abdominal aortic aneurysm: A systematic review of a genetic background. European journal of vascular and endovascular surgery : the official journal of the European Society for Vascular Surgery 2002;24:105–116 [DOI] [PubMed] [Google Scholar]
- 31.Siasos G, Mourouzis K, Oikonomou E, Tsalamandris S, Tsigkou V, Vlasis K, Vavuranakis M, Zografos T, Dimitropoulos S, Papaioannou TG, Kalampogias A, Stefanadis C, Papavassiliou AG, Tousoulis D. The role of endothelial dysfunction in aortic aneurysms. Current pharmaceutical design. 2015;21:4016–4034 [DOI] [PubMed] [Google Scholar]
- 32.Sabiston DC, Townsend CM. Sabiston textbook of surgery : The biological basis of modern surgical practice. Philadelphia, PA: Elsevier Saunders; 2012. [Google Scholar]
- 33.Tung WS, Lee JK, Thompson RW. Simultaneous analysis of 1176 gene products in normal human aorta and abdominal aortic aneurysms using a membrane-based complementary DNA expression array. Journal of vascular surgery. 2001;34:143–150 [DOI] [PubMed] [Google Scholar]
- 34.Nakahashi TK, Hoshina K, Tsao PS, Sho E, Sho M, Karwowski JK, Yeh C, Yang RB, Topper JN, Dalman RL. Flow loading induces macrophage antioxidative gene expression in experimental aneurysms. Arteriosclerosis, thrombosis, and vascular biology. 2002;22:2017–2022 [DOI] [PubMed] [Google Scholar]
- 35.Thompson RW, Absi T, Tung WS. Gene expression profiling of abdominal aortic aneurysms. Journal of vascular surgery. 2002;35:403–404 [DOI] [PubMed] [Google Scholar]
- 36.Zheng Y, Guan H, Li Y, Liu C, Liu B, Sheng Q, Miao S. [microarray in screening for differentially expressed genes of cellular cycle and apoptosis in abdominal aortic aneurysms]. Zhonghua Wai Ke Za Zhi. 2002;40:817–819 [PubMed] [Google Scholar]
- 37.Absi TS, Sundt TM 3rd, Tung WS, Moon M, Lee JK, Damiano RR Jr., Thompson RW. Altered patterns of gene expression distinguishing ascending aortic aneurysms from abdominal aortic aneurysms: Complementary DNA expression profiling in the molecular characterization of aortic disease. J Thorac Cardiovasc Surg. 2003;126:344–357; [DOI] [PubMed] [Google Scholar]
- 38.Kazi M, Zhu C, Roy J, Paulsson-Berne G, Hamsten A, Swedenborg J, Hedin U, Eriksson P. Difference in matrix-degrading protease expression and activity between thrombus-free and thrombus-covered wall of abdominal aortic aneurysm. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:1341–1346 [DOI] [PubMed] [Google Scholar]
- 39.Choke E, Thompson MM, Jones A, Torsney E, Dawson J, Laing K, Nasr H, Loftus IM, Cockerill GW. Gene expression profile of abdominal aortic aneurysm rupture. Ann N Y Acad Sci. 2006;1085:311–314 [DOI] [PubMed] [Google Scholar]
- 40.Lenk GM, Tromp G, Skunca M, Gatalica Z, Berguer R, Kuivaniemi H. Global expression profiles in human normal and aneurysmal abdominal aorta based on two distinct whole genome microarray platforms. Ann N Y Acad Sci. 2006;1085:360–362 [DOI] [PubMed] [Google Scholar]
- 41.Van Vickle-Chavez SJ, Tung WS, Absi TS, Ennis TL, Mao D, Cobb JP, Thompson RW. Temporal changes in mouse aortic wall gene expression during the development of elastase-induced abdominal aortic aneurysms. Journal of vascular surgery. 2006;43:1010–1020 [DOI] [PubMed] [Google Scholar]
- 42.Zhao HG, Fu SJ, Jiang ME. [gene expression difference analysis between abdominal aorta aneurysm and normal abdominal artery]. Zhonghua Wai Ke Za Zhi. 2008;46:691–693 [PubMed] [Google Scholar]
- 43.Rush C, Nyara M, Moxon JV, Trollope A, Cullen B, Golledge J. Whole genome expression analysis within the angiotensin ii-apolipoprotein e deficient mouse model of abdominal aortic aneurysm. BMC Genomics. 2009;10:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spin JM, Hsu M, Azuma J, Tedesco MM, Deng A, Dyer JS, Maegdefessel L, Dalman RL, Tsao PS. Transcriptional profiling and network analysis of the murine angiotensin ii-induced abdominal aortic aneurysm. Physiol Genomics. 2011;43:993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pahl MC, Derr K, Gabel G, Hinterseher I, Elmore JR, Schworer CM, Peeler TC, Franklin DP, Gray JL, Carey DJ, Tromp G, Kuivaniemi H. Microrna expression signature in human abdominal aortic aneurysms. BMC Med Genomics. 2012;5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinterseher I, Erdman R, Elmore JR, Stahl E, Pahl MC, Derr K, Golden A, Lillvis JH, Cindric MC, Jackson K, Bowen WD, Schworer CM, Chernousov MA, Franklin DP, Gray JL, Garvin RP, Gatalica Z, Carey DJ, Tromp G, Kuivaniemi H. Novel pathways in the pathobiology of human abdominal aortic aneurysms. Pathobiology. 2013;80:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheuk BL, Cheng SW. Identification and characterization of micrornas in vascular smooth muscle cells from patients with abdominal aortic aneurysms. Journal of vascular surgery. 2014;59:202–209 [DOI] [PubMed] [Google Scholar]
- 48.Longo GM, Xiong W, Greiner TC, Zhao Y, Fiotti N, Baxter BT. Matrix metalloproteinases 2 and 9 work in concert to produce aortic aneurysms. The Journal of clinical investigation. 2002;110:625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daugherty A, Cassis LA. Mouse models of abdominal aortic aneurysms. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:429–434 [DOI] [PubMed] [Google Scholar]
- 50.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, Leeper NJ, Raaz U, Schoelmerich AM, McConnell MV, Dalman RL, Spin JM, Tsao PS. Microrna-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Science translational medicine. 2012;4:122ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ, McConnell MV, Dalman RL, Spin JM, Tsao PS. Inhibition of microrna-29b reduces murine abdominal aortic aneurysm development. The Journal of clinical investigation. 2012;122:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakao T, Horie T, Baba O, Nishiga M, Nishino T, Izuhara M, Kuwabara Y, Nishi H, Usami S, Nakazeki F, Ide Y, Koyama S, Kimura M, Sowa N, Ohno S, Aoki H, Hasegawa K, Sakamoto K, Minatoya K, Kimura T, Ono K. Genetic ablation of microrna-33 attenuates inflammation and abdominal aortic aneurysm formation via several anti-inflammatory pathways. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2161–2170 [DOI] [PubMed] [Google Scholar]
- 53.Liu G, Huang Y, Lu X, Lu M, Huang X, Li W, Jiang M. Identification and characteristics of micrornas with altered expression patterns in a rat model of abdominal aortic aneurysms. The Tohoku journal of experimental medicine. 2010;222:187–193 [DOI] [PubMed] [Google Scholar]
- 54.Cassis LA, Rateri DL, Lu H, Daugherty A. Bone marrow transplantation reveals that recipient at1a receptors are required to initiate angiotensin ii-induced atherosclerosis and aneurysms. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:380–386 [DOI] [PubMed] [Google Scholar]
- 55.Lu H, Howatt DA, Balakrishnan A, Graham MJ, Mullick AE, Daugherty A. Hypercholesterolemia induced by a pcsk9 gain-of-function mutation augments angiotensin ii-induced abdominal aortic aneurysms in c57bl/6 mice-brief report. Arteriosclerosis, thrombosis, and vascular biology. 2016;36:1753–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Wang J, Li X, Liu X, Yu X, Tian Y. Microrna-145 mediates the formation of angiotensin ii-induced murine abdominal aortic aneurysm. Heart, lung & circulation. 2017;26:619–626 [DOI] [PubMed] [Google Scholar]
- 57.Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H. Prevention of abdominal aortic aneurysm by anti-microrna-712 or anti-microrna-205 in angiotensin ii-infused mice. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao T, Yao Y, Zhang B, Hao DC, Sun QF, Li JB, Yuan C, Jing B, Wang YP, Wang HY. Role of microrna-103a targeting adam10 in abdominal aortic aneurysm. BioMed research international. 2017;2017:9645874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle Anna M. Hotair forms an intricate and modular secondary structure. Molecular Cell. 2015;58:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens L, Rühle F, Stoll M. Lncrna secondary structure in the cardiovascular system. Non-coding RNA Research. 2017;2:137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartel DP. Micrornas: Target recognition and regulatory functions. Cell. 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boon RA, Dimmeler S. Micrornas and aneurysm formation. Trends Cardiovasc Med 2011;21:172–177 [DOI] [PubMed] [Google Scholar]
- 63.Leeper NJ, Maegdefessel L. Non-coding rnas: Key regulators of smooth muscle cell fate in vascular disease. Cardiovascular research. 2018;114:611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kin K, Miyagawa S, Fukushima S, Shirakawa Y, Torikai K, Shimamura K, Daimon T, Kawahara Y, Kuratani T, Sawa Y. Tissue- and plasma-specific microrna signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc. 2012;1:e000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, van Heijningen P, Essers J, Brandes RP, Zeiher AM, Dimmeler S. Microrna-29 in aortic dilation: Implications for aneurysm formation. Circulation research. 2011;109:1115–1119 [DOI] [PubMed] [Google Scholar]
- 66.Merk DR, Chin JT, Dake BA, Maegdefessel L, Miller MO, Kimura N, Tsao PS, Iosef C, Berry GJ, Mohr FW, Spin JM, Alvira CM, Robbins RC, Fischbein MP. Mir-29b participates in early aneurysm development in marfan syndrome. Circulation research. 2012;110:312–324 [DOI] [PubMed] [Google Scholar]
- 67.Okamura H, Emrich F, Trojan J, Chiu P, Dalal AR, Arakawa M, Sato T, Penov K, Koyano T, Pedroza A, Connolly AJ, Rabinovitch M, Alvira C, Fischbein MP. Long-term mir-29b suppression reduces aneurysm formation in a marfan mouse model. Physiol Rep. 2017;5:e13257–e13257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zampetaki A, Attia R, Mayr U, Gomes RS, Phinikaridou A, Yin X, Langley SR, Willeit P, Lu R, Fanshawe B, Fava M, Barallobre-Barreiro J, Molenaar C, So PW, Abbas A, Jahangiri M, Waltham M, Botnar R, Smith A, Mayr M. Role of mir-195 in aortic aneurysmal disease. Circulation research. 2014;115:857–866 [DOI] [PubMed] [Google Scholar]
- 69.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of micrornas after myocardial infarction reveals a role of mir-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rupaimoole R, Slack FJ. Microrna therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222 [DOI] [PubMed] [Google Scholar]
- 71.Greineder CF, Hood ED, Yao A, Khoshnejad M, Brenner JS, Johnston IH, Poncz M, Gottstein C, Muzykantov VR. Molecular engineering of high affinity single-chain antibody fragment for endothelial targeting of proteins and nanocarriers in rodents and humans. Journal of controlled release : official journal of the Controlled Release Society. 2016;226:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hood ED, Chorny M, Greineder CF, I SA, Levy RJ, Muzykantov VR. Endothelial targeting of nanocarriers loaded with antioxidant enzymes for protection against vascular oxidative stress and inflammation. Biomaterials. 2014;35:3708–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kiseleva RY, Glassman PM, Greineder CF, Hood ED, Shuvaev VV, Muzykantov VR. Targeting therapeutics to endothelium: Are we there yet? Drug delivery and translational research. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie A, Belcik T, Qi Y, Morgan TK, Champaneri SA, Taylor S, Davidson BP, Zhao Y, Klibanov AL, Kuliszewski MA, Leong-Poi H, Ammi A, Lindner JR. Ultrasound-mediated vascular gene transfection by cavitation of endothelial-targeted cationic microbubbles. JACC. Cardiovascular imaging. 2012;5:1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS, Wen JK. Exosome-mediated mir-155 transfer from smooth muscle cells to endothelial cells induces endothelial injury and promotes atherosclerosis. Molecular therapy : the journal of the American Society of Gene Therapy. 2017;25:1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Wang D, Deuse T, Stubbendorff M, Chernogubova E, Erben RG, Eken SM, Jin H, Li Y, Busch A, Heeger CH, Behnisch B, Reichenspurner H, Robbins RC, Spin JM, Tsao PS, Schrepfer S, Maegdefessel L. Local microrna modulation using a novel anti-mir-21-eluting stent effectively prevents experimental in-stent restenosis. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:1945–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maegdefessel L, Spin JM, Raaz U, Eken SM, Toh R, Azuma J, Adam M, Nakagami F, Heymann HM, Chernogubova E, Jin H, Roy J, Hultgren R, Caidahl K, Schrepfer S, Hamsten A, Eriksson P, McConnell MV, Dalman RL, Tsao PS. Mir-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. 2014;5:5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. Mir-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang K, Bao H, Yan ZQ, Wang L, Zhang P, Yao QP, Shi Q, Chen XH, Wang KX, Shen BR, Qi YX, Jiang ZL. Microrna-33 protects against neointimal hyperplasia induced by arterial mechanical stretch in the grafted vein. Cardiovascular research. 2017;113:488–497 [DOI] [PubMed] [Google Scholar]
- 80.Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. Microrna-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. The Journal of clinical investigation. 2015;125:4334–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. Mir-145 and mir-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of mir-143 and −145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009;16:1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through mirnas. Nat Cell Biol. 2012;14:249–256 [DOI] [PubMed] [Google Scholar]
- 84.Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. Tgfbeta triggers mir-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circulation research. 2015;116:1753–1764 [DOI] [PubMed] [Google Scholar]
- 85.Di Gregoli K, Mohamad Anuar NN, Bianco R, White SJ, Newby AC, George SJ, Johnson JL. Microrna-181b controls atherosclerosis and aneurysms through regulation of timp-3 and elastin. Circulation research. 2017;120:49–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hori D, Dunkerly-Eyring B, Nomura Y, Biswas D, Steppan J, Henao-Mejia J, Adachi H, Santhanam L, Berkowitz DE, Steenbergen C, Flavell RA, Das S. Mir-181b regulates vascular stiffness age dependently in part by regulating tgf-beta signaling. PLoS One. 2017;12:e0174108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Hunninghake GM, Vera MP, Registry M, Blackwell TS, Baron RM, Feinberg MW. Microrna-181b regulates nf-kappab-mediated vascular inflammation. The Journal of clinical investigation. 2012;122:1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wanhainen A, Mani K, Vorkapic E, De Basso R, Bjorck M, Lanne T, Wagsater D. Screening of circulating microrna biomarkers for prevalence of abdominal aortic aneurysm and aneurysm growth. Atherosclerosis. 2017;256:82–88 [DOI] [PubMed] [Google Scholar]
- 89.Ma X, Yao H, Yang Y, Jin L, Wang Y, Wu L, Yang S, Cheng K. Mir-195 suppresses abdominal aortic aneurysm through the tnf-alpha/nf-kappab and vegf/pi3k/akt pathway. International journal of molecular medicine. 2018;41:2350–2358 [DOI] [PubMed] [Google Scholar]
- 90.Liang B, Che J, Zhao H, Zhang Z, Shi G. Mir-195 promotes abdominal aortic aneurysm media remodeling by targeting smad3. Cardiovascular therapeutics. 2017;35. [DOI] [PubMed] [Google Scholar]
- 91.Wang YS, Wang HY, Liao YC, Tsai PC, Chen KC, Cheng HY, Lin RT, Juo SH. Microrna-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovascular research. 2012;95:517–526 [DOI] [PubMed] [Google Scholar]
- 92.Jin W, Reddy MA, Chen Z, Putta S, Lanting L, Kato M, Park JT, Chandra M, Wang C, Tangirala RK, Natarajan R. Small rna sequencing reveals micrornas that modulate angiotensin ii effects in vascular smooth muscle cells. J Biol Chem. 2012;287:15672–15683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartmann P, Zhou Z, Natarelli L, Wei Y, Nazari-Jahantigh M, Zhu M, Grommes J, Steffens S, Weber C, Schober A. Endothelial dicer promotes atherosclerosis and vascular inflammation by mirna-103-mediated suppression of klf4. Nature Communications. 2016;7:10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H. The atypical mechanosensitive microrna-712 derived from pre-ribosomal rna induces endothelial inflammation and atherosclerosis. Nat Commun. 2013;4:3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maegdefessel L, Azuma J, Tsao PS. Microrna-29b regulation of abdominal aortic aneurysm development. Trends Cardiovasc Med 2014;24:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. Gencode: The reference human genome annotation for the encode project. Genome research. 2012;22:1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quinn JJ, Chang HY. Unique features of long non-coding rna biogenesis and function. Nature reviews. Genetics. 2016;17:47–62 [DOI] [PubMed] [Google Scholar]
- 98.Palazzo AF, Lee ES. Non-coding rna: What is functional and what is junk? Frontiers in genetics. 2015;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding rnas; sequence, structure, function. Biochimica et biophysica acta. 2014;1840:1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang KC, Chang HY. Molecular mechanisms of long noncoding rnas. Mol Cell. 2011;43:904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding rnas. RNA Biol. 2013;10:925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lv J, Wang L, Zhang J, Lin R, Wang L, Sun W, Wu H, Xin S. Long noncoding rna h19-derived mir-675 aggravates restenosis by targeting pten. Biochem Biophys Res Commun. 2018;497:1154–1161 [DOI] [PubMed] [Google Scholar]
- 103.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncrna promoters, transcription and splicing. Nature. 2016;539:452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33:464–478 [DOI] [PubMed] [Google Scholar]
- 105.Boon RA, Hofmann P, Michalik KM, Lozano-Vidal N, Berghauser D, Fischer A, Knau A, Jae N, Schurmann C, Dimmeler S. Long noncoding rna meg3 controls endothelial cell aging and function: Implications for regenerative angiogenesis. Journal of the American College of Cardiology. 2016;68:2589–2591 [DOI] [PubMed] [Google Scholar]
- 106.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. Lincrna-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P, Backlund A, Eken SM, Roy J, Eriksson P, Dacken B, Ramanujam D, Dueck A, Engelhardt S, Boon RA, Eckstein HH, Spin JM, Tsao PS, Maegdefessel L. H19 induces abdominal aortic aneurysm development and progression. Circulation. 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding rna. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, Joshi N, Dweck MR, Miano JM, McBride MW, Newby DE, McDonald RA, Baker AH. Smooth muscle enriched long noncoding rna (smilr) regulates cell proliferation. Circulation. 2016;133:2050–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding rnas are regulated by angiotensin ii in vascular smooth muscle cells. Circulation research. 2013;113:266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microrna-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo X, Chang Q, Pei H, Sun X, Qian X, Tian C, Lin H. Long non-coding rna-mrna correlation analysis reveals the potential role of hotair in pathogenesis of sporadic thoracic aortic aneurysm. European journal of vascular and endovascular surgery : the official journal of the European Society for Vascular Surgery. 2017;54:303–314 [DOI] [PubMed] [Google Scholar]
- 113.Falak S, Schafer S, Baud A, Hummel O, Schulz H, Gauguier D, Hubner N, Osborne-Pellegrin M. Protease inhibitor 15, a candidate gene for abdominal aortic internal elastic lamina ruptures in the rat. Physiol Genomics. 2014;46:418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Y, Liu Y, Liu S, Wu F, Li S, Yang F, Gu Y, Wang G, Xu Z. Differential expression profile of long non-coding rnas in human thoracic aortic aneurysm. J Cell Biochem. 2018 [DOI] [PubMed] [Google Scholar]
- 115.He Q, Tan J, Yu B, Shi W, Liang K. Long noncoding rna hif1a-as1a reduces apoptosis of vascular smooth muscle cells: Implications for the pathogenesis of thoracoabdominal aorta aneurysm. Pharmazie. 2015;70:310–315 [PubMed] [Google Scholar]
- 116.Wang S, Zhang X, Yuan Y, Tan M, Zhang L, Xue X, Yan Y, Han L, Xu Z. Brg1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding rna hif1a-as1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446 [DOI] [PubMed] [Google Scholar]
- 117.Yu B, Liu L, Sun H, Chen Y. Long noncoding rna ak056155 involved in the development of loeys-dietz syndrome through akt/pi3k signaling pathway. Int J Clin Exp Pathol. 2015;8:10768–10775 [PMC free article] [PubMed] [Google Scholar]
- 118.Jones GT, Tromp G, Kuivaniemi H, Gretarsdottir S, Baas AF, Giusti B, Strauss E, Van’t Hof FN, Webb TR, Erdman R, Ritchie MD, Elmore JR, Verma A, Pendergrass S, Kullo IJ, Ye Z, Peissig PL, Gottesman O, Verma SS, Malinowski J, Rasmussen-Torvik LJ, Borthwick KM, Smelser DT, Crosslin DR, de Andrade M, Ryer EJ, McCarty CA, Bottinger EP, Pacheco JA, Crawford DC, Carrell DS, Gerhard GS, Franklin DP, Carey DJ, Phillips VL, Williams MJ, Wei W, Blair R, Hill AA, Vasudevan TM, Lewis DR, Thomson IA, Krysa J, Hill GB, Roake J, Merriman TR, Oszkinis G, Galora S, Saracini C, Abbate R, Pulli R, Pratesi C, Saratzis A, Verissimo AR, Bumpstead S, Badger SA, Clough RE, Cockerill G, Hafez H, Scott DJ, Futers TS, Romaine SP, Bridge K, Griffin KJ, Bailey MA, Smith A, Thompson MM, van Bockxmeer FM, Matthiasson SE, Thorleifsson G, Thorsteinsdottir U, Blankensteijn JD, Teijink JA, Wijmenga C, de Graaf J, Kiemeney LA, Lindholt JS, Hughes A, Bradley DT, Stirrups K, Golledge J, Norman PE, Powell JT, Humphries SE, Hamby SE, Goodall AH, Nelson CP, Sakalihasan N, Courtois A, Ferrell RE, Eriksson P, Folkersen L, Franco-Cereceda A, Eicher JD, Johnson AD, Betsholtz C, Ruusalepp A, Franzen O, Schadt EE, Bjorkegren JL, Lipovich L, Drolet AM, Verhoeven EL, Zeebregts CJ, Geelkerken RH, van Sambeek MR, van Sterkenburg SM, de Vries JP, Stefansson K, Thompson JR, de Bakker PI, Deloukas P, Sayers RD, Harrison SC, van Rij AM, Samani NJ, Bown MJ. Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circulation research. 2017;120:341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jeck WR, Sharpless NE. Detecting and characterizing circular rnas. Nat Biotechnol. 2014;32:453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lasda E, Parker R. Circular rnas: Diversity of form and function. RNA. 2014;20:1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng C, Niu H, Li M, Zhang H, Yang Z, Tian L, Wu Z, Li D, Chen X. Cyclic rna hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol Med Rep. 2015;12:6656–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zou M, Huang C, Li X, He X, Chen Y, Liao W, Liao Y, Sun J, Liu Z, Zhong L, Bin J. Circular rna expression profile and potential function of hsa_circrna_101238 in human thoracic aortic dissection. Oncotarget. 2017;8:81825–81837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Teng L, Chen Y, Chen H, He X, Wang J, Peng Y, Duan H, Li H, Lin D, Shao B. Circular rna hsa_circ_0021001 in peripheral blood: A potential novel biomarker in the screening of intracranial aneurysm. Oncotarget. 2017;8:107125–107133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for sequence-based mirna target prediction: What to choose? Int J Mol Sci. 2016;17:1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao Y, Zhao F. Computational strategies for exploring circular rnas. Trends Genet. 2018;34:389–400 [DOI] [PubMed] [Google Scholar]
- 126.Zhang Y, Wang Z, Gemeinhart RA. Progress in microrna delivery. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McLendon JM, Joshi SR, Sparks J, Matar M, Fewell JG, Abe K, Oka M, McMurtry IF, Gerthoffer WT. Lipid nanoparticle delivery of a microrna-145 inhibitor improves experimental pulmonary hypertension. Journal of controlled release : official journal of the Controlled Release Society. 2015;210:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kheirolomoom A, Kim CW, Seo JW, Kumar S, Son DJ, Gagnon MK, Ingham ES, Ferrara KW, Jo H. Multifunctional nanoparticles facilitate molecular targeting and mirna delivery to inhibit atherosclerosis in apoe(−/−) mice. ACS nano. 2015;9:8885–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haussecker D Current issues of rnai therapeutics delivery and development. Journal of controlled release : official journal of the Controlled Release Society. 2014;195:49–54 [DOI] [PubMed] [Google Scholar]
- 130.Jia C, Chen H, Wei M, Chen X, Zhang Y, Cao L, Yuan P, Wang F, Yang G, Ma J. Gold nanoparticle-based mir155 antagonist macrophage delivery restores the cardiac function in ovariectomized diabetic mouse model. International journal of nanomedicine. 2017;12:4963–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghosh R, Singh LC, Shohet JM, Gunaratne PH. A gold nanoparticle platform for the delivery of functional micrornas into cancer cells. Biomaterials. 2013;34:807–816 [DOI] [PubMed] [Google Scholar]
- 132.Ekin A, Karatas OF, Culha M, Ozen M. Designing a gold nanoparticle-based nanocarrier for microrna transfection into the prostate and breast cancer cells. The journal of gene medicine. 2014;16:331–335 [DOI] [PubMed] [Google Scholar]
- 133.Zhou Y, Zhou G, Tian C, Jiang W, Jin L, Zhang C, Chen X. Exosome-mediated small rna delivery for gene therapy. Wiley interdisciplinary reviews. RNA. 2016;7:758–771 [DOI] [PubMed] [Google Scholar]
- 134.Xitong D, Xiaorong Z. Targeted therapeutic delivery using engineered exosomes and its applications in cardiovascular diseases. Gene. 2016;575:377–384 [DOI] [PubMed] [Google Scholar]
- 135.Sutaria DS, Jiang J, Elgamal OA, Pomeroy SM, Badawi M, Zhu X, Pavlovicz R, Azevedo-Pouly ACP, Chalmers J, Li C, Phelps MA, Schmittgen TD. Low active loading of cargo into engineered extracellular vesicles results in inefficient mirna mimic delivery. Journal of extracellular vesicles. 2017;6:1333882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Silva M, Melo SA. Non-coding rnas in exosomes: New players in cancer biology. Current genomics. 2015;16:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, Lu L. Exosomes derived from mir-181–5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. Journal of cellular and molecular medicine. 2017;21:2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered exosome-mediated delivery of functionally active mir-26a and its enhanced suppression effect in hepg2 cells. International journal of nanomedicine. 2018;13:585–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, Shahar T, Yamashita S, Parker Kerrigan B, Fueyo J, Sawaya R, Lang FF. Mesenchymal stem cells as natural biofactories for exosomes carrying mir-124a in the treatment of gliomas. Neuro-oncology. 2018;20:380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacology & therapeutics. 2017;174:63–78 [DOI] [PubMed] [Google Scholar]
- 141.Biros E, Moran CS, Wang Y, Walker PJ, Cardinal J, Golledge J. Microrna profiling in patients with abdominal aortic aneurysms: The significance of mir-155. Clin Sci (Lond). 2014;126:795–803 [DOI] [PubMed] [Google Scholar]